Abstract
Mutants of Escherichia coli K-12 were isolated which lack the normal phosphotransferase system-dependent catabolic pathway for d-mannitol (Mtl). In some mutants the pts genes for the general proteins enzyme I and histidine protein of the phosphoenolpyruvate-dependent carbohydrate phosphotransferase systems were deleted. Other mutants expressed truncated mannitol-specific enzymes II (IIMtl) which lacked the IIAMtl or IIBAMtl domain(s), and the mtlA genes originated either from E. coli K-12 or from Klebsiella pneumoniae 1033-5P14. The dalD gene from Klebsiella oxytoca M5a1 was cloned on single-copy plasmids and transformed into the strains described above. This gene encodes an NAD-dependent d-arabinitol dehydrogenase (DalD) which converts d-arabinitol into d-xylulose and also converts d-mannitol into d-fructose. The different strains were used to isolate mutations which allow efficient transport of mannitol through the nonphosphorylated IIMtl complexes by selecting for growth on this polyhydric alcohol. More than 40 different mutants were analyzed to determine their ability to grow on mannitol, as well as their ability to bind and transport free mannitol and, after restoration of the missing domain(s), their ability to phosphorylate mannitol. Four mutations were identified (E218A, E218V, H256P, and H256Y); all of these mutations are located in the highly conserved loop 5 of the IIC membrane-bound transporter, and two are located in its GIHE motif. These mutations were found to affect the various functions in different ways. Interestingly, in the presence of all IIMtl variants, whether they were in the truncated form or in the complete form, in the phosphorylated form or in the nonphosphorylated form, and in the wild-type form or in the mutated form, growth occurred on the low-affinity analogue d-arabinitol with good efficiency, while only the uncoupled mutated forms transported mannitol at a high rate.
The bacterial phosphoenolpyruvate (PEP)-dependent mannitol phosphotransferase system (PTS) catalyzes the concomitant transport and phosphorylation of d-mannitol (Mtl) (14, 15, 46). Transfer of the phosphoryl group from PEP to the substrate is catalyzed in four reversible steps by a soluble PEP-dependent protein kinase designated enzyme I, a soluble histidine protein (HPr), and an mtlA-encoded d-mannitol-specific enzyme II (IIMtl) which also accepts, but with a lower affinity, d-glucitol and d-arabinitol (16, 50). IIMtl comprises two hydrophilic domains, IIAMtl and IIBMtl, which accept the phosphoryl group from HPr on His554 and then Cys384, respectively. A membrane-bound domain (IICMtl) constitutes the transporter proper. IICMtl (amino acids 1 to 346) forms six transmembrane segments which are joined by three short periplasmic loops and by two large intracellular loops (loop 3, residues 70 to 134; loop 5, residues 185 to 273) (19, 24, 37, 45). Loop 5, which comprises a conserved structure centered around a characteristic GIHE motif, has been postulated to be essential in substrate binding and translocation (16, 49). In the enteric bacteria, the three domains are fused in a large peptide consisting of 637 amino acids and function as a dimeric complex (37). The domains must, however, be relatively autonomous because artificial splitting and fusion at the natural linkers do not grossly affect their activities. Thus, after deletion of IIA and IIB, the IIC domain alone still seems to form a functional transporter which binds and discriminates the various substrates like the intact IIMtl complex (8, 22).
In mutants which lack the protein kinase enzyme I and HPr, enzymes II cannot be phosphorylated and are unable to transport substrates at a rate sufficient to support growth (34). ptsHI mutants of Salmonella enterica serovar Typhimurium and Escherichia coli K-12 have been isolated which have increased rates of transport through an intact IIGlc (30, 40). In such mutants, uptake of free glucose occurred via facilitated diffusion, and fermentation required an ATP-dependent glucokinase. Thus, translocation and phosphorylation of the substrate can be uncoupled. Sequencing revealed that all uncoupling mutations mapped in loop 5 close to the conserved GITE motif of IIGlc. All mutated IIGlc enzymes had drastically lower affinities for glucose (≥100-fold lower) but were still able to phosphorylate the substrates in a pts+ background. Revertants which exhibited high levels of binding but were still uncoupled could be isolated (38). Some of the mutants transported glucose at a higher rate and with a higher affinity in the phosphorylated state than in the dephosphorylated state (39). A higher affinity was also observed with wild-type IIMtl in E. coli in the presence of phosphorylated Cys384 but not in the presence of dephosphorylated Cys384 (23). These and other results (references 19 and 32 and references therein) suggest that in PTS transporters high-affinity substrates are tightly bound in the IIC domain and can be released efficiently into the cytoplasm only in the presence of a IIB domain in which Cys384 or the equivalent residue is phosphorylated. Sometime during uptake, the phosphoryl group is transferred to the substrate, and substrate phosphate is consequently released into the cell.
In this paper we describe isolation and characterization of more than 40 uncoupled mutants with mutations in IIMtl of E. coli and Klebsiella pneumoniae. Selection of these mutants required construction of an artificial metabolic pathway for intracellular free d-mannitol and then either an intact IIMtl in a ΔptsHI strain or a truncated IIMtl from which the IIAMtl domain or the IIBAMtl domains had been deleted. The resulting mutations all mapped in loop 5. These mutations had characteristic effects on transport, phosphorylation, substrate affinity, and substrate specificity and helped to identify amino acids that have a central role in transport via this enzyme II.
MATERIALS AND METHODS
Chemicals.
d-[1-14C]mannitol (60 mCi/mmol) and d-[1-14C]glucitol (307 mCi/mmol) were purchased from Amersham Biosciences Europe GmbH, Freiburg, Germany, and the latter compound was purified from d-mannitol as described previously (15). All other chemicals were of commercially available analytical grades.
Bacterial strains, plasmids, and media.
The origins and relevant phenotypes and/or genotypes of most of the strains used in this study are shown in Table 1; the exceptions are standard strains and plasmids, such as HB101, pACYC177, pUC18, etc., which have been described by Sambrook et al. (42). To construct the F′ plasmid carrying the dalD gene, a 2.7-kb HindIII-EcoRI subfragment was excised from pGHL3 which carried the dalD+K+T+ genes from Klebsiella oxytoca M5a1. This fragment included the dalDp promoter, the entire dalD gene, and a truncated dalK′ gene. It was cloned next into pHEX3 (10), excised as a ClaI-SacI fragment, and cloned into the multiple-cloning site of pTIM101 (51). This vector contains the inverted repeat regions of transposon Tn1721, and between these inverted repeat regions are the multiple-cloning site of pBluescript II SK(+) and the so-called Ω-element with an spc gene encoding spectinomycin and streptomycin resistance. The new construct pLSD101, carrying the dalD+K′ genes within the artificial transposon TnLSD101, was transformed into strain CSH36/F′lac (26) together with pPSO110 (47), which carries the gene for the Tn1721 transposase to activate transposition. After 3 days of incubation, the transformants were mated with the Lac− recipient strain S136-3 with subsequent selection for Lac+ Spcr Kanr exconjugants which were also Aps Cms (i.e., cured of pPSO110). One of these purified exconjugants (STL 136) was used to conjugate an F′lac::TnLSD110 dalD+K′ plasmid (designated F′DalD) into other strains. Strain LTK31-2 is a derivative of LGS31 (8) which carries a Δ(ptsHIcrr)::kan cassette (51), and it contains the F′DalD plasmid and conjugative plasmid pUR404 synthesizing a d-fructokinase ScrK constitutively (1).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Origin and/or relevant properties | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| JWL181 | mtlAp,o50(Con) gutA50 gatA50 | 17 |
| JWL191 | ptsI191 | 17 |
| LGS322 | JWL181 ΔmtlA55 mtlD+ Δ(gut-rec)63 Atls | 8 |
| LGS323 | LGS322 Atlr Crr− | 10 |
| LGS324 | LGS322 Atlr | This study |
| LGS31 | JWL181 ΔmtlA55 mtlD+gutA50 | 8 |
| LTK31-2 | LGS31 Δ(ptsHI crr)::kan/F′DalD/pUR404 | This study |
| STL136 | lac-85 malA1 mel-2 rpsL Δ(ptsHIcrr)::kan/F′DalD | This study |
| Other strains | ||
| 1033-5P14 | K. pneumoniae | 9 |
| M5a1 | K. oxytoca | 25 |
| Plasmids | ||
| pGJ9 | mtlA+E. coli (637 amino acids) | 8 |
| pGJ9Δ137 | mtlAΔ137 (deleted after bp 1530, amino acid 510) | 8 |
| pGJ9ΔSnaBI | mtlAΔSnaBI (deleted after bp 1128, amino acid 376) | 8 |
| pDW1 | mtlA+ (bp 1 to 1128 deleted, amino acids 1 to 376) | 50 |
| pSOL300 | pHEX3 mtlA+K. pneumoniae (635 amino acids) | 28 |
| pSOL110 | pHEX3 ΔmtlA′ (deleted after bp 929, amino acid 361) | 28 |
| pGHL3 | pACYC 177 dalD+K+T+K. oxytoca | G. Heilenmann |
| pASL14 | pUC18 dalD+ K′K. oxytoca (2.7-kb HindIII-EcoRI fragment) | This study |
| F′DalD | F′laclacI proA+B+::TnLSD101 dalD+K′K. oxytocaSpr | This study |
| pTIM101 | pIC-19H::TnLSD | 51 |
| pLSD101 | pTIM101 Apr::TnLSD101 (dalD+K′ spc) | This study |
| pUR404 | scr+ ScrR− Tcr | 1 |
Plasmids pGJ9 and pSOL300 carry the intact mtlA gene from E. coli K-12 and the intact mtlA gene from K. pneumoniae 1033-5P14, respectively, each of which encodes a complete IICBAMtl complex (637 and 635 amino acids, respectively), while pGJ9Δ137 expresses a truncated IICBMtl (510 amino acids) from E. coli (8) and pSOL110 expresses a truncated IICMtl (361 amino acids) from K. pneumoniae (28). Derivatives of these plasmids with the various uncoupling mutations were designated accordingly (e.g., pGJ9 E218A or pGJ9Δ137 E218A).
Cells were routinely grown in complex tryptone broth, in Lennox broth with or without glucose and CaCl2, or in minimal phosphate-buffered medium (14, 46) and were tested for substrate utilization on MacConkey indicator plates containing the carbohydrate to be tested at a concentration of 1% (wt/vol). In minimal media, amino acids were added at concentrations of 20 mg/liter and carbohydrates were added at concentrations of 2 g/liter. The antibiotic concentrations used were as follows: ampicillin, 100 mg/liter; chloramphenicol, 25 mg/liter; kanamycin, 25 mg/liter; spectinomycin, 100 mg/liter; and tetracycline, 10 mg/liter.
Isolation of plasmid DNA, restriction analyses, and cloning procedures.
All manipulations with recombinant DNA were performed by using standard procedures (2). Plasmid DNA was prepared by the standard phenol extraction procedure (42) or by using the JETstar DNA purification system (Genomed, Bad Oeynhausen, Germany). Restriction enzymes were purchased from New England Biolabs (Schwalbach, Germany) and were used according to the recommendations of the supplier. The oligonucleotides used for sequencing or PCRs were obtained from Interactiva (Ulm, Germany).
Site-directed mutagenesis and DNA sequencing.
DNA amplification of the various mtlA genes was performed as described by Saiki et al. (41), and site-directed mutagenesis was performed with the Altered Sites II in vitro mutagenesis system (Promega Corp., Mannheim, Germany) by using phosphorylated mutagenesis primers and single-stranded DNA replicated by using strain BMH71-18 according to the recommendations of the supplier. Plasmid pGJ9 was digested with HindIII, and the resulting 2.1-kb fragment carrying the 5′-terminal mtlA′ portion was cloned into pALTER-1. During mutagenesis, frequent deletions were observed and were found to originate from an 11-bp repeat (5′-GGCGTGCTGCT-3′) present in the mtlA gene (bp 676 to 686) and in the tetA gene (bp 876 to 886) of the vector. The primers were designed such that two bases were exchanged in the target codon (i.e., GAA to GCC for E218A, GAA to GTC for E218V, and CAC to CCA for H256P). The corresponding primers were 5′-CAATCTTCTTCCTGATTGCCGCTAACCCAGGTCCAGG-3′, 5′-CAATCTTCTTCCTGATTGTCGCTAACCCAGGTCCAGG-3′, and 5′-CTTCCTGGGGGGTATCCCAGAAATCTACTTCCCG-3′. The mutated sequences in the new vectors (designated pGAL3-1, pGAL3-2, and pGAL3-3) were confirmed by sequencing. PCR products were purified by using the Wizard PCR preps DNA purification system (Promega Corp.). All DNA sequencing reactions were performed by the dideoxy chain termination method by using an ALF express Auto Read or dATP labeling mix sequencing kit from Amersham Biosciences and determining the sequences of both strands. A computer analysis was done with the DNASIS sequence analysis software (Hitachi) and by using the BLAST programs and database services of the National Center for Biotechnology Information, Bethesda, Md.
Transport, binding, phosphorylation, and enzyme assays.
Transport of 14C-labeled d-mannitol, d-glucitol, or d-arabinitol (usually at a final concentration of 10 μM) was tested by using exponentially growing cells, and the activities were calculated from the initial uptake rates (0 to 30 s) as described previously (10, 44). The tests used for the d-mannitol 1-phosphate dehydrogenase (MtlD) and for the d-arabinitol dehydrogenase (DalD) have also been described previously in detail (15, 28).
Membrane vesicles from strain LGS322 with the various mtlA-carrying plasmids were prepared as described previously (22), and their PEP-dependent mannitol phosphorylation activity was determined as described by Robillard and Blaauw (36) by using saturating amounts of enzyme I and HPr and 500 μM d-[3H]mannitol unless indicated otherwise. Binding of d-[3H]mannitol was tested by using membrane vesicles after solubilization with 0.25% pentaethylene glycol monodecyl ether (dPEG) and the flow dialysis technique, which measures the free concentration of mannitol in equilibrium with the mannitol-enzyme complex. The total d-[3H]mannitol concentration used in the Scatchard analyses varied from 26 to 524 nM, the temperature was 25°C, and the tests were performed as described previously (22), with some modifications (48). Normal phosphorylation tests with purified membranes and cell extracts containing enzyme I, HPr, and d-[3H]mannitol at various concentrations were performed as described previously (15).
Cell extraction and chromatography.
Cells of the various strains growing exponentially in Lennox broth without glucose and CaCl2 were harvested, washed, and extracted as described previously (28). In essence, washed cells resuspended in 0.5 ml of minimal medium at a concentration of 1010 cells per ml at 25°C were incubated for 5 min with 6.4 × 105 cpm of d-[14C]mannitol at a concentration of 280 μM. After two careful washes with 0.5 ml of medium with the supernatants removed completely each time, the cells were resuspended in 50 μl of H2O and heated for 2 min in a boiling water bath, and the cell debris was removed by centrifugation. Portions (5 μl) of the supernatants were applied to Nono-Sil G plates (Macherey-Nagel, Düren, Germany) and developed in 1-butanol-ethanol-H2O (3:1:1). The spots were visualized by autoradiography and were identified by comparison to known substrates and intermediates, or they were excised from the plates and resuspended in scintillation fluid for quantitative determinations.
RESULTS
Construction of strains and plasmids for selection of uncoupled IIMtl mutants.
Strain E. coli K-12 lacks a degradative pathway for free intracellular d-mannitol (14, 46). Therefore, mutants of K-12 lacking either the d-mannitol (IIMtl) and d-glucitol (IIGut) PTS or the pts genes were unable to grow on the PTS carbohydrate d-mannitol (Table 2). A non-PTS catabolic pathway for this polyhydric alcohol has been described for various enteric bacteria (6, 15, 35, 46). This pathway relies on constitutively expressed genes and enzymes for d-arabinitol (Atl) catabolism. Free mannitol is taken up via an H+ symporter (the gene is designated atlT in E. coli C and dalT in Klebsiella) and is converted by a soluble NAD+-dependent d-arabinitol dehydrogenase (encoded by atlD or dalD) into d-fructose, while DalD converts d-arabinitol into d-xylulose (9). The d-xylulose is phosphorylated by an ATP-dependent kinase (encoded by dalK) to d-xylulose 5-phosphate, while fructose is phosphorylated in pts+ strains by the d-fructose PTS or the d-mannose PTS. Alternatively (e.g., in mutants lacking the pts genes), an ATP-dependent fructokinase (encoded by scrK) of sucrose metabolism may be used (1). Thus, uncoupled IIMtl mutants should grow on free mannitol when a transporter, the DalD dehydrogenase, and (where necessary) the ScrK kinase are supplied.
TABLE 2.
Properties of the selection strains
| Strain | Plasmid | Growth ona:
|
IIMtl phosphorylation activity (nmol/min/mg of total membrane protein)b | MtlD activity (nmol/min/mg of total protein)b | DalT activity (nmol/min/mg of total protein)b | DalD activity (nmol/min/mg of total protein)b | |
|---|---|---|---|---|---|---|---|
| Mtl | Atl | ||||||
| JWL181 mtl+gutA50 | None | 3+ | s | 9.8 | 2.20 | ≤0.1 | ≤0.01 |
| pASL14 | 3+ | 2+ | NT | NT | ≤0.1 | 1.20 | |
| LGS322 ΔmtlA55 Δ(gut-rec)63 Atls | None | − | s | ≤0.1 | 0.73 | ≤0.1 | ≤0.01 |
| pGHL3 | 2+ | 3+ | ≤0.1 | NT | 10.0 | 1.19 | |
| pASL14 | − | 2+ | ≤0.1 | NT | ≤0.1 | 1.18 | |
| F′DalD | − | + | ≤0.1 | NT | ≤0.1 | NT | |
| LGS323 ΔmtlA55 Δ(gut-rec)63 Crr− Atlr | None | − | − | ≤0.1 | 0.75 | ≤0.1 | ≤0.01 |
| pGHL3 | 2+ | 3+ | ≤0.1 | NT | 9.5 | 1.18 | |
| pASL14 | − | − | ≤0.1 | NT | ≤0.1 | 1.21 | |
| F′DalD | − | − | ≤0.1 | NT | ≤0.1 | NT | |
| JWL191 ptsI191 | None | − | − | ≤0.1 | 1.52 | ≤0.1 | NT |
| pGHL3 | 2+ | 3+ | ≤0.1 | NT | 3.5 | 0.45 | |
| pASL14 | − | 3+ | ≤0.1 | NT | ≤0.1 | 0.40 | |
Growth on d-mannitol (Mtl) and d-arabinitol (Atl) was tested on MacConkey indicator plates containing 1% carbohydrate. −, white colonies; +, pink colonies; 2+ or 3+, purple or deep purple colonies; s, sensitive.
Cells in the exponential growth phase on minimal glycerol (0.2%, vol/vol) medium were harvested and tested as described in Materials and Methods. The IIMtl phosphorylation activity was tested with 30 μM d-mannitol, the d-mannitol-1-phosphate dehydrogenase (MtlD) activity was tested with 2 mM d-mannitol, the d-arabinitol transporter (DalT) activity was tested with 500 μM d-[14C]mannitol, and the d-arabinitol dehydrogenase (DalD) activity was tested with 2 mM d-arabinitol. NT, not tested.
Strain JWL191 (ptsI191) lacks enzyme I of the PTS and hence all PTS-dependent transport activities. In contrast, strain LGS322 carries the ΔmtlA55 and Δ(gut-recA)63 deletions and consequently lacks IIMtl and IIGut (8). As expected, both of these strains were Mtl−, and upon transformation with pGHL3 (dalDp dalD+K+T+), which carries the dal genes from K. oxytoca M5al and allows constitutive expression of the d-arabinitol enzymes, they became Mtl+ and Atl+ (Table 2). After transformation with pASL14, a derivative of pGHL3 from which the dalT and dalK genes were deleted, the strains remained Mtl− but were Atl+. During this study it became clear that growth of these DalT− mutants on d-arabinitol required the presence of constitutively expressed IIMtl, which was caused by the mtlAp,o50(Con) allele present in JWL181 and its derivatives, JWL191 and LGS322 (17). Alternatively, the glucose PTS (IIGlc) may be used (4). These three strains also have a mutation in the ptsG locus which causes induction of IIGlc during growth in the presence of d-arabinitol (51). As discussed below, a substantial part of the transported substrate must be in the nonphosphorylated form. Free d-arabinitol can be converted by DalD into d-xylulose and by ATP-dependent kinases into d-xylulose 5-phosphate, thus allowing growth on the pentitol. In the absence of DalD, the accumulating d-arabinitol is apparently converted by the endogenous d-xylulose kinase XylK into toxic d-arabinitol 5-phosphate (35), thus causing the Atls phenotype of the strains (Table 2). Consequently, the IIMtl-negative mutant LGS322 still grew on d-arabinitol when it was provided with DalD, while the double mutant LGS323, which also lacked IIGlc, remained Atl−. The ptsI mutant JWL191 supplied with DalD was also Atl+, thus supporting the observation that the transported substrate (or parts of the transported substrate) was nonphosphorylated (4). The Atlr phenotype of this strain in the absence of DalD, however, may be caused either by a lower XylK kinase activity or by the lack of PTS-dependent phosphorylation during uptake. Most strains containing pASL14 were highly unstable during growth in the presence of mannitol or d-arabinitol; an exception was LGS323. Later in this study, we used a single-copy plasmid (F′DalD) which carried the dalD+K′-containing DNA fragment of pASL14, expressed the DalD activity at a normal level, and in LGS323 gave more stable transformants for selection of uncoupled IIMtl mutants.
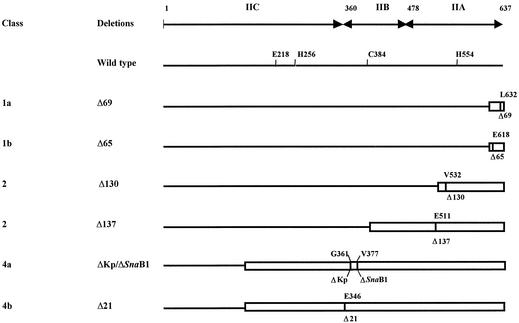
To identify more precisely the functions of the IIC, IIB, and IIA domains in substrate recognition, transport, and phosphorylation, the exact extents of several C-terminal deletions isolated previously (8) and their effects on the various functions were determined (Fig. 1 and Table 3). The class 1 deletions, including Δ69 and Δ65, ended between residues 637 and Leu611; the class 2 deletions, including Δ130, ended between residues Leu611 and Gln520; the class 3 deletions, including Δ137, ended between residues Gln520 and Met393; and the class 4 deletions, including ΔSnaBI and Δ21, ended between residues Met393 and Leu161. Thus, the class 1 and 2 deletions ended within the IIA domain, the class 3 deletions ended in the IIB domain, and the class 4 deletions ended beyond residue 360 in the IIC domain. The ΔKp deletion used for the K. pneumoniae constructs (pSOL110) ended at Gly361 within the linker joining the IIB and IIC domains (Fig. 1).
FIG. 1.
Deletions of the IIMtl complex of E. coli K-12. The locations of domains A, B, and C of IIMtl are indicated together with the locations of the essential residues E218, H256, C384, and H554 and the locations of the approximate borders between the domains. The longest deletions in the four deletion classes isolated previously are L611 to K637 in class 1, Q520 to K637 in class 2, M393 to K637 in class 3, and L161 to K637 in class 4. Representative deletions for each class are Δ69 (ΔL632-K637) and Δ65 (ΔE618-K637) for class 1, Δ130 (ΔV532-K637) for class 2, Δ137 (ΔE511-K637) for class 3, and ΔSnaBI (ΔV377-K637) and Δ21 (ΔE346-K637) for class 4. In K. pneumoniae, ΔKp (ΔG361-K635) was used as a representative of class 4 and corresponds to the allele present in pSOL110 (Table 1).
TABLE 3.
Transport and phosphorylation activities of various IIMtl deletion mutantsa
| Deletion | Growth on Atl | Growth on Mtl | Transport (nmol/min/mg of protein) | Phosphor- ylation (%) | Trans- phosphor- ylation | Binding |
|---|---|---|---|---|---|---|
| None (wild type) | 2+ | 3+ | 9.0 | 100 | + | + |
| Δ69 | 2+ | + | 6.0 | 20 | + | + |
| Δ65 | 2+ | − | ≤0.1 | 0 | + | + |
| Δ130 | 2+ | − | ≤0.1 | 0 | + | + |
| Δ137 | 2+ | − | ≤0.1 | 0 | + | + |
| ΔSnaBI | 2+ | − | ≤0.1 | 0 | − | + |
| ΔKp | 2+ | − | ≤0.1 | 0 | NTb | NT |
| Δ21 | − | − | ≤0.1 | 0 | − | − |
The deletions (Fig. 1) were transformed into strain LGS323/F′DalD to test for growth on d-arabinitol (Atl) and into strain LGS323 to test for growth on d-mannitol (Mtl) in MacConkey indicator plates with 1% carbohydrate. −, white colonies; +, pink colonies; 2+ or 3+, purple or deep purple colonies. d-Mannitol transport activity with intact cells and PEP-dependent phosphorylation activity (100% was equivalent to 11 nmol per min per mg of membrane protein) with crude cell extracts were tested with 25 and 30 μM d-mannitol, respectively. The data for transphosphorylation activity and d-mannitol binding activity were obtained from reference 8.
NT, not tested.
Deletion of only five C-terminal residues (Δ69) decreased transport and phosphorylation of mannitol, and both activities disappeared upon deletion of 19 residues (Δ65) or more residues (class 1b) (Table 3). In contrast, the transphosphorylation activity was inactivated only by deletions which removed both phosphorylation sites (e.g., ΔSnaBI), while binding of mannitol remained intact (8) until parts of a series of charged residues (residues 335 to 370) were deleted (e.g., Δ21). These residues link the IIC and IIB domains and may be an essential part of the second phosphorylation site which includes Cys384 (20). As noted above, the high-affinity substrate d-mannitol was not transported via a nonphosphorylated IIMtl at a rate sufficient to allow growth in the presence of DalD (strain JWL191/pASL14 in Table 2); i.e., the transporter remained in the closed state. Surprisingly, however, the low-affinity substrate d-arabinitol (Km, >500 μM) (16, 50) allowed fast growth (doubling time, about 85 min) in the presence of wild-type IIMtl, whether this complex was in the phosphorylated state or the nonphosphorylated state and also whether it was complete or truncated as long as the IIC domain remained complete (classes 1 to 4a) and DalD was present (Table 2). Furthermore, growth on d-arabinitol required neither an intact enzyme I of the PTS nor the kinase ScrK. Based on these results, we concluded that the isolated IIC domain not only bound mannitol normally, but it must have formed a functional complex which transported arabinitol efficiently.
Isolation and characterization of uncoupled IIMtl mutants of E. coli and K. pneumoniae.
To increase the variety of isolated mutations, different selection conditions and strains were used. Thus, the cells were plated on MacConkey Mtl indicator plates and on minimal media containing 5 to 50 mM d-mannitol as a single carbon source and were incubated at 30, 37, and 42°C. In later selections, the unstable multicopy plasmid pASL14 encoding DalD was replaced by the stable single-copy plasmid F′DalD. Analogous to the previous IIGlc studies (30), strain LTK31-2(ΔptsHIcrr::kan)/F′DalD, which expressed the ATP-dependent d-fructokinase ScrK from pUR404, was used first as a host. In addition, we speculated that mutant strains that lacked IIMtl and IIGut, including LGS322 and LGS323 [ΔmtlA55 Δ(gut-rec)63], but expressed the dehydrogenase DalD could also be used when they were transformed with plasmids encoding truncated forms of IIMtl which lack one (H554) or both (H554 and C384) phosphorylation sites. The advantage of this technique was that the faster-growing pts+ strains and mtlA genes from different enteric bacteria could be used. The corresponding plasmids were pGJ9Δ137 for the mtlA gene from E. coli K-12 and pSOL110 for K. pneumoniae (Fig. 1), which have 85% identical base pairs and whose IIMtl sequences have 95% identical amino acid residues (28).
Using nonmutagenized cells, we selected the following mutants from independent selection batches within 2 weeks after inoculation or plating: eight independent Mtl+ mutants of strain LTK31-2/F′DalD/pUR404, 33 Mtl+ mutants of LGS323/F′DalD/pGJ9Δ137, and 40 Mtl+ mutants of LGS323/F′DalD/pSOL110. After the Mtl+ colonies were purified, their small plasmids were isolated and retransformed into strains LGS323 and LGS323/F′DalD. All transformants in LGS323 remained Mtl−, while all LGS323/F′DalD transformants were Mtl+. The various uncoupling mutations thus must map on pGJ9Δ137 and pSOL110, respectively. Based on their phenotypes, which ranged from Mtl+ to Mtl3+ (Table 4), the 81 mutants were grouped into four classes. Sequencing of the corresponding mtlA fragments revealed the following mutations and amino acid exchanges: three times there was a CAT-to-TAT transition at nucleotide 765 causing an H256Y exchange in the K. pneumoniae gene; 21 times there was a CAT-to-CCT transversion at nucleotide 766 in the K. pneumoniae gene causing an H256P exchange in the GIHE motif; 13 times there was a CAC-to-CCC transversion at nucleotide 767 in the E. coli gene causing an H256P exchange in the GIHE motif; twice there was a GAA-to-GTA transversion at nucleotide 653 in the E. coli gene causing an E218V exchange; 26 times there was a GAA-to-GCA transversion at nucleotide 653 in the E. coli gene causing an E218A exchange; and 16 times there was a GAA-to-GCA transversion at nucleotide 653 in the K. pneumoniae gene causing an E218A exchange. Thus, despite various selections and the large number of mutants, only four different uncoupling mutations were found in the two bacteria; however, all of these mutations were located in loop 5, and two affected the highly conserved GIHE motif.
TABLE 4.
Uncoupling mutations obtained by localized mutagenesisa
| Mutation | Plasmid | LGS322
|
LGS322/F′DalD
|
||||
|---|---|---|---|---|---|---|---|
| Doubling time (min) on Mtl | Growth yield (OD600) on Mtl | Growth on Atl | Doubling time (min) on Mtl | Growth yield (OD600) on Mtl | Growth on Atl | ||
| None (wild-type E. coli) | pGJ9Δ137 | NG | NG | s | NG | NG | 2+ |
| pGJ9 | 85 | 8.0 | s | 85 | 7.7 | 2+ | |
| None (wild-type K. pneumoniae) | pSOL110 | NG | NG | s | NG | NG | 2+ |
| pSOL300 | 100 | 6.7 | s | 100 | 6.5 | 2+ | |
| E218A | pGJΔ137 | NG | NG | s | 200 | 3.5 | 2+ |
| pGJ9 | 190 | 4.2 | s | 117 | 7.0 | 2+ | |
| E218V | pGJ9Δ137 | NG | NG | s | 190 | 4.0 | 2+ |
| pGJ9 | 300 | 2.7 | s | 110 | 6.8 | 2+ | |
| H256P | pGJ9Δ137 | NG | NG | s | ≤300 | 2.5 | 2+ |
| pGJ9 | NG | NG | s | 280 | 3.9 | 2+ | |
| H267Y | pSOL114 | NG | NG | s | 250 | 2.5 | 2+ |
| pSOL314 | 105 | 7.0 | s | 85 | 6.9 | 2+ | |
The mutations were generated by localized mutagenesis in the complete or truncated mtlA gene of E. coli K-12 by exchanging 2 bp in the corresponding triplets to minimize reversions. The H256Y allele was obtained by selection in the mtlA gene of K. pneumoniae and was also present in the complete form (pSOL300) and in the deleted form (pSOL110). Cells of strains LGS322 and LGS322/F′DalD transformed with the plasmids were tested for growth in minimal d-mannitol (0.2%) medium (Mtl) and on MacConkey indicator plates containing d-arabinitol (Atl). s, sensitive; 2+, purple or deep purple colonies; OD600, optical density at 600 nm; NG, no growth.
Effect of uncoupling mutations in complete IIMtl complexes.
As stated above, transformants of strain LGS323/F′DalD containing plasmids p6J9ΔSnaBI and pSOL110, which lack both phosphorylation sites, were Mtl− but Atl+, while LGS323 transformants were Mtl− Atl− (Table 3). These large deletions can be complemented by pDWI, which expresses the IIBAMtl domains independently (50). Transformants of LGS323/pDWI containing either pG79Δ137 or pGJ9Δ137 E218A were Mtl+ (doubling time, about 90 min) in the absence of DalD (data not shown). Obviously, the wild-type and uncoupled IIC domains still catalyzed d-mannitol phosphorylation when they were complemented with intact IIBAMtl domains. However, cells containing several plasmids were highly unstable and prone to artifacts in tests. Therefore, plasmids encoding complete IICBAMtl transporters with the uncoupling mutations were constructed. The 5′-terminal AvaI-SnaBI fragments of the various mutants of E. coli encoding amino acid residues 1 to 377 were substituted for the wild-type fragment in pGJ9 to obtain, e.g., pGJ9 E218A, and similarly, these fragments were substituted for an EcoRV fragment comprising bp 13 to 1032 (amino acid residues 4 to 344) of mtlA from K. pneumoniae in pSOL300 to obtain plasmids pSOL312 to pSOL314. The correct orientations of all fragments and the mutations were ascertained by sequencing. The new constructs not only allowed analysis of their transport and phosphorylation activities but also comparisons of IIC activities in the presence and in the absence of the IIBA domains.
Besides being very unstable, some of the constructs had a strong tendency to revert to the wild type when they were cloned on high-copy-number plasmids. Furthermore, some mutations with identical substitutions seemed to differ in the variants of E. coli and K. pneumoniae. Therefore, double exchanges were introduced by localized mutagenesis into the mtlA gene from E. coli (in particular, GAA to GCC for the E218A mutation, GAA to GTC for the E218V mutation, and CAC to CCA for the H256P mutation). All double exchanges were introduced both into the truncated mtlA′ gene on pGJ9Δ137 and into the complete mtlA gene on pGJ9 and were verified by sequencing. Transformants of strain LGS322 with the new plasmids had phenotypes on mannitol plates similar to those of the mutants obtained through selection. Thus, only the phenotypes of the new mutants in LGS322 are shown in Table 4; in addition, the H256Y mutant (pSOL314) was found only in K. pneumoniae. This plasmid allowed growth on mannitol with nearly normal doubling times and yields which were not increased by the presence of DalD. The pGJ9 E218A transformants had somewhat shorter doubling times and lower growth yields on mannitol, the pGJ79 E218V transformant had a markedly shorter doubling time and lower growth yield, and the pGJ9 H256P transformants exhibited no detectable growth. In the presence of DalD, both E218 mutants had nearly normal doubling times and growth yields, and the H256P mutant grew on mannitol, although the growth was reduced. In the host LGS323 the growth behavior was nearly identical to that in LGS322, and in particular, all variants showed growth on d-arabinitol provided that DalD was also present (Table 4 and data not shown).
Further characterization of uncoupled mutants.
While the phenotypes of the mutants were for the most part similar regardless of whether they had been obtained through selection or through localized mutagenesis, the initial results of the transport and PEP-dependent phosphorylation tests were not as clear. Thus, LGS322 transformants with pGJ9 E218A consistently lacked transport activity for mannitol when concentrations in the micromolar range were used and showed only some transport and phosphorylation activity (about 15%) at higher substrate concentrations (the apparent Km was 750 μM, compared to 4 μM for pGJ9). In contrast, transformants with pSOL313 which carried the same E218A mutation in the K. pneumoniae gene had nearly normal transport activities and a Km that was approximately twice the wild-type Km (Table 5). Because the high-copy-number plasmid pGJ9 E218A was very unstable compared to the lower-copy-number plasmid pSOL313, the E. coli DNA was recloned into the same pHEX vector as pSOL313 and transformed into LGS323. After this, both the E. coli and K. pneumoniae E218A alleles conferred nearly normal transport activities to these constructs, and the apparent Km values were not more than twice the wild-type value. We assumed, therefore, that these values represent the real phenotype of the E218A uncoupled mutants, and only these values are given in Table 5. With the E218V allele from E. coli and the H256P alleles from both bacteria, no transport (tested at a concentration of 10 μM) and only low phosphorylation activity (about 10% at a concentration of 500 μM), if any, could be detected. The activities of the fourth allele (H256Y) were nearly normal compared to the activities of the wild type, except for the transport activity that had an apparent Km that was more-than-20-fold increased (58 μM) (Table 5).
TABLE 5.
Transport and related activities in various uncoupled mutantsa
| Allele of mtlA | Transport
|
Phosphor- ylation (%) | Binding constant (nM) | Free intracellular mannitol | Uncoupling | ||
|---|---|---|---|---|---|---|---|
| Activity (nmol/min/mg of total protein) | Km (μM) | Vmax (nmol/min/mg of total protein) | |||||
| E. coli K-12 | |||||||
| Wild type | 9.5 | 4 | 82 | 100 | 40 | No | No |
| E218A | 11.5 | 5 | 25 | 110 | —b | NT | Yes |
| E218V | ≤0.1 | 12 | No | NT | Yes | ||
| H256P | ≤0.1 | ≤1 | No | NT | Yes | ||
| K. pneumoniae | |||||||
| Wild type | 3.1 | 1.2 | 33 | 100 | NT | No | No |
| E218A | 3.4 | 2.9 | 59 | 100 | NT | No | Yes |
| H256P | ≤0.1 | 14 | NT | Yes | Yes | ||
| H256Y | NT | 58.0 | 31 | 100 | NT | No | Yes |
Cells of strain LGS324 transformed with plasmids carrying the complete mtlA gene either from E. coli K-12 on pGJ9 or from K. pneumoniae on pSOL300 in the wild-type and mutated forms were grown exponentially in minimal glycerol medium. Whole cells were used to determine the transport activity with 10 μM d-[14C]mannitol and to determine the apparent Km values and the corresponding Vmax values. Phosphorylation activities (100% corresponded to 278 nmol per min per mg of membrane protein) were determined with 500 μM d-[14C]mannitol or were estimated based on thin-layer chromatography tests as described in the text. The binding constants were determined by flow dialysis as described in Materials and Methods with membrane vesicles. The presence of free intracellular mannitol was determined by thin-layer chromatography (Fig. 2), and coupling was determined by measuring the growth on d-mannitol of transformants expressing the wild-type or mutated truncated IIC domains in the presence of DalD. NT, not tested.
—, see text.
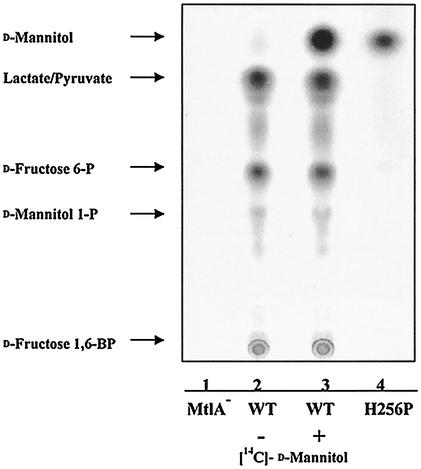
According to the model proposed by Lolkema et al. (21, 24), a complete and phosphorylated IIMtl system transports approximately 50% free mannitol, which is trapped immediately and phosphorylated in a subsequent step by other IIMtl transporters. Consequently, in wild-type cells only mannitol 1-phosphate is detected intracellularly. We wanted to confirm and extend these results, which were obtained with membrane vesicles, by using intact cells and uncoupled as well as wild-type IIMtl transporters. When cells of an MtlD− mutant lacking the mannitol 1-phosphate dehydrogenase were incubated with 14C-labeled mannitol (280 μM), the corresponding cell extracts invariably contained about 95% mannitol 1-phosphate and about 5% free intracellular mannitol as determined by using thin-layer chromatography plates (data not shown). The percentage of free mannitol increased somewhat with the boiling time used to lyse the cells, and this may indicate that there was some hydrolysis of mannitol 1-phosphate during boiling. Another possible explanation is that accumulating intracellular concentrations of mannitol 1-phosphate increasingly inhibited phosphorylation of the free intracellular mannitol. When the MtlD+ strain LGS323 was transformed with the wild-type mtlA allele present in pSOL300, mannitol 1-phosphate was metabolized almost quantitatively to fructose 6-phosphate and to C3 compounds, and no free intracellular mannitol was detected (Fig. 2). Similar results were obtained with the uncoupled alleles present on pSOL313 and pSOL314. However, not only did the mutant allele H256P present in pSOL312 result in drastically reduced transport and phosphorylation activity (Table 5), but most of the intracellular product found in cell extracts was free mannitol (Fig. 2). Thus, uncoupled mutants could be used to visualize transport of free mannitol via a complete IICBAMtl transporter provided that they had drastically reduced phosphorylation activity. Unfortunately, the technique could not be used with wild-type transporters and intact cells to prove or disprove the model of Lolkema et al. (21, 24) by direct tests. Nor could the ratio of d-arabinitol to d-arabinitol 5-phosphate be directly determined as these compounds accumulated intracellularly during transport via an intact and phosphorylated IIMtl, because radioactively labeled d-arabinitol of sufficient quality was not available.
FIG. 2.
Intracellular d-[14C]mannitol in cell extracts from wild-type and uncoupled mutant strains. Cells of strain LGS323 [ΔmtlA55 Δ(gut-rec)63] (lane 1) and cells of transformants of this strain with the wild-type allele of mtlA on pGJ9 (lanes 2 and 3) or with the uncoupled allele on pGJ9 H256P (lane 4) were incubated for 5 min with 280 μM d-[14C]mannitol at 25°C, washed, and extracted as described in Material and Methods. Portions (5 μl) of the supernatants were applied to Nono-Sil G plates and developed in 1-butanol-ethanol-water (3:1:1). In lane 3, 1.6 × 104cpm of d-[14C]mannitol was added to each of the cell extracts before application to the gel plates. Free d-mannitol and several metabolic derivatives of mannitol 1-phosphate in the wild type were visualized by radioautography. WT, wild type.
DISCUSSION
The substrate-specific enzymes II of the different PTSs catalyze concomitant transport and phosphorylation of their substrates during uptake. The two processes have long been considered tightly coupled, mostly because nonphosphorylated enzymes II were unable to transport their high-affinity substrates at rates sufficient to support efficient growth. However, early experiments on the role of enzymes II during induction, the efflux of PTS carbohydrates from cells, and the transport of steric analogues (e.g., the transport of nonphosphorylated d-galactose via the IIGlc and IIMan transporters in enteric bacteria) indicated that this coupling might not be absolute (reference 31 and references therein). The successful separation of the two phosphoryl transfer domains IIABMtl from a still active membrane-bound IICMtl domain provided further evidence that transport and phosphorylation should be considered two consecutive processes which may be uncoupled (8, 22). According to these data and the results described in this paper, the isolated IICMtl domain is an autonomous transporter that is able to distinguish and to bind its various substrates with normal affinity but is unable to translocate the high-affinity substrate d-mannitol (apparent Km, 1 to 4 μM) at a high rate. In contrast, the low-affinity C5 isomer d-arabinitol (apparent Km, ≥500 μM) (16, 50) was transported via the truncated but otherwise wild-type IICMtl transporter at a rate that allowed doubling times of up to 85 min for transformants (Tables 3 and 4). The major conclusions based on these results were that the IICMtl domain alone formed a stereospecific translocator that was able to transport d-arabinitol but not d-mannitol and that neither deletion of the IIA and IIB domains nor the absence of PTS-dependent phosphorylation was sufficient to convert IICMtl into an efficient transporter for this high-affinity substrate.
E. coli and S. enterica mutants in which glucose transport and phosphorylation via a complete IIGlc are uncoupled have been isolated (40). We extended these studies by using intact IIMtl complexes or truncated IICBMtl and IICMtl transporters from E. coli and K. pneumoniae and selecting for growth on d-mannitol through a non-PTS catabolic pathway. Despite different selection conditions and sequencing of many mutated mtlA variants of both organisms, only four unique mutations in two amino acid residues were found. Most of the mutations affected Glu218, a residue present in all functional IIMtl enzymes but not in the inactive IICmt enzyme, which has an E218A mutation. This mutation is also the most frequent mutation (52%) found among our uncoupled mutants The E218A allele from E. coli, as well as the E218A allele from K. pneumoniae in its truncated form, encoded an efficient transporter for free d-mannitol, as determined by growth in the presence of DalD (Tables 3 and 4). The effect on transport, phosphorylation, and substrate binding of the E218V mutation, which in terms of growth resembled E218A, was much more severe in the intact complex. Even more drastic were the differences for the H256P mutation, the second most frequent allele in both organisms. Finally, the isolocal mutation H256Y with nearly normal growth activity resulted in a markedly increased Km in transport tests and resembled the mutations in many IIGlc mutants that also show lower affinities (40). Obviously, residues E218 and H256 play essential roles in coupling transport and phosphorylation and perhaps also in binding of the substrates in the IIC transporter. Thus, alleles E218A, E218V, and H256P resulted in no binding of mannitol (at a concentration of 500 μM) in direct tests (Table 5). However, for allele E218A, which has high transport activity, the absence of binding was perhaps an artifact due to the plasmid segregation problems discussed above. The mutations of H256, a member of the GIHE motif, corroborate the hypothesis (16, 49) that the highly conserved amino acids have an essential role in transport and phosphorylation.
The model of Lolkema et al. (21, 24) states that mechanistically, the coupling between transport and phosphorylation is less than 50% even for wild-type IIMtl complexes but that free mannitol is captured and phosphorylated immediately at the inside of the membrane. This hypothesis coincides with our results which showed that there was free intracellular mannitol only in the uncoupled mutants with negligible phosphorylation activity (Fig. 2). The model states further that only phosphorylated IIMtl catalyzes facilitated diffusion at a high rate; i.e., the phosphorylation state of the IIBAMtl domains modulates the activity of the translocator IIC (e.g., by drastically lowering the activation energy for the translocation step). This again coincides with our results showing that the wild-type IIC translocator does not translocate d-mannitol efficiently. In the uncoupled mutants, neither phosphorylation nor the presence of the IIBAMtl domains is needed for efficient mannitol transport. Possibly, the mutations activate IIC, similar to phosphorylation in the wild type. In this context it should be pointed out that based on growth experiments, the low-affinity analogue d-arabinitol is transported in the nonphosphorylated form at a rate sufficient to allow fast growth via wild-type and uncoupled IICMtl transporters whether the transporters are complete or lack IIBAMtl and whether they are phosphorylated or not phosphorylated (Table 4). Based on similar growth experiments, other non-PTS substrates which are low-affinity stereoisomers of natural PTS substrates are also transported in the nonphosphorylated form via enzymes II; for example, d-galactose and trehalose are transported via IIMan in S. enterica (29, 33), d-galactose, d-arabinitol, ribitol, and d-fructose are transported via IIGlc in E. coli (4, 11, 12, 51), and d-arabinitol and xylitol are transported via IIGat (our unpublished results). Together with the observation that uncoupling mutations often affect the binding and lower the affinity of the transporter for its natural substrates (40) (Table 5), this seems to indicate that binding efficiency and the coupling between transport and phosphorylation may be related (19). According to the model, substrates that enter IICMtl from the periplasmic site and bind with high affinity lock the translocator in a closed state. In wild-type enzymes II, a locked-in IIC domain with its trapped substrate molecule is converted by means of a phosphorylated IIB domain into the open form, thus allowing efficient translocation of the substrate into the cell. Low-affinity substrates in wild-type IIC transporters and high-affinity substrates in uncoupled mutants apparently do not lock in. In uncoupled mutants with decreased binding affinity, mannitol is a low-affinity substrate; i.e., it fails to lock in. Uncoupled mutants with nearly normal binding properties cannot be explained in this way. However, in all these cases substrates are transported at an increased rate by facilitated diffusion even in the absence of the phosphorylated IIB domain. In accordance with these observations and the results obtained for IIGlc (27), a C384S exchange did not allow facilitated diffusion of mannitol via an otherwise wild-type IIMtl system into the cell, as measured by growth in strain LGS323/F′DalD (our unpublished results). This contrasts with the results of Lolkema et al. (23, 24) obtained with vesicles. Finally, the intracellular ratio of free mannitol to mannitol 1-phosphate is very low in wild-type strains because the free mannitol molecules are recaptured and phosphorylated immediately. This ratio thus depends primarily on the affinity of the free substrate for the intracellular substrate binding sites of the IIMtl complexes and on the properties of the d-mannitol 1-phosphate dehydrogenase MtlD. Accordingly, free intracellular mannitol could be found in uncoupled mutants only if the phosphorylation activity was low, as it is in mutant H256P (Table 5). The C5 isomer d-arabinitol was apparently translocated more efficiently by all IICMtl variants. Because it lacks C-1 and hence the normal phosphorylation site for IIMtl substrates, this compound should accumulate intracellularly in the free form and thus allow growth in the presence of DalD. In contrast, 75% of the low-affinity C2 isomer d-glucitol (apparent Km, 2 mM) (16) was found as d-glucitol 6-phosphate when it was transported via IIMtl (our unpublished results). Similarly, the differences between mutants E218A and E218V and between mutants H256P and H256Y suggest that binding, translocation, and phosphorylation are affected in different ways and to different degrees.
The PTSs have traditionally been classified as a third group of transporters which mechanistically resemble neither ion-driven symporters and antiporters nor ATP-driven ABC transporters. In view of the results described in this paper, more analogies to substrate-specific uniporters which catalyze facilitated diffusion, and even to ABC transporters, become obvious (reference 19 and references therein). In the latter case, the free energy of ATP is apparently used to lower the energy barrier for a substrate molecule bound in a high-affinity binding site, thus causing accumulation of free substrate in the cell following the release of ADP and inorganic phosphate (7). In enzymes II, the free energy of either a high-energy P∼His or a P∼Cys could also be used to lower the energy barrier for substrates trapped in the IIC transporter. In the most economical process, however, the phosphoryl group is not transferred to an H2O molecule as it is in the ABC transporters but is used to phosphorylate the substrate. This group translocation or vectorial phosphorylation then causes accumulation of substrate phosphate molecules in the cell in a process which under normal conditions is nearly irreversible. A topological model for IIMtl has been proposed (16, 19), and according to this model six to eight transmembrane helices form an integral membrane structure (channel) into which the large hydrophilic loop 5 folds back. Together with the highly conserved amino acids surrounding C384 (3, 48), this loop could form at the interface of the membrane the catalytic center for substrate recognition, binding, and phosphorylation which couples transport and phosphorylation. We postulated, furthermore, that H195 and the GIHE motif are central to these functions. In agreement with this model, all uncoupling mutations found in IIMtl (this study) and in IIGlc (5, 40) affected highly conserved residues in loop 5 and the GIHE (or GITE) motif. Furthermore, the multiple residues exchanged over the years by site-directed mutagenesis and found to be essential in transport-related functions also mapped almost exclusively in loop 5 or its immediate surroundings (5, 13, 43, 49).
Further exchange and cross-linking studies are in progress, and these studies should help us obtain a more detailed model of this interesting class of carbohydrate transporters which have an interesting dual role in transport and signal transduction (18).
Acknowledgments
We thank G. Robillard and his coworkers for help with the work with membrane vesicles performed in their laboratories, G. Sucher-Heilenmann for the gift of pGHL3, K. Jahreis and K. Schmid for critical reading of the manuscript and helpful discussions, and L. Schmieding for help with preparing the manuscript.
We thank the Deutsche Forschungsgemeinschaft for financial support through Sonderforschungsbereiche SFB171 and SFB431. S.O. was also supported by a grant from the III Hochschulsonderprogramm-Spezielle frauenfördende Maßnahmen
REFERENCES
- 1.Aulkemeyer, P., R. Ebner, G. Heilenmann, K. Jahreis, K. Schmid, S. Wrieden, and J. W. Lengeler. 1991. Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol. Microbiol. 5:2913-2922. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, N.Y.
- 3.Boer, H., R. H. ten Hoeve-Duurkens, and G. T. Robillard. 1996. Relation between the oligomerization state and the transport and phosphorylation function of the Escherichia coli mannitol transport protein: interaction between mannitol-specific enzyme II monomers studied by complementation of inactive site-directed mutants. Biochemistry 35:12901-12908. [DOI] [PubMed] [Google Scholar]
- 4.Budde, A. 1991. Genetische and biochemische Analyse der Pentitol- und Hexitoltransport-systeme und -stoffwechselwege in Stämmen von Escherichia coli. Diploma thesis. Universität Osnabrück, Osnabrück, Germany.
- 5.Buhr, A., G. A. Daniels, and B. Erni. 1992. The glucose transporter of Escherichia coli. Mutants with impaired translocation activity that retain phosphorylation activity. J. Biol. Chem. 267:3847-3851. [PubMed] [Google Scholar]
- 6.Charnetzky, W. T., and R. P. Mortlock. 1974. Close genetic linkage of the determinants of the ribitol and d-arabitol catabolic pathways in Klebsiella aerogenes. J. Bacteriol. 119:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson, A. 2002. Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J. Bacteriol. 184:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grisafi, P. L., A. Scholle, J. Sugiyama, C. Briggs, G. R. Jacobson, and J. W. Lengeler. 1989. Deletion mutants of the Escherichia coli K-12 mannitol permease: dissection of transport-phosphorylation, phospho-exchange, and mannitol-binding activities. J. Bacteriol. 171:2719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heuel, H., A. Shakeri-Garakani, S. Turgut, and J. W. Lengeler. 1998. Genes for d-arabinitol and ribitol catabolism from Klebsiella pneumoniae. Microbiology 144:1631-1639. [DOI] [PubMed] [Google Scholar]
- 10.Heuel, H., S. Turgut, K. Schmid, and J. W. Lengeler. 1997. Substrate recognition domains as revealed by active hybrids between the d-arabinitol and ribitol transporters from Klebsiella pneumoniae. J. Bacteriol. 179:6014-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornberg, H. L., T. M. Lambourne, and A. A. Sproul. 2000. Facilitated diffusion of fructose via the phosphoenolpyruvate/glucose phosphotransferase system of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:1808-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg, H. L., and C. Riordan. 1976. Uptake of galactose into Escherichia coli by facilitated diffusion. J. Gen. Microbiol. 94:75-89. [DOI] [PubMed] [Google Scholar]
- 13.Lanz, R., and B. Erni. 1998. The glucose transporter of the Escherichia coli phosphotransferase system. J. Biol. Chem. 273:12239-12243. [DOI] [PubMed] [Google Scholar]
- 14.Lengeler, J. W. 1975. Mutations affecting transport of the hexitols d-mannitol, d-glucitol and galactitol in Escherichia coli K-12: isolation and mapping. J. Bacteriol. 124:26-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lengeler, J. W. 1975. Nature and properties of hexitol transport systems in Escherichia coli. J. Bacteriol. 124:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lengeler, J. W. 1990. Molecular analysis of the enzyme II-complexes of the bacterial phosphotransferase system (PTS) as carbohydrate transport systems. Biochim. Biophys. Acta 1018:155-159. [Google Scholar]
- 17.Lengeler, J. W., A.-M. Auburger, R. Mayer, and A. Pecher. 1981. The phosphoenolpyruvate-dependent carbohydrate:phosphotransferase system enzymes II as chemoreceptors in chemotaxis of Escherichia coli K-12. Mol. Gen. Genet. 183:163-170. [DOI] [PubMed] [Google Scholar]
- 18.Lengeler, J. W., and K. Jahreis. 1996. Phosphotransferase systems or PTSs as carbohydrate transport and as signal transduction systems, p. 573-598 In W. N. Konings, H. R. Kaback, and J. S. Lolkema (ed.), Transport processes in eukaryotic and prokaryotic organisms. Handbook of biological physics, vol. 2. Elsevier Science, Amsterdam, The Netherlands.
- 19.Lengeler, J. W., K. Jahreis, and U. F. Wehmeier. 1994. Enzymes II of the phosphoenol-pyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim. Biophys. Acta 1188:1-28. [DOI] [PubMed] [Google Scholar]
- 20.Lengeler, J. W., F. Titgemeyer, A. P. Vogler, and B. M. Wöhrl. 1990. Structure and homologies of carbohydrate phosphotransferase system (PTS) proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326:489-504. [DOI] [PubMed] [Google Scholar]
- 21.Lolkema, J. S., D. S. Dijkstra, and G. T. Robillard. 1992. Mechanics of solute translocation catalysed by enzyme IImtl of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. Biochemistry 31:5514-5521. [DOI] [PubMed] [Google Scholar]
- 22.Lolkema, J. S., D. S. Dijkstra, R. H. ten Hoeve-Duurkens, and G. T. Robillard. 1990. The membrane-bound domain of the phosphotransferase enzyme IImtl of Escherichia coli constitutes a mannitol translocating unit. Biochemistry 29:10659-10663. [DOI] [PubMed] [Google Scholar]
- 23.Lolkema, J. S., D. S. Dijkstra, R. H. ten Hoeve-Duurkens, and G. T. Robillard. 1991. Interaction between the cytoplasmic and membrane-bound domains of enzyme IImtl of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system. Biochemistry 30:6721-6726. [DOI] [PubMed] [Google Scholar]
- 24.Lolkema, J. S., R. H. ten Hoeve-Duurkens, D. S. Dijkstra, and G. T. Robillard. 1991. Mechanistic coupling of transport and phosphorylation activity by enzyme IIMtl of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system. Biochemistry 30:6716-6721. [DOI] [PubMed] [Google Scholar]
- 25.Mahl, M. C., P. W. Wilson, M. A. Fife, and H. W. Ewing. 1965. Nitrogen fixation by members of the tribe Klebsielleae. J. Bacteriol. 89:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Nuoffer, C., B. Zanolari, and B. Erni. 1988. Glucose permease of Escherichia coli. The effect of cysteine to serine mutations on the function, stability, and regulation of transport and phosphorylation. J. Biol. Chem. 263:6647-6655. [PubMed] [Google Scholar]
- 28.Otte, S., and J. W. Lengeler. 2001. The mtl genes and the mannitol-1-phosphate dehydrogenase from Klebsiella pneumoniae KAY2026. FEMS Microbiol. Lett. 194:221-227. [DOI] [PubMed] [Google Scholar]
- 29.Postma, P. W. 1976. Involvement of the phosphotransferase system in galactose transport in Salmonella typhimurium. FEBS Lett. 61:49-53. [DOI] [PubMed] [Google Scholar]
- 30.Postma, P. W. 1981. Defective enzyme II-BGlc of the phosphoenolpyruvate:sugar phosphotransferase system leading to uncoupling of transport and phosphorylation in Salmonella typhimurium. J. Bacteriol. 147:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postma, P. W., and J. W. Lengeler. 1985. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 49:232-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phosphoenolpyruvate:carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D. C.
- 33.Postma, P. W., H. G. Keizer, and P. Koolwijk. 1986. Transport of trehalose in Salmonella typhimurium. J. Bacteriol. 168:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postma, P. W., and J. B. Stock. 1980. Enzymes II of the phosphotransferase system do not catalyze sugar transport in the absence of phosphorylation. J. Bacteriol. 141:476-484. [DOI] [PMC free article] [PubMed]
- 35.Reiner, A. M. 1975. Genes for ribitol and d-arabitol catabolism in Escherichia coli: their loci in C strains and absence in K-12 and B strains. J. Bacteriol. 123:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robillard, G. T., and M. Blaauw. 1987. Enzyme II of the Escherichia coli phosphoenol-pyruvate-dependent phosphotransferase system: protein-protein and protein-phospholipid interactions. Biochemistry 26:5796-5803. [DOI] [PubMed] [Google Scholar]
- 37.Robillard, G. T., H. Boer, P. I. Haris, W. Meijberg, D. Swaving-Dijkstra, G. K. Scuurman-Wolters, R.H. ten Hoeve-Duurkens, D. Chapman, and J. Boos. 1996. Domain and subunit interactions and their role in the function of the E. coli mannitol transporter EIIMtl, p. 549-572. In W. N. Konings, H. R. Kaback, and J. S. Lolkema (ed.), Transport processes in eukaryotic and prokaryotic organisms. Handbook of biological physics, vol. 2. Elsevier Science, Amsterdam, The Netherlands.
- 38.Ruijter, G. J. G., P. W. Postma, and K. van Dam. 1990. Adaptation of Salmonella typhimurium mutants containing uncoupled enzyme IIGlc to glucose-limited conditions. J. Bacteriol. 172:4783-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruijter, G. J. G., P. W. Postma, and K. van Dam. 1991. Energetics of glucose uptake in Salmonella typhimurium mutants containing uncoupled enzyme IIGlc. Arch. Microbiol. 155:234-237. [DOI] [PubMed] [Google Scholar]
- 40.Ruijter, G. J. G., G. van Meurs, M. A. Verwey, P. W. Postma, and K. van Dam. 1992. Analysis of mutations that uncouple transport from phosphorylation in enzyme IIGlc of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system. J. Bacteriol. 174:2843-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saiki, R., S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Ehrlich, and M. Arnheim. 1985. Enzymatic amplification of β-globin genomic sequences and restriction sites of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Saraceni-Richards, C. A., and G. R. Jacobson. 1997. A conserved glutamate residue, Glu-257, is important for substrate binding and transport by the Escherichia coli mannitol permease. J. Bacteriol. 179:1135-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid, K., R. Ebner, K. Jahreis, J. W. Lengeler, and F. Titgemeyer. 1991. A sugar-specific porin, ScrY, is involved in sucrose uptake in enteric bacteria. Mol. Microbiol. 5:941-950. [DOI] [PubMed] [Google Scholar]
- 45.Sugiyama, J. E., S. Mahmoodian, and G. R. Jacobson. 1991. Membrane topology analysis of Escherichia coli mannitol permease by using a nested-deletion method to create mtlA-phoA fusions. Proc. Natl. Acad. Sci. USA 88:9603-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, S., S. A. Lerner, and E. C. C. Lin. 1967. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J. Bacteriol. 93:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulmke, C., J. Kreth, J. W. Lengeler, W. Welte, and K. Schmid. 1999. Site-directed mutagenesis of loop L3 of sucrose porin ScrY leads to changes in substrate selectivity. J. Bacteriol. 181:1920-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Montfort, B. A., G. K. Schuurman-Wolters, R. H. Duurkens, R. Mensen, B. Poolman, and G. T. Robillard. 2001. Cysteine cross-linking defines part of the dimer and B/C domain interface of the Escherichia coli mannitol permease. J. Biol. Chem. 276:12756-12763. [DOI] [PubMed] [Google Scholar]
- 49.Weng, Q.-P., J. Elder, and G. R. Jacobson. 1992. Site-specific mutagenesis of residues in the Escherichia coli mannitol permease that have been suggested to be important for its phosphorylation and chemoreception functions. J. Biol. Chem. 267:19529-19535. [PubMed] [Google Scholar]
- 50.White, D. W., and G. R. Jacobson. 1990. Molecular cloning of the C-terminal domain of Escherichia coli d-mannitol permease: expression, phosphorylation, and complementation with C-terminal permease deletion proteins. J. Bacteriol. 172:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeppenfeld, T., C. Larisch, J. W. Lengeler, and K. Jahreis. 2000. Glucose transporter mutants of Escherichia coli K-12 with changes in substrate recognition of IICBGlc and induction behavior of the ptsG gene. J. Bacteriol. 182:4443-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]