Abstract
OxyR is a DNA binding protein that differentially regulates a cell's response to hydrogen peroxide-mediated oxidative stress. We previously reported that the reduced form of OxyR is sufficient for repression of transcription of agn43 from unmethylated template DNA, which is essential for deoxyadenosine methylase (Dam)- and OxyR-dependent phase variation of agn43. Here we provide evidence that the oxidized form of OxyR [OxyR(ox)] also represses agn43 transcription. In vivo, we found that exogenous addition of hydrogen peroxide, sufficient to oxidize OxyR, did not affect the expression of agn43. OxyR(ox) repressed in vitro transcription but only from an unmethylated agn43 template. The −10 sequence of the promoter and three Dam target sequences were protected in an in vitro DNase I footprint assay by OxyR(ox). Furthermore, OxyR(ox) bound to the agn43 regulatory region DNA with an affinity similar to that for the regulatory regions of katG and oxyS, which are activated by OxyR(ox), indicating that binding at agn43 can occur at biologically relevant concentrations. OxyR-dependent regulation of Ag43 expression is therefore unusual in firstly that OxyR binding at agn43 is dependent on the methylation state of Dam target sequences in its binding site and secondly that OxyR-dependent repression appears to be independent of hydrogen-peroxide mediated oxidative stress and the oxidation state of OxyR.
Bacteria have developed a multitude of means that allow them to survive and grow in a constantly changing environment. The formation of biofilms and cellular aggregates can, for example, facilitate resistance to certain environmental stress factors. In different bacterial species a variety of protein factors are being identified that promote the formation of biofilms and bacterial aggregates. In Escherichia coli, one of these factors is the outer membrane protein Ag43 (8, 9, 13, 14, 17, 24-26). Ag43 is a member of the autotransporter family and is encoded by the agn43 gene, originally identified as the flu locus (9, 18, 24, 25). The expression of Ag43 is under the control of phase variation, so that an individual cell either expresses the protein (ON) or does not (OFF). Thus, in a given population of cells only a percentage of individual cells will be expressing Ag43. The switch frequency of Ag43 expression, that is, the frequency with which the expression state changes, is approximately once per 1,000 cells per generation (12, 25).
Phase variation of Ag43 is regulated at the transcriptional level by OxyR and by the DNA-methylating enzyme deoxyadenosine methylase (Dam) (12, 17, 18). Dam is a maintenance methylase of E. coli that specifically methylates the adenine residue of GATC sequences. When three GATC sequences in the agn43 regulatory region are unmethylated, OxyR can bind and repress transcription, resulting in the OFF phase, but, when these GATC sequences are methylated, OxyR does not bind and expression is ON (12, 17, 18, 36, 37). Binding of OxyR prevents Dam from accessing the target sequences in the binding site, allowing them to remain unmethylated throughout the cell cycle, even in the presence of Dam (7). Thus, this DNA methylation state can be passed on from mother to daughter cell, which results in inheritance of the Ag43 OFF expression state. By definition, this regulation is epigenetic, since it is heritable and reversible and does not involve a change in DNA sequence.
OxyR is a peroxide-sensing, global transcriptional regulator (reviewed in references 11, 16, 23, and 31). Most data support a model in which the peroxide-mediated oxidative stress signal is transduced by oxidation of two cysteine residues in OxyR, which leads to the formation of an intramolecular disulfide bridge. This is accompanied by a protein conformation change and a change in protein-DNA contact sites (6, 20, 34, 38). The oxidized form of OxyR is a transcriptional activator of a multitude of genes that assist in protecting the cell from oxidative damage. These include regulatory factors, such as the small RNA oxyS, enzymes of the two main disulfide reduction pathways (trxC and grxA), and peroxide-metabolizing enzymes like catalase (katG) and alkyl hydroperoxide reductase (ahpC) (reviewed in references 23 and 31). The cellular response to peroxide that is mediated by oxidized OxyR is transient, since the inducing agent, peroxide, is enzymatically degraded rapidly and, in addition, since oxidized OxyR in the cell is converted back to the reduced state within 30 min (2). In the absence of oxidative stress, OxyR remains mainly, if not exclusively, in the reduced form, since the cytoplasm is a reducing environment (2). A recent report by Kim et al. suggests that other modifications of OxyR may also occur (16, 19). A mutant of OxyR, OxyR(C199S), is locked in the reduced form and has been shown to be sufficient for obtaining repression of the Ag43-encoding gene agn43 (12, 18, 20).
A key question in OxyR- and Dam-dependent phase variation of Ag43 is whether the switch in expression phase requires a specific cell cycle or metabolic event. It has been suggested that the oxidized form of OxyR may not mediate repression of agn43 (18, 28). If this were the case, it would not only elucidate the nature of the switch but would also implicate Ag43-dependent biofilm formation and autoaggregation as a survival mechanism to oxidative stress. However, the results presented here indicate that both the reduced and the oxidized forms of OxyR can repress agn43 transcription when the agn43 GATC sites are unmethylated.
MATERIALS AND METHODS
Medium and growth conditions.
Luria-Bertani (LB) broth, LB agar, M9 media, and agar were prepared as described earlier (22, 30). Glycerol was added as a carbon source to M9 medium at 0.2% (vol/vol), unless mentioned otherwise. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added as required. Catalase was added to plates with MV462 and MV463 to enhance growth of individual colonies. Antibiotics were added to the media at the following concentrations: kanamycin, 30 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 34 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 15 μg/ml.
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. The single-copy agn′-lacZ reporter plasmid, pMV169, was constructed by cloning a 740-bp fragment of the agn43 regulatory region (nucleotides [nt] 8785 to 9525 of AE000291 [4]) at the NotI and HindIII site of pNN387 (10). This is the same region of agn43 present in the previously described strain MV198 and its derivatives (7, 12, 37). MC4100 containing pMV169 (MV577) was used for oxidative stress experiments. Standard genetic manipulations and techniques were performed as described previously (3, 21, 22).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or Source |
|---|---|---|
| Strains | ||
| MC4100 | F−araD ΔlacU169 rpsL thi-1 | 5 |
| DHB4 | F−lac-pro lacIq/Δ(ara-leu)7697 araD139 ΔlacX74 galE galK rpsL phoR Δ(phoA)PvuII ΔmalF3 thi | 2 |
| FA369 | DHB4(katG::Tn10[Tet], ΔahpCF::Kan) | 2 |
| MV201 | MC4100 oxyR::Ω(Spec) λMV195(agn′-lacZ) lysogen | 12 |
| MV247 | MV243 oxyR::Ω(Spec) containing pMV107(oxyR) | 12 |
| MV462 | MV201 containing pAQ5(oxyR) | This study; 20 |
| MV463 | MV201 containing pGS058[oxyR(A233V)] | This study; 20 |
| MV577 | MC4100 containing pMV169(agn′-lacZ) | This study |
| MV612 | MC4100 containing pAQ24(katG′-lacZ) | This study; 33 |
| MV651 | FA369 containing pMV169 | This study |
| MV764 | MV201 containing pGS054[oxyR(H198R)] | This study; 20 |
| Plasmids | ||
| pMV151 | pRLG770 with an agn(8785-9389) insert | 37 |
| pMV169 | pNN387 with a 740-bp agn(8785-9525) NotI-HindIII fragment | This study; 10 |
Induction of oxidative stress.
An overnight culture of MV577 in M9 media was diluted 1:100, subcultured in the same media, and grown to an A600 of 0.200. The culture was aliquoted in three parts. One part was left untreated as a control for the β-galactosidase assay and for determining the percentage of ON cells. The other two parts (25 ml) were treated with hydrogen peroxide. To one of these, 2.5 μl of H2O2 (2 M stock) was added once for transient stress, and to the other 2.5 μl of H2O2 was added 15 times at 6-min intervals to obtain a sustained oxidative stress. The treated and untreated cells were plated after 10 min and 2 h to determine the percentage of Lac+ cells. Cultures were vigorously vortexed before diluting and plating to ensure the absence of aggregates.
EMSA and DNase I footprint analysis.
Electrophoretic mobility shift assays (EMSA) were performed as described previously (12), with either purified OxyR or crude cell extracts, as specified. Wild-type OxyR was purified as described previously for OxyR(C199S) (7) from MV247. This oxyR strain carries plasmid pMV107, which contains the wild-type oxyR gene behind an inducible promoter (7, 12). Purified, wild-type OxyR is in the oxidized form, since it was purified under aerobic conditions. To analyze the binding of mutant forms of OxyR at agn43 (20) crude cell extracts of MV462 and MV463 and of MV764 containing pAQ5(oxyR), pGS058[oxyR(A233V)], and pGS054[oxyR(H198R)], respectively, were used in an EMSA with radiolabeled, unmethylated agn43 DNA as a probe (20). The oxyR gene in these plasmids is contained within a larger insert on pACYC184. Transcription of oxyR does not require induction. Cell extracts were prepared from exponential-phase, LB- grown cultures. All these isolates contain an insertional mutation in the chromosomal copy of the oxyR gene and a single-copy agn′-lacZ reporter fusion. The protein concentration was determined, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was run to confirm that equal concentrations of protein were used in the EMSA.
DNA probes were obtained by amplifying the regulatory regions from chromosomal DNA of MC4100 by using PCR and were radiolabeled for EMSA and DNase I footprint analyses. The agn43 probe consisted of nt 9129 to nt 9389 of AE000291, the katG probe of nt 5252 to 5467 of AE000468, and the oxyRS probe of nt 55 to 311 of X52666.
The DNase I footprinting assay was performed as previously described on PCR-derived fragments of the agn43 regulatory region and of oxyRS described above by using purified protein (7).
In vitro transcription.
Multiple rounds of in vitro transcription reactions were performed with pMV151 as a template that contains agn43 regulatory region DNA, as described earlier (37). Both methylated and unmethylated pMV151 DNA was used as template for transcription. The effect of oxidized OxyR on in vitro transcription was studied by the addition of 9.5 and 19 nM purified OxyR to the reaction mix.
Southern blot analysis.
The methylation of agn43 GATC sequences in oxidative stress strain MV651 was determined using by using Southern blot analysis as described previously (12).
EMSA, DNase I footprint, and in vitro transcription data that were all obtained with radioactive DNA probes were visualized with a Storm phosphorimager and were quantified by using ImageQuant (Molecular Dynamics).
RESULTS
Mutant OxyR and agn43 regulation.
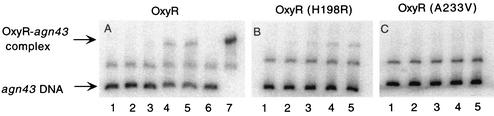
It was previously shown that oxidized OxyR binds to unmethylated agn43 DNA in vitro (7, 12), but its role in agn43 regulation was not addressed further. One approach for studying this is to use mutants that mimic the biochemical properties and biological activity of oxidized OxyR in normal, reduced cytoplasm. We analyzed the mutants OxyR(A233V) and OxyR(H198R), which constitutively activate transcription of oxyS (20). OxyR(H198R) and OxyR(A233V) differ in the degree of activation and their DNA binding properties (20). Henderson et al. previously showed that OxyR(A233V) does not complement the oxyR null phenotype, in that agn43 transcription was not repressed (18). MV201(agn′-lacZ; oxyR) was transformed with pAQ5 carrying the wild-type oxyR gene (MV462), pGS058[oxyR(A233V)] resulting in MV463, or pGS054[oxyR(H198R)] resulting in MV764. These isolates differed from each other in the Lac phenotype of individual colonies (Table 2). The predominance of Lac− colonies of MV462(oxyR) confirms that wild-type OxyR, which is mainly if not exclusively in the reduced form, represses agn43 transcription. An isolate with a chromosomal copy of oxyR phase varies, but in this isolate phase variation is absent as a result of the relative increase in level of cellular OxyR. Conversely, the Lac+ phenotype of the colonies of MV463 confirms that OxyR(A233V) does not repress agn43 transcription. In contrast, MV764[oxyR(H198R)] resulted in a mixed Lac phenotype, which indicates that this mutant can repress agn43 transcription but less efficiently than the wild-type protein. The latter result could be interpreted as the oxidized form of OxyR being able to repress agn43. However, these differences in repression at the population level can also be the result of differences in the binding affinity of the OxyR proteins for agn43 DNA. In Fig. 1 the results of EMSA are shown with the use of extracts prepared from these three isolates, which suggests that this is indeed the case. Binding was observed with wild-type OxyR (Fig. 1A), whereas no binding was observed with OxyR(A233V) (Fig. 1C). This indicates that the affinity of OxyR(A233V) for the unmethylated agn43 binding site was at least fivefold lower than that of wild-type OxyR. In contrast, and consistent with the observed repression in vivo, binding was observed with OxyR(H198R) (Fig. 1B). However, from this and a duplicate experiment (results not shown), it appears that the affinity may be lower than obtained with wild-type OxyR. Taken together, these results suggest that OxyR(A233V) and OxyR(H198R) may not accurately reflect the role of oxidized OxyR in agn43 regulation. Therefore, we directly address the role of oxidized OxyR on Ag43 phase variation by using both in vivo and in vitro approaches.
TABLE 2.
Lac phenotype of colonies of isolates containing an agn′-lacZ fusion and plasmids encoding wild-type or mutant OxyR
| Strain | oxyR genotypea | Lac phenotype (original colony) | No. of colonies examined | % Lac− (OFF) in daughtersb |
|---|---|---|---|---|
| MV462 | oxyR | Lac− (OFF) | 1,622 | >99.9c |
| 1,960 | >99.9 | |||
| MV463 | oxyR(A233V) | Lac+ (ON) | 715 | <0.2d |
| 967 | <0.2 | |||
| MV764 | oxyR(H198R) | Lac+ (ON) | 274 | 23 |
| 387 | 24 | |||
| Lac− (OFF) | 1,090 | 95.5 | ||
| 1,839 | 95.5 |
This is the genotype of the plasmid-encoded oxyR allele. The chromosomal oxyR gene is inactivated in all isolates.
A colony with a specific Lac phenotype (original colony) was resuspended, diluted, and plated. The phenotype of these latter, daughter colonies is given.
Some colonies showed blue sectors, but no Lac+ (ON) colonies could be obtained.
A Lac− (OFF) colony of this isolate was never observed in any experiment.
FIG. 1.
Different binding properties of wild-type and mutant OxyR to unmethylated agn43 regulatory region. Results obtained from EMSA with cell extracts containing OxyR are shown in panel A (lanes 2 to 5), with OxyR(H198R) in panel B, and with OxyR(A233V) in panel C. Amounts of total protein added were 0 μg (lanes 1 and 6), 0.42 μg (lanes 2), 0.85 μg (lanes 3), 2.55 μg (lanes 4), and 4.25 μg (lanes 5). Also shown is the shift obtained with 7 ng of purified, oxidized OxyR (A, lane 7).
The OFF phase is maintained during oxidative stress.
To study the effect of oxidized OxyR on expression of agn43, we constructed an agn′-lacZ reporter fusion in a single-copy plasmid (pMV169) and introduced it into a lacZ mutant background (MV577). The expression of lacZ in the strain MV577 was phase variable. The switch frequencies were 3.9 × 10−3 for OFF to ON and 3.2 × 10−2 for ON to OFF. This is a few times lower than previously described for the agn′-lacZ reporter fusion on the chromosome in MV198 (12). The plasmid-based approach avoids complications that can arise if induction of the agn′-lacZ containing lambda prophage in MV198 occurs as a result of oxidative stress.
OxyR can be oxidized in vivo by increasing intracellular hydrogen peroxide levels (2). Thus, the effect of hydrogen peroxide addition to a culture of MV577 (OFF phase) in early logarithmic growth phase was determined. β-Galactosidase levels were measured to observe an immediate change in transcription levels. In addition, aliquots of MV577 culture were plated 10 min and 2 h after addition of H2O2 to determine whether a heritable change in the expression phase had occurred, as determined by the percentage of Lac+ colonies. The addition of peroxide altered neither the β-galactosidase activity (not shown) nor the percentage of ON cells (Table 3). Transcription from a katG′-lacZ fusion increases in the presence of oxidized OxyR (33). When the same protocol with MV612 containing a multicopy katG′-lacZ fusion was used, a 1.5-fold increase in LacZ expression was obtained at 10 min, indicating that at least part of the cellular OxyR was converted to the oxidized form by using this approach.
TABLE 3.
Transient and sustained peroxide-mediated oxidative stress does not affect phase variation of agn43
An aliquot of H2O2 was added to a growing culture of MV577 (see Materials and Methods).
Aliquots of the culture were plated at the time indicated, relative to peroxide addition.
Percentage of Lac+ colonies.
Untreated MV577 culture.
After an oxidative pulse like the one described above, OxyR is completely converted to the oxidized form within 30 s and is converted back to the reduced form quite rapidly as well, with a predicted half-life that, dependent on the cell density, varies from 1.6 to 17 min (2). The transition to the ON phase of Ag43 not only requires dissociation of OxyR from the DNA but also methylation of the three target GATC sequences in the agn43 OxyR binding site to prevent reassociation of OxyR (7). In the experiment above, the OFF state could have been maintained either by binding of oxidized OxyR or by a lack of methylation during the brief period during which OxyR(ox) was dissociated. The latter would allow OxyR after conversion back to the reduced form to readily reassociate with the DNA. To address the latter possibility, we tested whether maintaining oxidized OxyR for a prolonged period would affect Ag43 regulation. Hydrogen peroxide was added 15 times to a culture of MV577 at intervals of 6 min as described in Materials and Methods. This prolonged peroxide-mediated oxidative stress also did not result in a change in the percentage of Lac+ cells in the progeny (Table 3). It must be noted that growth of the culture ceased after five additions (30 min). Because of this concern, we took additional approaches to examine the effect of sustained oxidative stress. Paraquat, a superoxide-generating compound, indirectly activates the conversion to oxidized OxyR by generating hydrogen peroxide at low levels (40). Various concentrations of paraquat, in the range of 0.01 to 1.0 mM, were added to a culture of MV577. At these concentrations the culture continued to grow, even though the growth rate decreased. The addition of paraquat also did not result in a change in the number of Lac+ cells (data not shown).
A high level of oxidized OxyR in vivo can also be achieved by introducing mutations that increase intracellular H2O2 concentration. This is the case in an isolate containing mutations in both ahpCF and katG (2). The single-copy plasmid with the agn′-lacZ fusion, pMV169, was introduced into the double mutant FA369 (ahpCF, katG) (MV651). We were not able to analyze agn43 phase variation based on Lac phenotype of the colonies due to the very slow growth of MV651. As an indicator for the Ag43 expression state, the agn43 DNA methylation state of three independent cultures of MV651 was determined, and in each case over 95% of the agn43 regulatory region of pMV169 was sensitive to restriction with MboI, indicating that the GATC sequences were unmethylated (data not shown). This is consistent with methylation protection at agn43 by bound OxyR in this strain, which suggests that OxyR can be bound to the agn43 regulatory region in vivo under persistent oxidative stress.
Taken together, the lack of an effect of peroxide-mediated oxidative stress on Ag43 expression indicates that, in cells with OxyR mainly or completely in the oxidized form, agn43 transcription is repressed. To determine whether the observed repression in the OFF cells can be mediated by oxidized OxyR, we examined in vitro the interactions of oxidized OxyR with agn43 regulatory region DNA.
Oxidized OxyR represses agn43 transcription in vitro.
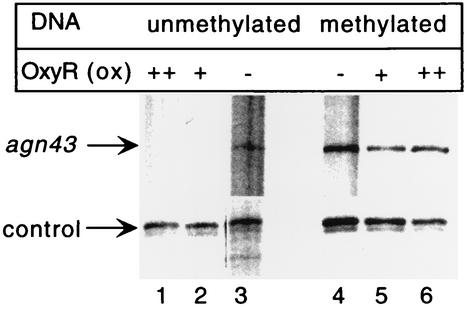
Previously, we showed that OxyR(C199S), which has the biological and biochemical properties of the reduced form, is able to repress in vitro transcription of agn43 from an unmethylated DNA template. We carried out an in vitro transcription assay with agn43 DNA in the presence of oxidized OxyR. Oxidized OxyR completely repressed transcription from an unmethylated template (Fig. 2, compare lanes 1 and 2 to lane 3) but not from a methylated template (Fig. 2, compare lane 4 to lanes 5 and 6). These results are in agreement with binding of oxidized OxyR to the unmethylated but not methylated agn43 regulatory region (12). No additional products were observed in the presence of OxyR(ox), indicating that it was also not acting as an activator for an alternative agn43 promoter. This repression in vitro is consistent with the interpretation that in vivo under hydrogen peroxide-induced oxidative stress agn43 repression is mediated by oxidized OxyR.
FIG. 2.
Oxidized OxyR represses transcription from an unmethylated agn43 template in vitro. Template DNA was either unmethylated (lanes 1 to 3) or methylated (lanes 4 to 6). OxyR(ox) was added as indicated before RNA polymerase to a final concentration of 9.5 nM (lanes 2 and 5) or 19 nM (lanes 1 and 6). No OxyR was added to reactions shown in lanes 3 and 4. “Control” refers to a transcript obtained from a vector-derived promoter. The size of the agn43 transcription product is 220 nt. Lane 2 was obtained from a different exposure of the same gel.
The binding region of oxidized OxyR at agn43.
We examined the binding of oxidized OxyR to the unmethylated agn43 regulatory region DNA by in vitro footprinting with DNase I. A hallmark of the members of the OxyR regulon is that either the reduced or the oxidized form of OxyR binds to the regulatory region or that, if both forms bind, the DNA-protein contact sites differ (34). For example, the DNase I footprints of the two forms of OxyR at the oxyRS intergenic region differ (Fig. 3, lanes 4 to 6) in accordance with previously published results by Toledano et al. (34). Specifically, OxyR(C199S), which has the properties of the reduced form, resulted in an extended footprint with a hypersensitive site (Fig. 3, lane 6). This hypersensitive site was absent in the shorter footprint obtained with the oxidized form (Fig. 3, lane 4) (34).
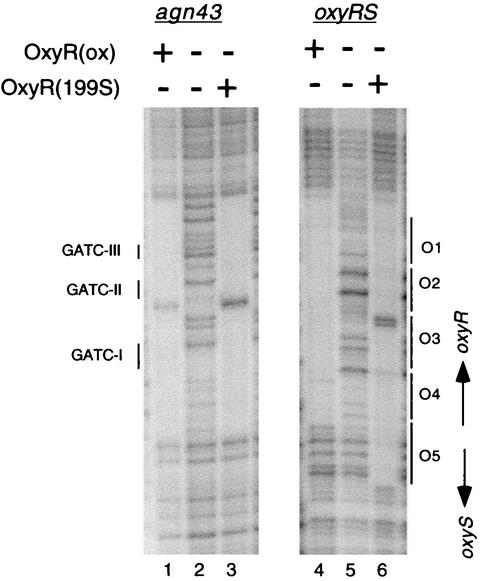
FIG. 3.
The DNase I footprint of oxidized OxyR is similar to that of OxyR(C199S) at agn43 DNA. Footprints were obtained using a final concentration of 12 nM OxyR(ox) (lanes 1 and 4) or 10 nM OxyR(C199S) (lanes 3 and 6). Template DNA was unmethylated agn43 DNA (lanes 1 to 3) and oxyRS DNA (lanes 4 to 6). For the agn43 sequence, the position of the three GATC sequences is shown. The direction of transcription of oxyR and oxyS is shown for the oxyRS footprint, as well as the position of the OxyR binding motifs O1 through O5, as designated previously by Toledano et al. (34).
The DNase I-protected region of unmethylated agn43 DNA in the presence of oxidized OxyR included the −10 sequence of the agn43 promoter and the three GATC sequences (Fig. 3, lane 1). The protected region at this agn43 DNA was the same as that protected with OxyR(C199S) (Fig. 3, lane 3). Binding by both oxidized OxyR and OxyR(C199S) resulted in a hypersensitive site. The footprint obtained with OxyR(C199S) is consistent with our previously reported results (37).
The affinity of oxidized OxyR for agn43 DNA is biologically relevant.
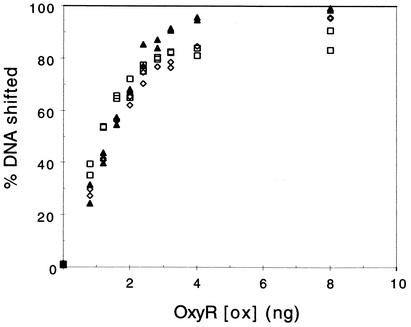
The results described above show that the oxidized form of OxyR binds to unmethylated agn43 DNA and represses agn43 transcription in vitro. To address the potential biological relevance of this in vitro activity, we compared the relative affinity of oxidized OxyR for unmethylated agn43 DNA with that for the regulatory regions of katG and oxyRS. Binding of oxidized OxyR at these latter regions activates transcription of katG and oxyS, respectively (33). In Fig. 4 the results of EMSA with oxidized OxyR are shown. The results indicate that the affinities of OxyR(ox) for the regulatory regions of agn43, oxyRS, and katG differ by less than 20%. This suggests that, under peroxide-mediated stress, when oxidized OxyR is known to bind to the katG and oxyRS regions and activate transcription, it can also be bound to the unmethylated agn43 regulatory region and thus repress agn43 transcription.
FIG. 4.
The affinity of oxidized OxyR for unmethylated agn43 DNA is similar to that for katG and oxyRS DNA. The percent shifted DNA in an EMSA with increasing amounts of purified wild-type OxyR is shown. Eight nanograms of OxyR corresponds to a final concentration of 12 nM. Template DNA consisted of oxyRS (open diamonds), katG (open squares), and unmethylated agn43 regulatory regions (closed triangles). Data from two independent series of experiments are shown.
DISCUSSION
The data presented here strongly suggest that phase variation of Ag43 is not affected in cells experiencing peroxide-mediated oxidative stress and furthermore that under these conditions the OFF phase can be a result of repression by oxidized OxyR. Firstly, the absence of a change in the level of agn′-lacZ transcription and in the percentage of ON cells after a transient oxidative stress indicates that a change in the oxidation state of OxyR does not result in an alleviation of repression and methylation of the GATC sequences (Table 3). Previously, global analysis of transcription also showed that the level of transcription from agn43 (flu) was not altered after addition of hydrogen peroxide (40). These results suggest that OxyR, which is mainly in the oxidized form, was bound and repressed agn43 transcription. Alternatively, oxidation of OxyR resulted in abrogation of repression, but a lack of rapid methylation allowed OxyR to rebind after being converted back to the reduced form. However, a constantly applied peroxide-mediated oxidative stress also did not result in a change in the percentage of ON cells (Table 3). The cells continued to grow until the fifth addition of peroxide, indicating that metabolic activity including DNA methylation was active during this period. Nevertheless, the percentage of ON cells did not change, indicating that the OFF phase had been maintained throughout the oxidative stress period. Furthermore, in the presence of paraquat the culture grew continuously, even though the growth rate was affected. The lack of conversion from the OFF to the ON phase in these cells under oxidative stress also is consistent with OxyR(ox) binding and repressing. Finally, the presence of unmethylated regulatory region in the ahpCF katG mutant is consistent with methylation protection by bound OxyR. This binding would result in repression of agn43 transcription in these cell that are under sustained oxidative stress.
Our in vivo data show that repression of agn43 occurs when the cellular pool of OxyR is mainly in the oxidized form, indicating that the oxidized form of OxyR is functioning as a repressor. However, neither the presence of a low level of reduced OxyR nor the possibility that DNA methylation was slightly affected can be fully excluded. Furthermore, the level of oxidative stress that is required to maintain OxyR(ox) will inherently affect growth and related metabolic processes. Even though our results obtained with OxyR(H198R) support our conclusion, we show that analysis of the effect of OxyR mutants on agn43 expression does not necessarily reflect the role of oxidized OxyR (Fig. 1; Table 2). The absence of and decrease in percentage of ON colonies in isolates with OxyR(A233V) and OxyR(H198R), respectively, appear to be a result of the decreased binding affinity of these mutants for agn43 DNA and not to be related to the redox state of OxyR (Fig. 1). Recently, Schembri et al. presented data indicating that OxyR(H198R) does not repress agn43 transcription (27). The difference between our results and theirs can be reconciled by taking into consideration that different expression systems for OxyR were used, which can lead to different cellular levels of OxyR. A decrease in concentration of OxyR, like a decrease in affinity, will affect the competition with Dam for the agn43 binding site, which leads to decreased OxyR binding, and thus results in an increased incidence of the ON phase.
Our in vitro analyses with purified, oxidized OxyR directly support the conclusion that the repression of agn43 transcription, which occurs under oxidative stress, is a direct result of binding of oxidized OxyR. The affinity of oxidized OxyR for unmethylated and hemimethylated agn43 DNA is the same as the affinity of OxyR(C199S), which behaves like the reduced form (12, 20). Furthermore, it was previously shown that methylation blocks binding of OxyR(ox) in vitro and conversely that binding of OxyR(ox) protects agn43 DNA from Dam-dependent methylation, which are both essential features of Ag43 phase variation (7, 12). Consistent with this, the region protected by OxyR(ox) from DNase I includes the GATC sequences. In addition, part of the agn43 promoter sequence was protected, which is consistent with the data showing that oxidized OxyR repressed agn43 in vitro transcription (Fig. 2 and 3). Taken together, the data support a model in which the ON state for Ag43 expression is obtained by DNA methylation-dependent abrogation of OxyR binding and in which the OFF phase is a result of repression mediated by binding of either the reduced or the oxidized form of OxyR to unmethylated agn43 DNA (Fig. 5).
FIG. 5.
Schematic model for regulation of agn43 transcription. Vertical lines indicate the GATC sequences, and methylation is indicated by CH3. The oxidized and the reduced forms of OxyR are as indicated, and the transcription start site is indicated by an arrow. The ON phase (A) is obtained if the three GATC sequences in regulatory region are methylated. The OFF phase is obtained by OxyR-dependent repression. Both the oxidized (C) and reduced forms (B) of OxyR repress transcription from the unmethylated agn43 template. Neither the oxidized nor reduced form of OxyR could bind to the methylated agn43 template.
At the agn43 regulatory region, the oxidized form has a DNase I footprint that resembles the reduced form and protects the same region (Fig. 3) (37). This is in contrast to the distinctive differences in the DNase I footprints obtained by the reduced and oxidized forms at the katG and oxyRS sites (Fig. 3) (34, 39). Similar DNase I footprints for both forms of OxyR have, however, also been observed at a mutated ahpC regulatory region (34). The relative intensity of the hypersensitive site in the footprint of OxyR(ox) at agn43 is lower than that obtained with OxyR(C199S), which indicates that there is some difference in the protein-DNA interaction between the wild-type, oxidized form and OxyR(C199S).
The affinity of oxidized OxyR for unmethylated agn43 DNA was similar not only to OxyR(C199S) for agn43 DNA but also to the oxyRS and katG regulatory regions (Fig. 4). Since these genes are immediately activated by oxidized OxyR upon oxidative stress, this indicates that binding of oxidized OxyR to agn43 DNA will occur in a cell under oxidative stress as long as this site is unmethylated. Interestingly, a high binding affinity of OxyR for agn43 DNA had been predicted by Zheng et al., based on analysis of a binding sequence with contacts at four adjacent major grooves (39). Our data suggest that the OxyR-agn43 contacts are different but that the affinity is nevertheless relatively high, which supports these authors' conclusion that DNA binding of OxyR and OxyR-dependent regulation are versatile (6, 39).
OxyR is known to repress several genes. In the reduced form it is a repressor of its own transcription, of agn43, and of the phage Mu mom gene (12, 32). In the oxidized form, it is a repressor of itself, of fhuF, a putative iron reductase, and, as we show here, of agn43 (39, 40). The oxidized form also binds to mom DNA (15, 32). However, differences between binding of OxyR and the mechanism of repression of mom and agn43 make it difficult to extrapolate our results to the role of oxidative stress in regulation of mom.
Our conclusion that oxidized OxyR as well as reduced OxyR can repress agn43 transcription has implications for a model proposed by Schembri et al., which addresses the coordinated regulation between fimbrial production and Ag43 expression. These authors proposed that fimbrial production leads to a change in the environmental redox potential that would result in the conversion of cytoplasmic OxyR to the oxidized form. In their model, this conversion results in abrogation of OxyR-dependent repression (27-29). In contrast, our results indicate that oxidized OxyR can efficiently repress agn43 transcription, which suggests that the coordinate regulation is not mediated by the differential oxidation state of OxyR at the level of agn43 transcription. However, processes that affect the cellular or environmental redox state will affect other pathways that are involved in responding to the redox potential, including those that affect protein secretion, stability, and proteolytic cleavage. These changes may affect the presence of processed Ag43 on the cell surface, as was shown to occur upon the addition of dithiothreitol (27).
To summarize, our data indicate that agn43 is an unusual member of the OxyR regulon in that its OxyR-dependent regulation is dependent on the DNA methylation state of the regulatory region but is apparently independent of the oxidation state of OxyR. Thus, peroxide-mediated stress does not alter the regulation of this specific member of the OxyR regulon. Ag43-mediated biofilm formation or aggregation may nevertheless confer a survival advantage under oxidative stress (27). It remains to be determined if environmental signals exist that lead to differential expression patterns of Ag43.
A family of agn43-like genes has been identified in E. coli isolates and in Shigella flexneri (1, 26). The function of this extended family of proteins is not clear, even though it was recently shown that the Ag43-like Cah protein in E. coli O157:H7 confers the characteristic Ag43-dependent autoaggregation and biofilm formation but also binds calcium (35). The regulatory region of this cah gene, as well as of all other members of this family for which sequence is available, has retained the essential features for OxyR- and Dam-dependent phase variation (37). Thus, it is very likely that the basic principles underlying Ag43 phase variation will be applicable to other members of the family of agn43-like genes.
Acknowledgments
This work was supported by grant MCB-0077501 from the National Science Foundation.
We thank Emmanuelle Binet and Sol Goodgal for critical reading of the manuscript and Gisela Storz for helpful discussions and strains.
REFERENCES
- 1.Al-Hasani, K., K. Rajakumar, D. Bulach, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Åslund, F., M. Zheng, J. Beckwith, and G. Storz. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 96:6161-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 4.Blattner, F. R., G. R. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. 1976. Transposition and fusion of the lac genes to selected promoters in E. coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 6.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103-113. [DOI] [PubMed] [Google Scholar]
- 7.Correnti, J., V. Munster, T. Chan, and M. van der Woude. 2002. Dam-dependent phase variation of Ag43 in E. coli is altered in a seqA mutant. Mol. Microbiol. 44:521-532. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., L. A. Pratt, S. Dove, and R. Kolter. 2000. The outer-membrane protein, Ag43, mediates cell-to-cell interactions within E. coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 9.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elledge, S. J., and R. W. David. 1989. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3:185-197. [DOI] [PubMed] [Google Scholar]
- 11.Georgiou, G. 2002. How to flip the (redox) switch. Cell. 111:607-610. [DOI] [PubMed] [Google Scholar]
- 12.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in E. coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 13.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasman, H., M. A. Schembri, and P. Klemm. 2000. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K12. J. Bacteriol. 182:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattman, S., and W. Sun. 1997. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 25:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D. 5 November 2002, posting date. OxyR: a molecular code for redox sensing? Sci. STKE 2002:PE46. [Online.] http://stke.sciencemag.org/cgi/content/full/sigtrans;2002/157/pe46.. [DOI] [PubMed]
- 17.Henderson, I. R., M. Meehan, and P. Owen. 1997. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli, p. 349-355. In P. S. Paul et al. (ed.), Mechanisms in the pathogenesis of enteric diseases. Proceedings of the First International Rushmore Conference Held in Rapid City, South Dakota, Sept. 28-30, 1995. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 18.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S. O., K. Merchant, R. Nudelman, W. F. Beyer, Jr., T. Keng, J. DeAngelo, A. Hausladen, and J. S. Stamler. 2002. OxyR: a molecular code for redox-related signaling. Cell 109:383-396. [DOI] [PubMed] [Google Scholar]
- 20.Kullik, I., M. B. Toledano, L. A. Tartaglis, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 24.Owen, P., P. Caffrey, and L.-G. Josefsson. 1987. Identification and partial characterization of a novel bipartite protein antigen associated with the outer membrane of Escherichia coli. J. Bacteriol. 169:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen, P., M. Meehan, H. de Loughry-Doherty, and I. Henderson. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16:63-76. [DOI] [PubMed] [Google Scholar]
- 26.Roche, A., J. McFadden, and P. Owen. 2001. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology 147:161-169. [DOI] [PubMed] [Google Scholar]
- 27.Schembri, M. A., L. Herrild, M. Gjermansen, and P. Klemm. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 28.Schembri, M. A., and P. Klemm. 2001. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 20:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schembri, M. A., D. W. Ussery, C. Workman, H. Hasman, and P. Klemm. 2002. DNA microarray analysis of fim mutations in Escherichia coli. Mol. Genet. Genomics 267:721-729. [DOI] [PubMed] [Google Scholar]
- 30.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:88-94. [DOI] [PubMed] [Google Scholar]
- 32.Sun, W., and S. Hattman. 1996. Escherichia coli OxyR protein repressses the unmethylated bacteriophage Mu mom operon without blocking binding of the transcriptional activator C. Nucleic Acids Res. 24:4042-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tartaglia, L. A., G. Storz, and B. N. Ames. 1989. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 210:709-719. [DOI] [PubMed] [Google Scholar]
- 34.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 35.Torres, A. G., N. T. Perna, V. Burland, A. Ruknudin, F. R. Blattner, and J. B. Kaper. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45:951-966. [DOI] [PubMed] [Google Scholar]
- 36.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 37.Wallecha, A., V. Munster, J. Correnti, T. Chan, and M. van der Woude. 2002. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, M., X. Wang, B. Doan, K. A. Lewis, T. D. Schneider, and G. Storz. 2001. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 183:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]