Abstract
In Bacillus subtilis, RNA polymerase becomes concentrated into regions of the nucleoid called transcription foci. With green fluorescent protein-tagged RNA polymerase, these structures are only observed at higher growth rates and have been shown to represent the sites of rRNA synthesis. There are 10 rRNA (rrn) operons distributed around nearly half of the chromosome. In this study we analyzed the rrn composition of transcription foci with fluorescently tagged loci and showed that they comprise the origin-proximal operon rrnO but not the more dispersed rrnE or rrnD. This suggests that transcription foci comprise only the seven origin-proximal operons rrnO, rrnA, rrnJ, rrnW, rrnI, rrnH, and rrnG. These results have important implications for our understanding of microbial chromosome structure.
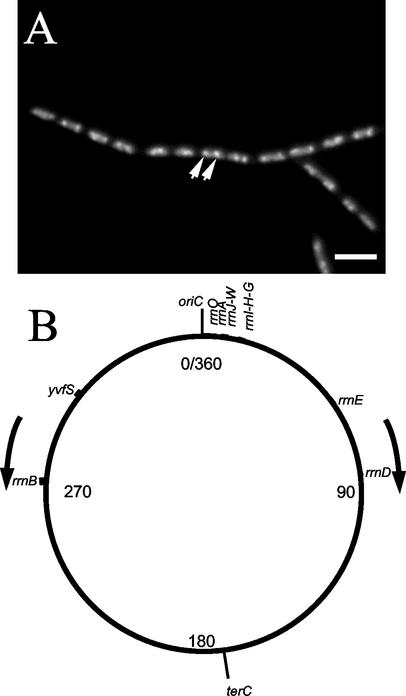
As the growth rate of a bacterial cell increases, so does the cellular demand for ribosomes (16). As a result, transcription of rRNA operons (rrns) increases with growth rate, so that at very high rates, nearly 80% of the cellular RNA is rRNA (5). The localization of RNA polymerase in the gram-positive bacterium Bacillus subtilis has recently been described (25). RNA polymerase was found to lie primarily within the central region of the nucleoid and to concentrate into subnucleoid regions termed transcription foci at higher growth rates (25) (Fig. 1A).
FIG. 1.
Transcription focus appearance and rrn distribution in the Bacillus subtilis chromosome. Panel A shows representative cells of strain 1048 carrying an rpoC-gfp fusion and grown in CH medium, displaying transcription foci (arrows) Scale bar, 5 μm. Panel B shows the locations of rrn operons and the control yvfS locus in the B. subtilis chromosome. Numbers represent coordinates in degrees on the 360-degree map of the chromosome. oriC and terC represent the origin and terminus of chromosome replication, respectively. Arrows indicate the direction of movement of replication forks.
On induction of the stringent response, transcription foci rapidly disappeared, suggesting that they represent the sites of rRNA synthesis within the cell. The stringent response occurs when ribosomes encounter uncharged tRNA molecules and is a starvation response that results in the production of the alarmone (p)ppGpp (6, 16). There is mounting evidence that (p)ppGpp exerts its pleiotropic effects on gene expression through direct interaction with RNA polymerase (7). In Escherichia coli, ribosomal genes have promoters which are sensitive to (p)ppGpp levels (11). When these levels rise within the cell, there is a concomitant downregulation of rRNA (rrn) transcription (16). It has also been long established that due to the strength of rrn promoters, there is a disproportionate loading of RNA polymerase onto rrn genes (14). Therefore, transcription foci in B. subtilis were interpreted to represent the sites of rrn transcription within the cell and presented a dynamic illustration of transcriptional activity in vivo. Transcription foci are only rarely observed at slow growth rates (25). This is consistent with the observation that ribosome number is directly related to growth rate, and at low rates, the cell requires relatively few ribosomes. Therefore, at low growth rates, the level of rrn transcription would also be relatively low (9, 16).
There are 10 rrn operons in the B. subtilis chromosome (21). The number and chromosomal location of rrns appear to be directly related to the growth rate of organisms (1). For example, the slow-growing Mycobacterium tuberculosis (doubling time, about 24 h) contains a single rrn operon located 1.5 Mb away from the origin region (about 120° on a 360° circular chromosomal map [8]), whereas rapidly growing organisms like Escherichia coli (doubling time, ≤20 min) contain seven operons, all in the origin half of the chromosome (within 90° on both sides of oriC on a circular map [4]). Vibrio natriegens, which can grow with a doubling time of around 10 min, has at least 13 rrn operons per genome (1).
The 10 rrns in B. subtilis are arranged in four groups (Fig. 1B). The first group contains seven rrn operons arranged in four clusters within 175 kb of the origin region. The remaining three operons are located at 54° (rrnE), 81° (rrnD), and 275° (rrnB). These origin-distal rrns are associated with large numbers of tRNA (trn) genes (21). Therefore, these operons are likely to be highly transcribed and important within the cell. It is known that the number of rrns in B. subtilis can vary due to deletion and recombination events, but this normally occurs in the rrnI-H-G and rrnJ-W clusters close to oriC (15, 37).
Transcription foci appear to be well-defined structures with a very regular appearance within the nucleoid (see Fig. 1A). Given that transcription foci represent the sites of rrn synthesis, we wanted to investigate their genetic composition. Do they contain all or just some of the rrn operons? If so, which ones? The clustering of origin-distal rrns into transcription foci would represent significant chromosomal reorganization during rapid growth. If this clustering occurs, is it also observed at low growth rates, when transcription foci are not observed due to the low level of loading of RNA polymerase onto rrns? If transcription foci contain origin-distal rrns, does incorporation occur following replication of the locus, in a cell cycle-dependent manner, or as a single-step event following duplication of all rrns? Previous studies have elegantly shown that the linear arrangement of genes on the chromosomes appears to be spatially retained in vivo (35). Therefore, any clustering of loci into transcription foci would most likely be specific for highly transcribed genes such as the rrns and imply the formation of structures akin to eukaryotic nucleoli where such clustering of rRNA genes does occur (33).
In this study, rrns were individually labeled with the system developed by Webb et al. (36), in which a green fluorescent protein (GFP)-LacI fusion binds to tandem repeats of the lacO site inserted at the desired chromosomal locus. These fusions were colocalized with origin regions with a Spo0J-cyan fluorescent protein (CFP) fusion. Spo0J foci have previously been shown to colocalize with transcription foci (25), and so it could also be used as a marker for rrn clustering at low growth rates, when transcription foci are not observed. A strong correlation between origin regions and the origin-proximal rrnO was observed at both low and medium growth rates. However, no evidence for clustering was observed for origin-distal rrns at either growth rate. These results indicate that transcription foci represent the loading of RNA polymerase onto the seven origin-proximal rrns and that all 10 rrns in B. subtilis do not aggregate to form a bacterial nucleolus.
In addition, we have shown that the rrn loci tested all had functional, active promoters, indicating that lack of clustering of origin-distal loci was not due to any lack of transcription of those genes. No change in colocalization frequencies was observed on induction of the stringent response. These results confirm that the cell is required to maintain the linear order of genes on the chromosome and that complex chromosome remodeling does not occur as a normal part of the cell cycle. The maintenance of chromosome structure throughout the cell cycle has important implications for our understanding of chromosome segregation.
MATERIALS AND METHODS
Bacterial strains and media.
All cloning was carried out in Escherichia coli DH5α (Gibco-BRL). The Bacillus subtilis strains used and constructed in this study are listed in Table 1. Transformation of B. subtilis was carried out by the method of Anagnostopolous and Spizizen (2), as modified by Jenkinson (20). B. subtilis transformants were selected on nutrient agar containing the appropriate antibiotic (chloramphenicol, 5 μg/ml; spectinomycin, 50 μg/ml; kanamycin, 5 μg/ml). Double-crossover integrants into the amyE locus were selected as described previously (24). S medium, used for slow growth (doubling time, 70 min at 37°C), and CH medium, used for medium growth (doubling time, 45 min at 37°C), have been described previously (34).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or construction |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | C. Anagnostopoulos |
| 1501 | trpC2 chr::pSG1517 (spo0J-gfp cat) | 13 |
| 1063 | trpC amyE::(spec Pxyl-gfp-lacI) | 168 transformed with pSG1189 |
| 1064 | trpC2amyE::(spec Pxyl-gfp-lacI) chr::pSG1196 (rrnD-lacO cat) | 1063 transformed with pSG1196 |
| 1067 | trpC2 chr pSG1197 (spo0J-cfp kan) | 168 transformed with pSG1197 |
| 105 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pSG1196 (rrnD-lacO cat) chr::pSG1197 (spo0J-cfp kan) | 1064 transformed with chromosomal DNA of 1067 |
| 106 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pSG1201 (rrnO-lacO cat) | 1063 transformed with pSG1201 |
| 107 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pNG105 (rrnE-lac0 cat) | 1063 transformed with pNG105 |
| 108 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pNG106 (yvfs-lacO cat) | 1063 transformed with pNG106 |
| 109 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pSG1201 (rrn0-lacO) chr::pSG1197 (spo0J-cfp kan) | 106 transformed with chromosomal DNA of 1067 |
| 110 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pNG105 (rrnE-lacO) chr::pSG1197 (spo0J-cfp kan) | 107 transformed with chromosomal DNA of 1067 |
| 111 | trpC2 amyE::(spec Pxyl-gfp-lacI) chr::pNG106 (yvfS-lacO) chr::pSG1197 (spo0J-cfp kan) | 108 transformed with chromosomal DNA of 1067 |
| 1285-05 | trpC2 lys1 aprEΔ3 nprR2 nprE18 amyE::PrrnO-bgaB cat | F. Kawamura (Rikkyo [St. Paul's] University, Japan) |
| 1285-DN1 | trpC2 lys1 aprEΔ3 nprR2 nprE18 amyE::PrrnD-bgaB cat | F. Kawamura |
| 1285-EN1 | trpC2 lys1 aprEΔ3 nprR2 nprE18 amyE::PrrnE-bgaB cat | F. Kawamura |
| Plasmids | ||
| pAT12 | bla lacO cat | 35 |
| pSG1170 | bla Pspac-gfpUV Ppen-lacI cat | 24 |
| pSG1729 | bla amyE3′ spc Pxyl-′gfp amyE5′ | 24 |
| pSG1192 | bla amyE3′ spc Pxyl-cfp′ amyE5′ | 12 |
| pSG1189 | bla amyE3′ spc Pxyl-gfp-lacI amyE5′ | BamHI-XhoI digest of lacI (Δ C-terminal 11 amino acids) PCR product from pSG1170 into pSG1729 cut with BamHI-XhoI |
| pSG1195 | bla amyE3′ spc Pxyl-spo0J-cfp amyE5′ | PCR spo0J from strain 1510 DNA, cut with XhoI-NcoI and insert into XhoI-NcoI-cut pSG1192 |
| pSG1196 | bla rrnD-lacO cat | PCR of region immediately downstream of rrnD cut with XhoI-BamHI into XhoI-BamHI-cut pAT12 |
| pSG1197 | bla spo0J-cfp kan | Acc651-XbaI-digested spo0J-cfp into Acc651-XbaI pUK19 |
| pSG1201 | bla rrnO-lacO cat | PCR of region immediately downstream of rrnO operon cut with XhoI-BamHI-cut pAT12 |
| pNG105 | bla rrnE-lacO cat | PCR of region immediately downstream of rrnE operon, cut with XhoI-BamHI into XhoI-BamHI-cut pAT12. |
| pNG106 | bla yvfS-lacO cat | PCR of region immediately downstream of yvfS gene cut with XhoI-BamHI into XhoI-BamHI-cut pAT12 |
DNA manipulations.
Cloning steps were performed as detailed before (32). Oligonucleotide primers used for PCR are detailed in Table 2, and details of plasmid construction are given in Table 1.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| lacI forward | 5′-GATCGGATCCACGTGAAACCAGTAACGTTA-3′ |
| lacI reverse | 5′-AACCTCCTCGAGCAGCTGCATTAATGAATC-3′ |
| rrnD forward | 5′-CTTACTCAACTCGAGGGTCTAGGATCTCTT-3′ |
| rrnD reverse | 5′-CTTACGTATATCTGTGGATCCAAAAGAAAA-3′ |
| rrnE forward | 5′-CCATTAATGCTCGAGAAACGATCATGT-3′ |
| rrnE reverse | 5′-GCTTTCAACGGATCCAATTGAAACACA-3′ |
| rrnO forward | 5′-GAATGATGTCTCGAGTGTTATCTAGTTTTG-3′ |
| rrnO reverse | 5′-TTATAAAGGATCCGCATATTCTCCCTTAGA-3′ |
| yvfS forward | 5′-GTGTAATACCTCGAGAAGGCAATATCT-3′ |
| yvfS reverse | 5′-GTG CTT GAT GGA TCC AGT CAC GGC TTC-3′ |
| spo0J forward | —a |
| spo0J reverse | 5′-TTCTCCTTTACTCACCATGGGCAGGAATTC-3′ |
Glaser et al. (13).
Cell cycle modeling.
Detailed cell cycle parameters for B. subtilis grown in S and CH media have been determined (34). Cell length measurements were made with MetaMorph (version 4.6; Universal Imaging Corporation) and used to determine the most likely stage of the cell cycle for each individual cell measured with the cell cycle models of Sharpe et al. (34). The point in the cell cycle at which each locus would be duplicated was calculated and placed onto the models so that cells could be classified as having oriC-locus ratios of either 1:1 or 2:1. Cells in which the ratio was 1:1 were classed as cells in which colocalization was most likely to occur if transcription foci contained clustered loci.
rrn-bgaB fusions and assays.
rrn promoter activity was determined with strains 1285-O5, 1285-DN1, and 1285-EN1 (Table 1), kindly provided by F. Kawamura (Rikkyo [St. Paul's] University, Japan). These strains utilize the thermostable β-galactosidase gene from Bacillus stearothermophilus (19). Assays were carried out at 62°C as described previously (19, 29).
Induction of stringent response.
Cultures in CH medium were grown to mid-exponential phase (A600 0.4) and then split into two. To one half, arginine hydroxamate was added to a final concentration of 500 μg/ml (25) to induce the stringent response. The other half of the culture was used as a positive control. For β-galactosidase assays, samples were taken at 10-min intervals and assayed as detailed above. For microscopic analysis, cultures were placed onto an agarose slide 10 min after the addition of arginine hydroxamate to the culture, and images were acquired for the following 20 min. Images were processed as detailed below.
Microscopy, image acquisition, and analysis.
Cultures were grown in S and CH media as detailed previously (34). Image data for all strains were collected from cells grown to mid-exponential phase in S and CH media at 30°C and 25°C, respectively. Strains required the addition of 0.2% xylose in S medium and 0.1% xylose in CH medium for the efficient expression of gfp-lacI. Cells were viewed on 1.2% agarose slides (17). All images were acquired with a Zeiss Axioskop 2 fluorescence microscope and Photometrics Quantix 1401E cooled charge-coupled device camera. GFP fluorescence was visualized with filter set 41018, and CFP fluorescence was visualized with filter set 31044v2 (Chroma Technology, Brattleboro, Vt.). Image analysis was performed with MetaMorph software (version 4.6; Universal Imaging Corporation), and figures were prepared for publication with Adobe Photoshop.
RESULTS
Strain construction and determination of signal crossover.
Since rrn clustering could happen at both slow and fast growth rates but transcription foci are only visible at higher growth rates, it was not possible to colocalize rrn loci directly with RNA polymerase. Instead, rrn loci were colocalized with a Spo0J fusion that marked the origin regions of the cell (see below) and has been shown previously to colocalize with transcription foci (25). Dual-labeled strains containing Spo0J-CFP and GFP-LacI fusions were constructed. The GFP-LacI fusion was able to bind to lacO sequences inserted at a variety of places in the chromosome (Table 1). The lac operator inserts contain 256 tandem repeats of lacO (31), and so if these sites are saturated with GFP-LacI, they could represent a significant block to an advancing DNA replication fork, slowing the natural growth rate. Strains 105, 109, 110, and 111 containing Spo0J-CFP, GFP-LacI, and the lac operators downstream of rrnD, rrnO, rrnE, and yvfS, respectively (Table 1), were grown in S and CH media in the presence of 0.2 and 0.1% (wt/vol) xylose (see below), and their growth rates and cell size distributions were compared with that of the wild-type strain 168. These were found to be identical to 168 (not shown). Therefore, under the growth conditions used in this study, it is reasonable to assume that the cell cycle parameters of the labeled strains will be the same as that of the wild-type control strain.
Fluorescent protein fusions have quite broad excitation and emission spectra (18), and so considerable fluorescence crossover would be expected to occur between the cyan and green fluorescent fusion proteins. If this occurred, it would not be possible to assign signal definitively to one fusion or the other when observing fluorescence in either the cyan or green channel. Nevertheless, since the fluorescence fusions used in this study were relatively weak, the small percentage of signal that crossed over into another channel represented a very small proportion of the total amount of signal present in that channel. Strains containing just spo0J-cfp (1067; Table 1) or rrnD-lacO and gfp-lacI (1064; Table 1) fusions were grown, and images were obtained in the green and cyan fluorescence channels. Strain 1064, containing a xylose-inducible copy of gfp-lacI, was grown in both S and CH media containing different levels of xylose to determine the optimum levels required for good fluorescence in the green channel and minimal crossover into the cyan channel. Strains grown in the presence of 0.2% (wt/vol) xylose in S and 0.1% (wt/vol) xylose in CH medium were found to display good fluorescence in the green channel with virtually no crossover into the cyan channel (not shown). The weakly fluorescent Spo0J-CFP fusion strain 1067 gave no observable crossover into the green channel (not shown).
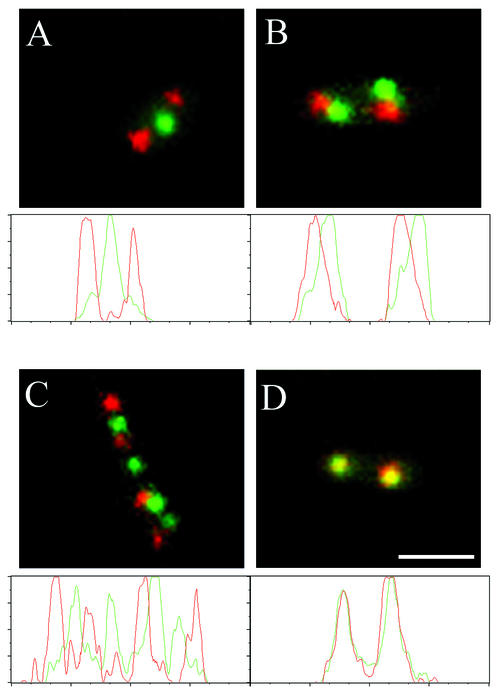
Dual-labeled strains were constructed, and cyan and green fluorescence was observed. In strain 105 (Spo0J-CFP rrnD-lacO GFP-LacI; Table 1), the oriC-rrnD ratio will be 2:1 for nearly half a cell cycle, as rrnD is distal to the origin (Fig. 1B; see below), and so around half the cell population should contain readily distinguishable cyan and green foci. This was observed and is illustrated in Fig. 2A. Spo0J-CFP foci have been pseudocolored red, and GFP-LacI foci have been pseudocolored green. The image overlay in Fig. 2A shows a single cell containing two Spo0J foci (red) and a single LacI focus (green), with no observable overlap between the signals. This is confirmed by the line scan of the cells shown directly below the overlay. The peak corresponding to GFP-LacI is mutually exclusive to the Spo0J-CFP peaks, confirming that there is essentially no signal crossover between the fusions and that it is possible to localize these green and cyan fluorescent protein fusions in the same cells with confidence.
FIG. 2.
Representative images and line scans of dual-labeled cells. Panels A, B, and D show single cells, whereas a pair of cells is shown in panel C. Line scans through the cells are shown below the images. Cells from strain 105 (rrnD) are shown in panels A and C, from strain 110 (rrnE) in panel B, and from strain 109 (rrnO) in panel D. Spo0J-CFP foci have been pseudocolored red, whereas GFP-LacI foci have been pseudocolored green. These color assignments have been conserved in the line scans below the images. Bar, 2 μm.
Colocalization of oriC with rrns.
Once the level of signal crossover had been determined, it was important to establish how images would be scored to ascertain which rrns cluster into transcription foci. A Spo0J-CFP fusion was used to mark the location of the origin region as it binds to multiple sites and condenses approximately 400 kb of the chromosome around oriC that also encompasses part of the rrnO-rrnG operon cluster (23, 27, 28). Thus, there should be a strong correlation between the subcellular localization of the oriC region and the rrnO-rrnG cluster.
Image overlays of Spo0J-CFP with different GFP-decorated rrns are shown in Fig. 2. In Fig. 2A, an image is shown containing two Spo0J foci and a single rrnD focus. The line scan shown below the image clearly shows that the signals are mutually exclusive (the peak of one signal corresponds to the trough of the other) and as such would be scored as noncoincident signals. Figures 2B, C, and D shows cells containing equal numbers of Spo0J and rrn foci. In Fig. 2B (rrnE) and 2C (rrnD), the foci lie either adjacent to or between each other. The line scans below the images show that peaks either do not coincide (they are offset from each other; Fig. 2B) or are mutually exclusive (Fig. 2C) and so would also be scored as noncoincident signals. In Fig. 2D (rrnO), the signals are perfectly coincident, and this was confirmed by the line scan. Only these signal patterns were scored as coincident signals.
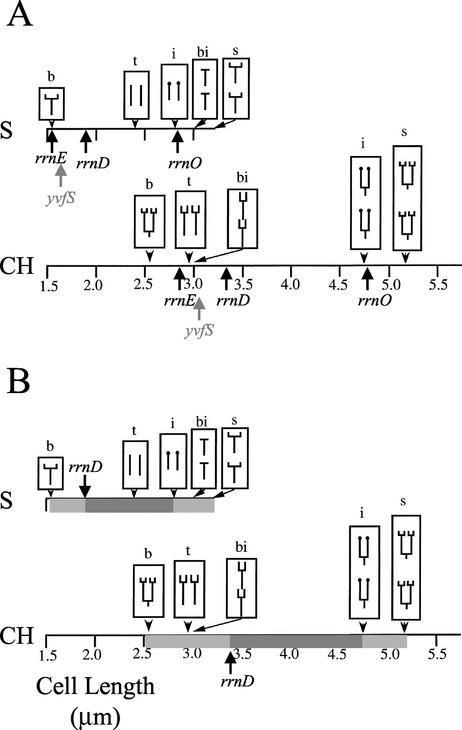
It was also necessary to take into account the stage of the cell cycle for individual cells being examined. If chromosome replication was under way and the origin regions duplicated but replication forks had not yet moved through an rrn locus (e.g., Fig. 2A), the ratio of oriC to rrn would be 2:1. Therefore, the maximum possible level of signal colocalization for that locus would be 50%. Detailed cell cycle models have been produced describing the growth of B. subtilis under a range of defined conditions in previous studies (34). Use was made of these models to assign the cell cycle stage of individual cells in this study. Cells were grown in S medium, in which transcription foci are not observed, and CH medium, in which they are. Cell cycle models for cultures grown in these media are shown in Fig. 3. The points at which individual loci are duplicated were calculated and mapped onto the models (Fig. 3A). For example, rrnD is situated at 81° on the chromosome and so will not be duplicated until the replication cycle is nearly 50% complete (forks situated at 90° and 270° on a circular map have duplicated 50% of the chromosome). Therefore, the oriC-rrnD ratio will be 2:1 for nearly half a cell cycle.
FIG. 3.
Cell cycle models for B. subtilis. The figures shown in both panels A and B are derived from those developed previously (34). S and CH represent S and CH medium, respectively. The letters i, bi, s, b, and t above the cartoon cells correspond to initiation of chromosome replication, binucleation, chromosome segregation, cell birth, and termination of the chromosome replication phases of the cell cycle, respectively. In panel A, the points in the cell cycle at which replication forks move through rrnO, rrnE, yvfS, and rrnD are labeled and marked by arrows under the figures. In panel B, the light grey boxes highlight the cell length distributions in which the oriC-rrnD ratio is 2:1, and the dark grey boxes highlight the cell length distributions in which the oriC-rrnD ratio is 1:1. Cell length is indicated on the scales below the figures. Adapted from reference 34 with permission.
The cell size distribution for this part of the cell cycle is indicated by the light gray boxes in Fig. 3B. Since the oriC-rrnD ratio is 2:1 in this portion of the cell cycle, the maximum level of colocalization of signals would be 50%. When replication forks pass through the rrnD locus, the ratio of signals will become 1:1, and so up to 100% colocalization of signals could be expected in this size range (dark gray boxes, Fig. 3B). In addition to rrnO, rrnE, and rrnD, the colocalization frequency of the control locus yvfS with oriC was also determined. yvfS is the last gene in an operon containing a proposed ABC transporter, is located at 299° on the B. subtilis chromosome, and is approximately equivalent to the location of rrnE but on the opposite arm of the chromosome (Fig. 1B). Therefore, if rrn locus clustering was found to occur, we could determine whether it was specific to those loci or a general feature of the origin half of the chromosome.
At least 400 cells of each strain in both S and CH media were scored to determine the level of colocalization of signals, and the results are presented in Table 3. The theoretical size range in which transcription foci could contain each locus (oriC-locus ratio of 1:1) was determined from the cell cycle models shown in Fig. 3 and detailed in reference 34 and is presented in the left-hand column for each of the media used. The following two columns present the percentage signal colocalization observed in cells that fell into the predicted size range in which transcription foci could be observed for that strain (in range), or that fell outside the predicted size range (ex range).
TABLE 3.
Colocalization of oriC regions with rrn loci
| Marker | S medium
|
CH medium
|
Frequency of colocalization in CH medium + arginine hydroxamate (500 μg/ml) (%)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Theoretical transcription focus size range (μm) | Frequency of colocalization (%)
|
Theoretical transcription focus size range (μm) | Frequency of colocalization (%)
|
|||||
| In range | Ex range | In range | Ex range | In range | Ex range | |||
| rrnO | All | 74.4 | All | 75.5 | 75 | |||
| rrnE | 1.70-2.80 | 46.9 | 43.4 | 2.85-4.80 | 39.4 | 34.3 | NDa | ND |
| rrnD | 1.95-2.80 | 33.3 | 20.5 | 3.35-4.80 | 50.2 | 36.4 | 38 | 35 |
| yvfS | 1.75-2.80 | 24.8 | 18.1 | 3.05-4.80 | 34.7 | 40.0 | ND | ND |
ND, not determined.
Strain 109 (Table 1), in which rrnO is labeled, lies so close to oriC (9.8 kb) and within the region of the chromosome that contains utilized Spo0J binding sites (28) that all of the cells were considered to lie within the range in which the locus would be able to form transcription foci. In both S and CH media, a high level of colocalization of rrnO and Spo0J signals was observed (74.4% and 75.5%, respectively; Table 3). This level of colocalization is in close agreement with the level of Spo0J and transcription focus colocalization previously measured with a strain carrying RNA polymerase-GFPuv and Spo0J-GFP fusions (79%) (25). Therefore, it is highly likely that rrnO is a component of transcription foci in vivo.
For the remaining strains used in this study, if a specific locus was incorporated into a transcription focus, the frequency of colocalization should be much higher for cells that fall into the in range category than those falling into the ex range category. The level of colocalization in the ex range category would represent the random colocalization of signals observed due to the orientation of cells on the slide, the resolving limitations of the light microscope, and the movement of DNA-protein-fluorescent protein complexes in the cell due to random diffusion. In addition, if transcription foci represent the exclusive clustering of rrn operons, the frequency of in range colocalization should be much higher for rrn loci than for the control yvfS locus. Results for colocalization of the loci are listed in Table 3. For all of the remaining loci, there was no significant difference in the level of colocalization of signal between cells falling into the in range and ex range classes. In addition, the levels of colocalization observed were similar for the control yvfS locus and the rrn loci. Overall, these results indicate that rrnE and rrnD are not specifically recruited into transcription foci in either S or CH medium.
Nevertheless, some differences in colocalization frequency between the media were observed. The level of colocalization was higher and less variable in CH medium than in S medium (Table 3). This may be due to the DNA content of the cells at higher growth rates. Under the conditions used, the large majority of cells growing in S medium contained two origins and one terminus (34) (Fig. 3). By contrast, the large majority of cells grown in CH medium contained four origins and one or two terminus regions. The increase in the number of loci per cell in CH medium might simply lead to a higher chance of random colocalization being observed in cells, accounting for the differences noted in Table 3.
rrn promoter activity.
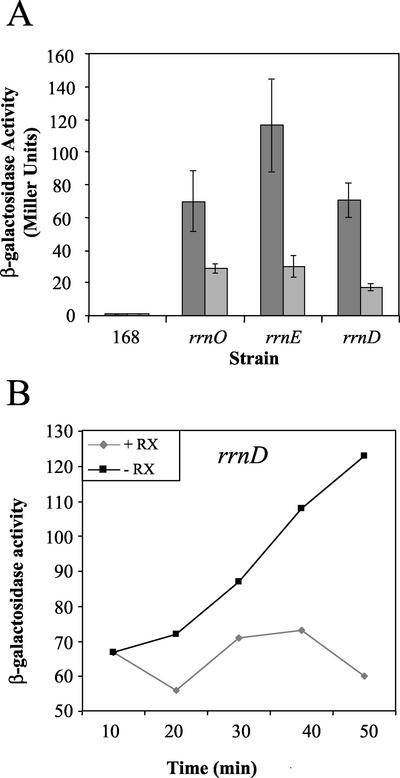
Our colocalization studies indicated that rrnE and rrnD were not recruited into transcription foci. However, it was not known whether these operons were actually being transcribed under the experimental conditions used, and very little data are available on the transcriptional activity of rrn promoters in B. subtilis. Therefore, we decided to check the promoter activity of rrnO, rrnE, and rrnD in S and CH media. β-Galactosidase gene (lacZ) fusions to rrn promoters were kindly donated by F. Kawamura (Rikkyo [St. Paul's] University, Japan) and are listed in Table 1. The strains were grown in both S and CH media, and assays were performed as detailed in Materials and Methods. The results of the assays are shown in Fig. 4A.
FIG. 4.
Panel A shows rrn promoter activity measured as β-galactosidase activity in both CH (dark grey) and S (pale grey) media. Error bars indicate standard deviations. The promoter fusions are indicated on the x axis. β-galactosidase activity (Miller units) is shown on the y axis. Panel B shows a plot of a time course assay for rrnD promoter activity in CH medium in the presence (grey diamonds) and absence (black squares) of arginine hydroxamate (RX).
It is clear that all the promoters were active in both media. As expected, the level of activity was approximately two- to fourfold greater in CH medium than in S medium. This would be consistent with the requirement for greater numbers of ribosomes for rapidly growing cells in rich media (5, 16). There were other slight variations with the level of activity of the promoters in different media. The level of expression of rrnD was approximately two-thirds that of rrnO and rrnE in S medium, while the level of rrnE expression was about 50% greater than that of rrnO and rrnD in CH medium (Fig. 4A). However, these differences are relatively modest, and quite large standard deviations were obtained during the assays (n = 5; Fig. 4A). Nevertheless, it is clear that all the rrn promoters tested showed considerable and similar levels of activity, indicating that they would all be transcribed under the experimental conditions used to determine transcription focus composition.
Colocalization patterns on induction of stringent response.
Transcription foci were shown to be the sites of rrn transcription, as transcription foci disappear on induction of the stringent response and subsequent downregulation of rrn transcription (25). In order to ensure that all of the rrn promoters used in this study functioned correctly and were under stringent control, they were tested as detailed in Materials and Methods. Within 10 min of the addition of the stringent response inducer arginine hydroxamate, there was a significant decrease in the level of transcription from PrrnD (Fig. 4B). Similar results were also obtained for PrrnO and PrrnE (not shown). The low level of expression from the promoters in the presence of arginine hydroxamate continued throughout the time course and showed that they behaved in the expected manner on induction of the stringent response.
Although there was no evidence to suggest clustering of rrns outside of the origin region into transcription foci, an additional check was performed to determine if any changes in colocalization of labeled loci were observed upon induction of the stringent response. Colocalization frequencies of CFP and GFP signals in strains 109 (rrnO) and 105 (rrnD) (Table 1) were determined as detailed in Materials and Methods, and the results are presented in Table 3. The level of colocalization of both rrnO and rrnD with Spo0J was similar to that observed in CH medium during normal exponential growth (compare rrnO and rrnD colocalization data in CH medium in Table 3). The in range level of rrnD colocalization was slightly lower than that observed in normal growing cells, and this may be due to a slight reduction in the number of rrnD loci on induction of the stringent response due to the activity of Ster sites (see Discussion).
Ster sites are located approximately 200 kb either side of oriC and act as reversible replication termination sites on induction of the stringent response (3). The rrnD operon is located nearly 1 Mb from oriC, and so on activation of Ster sites, replication forks will not be able to move beyond 200 kb of oriC and the rrnD locus will not be duplicated. Therefore, the proportion of cells with an oriC-rrnD ratio of 1:1 will drop on induction of the stringent response. Since the rrnO operon lies within 10 kb of oriC, the proportion of cells with an oriC-rrnO ratio of 1:1 should not be affected on induction of the stringent response. Since image acquisition was carried out within 30 min of induction of the stringent response, Ster termination would have had only a modest effect on the ratios of loci in our samples. Our results confirm that it is extremely unlikely that there is any chromosomal refolding involved in clustering of distantly located rrn operons to form transcription foci. Rather, the formation of transcription foci is most likely to be exclusively due to the increased loading of RNA polymerase onto the seven rrn operons clustered adjacent to oriC.
DISCUSSION
In this work, we characterized the structural composition and nature of transcription foci in Bacillus subtilis. In addition to the increased level of loading of RNA polymerase onto rrn genes at high growth rates (14), it was also possible that some or all of the 10 rrns distributed around nearly half of the B. subtilis chromosome could be involved in the formation of these structures. We analyzed the level of clustering of several rrn operons at both slow growth rates, when transcription foci are not observed, and high growth rates, when they are (Table 3). In addition, we also analyzed any dynamic changes in transcription focus structure that occurred on induction of the stringent response and subsequent downregulation of rrn expression (Table 3).
Our results clearly show that rrnO located adjacent to oriC is involved in transcription focus formation. Likewise, there is no evidence to suggest that rrnE and rrnD are involved in the formation of transcription foci (Table 3). By inference, it is also extremely unlikely that the remaining origin-distant operon, rrnB, would be involved in transcription focus formation either.
We also determined the transcriptional activity of the promoters for all the rrn operons used in this study in order to determine their activity under our experimental conditions. All the promoters were active to a similar level, indicating that all of the operons were being transcribed. This result indicates that while oriC-distal rrn operons are transcribed, transcriptional activity does not require translocation to a nucleolus-like subregion of the bacterial nucleoid dedicated to rrn transcription and ribosome assembly. It should be noted that transcriptional activity of rrn promoters was determined from β-galactosidase gene fusions at the amyE locus rather than from the chromosomal location. Therefore, our results reflect a direct comparison of the transcriptional activity of these promoters in the cell independent of gene dosage effects. The level of expression of the strains used in this study was similar to that of constructs with β-galactosidase fusions inserted at the chromosomal locus of the rrn operons (R. Rudner, New York University, New York, N.Y., personal communication). Nevertheless, it was important to check that these promoter sequences behaved as would be expected for sequences located at their normal chromosomal sites. On induction of the stringent response, the activity of all the promoters was significantly reduced, indicating that Prrn activity was attenuated, as would be expected. No significant change in colocalization frequency was observed for labeled loci on induction of the stringent response, confirming that transcription focus disappearance is not due to disaggregation of clustered rrn loci and is due to the reduced level of loading of RNA polymerase onto the seven origin-proximal rrn operons.
Therefore, we conclude that transcription foci comprise the seven rrn operons that are located within 175 kb of oriC, rrnO, rrnA, rrnJ, rrnW, rrnI, rrnH, and rrnG. Rather than specifically clustering, we suggest that transcription foci purely represent the additional loading of RNA polymerase that occurs at rrns when cells are growing rapidly (14, 16). Although we have shown that all the rrns tested in this study are transcribed at similar levels, we cannot detect additional foci that represent RNA polymerase loading onto the more dispersed sites rrnE and rrnD at high growth rates with any confidence (not shown). If we assume that the rate of transcription from each rrn promoter is approximately equal, the proportion of RNA polymerase involved in transcription of the three origin-distal loci at high growth rates represents not much more than 10% of the level of RNA polymerase involved in total rrn transcription when gene dosage is taken into account, and so it is unlikely that we would be able to detect specific signals above the background RNA polymerase level.
Our results help confirm an emerging model of chromosome structure and organization within living bacterial cells. While it had been assumed for years that there must be some level of structural organization of bacterial chromosomes, it is only relatively recently that we have been able to start probing these delicate structures in vivo (26). In both E. coli and B. subtilis, a number of studies have shown that origin regions tend to be oriented towards the cell poles, whereas terminus regions tend to localize to the cell center around the site where the new division septum will form. This orientation appears to be important both with respect to providing a polarity for chromosome segregation (13, 36) and for the faithful decatenation of chromosomes on completion of DNA replication (10, 22). More detailed studies on chromosome structure in E. coli indicate that genes in the origin half of the chromosome cluster into an origin domain, whereas those in the terminal half cluster in to a terminus domain (30). Other studies in B. subtilis imply that the linear order of genes observed in the chromosomal sequence is preserved in vivo (35). Therefore, a gene that lies closer to oriC on the linear DNA sequence of the chromosome will also lie closer to it in situ than a more distant gene.
Despite these data, it was still possible that certain sequences, such as those involved in the coordinated large-scale production of rRNA, might prove exceptions to this rule and be involved in the formation of a higher-order structure that increased the efficiency of rRNA synthesis. Our results suggest that this is not the case and are most consistent with the individual rrn genes' retaining their positions relative to other genes within the linear order of the chromosome. This, in turn, suggests that maintenance of chromosome structure is important throughout the cell cycle and at different growth rates and is likely to ensure accurate segregation of daughter nucleoids upon cell division.
Acknowledgments
This work was supported by an ARC grant to P.L. (A00105185).
We are indebted to F. Kawamurra for the supply of Prrn-bgaB strains prior to publication, R. Rudner for helpful discussions and providing unpublished data on Prrn activity, and E. Harry for comments on the manuscript.
REFERENCES
- 1.Aiyar, S. E., T. Gaal, and R. L. Gourse. 2002. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 184:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopolous, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autret, S., A. Levine, F. Vannier, Y. Fujita, and S. J. Seror. 1999. The replication checkpoint control in Bacillus subtilis: identification of a novel RTP-binding sequence essential for the replication fork arrest after induction of the stringent response. Mol. Microbiol. 31:1665-1679. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bremmer, H., and P. D. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt et al. (ed.). Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p.1458-1496. In F. C. Neidhardt et al. (ed.). Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 7.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signalling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Condon, C., J. Philips, Z. Y. Fu, C. Squires, and C. L. Squires. 1992. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 11:4175-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duggin, I., and R. G. Wake. 2002. Termination of chromosome replication, p. 87-96. In A. L. Sonenshine et al. (ed.), Bacillus subtilis and its closest relatives. American Society for Microbiology, Washington, D.C.
- 11.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feucht, A., and P. J. Lewis. 2001. Improved vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene 264:289-297. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behaviour of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 14.Gotta, S. L., O. L. Miller, Jr., and S. L. French. 1991. rRNA transcription rate in Escherichia coli. J. Bacteriol. 173:6647-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb, P., G. LaFauci, and R. Rudner. 1985. Alterations in the number of rRNA operons within the Bacillus subtilis genome. Gene 33:259-268. [DOI] [PubMed] [Google Scholar]
- 16.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 17.Harry, E. J., and P. J. Lewis. 2003. Early targeting of Min proteins to the cell poles in germinated spores of Bacillus subtilis: evidence for division apparatus-independent recruitment of Min proteins to the division site. Mol. Microbiol. 47:37-48. [DOI] [PubMed]
- 18.Haseloff, J. 1999. GFP variants for multispectral imaging of living cells, p. 139-152. In K. F. Sullivan and S.A. Kay (ed.), Green fluorescent proteins. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 19.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of β-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkinson, H. F. 1983. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J. Gen. Microbiol. 129:1945-1958. [DOI] [PubMed] [Google Scholar]
- 21.Kunst, F., et al. 1997. The complete genome sequence of the Gram positive Bacillus subtilis bacterium. Nature 390:249-256. [DOI] [PubMed]
- 22.Lemon, K. P., I. Kurtser, and A. D. Grossman. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis, P. J., and J. Errington. 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and colocalisation with the Spo0J partitioning protein. Mol. Microbiol. 25:945-954. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101-109. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, P. J., S. D. Thaker, and J. Errington. 2000. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, P. J. 2001. Bacterial chromosome segregation. Microbiology 147:519-526. [DOI] [PubMed] [Google Scholar]
- 27.Lin, D. C. H., P. A. Levin, and A. D. Grossman. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 94:4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, D. H. C., and A. D. Grossman. 1998. Identification and characterisation of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 29.Nanamiya, H., Y. Ohashi, K. Asai, et al. 1998. ClpC regulates the fate of a sporulation initiation sigma factor, σH protein, in Bacillus subtilis at elevated temperatures. Mol. Microbiol. 29:505-513. [DOI] [PubMed] [Google Scholar]
- 30.Niki, H., Y. Yamaichi, and S. Hiraga. 2000. Dynamic organisation of chromosomal DNA in Escherichia coli. Genes Dev. 14:212-223. [PMC free article] [PubMed] [Google Scholar]
- 31.Robinett, C. C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A. S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization with lac operator/repressor recognition. J. Cell Biol. 135:1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Scheer, T. M., and G. Goessens. 1991. Localisation of nucleolar chromatin by immunocytochemistry and in situ hybridisation at the electron microscopic level. Electron Microsc. Rev. 4:85-110. [DOI] [PubMed] [Google Scholar]
- 34.Sharpe, M. E., P. M. Hauser, R. G. Sharpe, and J. Errington. 1998. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J. Bacteriol. 180:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teleman, A., P. L. Graumann, D. H. C. Lin, A. Grossman, and R. Losick. 1998. Chromosome arrangement within a bacterium. Curr. Biol. 8:1102-1109. [DOI] [PubMed] [Google Scholar]
- 36.Webb, C. D., A. Teleman, S. Gordon, et al. 1997. Bipolar localisation of the replication origin region of chromosomes in vegetative and sporulating cells of Bacillus subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 37.Widom, R. L., E. D. Jarvis, G. LaFauci, and R. Rudner. 1988. Instability of rRNA operons in Bacillus subtilis. J. Bacteriol. 170:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]