Abstract
bldN is one of a set of genes required for the formation of specialized, spore-bearing aerial hyphae during differentiation in the mycelial bacterium Streptomyces coelicolor. Previous analysis (M. J. Bibb et al., J. Bacteriol. 182:4606-4616, 2000) showed that bldN encodes a member of the extracytoplasmic function subfamily of RNA polymerase σ factors and that translation from the most strongly predicted start codon (GTG1) would give rise to a σ factor having an unusual N-terminal extension of ca. 86 residues. Here, by using a combination of site-directed mutagenesis and immunoblot analysis, we provide evidence that all bldN translation arises from initiation at GTG1 and that the primary translation product is a proprotein (pro-σBldN) that is proteolytically processed to a mature species (σBldN) by removal of most of the unusual N-terminal extension. A time course taken during differentiation of the wild type on solid medium showed early production of pro-σBldN and the subsequent appearance of mature σBldN, which was concomitant with aerial mycelium formation and the disappearance of pro-σBldN. Two genes encoding members of a family of metalloproteases that are involved in the regulated proteolytic processing of transcription factors in other organisms were identified in the S. coelicolor genome, but their disruption did not affect differentiation or pro-σBldN processing.
The regulated proteolysis of transcription factors is an important theme in the control of gene expression in both eukaryotes and bacteria. Well-characterized examples include the sterol regulatory element binding protein (SREBP) in mammals (3) and σE and σK in the bacterium Bacillus subtilis (30). SREBP, which activates genes involved in cholesterol biosynthesis and uptake, has an N-terminal transcription activation domain and a C-terminal regulatory domain separated by two transmembrane helices. The protein is inserted into the membrane of the endoplasmic reticulum and the nuclear envelope in a hairpin fashion such that the N-terminal and C-terminal domains are in the cytoplasm and the short “luminal loop” between the two transmembrane helices projects into the lumen of the organelle (18, 36). In order to activate its target genes, the N-terminal domain has to be released from the membrane in a two-step proteolytic process that is activated by a drop in sterol levels (35, 38). In the first step, SREBP is cleaved at site 1 within the luminal loop in a sterol-dependent manner, separating the N-terminal and C-terminal domains but leaving both still anchored in the membrane (10). In the second step, which is not regulated by sterols but requires prior cleavage at site 1, a second protease cleaves at site 2 within the first transmembrane helix (11). This releases the mature N-terminal domain into the cytosol, from which it rapidly enters the nucleus and activates gene expression to bring about an increase in cellular sterol levels (3).
In the gram-positive bacterium B. subtilis, the mother cell-specific σ factors, σE and σK, play central roles in the control of gene expression during endospore formation. σE and σK are synthesized as inactive, membrane-associated pro-σ factors that are subsequently activated and released into the mother cell cytoplasm through proteolysis of the N-terminal 27 to 29 and 20 amino acids, respectively, in reactions catalyzed by membrane-localized proteases. The prosequences of both pro-σE and pro-σK are responsible for promoting membrane association, and the mature forms of these σ factors are found in the cytoplasm associated with core RNA polymerase (13, 16, 19, 20, 43). In both cases, the activation of pro-σ processing in the mother cell is triggered by signals derived from the forespore; in the case of σK, pro-σ processing is triggered by a putative signaling protein (SpoIVB), which is produced in the forespore and is secreted either into the mother cell or the space between the inner and outer forespore membranes (14, 15). These intercellular signaling pathways are critical for coordination of the divergent programs of gene expression between the two cells. Thus, for example, the engineered removal of the DNA encoding the prosequence from the gene encoding σK results in the premature appearance of σK activity in the mother cell and the consequent production of highly defective spores (8).
σBldN is an extracytoplasmic function (ECF) σ factor that carries an unusual N-terminal extension of ca. 86 amino acids that is absent from other σ factors. σBldN was discovered following the isolation of two NTG-induced point mutants in the bldN gene that affected morphological differentiation in the mycelial bacterium Streptomyces coelicolor (1, 34). bldN-null mutants cannot develop the specialized aerial hyphae that give Streptomyces colonies their characteristic fuzzy appearance and which ultimately differentiate to form chains of exospores (1, 6, 21).
bldN expression is developmentally regulated. The bldN transcript is almost undetectable during vegetative growth but its abundance increases dramatically during aerial mycelium formation (1). Three other bld loci have been implicated in this transcriptional regulation. Activation of bldN transcription depends, directly or indirectly, on both bldG and bldH (1), and BldD directly represses bldN transcription by binding to the bldN promoter at two sites, one at either side of the transcription start site (12). In addition to bldN, BldD also negatively regulates two other σ factor genes, whiG and sigH, and is thought specifically to repress transcription of these genes prior to aerial mycelium formation (12, 23).
σBldN, in turn, directly activates transcription of another gene required for aerial mycelium formation, bldM (1, 27). bldM has two promoters, a strong σBldN-dependent promoter (bldMp1) and a weak σBldN-independent promoter (bldMp2). As a consequence, bldM mRNA and protein levels are very low in a bldN mutant (1; M. Elliot, unpublished data).
The bldN orthologue of Streptomyces griseus, adsA, has also been characterized, and the unusual N-terminal extension that is present in σBldN is conserved in σAdsA (40). Like bldN, adsA is required for aerial mycelium formation and is developmentally regulated at the level of transcription. In S. griseus, the γ-butyrolactone signaling molecule A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) triggers a regulatory cascade required for both aerial mycelium formation and production of the antibiotic streptomycin (17). adsA is under the immediate control of the A-factor cascade, being directly activated by AdpA (A-factor-dependent protein A), a transcriptional activator required for both differentiation and streptomycin production (28).
Here we provide evidence that σBldN, in addition to being developmentally regulated at the transcriptional level, is synthesized as a proprotein (pro-σBldN) that is processed to a mature species (σBldN) through the proteolytic removal of the unusual N-terminal extension.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, transformation, and conjugal plasmid transfer from Escherichia coli to Streptomyces.
S. coelicolor strains (Table 1) were cultured on R2YE, on minimal medium containing 0.5% (wt/vol) mannitol as a carbon source, on MS agar (mannitol plus soya flour), or in liquid YEME medium (24). To bypass the methyl-specific restriction system of S. coelicolor during conjugation from E. coli, unmethylated plasmids were transferred by conjugation from the dam dcm hsdS E. coli strain ET12567 (26) as described by Ryding et al. (34). Streptomyces protoplast transformation was as described by Kieser et al. (24). E. coli BL21λ(DE3)(pLysS) (37) was used to express σBldN, and DH5α was the host strain for standard manipulations. The plasmids used were pET11a (Novagen), pSK+ (Stratagene), pSET152 (2), pRSET (Invitrogen), pIJ487 (39), and pIJ6650 (24).
TABLE 1.
Derivatives of S. coelicolor A3(2) used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| M145 | SCP1− SCP2− | 24 |
| J1501 | hisA1 uraA1 strA1 Pgl− SCP1− SCP2− | 7 |
| J1915 | ΔglkA119 SCP1− SCP2− | 22 |
| J2177 | ΔglkA119 bldN::hyg SCP1− SCP2− | 1 |
| J2178 | ΔglkA119 bldN::his SCP1− SCP2− | This work |
| J2180 | ΔglkA119 SCO2260::hyg SCP1− SCP2− | This work |
| J2182 | SCO5695::apr SCP1− SCP2− | This work |
| J2183 | ΔbldN::aadA SCP1− SCP2− | This work |
| J2184 | hisA1 uraA1 strA1 ΔbldN::aadA Pgl− SCP1− SCP2− | This work |
| J2188 | ΔglkA119 SCO2260::hyg SCO5695::apr SCP1− SCP2− | This work |
| R112 | bldN112 SCP1− SCP2− | 34 |
| R650 | bldN650 SCP1− SCP2− | 34 |
| WC103 | bldG103 hisA1 uraA1 strA1 Pgl− SCP1− SCP2− | 5 |
| WC109 | bldH109 hisA1 uraA1 strA1 Pgl− SCP1− SCP2− | 5 |
PCR-based site-directed mutagenesis.
A 1.2-kb BamHI-EcoRI fragment containing bldN was isolated from pIJ6715 (1) and cloned into pSK+ digested with BamHI and EcoRI to give pIJ6723. Pairs of abutting oligonucleotides were used to amplify the entire pIJ6723 plasmid, simultaneously introducing specific mutations. The oligonucleotides used were pIJ6724 (GTG1 to GTC; 5′-CTACCCACACGTCGGGGTTGA-3′ and 5′-ACGGGACTCCCAGAGGCAGAG-3′), pIJ6725 (GTG2 to GTC; 5′-CCCCGCCGGTCCGTGCTACGCA-3′ and 5′-ACGGCGGCCGCGGCGAGGGCG-3′), pIJ6726 (TTG to TGA1; 5′-TGACGCGGCTTCGTCCCCACCGCG and 5′-CAGGTCTTGCGCTGATTTGAC-3′), pIJ6727 (GGA to TGA2; 5′-TGAAGCGCCGTCGTCGGCAGAC-3′ and 5′-TTCGGCCAGTGCGTAGCACGG-3′), and pIJ6728 (AGTATG to GCGGCG; 5′-GCGGAGCTGGTCGAGCGGGCCCAG-3′ and 5′-CGCGCGGGCGCTGTCGCTGTCCGC-3′). After phosphorylation of the oligonucleotides, the PCR conditions used were 10 cycles of 96°C for 1 min, 60°C for 45 s, and 72°C for 8 min 30 s; followed by 10 cycles of 96°C for 1 min, 60°C for 45 s, and 72°C for 12 min 30 s; followed by an extension reaction at 72°C for 15 min. In each case the PCR product was self-ligated to recreate a circular plasmid, and the resulting bldN allele was sequenced over its entire length to ensure that only the desired mutation had been introduced. Lastly, each bldN allele was isolated as a BamHI-EcoRI fragment and ligated into pSET152 cut with the same enzymes. The plasmid numbers given above refer to the final pSET152 clones.
Immunoblot analysis.
Streptomyces liquid cultures were harvested by centrifugation, whereas surface-grown cultures were scraped from cellophane-covered R2YE plates. Approximately 500 mg of mycelium from each culture was resuspended in 1 ml of Complete protease inhibitor buffer (Roche Diagnostics) in a 1.5-ml Eppendorf tube, and samples were sonicated at half power for four cycles of 10 s, with cooling on ice in between. Cell debris was removed by centrifugation at 14,000 rpm for 2 min, and the protein concentration of the supernatant was estimated by using Bradford reagent (Bio-Rad). Samples (20 μg) were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to Hybond-C nylon membrane (Amersham Pharmacia Biotech), and probed with a 1:2,500 dilution of rabbit anti-BldN antibody. Horseradish peroxidase-coupled secondary antibody (Amersham Pharmacia Biotech) was used at a 1:5,000 dilution and detected by chemiluminescence by using ECL Western blotting detection reagents (Amersham Pharmacia Biotech).
C-terminally His-tagged σBldN.
A 5′-truncated allele of bldN carrying seven histidine codons at the 3′ end was generated by PCR by using pIJ6714 (1) as the template and the oligonucleotides 5′-GCCGGATCCGCGCGGCTTCGTCCCCACCGC-3′ and 5′-CGGGGATCCTCAGTGGTGGTGGTGGTGGTGGTGGCGGGCGTCGTCGGGGAGCAG-3′. The PCR conditions consisted of 20 cycles of 96°C for 1 min, 60°C for 50 s, and 72°C for 50 s, followed by 72°C for 10 min. The resulting PCR fragment was cut with BamHI and cloned into BamHI-cut pIJ6650 to create pIJ6729. The sequence of the bldN allele was confirmed in pIJ6729, which was introduced by conjugation into S. coelicolor J1915, selecting for apramycin resistance. A single crossover event between the full-length (chromosomal) and truncated copies of bldN to give strain J2178 was confirmed by PCR with an oligonucleotide complementary to the His tag sequence (5′-TCAGTGGTGGTGGTGGTGGTG-3′) and one within the bldN gene, upstream of the truncated version of bldN (5′-GCTACGACGGGTGAATGGTTC-3′).
Construction of a SCO2260 (ORF6 on cosmid C75A) null mutant.
A SCO2260 (putative metalloprotease gene) null mutant derivative of J1915, a plasmid-free, glkA derivative of the wild-type strain, was constructed by using the method of Buttner et al. (4).
A 2.8-kb SmaI fragment (nucleotides 7275 to 10058 from the Sanger Centre sequence) carrying SCO2260, isolated from cosmid C75A, was cloned into SmaI-digested pUC19 to give pIJ6735, and a 1.8-kb BglII hyg cassette (conferring resistance to hygromycin) (42) was cloned into the unique BclI site (at nucleotide 8515) internal to SCO2260 to give pIJ6736. pIJ6736 was digested with NdeI, end filled, digested with HindIII, and cloned into EcoRV- and HindIII-digested pIJ6650 to give pIJ6737.
pIJ6737 was introduced into S. coelicolor J1915 (ΔglkA119) by conjugation from E. coli and exconjugants in which the plasmid had presumptively integrated at the SCO2260 locus by single-crossover homologous recombination were selected with 50 μg of apramycin/ml. After we checked for apramycin and hygromycin resistance and 2-deoxyglucose sensitivity, one such isolate, J2179, was grown nonselectively through four rounds of sporulation and putative SCO2260::hyg mutants in which the delivery plasmid had been lost were selected on minimal medium containing 100 mM 2-deoxyglucose and 50 μg of hygromycin/ml. The structure of one null mutant was confirmed by PCR (using the oligonucleotides 5′-TGTCCACCGCCACCGCCCGTC-3′ from SCO2260 and 5′-GCGACGGTGTACGCCACAGCTTG-3′ from the hyg cassette) and by Southern hybridization, and the strain was designated J2180.
Construction of J2183 and J2184 (bldN::aadA).
bldN-null mutants, in which the entire bldN (SCO3323) coding sequence was precisely replaced by aadA (conferring resistance to both spectinomycin and streptomycin), were constructed by the PCR-targeted method of Gust et al. (B. Gust, G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater, unpublished data). In this method, S. coelicolor genes carried on cosmids in E. coli are replaced with a selectable marker generated by PCR with primers with 39-nucleotide gene-specific extensions. The selectable marker cassette also includes an oriT which permits the direct transfer of the mutagenized cosmid into S. coelicolor by conjugation (Gust et al., unpublished). Cosmid E68 was introduced into E. coli BW25113 (9), and bldN was disrupted by electroporation with an oriT/aadA cassette that had been amplified with oligonucleotides that carried bldN-specific extensions: 5′-CGTACTGCACGTGATGGAAGCTCTGCCTCTGGGAGTCCCATTCCGGGGATCCGTCGACC-3′ (forward) and 5′-CTTGGGGAACACGAAGGGTGAGCGCCTCTGTGGCGTCTCTGTAGGCTGGAGCTGCTTC-3′ (reverse). The resulting cosmid was introduced into S. coelicolor M145 and J1501 by conjugation. Three bldN-null mutant derivatives of each strain, generated by double crossing over, were identified by their spectinomycin-resistant, kanamycin-sensitive, and bald phenotypes. Their structures were confirmed by Southern hybridization, and representative bldN-null mutant derivatives of M145 and J1501 were designated J2183 and J2184, respectively.
Construction of a SCO5695 (ORF19 on cosmid 5H4) null mutant.
A SCO5695 (putative metalloprotease gene) null mutant was constructed by the PCR-directed method of Gust et al. (unpublished), by using an oriT/apr (apramycin resistance) cassette amplified with oligonucleotides with SCO5695-specific extensions: 5′-GGCGGCACAGACGGCGGAGGCCCGTGCATGACGACCCTGATTCCGGGGATCCGTCGACC-3′ (forward) and 5′-GTCCGGAGGGACAGGCTCTCCGGATCGGGCTTCTCGTCGTGTAGGCTGGAGCTGCTTC-3′ (reverse). The resulting SCO5695::apr derivative of cosmid 5H4 was introduced into S. coelicolor M145 by conjugation from E. coli, and two SCO5695 null mutants, generated by double crossing over, were identified by their apramycin-resistant, kanamycin-sensitive phenotype. After their structures were confirmed by Southern hybridization, one was designated J2182. A SCO5695::apr SCO2260::hyg double mutant (J2188) was constructed by repeating the disruption of SCO5695 in J2180.
Expression of σBldN in E. coli.
A truncated form of σBldN (starting at ATG3/Met-87) was overexpressed in E. coli by using the pRSET derivative pIJ6722 (1) and purified as described previously (1). Then, 2 mg of this protein was used to raise a polyclonal antiserum in rabbit (Genosys). In addition, two derivatives of pET11a were constructed to express bldN from GTG1 (Met-1) or ATG4 (Met-88). Pairs of abutting oligonucleotides were used to amplify the entire pIJ6723 plasmid, simultaneously introducing appropriately positioned NdeI sites. The PCR conditions were as described above (PCR-based site-directed mutagenesis), and the oligonucleotides used were as follows: 5′-CATATGTACCCACACGTCGGGGTTG-3′ and 5′-GGGACTCCCAGAGGCAGAGC-3′ (GTG1/Met-1) and 5′-ATGGAGCTGGTCGAGCGGGCC-3′ and 5′-ATGCGGGCGCTGTCGCTGTCC-3′ (ATG4/Met-88). The resulting plasmids, pIJ6731 and pIJ6733, were confirmed by sequencing, and the bldN alleles were removed as NdeI-BamHI fragments and ligated to NdeI-BamHI-digested pET11a to create pIJ6732 (GTG1/Met-1) and pIJ6734 (ATG4/Met-88).
RESULTS AND DISCUSSION
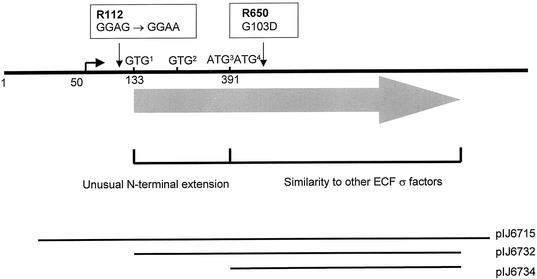
The cloning and characterization of bldN raised important questions about its translation (1). bldN has four in-frame ATG or GTG potential start codons (GTG1, GTG2, ATG3, and ATG4; Fig. 1), only one of which, GTG1, is preceded by a credible ribosome-binding site (GGAG). Translation from GTG1 would give rise to a σ factor with an unusual N-terminal extension of ca. 86 amino acids that is absent from the other σ factors. The discovery that R112, the more severe of the two point mutants that originally defined the bldN locus, had a wild-type protein coding sequence but carried a GGAG→GGAA mutation in the putative ribosome-binding site strongly implied that at least some bldN translation initiated at GTG1 (1). However, although the two adjacent ATG codons lack a recognizable ribosome-binding site, translation initiation at either codon would give rise to a σ factor approximately co-N-terminal with other σ factors (Fig. 1) (1). Moreover, these two ATG codons are conserved within adsA, the orthologue of bldN in S. griseus (40). These considerations raised the possibility that there might be two independent bldN primary translation products, differing in length by 86 or 87 amino acids.
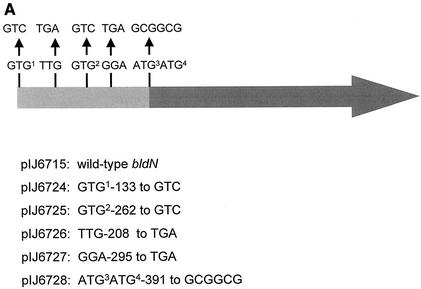
FIG. 1.
Genetic organization of the bldN locus, showing the four in-frame potential start codons, the nature and positions of the two point mutations (R112 and R650) that originally defined the bldN locus, and the extent of DNA carried in various bldN clones. The numbering of the nucleotides is taken from Bibb et al. (1). pIJ6715 is pSET152 carrying a FokI fragment containing bldN; pIJ6732 and pIJ6734 are pET11a-derived constructs expressing bldN from GTG1 and ATG4, respectively.
Two proteins of 28 and 35 kDa are encoded by bldN.
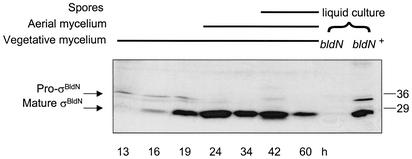
To examine bldN translation experimentally, a polyclonal antiserum was raised against purified protein expressed in E. coli, initiating at ATG3, and used in immunoblots against S. coelicolor extracts. A time course taken during differentiation of the wild type on R2YE solid medium (Fig. 2) showed early production of a cross-reacting species of apparent molecular mass of 35 kDa and the subsequent appearance of a second species of apparent molecular mass of 28 kDa, which was concomitant with aerial mycelium formation and the disappearance of the larger species. In liquid culture, conditions under which S. coelicolor does not sporulate, the 35-kDa species was more persistent (Fig. 2).
FIG. 2.
Immunoblot analysis of σBldN expression during a developmental time course of S. coelicolor M145 grown on R2YE solid medium. The time points at which mycelium was harvested, and the presence of vegetative mycelium, aerial mycelium, and spores, as judged by microscopic examination, are indicated. Control extracts from the bldN-null mutant (J2177) and its congenic bldN+ parent (J1915), both grown in YEME liquid medium, are shown. The positions of the molecular mass markers are indicated on the right in kilodaltons.
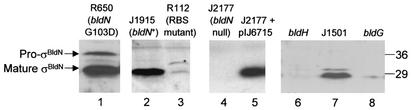
The presence of σBldN was also investigated in a variety of different genetic backgrounds. Immunoblots against single, late time points, in which only the 28-kDa species was present in the bldN+ parent strain J1915 (Fig. 3, lane 2), showed that the 28-kDa species was absent from the constructed bldN-null mutant, J2177 (Fig. 3, lane 4), and was barely detectable in R112, the bldN strain carrying the point mutation in the RBS associated with GTG1 (Fig. 3, lane 3). In R650, a second bldN point mutant carrying a G103D substitution in region 2.1 (Fig. 1) (1), the 35-kDa species was readily detected, despite the late time point (Fig. 3, lane 1; the persistence of the 35-kDa species in R650 was reproducible). Complemention of the null mutant (J2177) with the bldN clone pIJ6715 restored normal levels of the 28-kDa species (Fig. 3, lane 5).
FIG. 3.
Immunoblot analysis of σBldN expression in different genetic backgrounds. The following strains were grown on R2YE solid medium, and late (48 h) samples of mycelium were harvested for analysis: the bldN-null mutant J2177 (lane 4); its congenic bldN+ parent, J1915 (lane 2); the bldN-null mutant J2177 complemented with pIJ6715 (lane 5); the bldN point mutants, R112 (lane 3) and R650 (lane 1); the bldG (lane 8) and bldH (lane 6) mutants and their congenic parent, J1501 (lane 7). The positions of the molecular mass markers are indicated on the right in kilodaltons.
The previously observed transcriptional dependence of bldN on bldG and bldH (1) was confirmed and extended at the protein level. σBldN protein was absent in the bldH mutant, WC109 (Fig. 3, lane 6), and was only faintly detectable in the bldG mutant, WC103 (Fig. 3, lane 8). In contrast, the 28-kDa species was readily detectable in J1501, the congenic parent of both these strains (Fig. 3, lane 7).
To confirm that both the 28-kDa and the 35-kDa cross-reacting species were products of the bldN locus, a strain expressing C-terminally His-tagged BldN was constructed. pIJ6729, a suicide plasmid carrying a 5′-truncated version of bldN with a 3′ tag of seven histidine codons, was introduced into M145 by conjugation. Homologous recombination between the two copies of bldN resulted in a strain (J2178) in which transcription from the bldN promoter gave rise to expression of C-terminally His-tagged BldN. The chromosomal structure of J2178 was confirmed by PCR and sequencing. Immunoblot analysis showed that the His tag caused a decrease in mobility of both the 28- and 35-kDa cross-reacting species, confirming that they are bldN encoded and co-C-terminal (data not shown). J2178 showed reduced aerial mycelium formation relative to M145, implying that the His tag interfered with σ factor function.
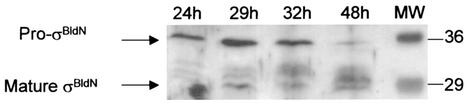
The 35-kDa species was detected only transiently in early time points during differentiation on plates but was more persistent in samples isolated from liquid culture (Fig. 2). To better visualize this species, bldN was cloned into the multicopy vector pIJ487 to give pIJ6739. When pIJ6739 was introduced into S. coelicolor, it caused only moderate overexpression. Despite this, not only was the 35-kDa species readily detectable in liquid cultures, it was the predominant species in early time points (Fig. 4). The presence of multicopy bldN had no obvious phenotypic consequences.
FIG. 4.
Immunoblot analysis of σBldN expression during growth in YEME liquid medium of S. coelicolor carrying bldN on a multicopy plasmid (pIJ6739). Molecular weight markers (MW) are shown in the rightmost lane in kilodaltons.
σBldN is synthesized as a proprotein.
The 28-kDa cross-reacting species could have resulted from posttranslational processing of the 35-kDa species or from translation initiation at a second, independent start site. To distinguish between these two possibilities, site-directed mutagenesis was used to remove each potential start codon in turn and to introduce internal stop codons within bldN. Each of the resulting bldN alleles was cloned into the attP+ integrative vector, pSET152, to generate a series of plasmids (pIJ6724-6728) identical to pIJ6715 (carrying wild-type bldN) except for the mutation indicated in Fig. 5A.
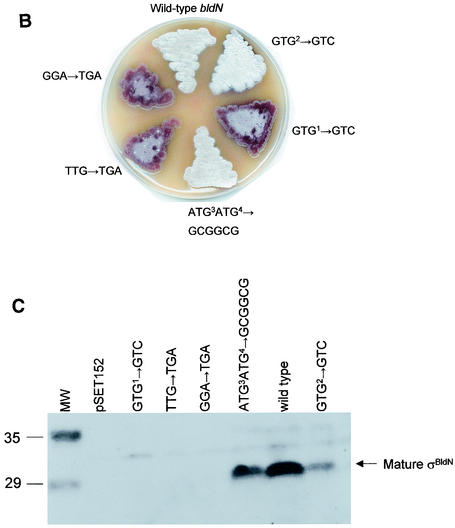
FIG. 5.
Mutational analysis of bldN translation. (A) Positions of the mutations introduced into bldN and the numbers of the plasmids carrying each allele. The numbering of the nucleotides is taken from Bibb et al. (1). (B) Phenotype of the bldN-null mutant (J2184) after the introduction of each bldN allele, grown on R2YE. (C) Immunoblot analysis of σBldN expression in the bldN-null mutant (J2184) after the introduction of each bldN allele. Mycelium was harvested at 48 h from cultures grown on R2YE solid medium. Molecular weight markers (MW) are shown in the leftmost lane in kilodaltons.
Each construct was introduced by conjugation into the bldN-null mutant, J2177. Although the phenotypes of J2177 carrying pIJ6724-6728 were unambiguous after the primary introduction of the plasmids, on repeated subculture the isolates carrying noncomplementing plasmids began progressively to take on a morphologically wild-type appearance. Because the allele carried by the null mutant J2177 replaced a 66-bp XhoI fragment internal to bldN with a hyg cassette (1) but left the remainder of the gene present (including GTG1, GTG2, ATG3, and ATG4), we speculated that the progressive change in phenotype might be caused by homogenotization. To prevent homogenotization, a new null mutant (J2184) was constructed in which the entire bldN coding sequence was replaced by an aadA cassette. The phenotypes of J2184 carrying pIJ6724-6728 were stable, suggesting that homogenotization was indeed the cause of the phenotypic instability associated with J2177 carrying pIJ6724-6728.
pIJ6725, carrying a mutation of GTG2 to GTC, fully complemented the bldN-null mutant J2184 (Fig. 5B) and restored production of the 28-kDa species (Fig. 5C), demonstrating that GTG2 is not an alternative initiation codon. In contrast, mutation of GTG1 to GTC (pIJ6724) completely abolished complementation of the bldN-null mutant phenotype (Fig. 5B) and no σBldN protein was detectable by immunoblot (Fig. 5C). Moreover, introduction of a TGA stop codon, either between GTG1 and GTG2 or between GTG2 and ATG3, also resulted in clones (pIJ6726 and pIJ6727, respectively) that could no longer complement the bldN-null mutant (Fig. 5B) and produced no detectable σBldN protein (Fig. 5C). The absence of σBldN protein in these strains suggested either (i) that the 28-kDa σBldN species arose from posttranslational processing of a 35-kDa pro-σBldN species, or (ii) that both the mutation of GTG1 and the introduced stop codons were blocking translation initiation at ATG3 or ATG4 through polar effects. To distinguish between these two alternatives, ATG3 and ATG4 were mutated to two alanine codons (GCGGCG), resulting in pIJ6728 (Fig. 5A). pIJ6728 fully complemented the bldN-null mutant (Fig. 5B) and restored wild-type levels of the 28-kDa σBldN species (Fig. 5C). These data are therefore consistent with the 28-kDa species (mature σBldN) arising by proteolytic processing of the 35-kDa species (pro-σBldN).
Pro-σBldN processing.
Translation initiation at GTG1 would give rise to a primary translation product of predicted molecular mass of 28.6 kDa, whereas translation initiation at ATG4 would give rise to one of 20.16 kDa. However, because many σ factors have highly aberrant mobilities on SDS-polyacrylamide gels, appearing larger than their actual size, it was not immediately possible to correlate predicted primary translation products with the experimentally observed bldN-encoded 28- and 35-kDa apparent molecular masses of the species. Two forms of σBldN were therefore expressed in E. coli and then purified, and their mobilities were compared with the species detected in S. coelicolor.
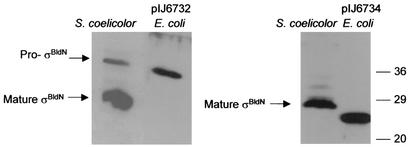
σBldN protein expressed from GTG1 (pIJ6732) had the same mobility as the large (35-kDa) species observed in S. coelicolor, confirming that pro-σBldN arises from translation initiation at GTG1 (Fig. 6). σBldN protein expressed from ATG4 (pIJ6734) had an apparent mass of ∼26 kDa, 2 kDa smaller than the mature (28-kDa) σBldN species observed in S. coelicolor (Fig. 6). This result suggested that the processing event that gives rise to mature σBldN occurs close to, but on the N-terminal side of, Met-87 (corresponding to ATG4; 2 kDa would correspond to ca. 18 residues). Repeated attempts to use nickel affinity chromatography to purify sufficient C-terminally His-tagged, mature σBldN to determine its N-terminal sequence were unsuccessful.
FIG. 6.
Immunoblot comparison of the pro-σBldN and mature σBldN species observed in S. coelicolor with recombinant σBldN protein expressed in E. coli from GTG1 (pIJ6732) or ATG4 (pIJ6734). The positions of the molecular mass markers are indicated on the right in kilodaltons.
Attempts to identify the gene encoding the pro-σBldN processing enzyme.
SpoIIGA and SpoIVFB, the proteases in B. subtilis responsible for processing pro-σE and pro-σK, respectively, have been characterized (25, 29, 30, 32). Database searches of the S. coelicolor genome failed to identify full-length homologues of these enzymes. However, Rudner et al. (33) and Yu and Kroos (41) assigned SpoIVFB and Site-2 protease (S2P), the enzyme responsible for cleavage of mammalian SREBP at site 2 (31), to a novel family of metalloproteases containing two signature motifs, HEXXH and NXXPXXXXDG. A search of the S. coelicolor genome database (http://www.sanger.ac.uk/Projects/S_coelicolor/) revealed two putative metalloproteases containing both motifs: SCO2260 (ORF6 from cosmid C75A) and SCO5695 (ORF19 from cosmid 5H4). To investigate the possibility that one or both of these proteins might be responsible for pro-σBldN processing, SCO2260 and SCO5695 were disrupted to yield J2180 and J2182, respectively, and a double mutant, J2188, was also constructed. All three strains were unaffected in pro-σBldN processing and were morphologically wild type (data not shown).
Conclusions.
Using a combination of site-directed mutagenesis and immunoblot analysis, we have provided evidence that all bldN translation arises from initiation at GTG1, and that the primary translation product is a proprotein (pro-σBldN) that is proteolytically processed to mature σBldN by removal of most of an unusual ∼86-residue N-terminal extension. In the absence of the N-terminal sequence of the 28-kDa species (mature σBldN), it is still possible that the change in SDS-polyacrylamide gel migration could reflect intein processing or a chemical modification that dramatically alters the mobility of the primary translation product, although we consider both of these possibilities unlikely. During differentiation on solid medium, pro-σBldN is detected in early time points and mature σBldN appears subsequently, concomitant with aerial mycelium formation and the disappearance of pro-σBldN. Thus, σBldN is developmentally regulated at at least two levels: transcription initiation and posttranslational processing. This is the first report of pro-σ factor processing in a bacterium outside of the genus Bacillus.
Placing bldN on a multicopy plasmid caused only mild overexpression of the protein, perhaps implying titration of the factors under the control of bldG and bldH that are required for activation of its transcription. Despite this, the low level of overexpression caused pro-σBldN to accumulate, suggesting that the processing enzyme is readily saturated. Pro-σBldN also persisted in the bldN point mutant R650, showing that the σBldN G103D substitution in this strain has a weak negative influence on pro-σBldN processing. Growth in liquid culture, conditions under which S. coelicolor does not sporulate, also led to increased persistence of pro-σBldN.
We identified two S. coelicolor genes encoding putative members of a family of metalloproteases that includes SpoIVFB and S2P, the proteases responsible for cleavage of pro-σK and SREBP Site-2, respectively. Both genes were dispensable for differentiation and pro-σBldN processing. However, the proteases involved in the regulated proteolysis of transcription factors are likely to be diverse, since SpoIVFB and S2P are not related to the pro-σE processing enzyme, SpoIIGA. There are no homologues of SpoIIGA encoded by S. coelicolor.
In other systems, transcription factor processing serves to coordinate transcription factor activity with other events. It will be interesting in the future to identify mutants that are blocked in pro-σBldN processing and to use these mutants to begin to understand how pro-σBldN processing is controlled during differentiation.
Acknowledgments
We thank Mark Paget for helpful discussion and Keith Chater and David Hopwood for helpful comments on the manuscript.
This work was supported by a grant-in-aid to the John Innes Centre from the BBSRC.
REFERENCES
- 1.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 4.Buttner, M. J., K. F. Chater, and M. J. Bibb. 1990. Cloning, disruption and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J. Bacteriol. 172:3367-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chater, K. F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F., C. J. Bruton, A. A. King, and J. E. Suarez. 1982. The expression of Streptomyces and Escherichia coli drug resistance determinants cloned into the Streptomyces phage φC31. Gene 19:21-32. [DOI] [PubMed] [Google Scholar]
- 8.Cutting, S., V. Oke, A. Driks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell 62:239-250. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan, E. A., M. S. Brown, J. L. Goldstein, and J. Sakai. 1997. Cleavage site for sterol-regulated protease localized to a Leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J. Biol. Chem. 272:12778-12785. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, E. A., U. P. Davé, J. Sakai, J. L. Goldstein, and M. S. Brown. 1998. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273:17801-17809. [DOI] [PubMed] [Google Scholar]
- 12.Elliot, M. A., M. J. Bibb, M. J. Buttner, and B. K. Leskiw. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:257-269. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 43:27-38. [DOI] [PubMed] [Google Scholar]
- 14.Gomez, M., S. Cutting, and P. Stragier. 1995. Transcription of spoIVB is the only role of σG that is essential for pro-σK processing during spore formation in Bacillus subtilis. J. Bacteriol. 177:4825-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoa, N. T., J. A. Brannigan, and S. M. Cutting. 2002. The Bacillus subtilis signaling protein SpoIVB defines a new family of serine peptidases. J. Bacteriol. 184:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmeister, A. 1998. Activation of the proprotein transcription factor pro-σE is associated with its progression through three patterns of subcellular localization during sporulation in Bacillus subtilis. J. Bacteriol. 180:2426-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 18.Hua, X., J. Sakai, Y. K. Ho, J. L. Goldstein, and M. S. Brown. 1995. Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J. Biol. Chem. 270:29422-29427. [DOI] [PubMed] [Google Scholar]
- 19.Ju, J., and W. G. Haldenwang. 1999. The “pro” sequence of the sporulation-specific σ transcription factor σE directs it to the mother cell side of the sporulation septum. J. Bacteriol. 181:6171-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju, J., T. Luo, and W. G. Haldenwang. 1997. Bacillus subtilis pro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J. Bacteriol. 179:4888-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelemen, G. H., and M. J. Buttner. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1:656-662. [DOI] [PubMed] [Google Scholar]
- 22.Kelemen, G. H., K. A. Plaskitt, C. G. Lewis, K. Findlay, and M. J. Buttner. 1995. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 17:221-230. [DOI] [PubMed] [Google Scholar]
- 23.Kelemen, G. H., P. H. Viollier, J. Tenor, L. Marri, M. J. Buttner, and C. J. Thompson. 2001. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40:804-814. [DOI] [PubMed] [Google Scholar]
- 24.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 25.Londono-Vallejo, J. A. 1997. Mutational analysis of the early forespore/mother-cell signalling pathway in Bacillus subtilis. Microbiology 143:2753-2761. [DOI] [PubMed] [Google Scholar]
- 26.MacNeil, D. J., J. L. Occi, K. M. Gewain, T. MacNeil, P. H. Gibbons, C. L. Ruby, and S. L. Danis. 1992. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115:119-125. [DOI] [PubMed] [Google Scholar]
- 27.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene for the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 29.Peters, H. K., and W. G. Haldenwang. 1991. Synthesis and fractionation properties of SpoIIGA, a protein essential for pro-σE processing in Bacillus subtilis. J. Bacteriol. 173:7821-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piggot, P. J., and R. Losick. 2001. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 31.Rawson, R. B., N. G. Zelenski, D. Nijhawan, J. Ye, J. Sakai, M. T. Hasan, T. Y. Chang, M. S. Brown, and J. L. Goldstein. 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1:47-57. [DOI] [PubMed] [Google Scholar]
- 32.Resnekov, O., and R. Losick. 1998. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1998 95:3162-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudner, D. Z., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 96:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryding, N. J., M. J. Bibb, V. Molle, K. C. Findlay, K. F. Chater, and M. J. Buttner. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J. Bacteriol. 181:5419-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai, J., E. A. Duncan, R. B. Rawson, X. Hua, M. S. Brown, and J. L. Goldstein. 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85:1037-1046. [DOI] [PubMed] [Google Scholar]
- 36.Sato, R., J. Yang, X. Wang, M. J. Evans, Y. K. Ho, J. L. Goldstein, and M. S. Brown. 1994. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1). J. Biol. Chem. 269:17267-17273. [PubMed] [Google Scholar]
- 37.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53-62. [DOI] [PubMed] [Google Scholar]
- 39.Ward, J. M., G. R., Janssen, T. Kieser, M. J. Bibb, M. J. Buttner, and M. J. Bibb. 1986. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468-478. [DOI] [PubMed] [Google Scholar]
- 40.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, Y. T., and L. Kroos. 2000. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zalacaín, M., A. González, M. C. Guerrero, R. J. Mattaliano, F. Malpartida, and A. Jiménez. 1986. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 14:1565-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, B., A. Hofmeister, and L. Kroos. 1998. The prosequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 180:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]