Abstract
Microorganisms in the family Geobacteraceae are the predominant Fe(III)-reducing microorganisms in a variety of subsurface environments in which Fe(III) reduction is an important process, but little is known about the mechanisms for electron transport to Fe(III) in these organisms. The Geobacter sulfurreducens genome was found to contain a 10-kb chromosomal duplication consisting of two tandem three-gene clusters. The last genes of the two clusters, designated omcB and omcC, encode putative outer membrane polyheme c-type cytochromes which are 79% identical. The role of the omcB and omcC genes in Fe(III) reduction in G. sulfurreducens was investigated. OmcB and OmcC were both expressed during growth with acetate as the electron donor and either fumarate or Fe(III) as the electron acceptor. OmcB was ca. twofold more abundant under both conditions. Disrupting omcB or omcC by gene replacement had no impact on growth with fumarate. However, the OmcB-deficient mutant was greatly impaired in its ability to reduce Fe(III) both in cell suspensions and under growth conditions. In contrast, the ability of the OmcC-deficient mutant to reduce Fe(III) was similar to that of the wild type. When omcB was reintroduced into the OmcB-deficient mutant, the capacity for Fe(III) reduction was restored in proportion to the level of OmcB production. These results indicate that OmcB, but not OmcC, has a major role in electron transport to Fe(III) and suggest that electron transport to the outer membrane is an important feature in Fe(III) reduction in this organism.
Microbial oxidation of organic matter coupled to the reduction of Fe(III) plays an important role in the degradation of natural and contaminant organic matter in aquatic sediments and the subsurface, and this process influences the mobility of a variety of trace metals in these environments (for reviews, see references 23 and 24). Insoluble Fe(III) oxides, which are the primary form of Fe(III) in most soils and sediments, are unlike commonly considered electron acceptors such as oxygen, nitrate, fumarate, sulfate, and carbon dioxide, which are soluble and can readily enter a cell to be reduced. If, as is generally considered to be the case (23, 43), Fe(III)-reducing microorganisms reduce extracellular Fe(III), then electron transport to Fe(III) is likely to proceed via mechanisms significantly different from those for the reduction of soluble electron acceptors.
Furthermore, phylogenetically distinct Fe(III) reducers might reduce Fe(III) oxides via significantly different mechanisms. For example, Geothrix fermentans and Shewanella species produce soluble electron shuttles and/or Fe(III)-chelating compounds which alleviate the need for contact between cells and insoluble Fe(III) for Fe(III) reduction (38-40, 54). Others, such as Geobacter metallireducens, do not produce electron shuttles or Fe(III) chelators (37) but utilize appendages such as flagella and pili to search for or to make direct contact with insoluble Fe(III) (6).
The mechanisms for Fe(III) reduction in Geobacter species are of particular interest, because molecular analysis of microbial community structure has demonstrated that microorganisms in the Geobacteraceae family are the predominant Fe(III)-reducing microorganisms in a wide variety of sedimentary environments in which Fe(III) reduction is the terminal electron-accepting process (18, 22, 44, 45, 51, 52). Early studies with G. metallireducens demonstrated that the Fe(III) reductase activity in G. metallireducens was localized in the membrane (15), and this was subsequently confirmed in studies with G. sulfurreducens (14, 29). A membrane-bound NADH-dependent Fe(III) reductase complex, consisting of at least six different proteins, including an 89-kDa c-type cytochrome designated FerA, was partially purified from G. sulfurreducens and characterized (29, 30). These results suggested that FerA is involved in the final step of Fe(III) reduction. However, the fact that many proteins with redox activity can nonspecifically transfer electrons to Fe(III) (19) suggested that further evaluation of the in vivo role of these proteins was required. Such studies are now possible due to the development of a genetic system for G. sulfurreducens (9).
A search of the G. sulfurreducens genome revealed two genes similar to ferA, designated omcB and omcC. Both genes encode polyheme c-type cytochromes, which are predicted to be localized in the outer membrane. The omcB and omcC genes, which are 79% identical, are within a 10-kb chromosomal duplication. Here we report on the roles of OmcB and OmcC in Fe(III) reduction in G. sulfurreducens.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
Escherichia coli strain DH5α [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (17, 55) was used for DNA manipulations. Targeted gene disruption experiments were performed on Geobacter sulfurreducens strain DL1 (4) to produce strains DL5 (omcC::kan) and DL6 (omcB::cam). G. sulfurreducens strains were routinely cultured anaerobically in either acetate-fumarate or acetate-Fe(III) citrate medium as previously described (9).
DNA manipulations and reverse transcriptase (RT) PCR conditions.
Genomic DNA was extracted using the MasterPure Complete DNA & RNA purification kit (Epicentre Technologies, Madison, Wis.). Plasmid DNA purification and PCR product purification were carried out using Mini Plasmid purification kits and PCR purification kits, respectively (Qiagen Inc., Valencia, Calif.). DNA cloning and other manipulations were carried out according to the methods outlined by Sambrook et al. (47). Restriction enzymes and other DNA-modifying enzymes were purchased from New England Biolabs, Beverly, Mass. Using the Multiprime DNA labeling system (Amersham Pharmacia Biotech, Piscataway, N.J.), probes for Southern blot analysis were labeled with [α-32P]dCTP. [α-32P]dCTP was purchased from New England Nuclear (Boston, Mass.). Unless otherwise stated, all primers used to amplify G. sulfurreducens sequences were designed using the preliminary sequence of the G. sulfurreducens genome (available at www.tigr.org). Taq DNA polymerase (Qiagen Inc.) was used for all PCR amplifications.
RT PCR was utilized to determine whether omcB and omcC were expressed during growth in the presence of two electron acceptors, fumarate and Fe(III) citrate. Due to the identical 5′ ends of the omcB and omcC genes, only one sense primer, 8907 (5′-CCGAAAACAAGGAGGGGACGG-3′), was designed. The most divergent regions of the omcB and omcC genes were utilized to design antisense primers specific for either omcC (89WR-2 [5′-GTTAGGCCGGTCCGCTCCG-3′]) or omcB (8908-2 [5′-GGTCAGCAGGCCACCGG-3′]). Using RNeasy Mini kits (Qiagen Inc.) followed by treatment with RNase-free DNase (Ambion, Inc., Austin, Texas), total RNA was purified from mid-log cultures. RT PCR was performed using the Thermoscript RT PCR system (Life Technologies, Rockville, Maryland) according to the manufacturer's instructions.
Disruption of the omcB and omcC genes via single-step gene replacement.
Single-step gene replacement was performed according to the protocols described by Lloyd et al. (21). To disrupt the omcC gene, a linear DNA fragment of 2.3 kb, containing the kanamycin resistance (Kanr) marker flanked by the sequence upstream of omcC and the 3′ end of omcC (Fig. 1A), was generated by recombinant PCR (21, 33). Three primary PCRs were carried out as follows: (i) primers 8904 (5′-CCGCAACCTGCAGTCGTGCC-3′) and 8903-2 (5′-GGCATGGTTGCCGTTGACG-3′) were used to amplify the upstream sequence of the omcC gene (594 bp from −209 to +385); (ii) primers 8902 (5′-CCTATCCTGATATCAAAGGG-3′) and 8901 (5′-CCCATGACATCTTGCCAGAGG-3′) were used to amplify the 3′ end of the omcC gene (606 bp from +1471 to +2077); and (iii) primers 8905 (5′-AGACCCTTTGATATCAGGATAGGATGGCAGGTTGGGCGTCGC-3′) and 8906 (5′-CGTCAACGGCAACCATGCCACCTGGGATGAATGTCAGC-3′) were used to amplify the Kanr cassette (1,114 bp), with pBBR1MCS-2 (20) as the template. Using the three PCR products as templates, recombinant PCR was carried out with distal primers 8901 and 8904. PCR conditions were similar to those described by Lloyd et al. (21), except that the annealing temperature was 55°C.
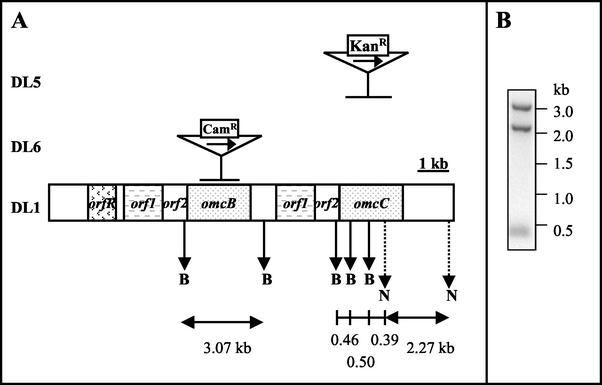
FIG. 1.
Organization of the gene duplication in G. sulfurreducens and mutation schemes. (A) Two gene clusters, orf1/orf2/omcB and orf1/orf2/omcC, were identified within a 10-kb duplication region in G. sulfurreducens (DL1). The central part of either omcC or omcB was replaced with a kanamycin or a chloramphenicol cassette in the mutant DL5 (omcC::kan) or DL6 (omcB::cam), respectively. Gene replacement is indicated by a horizontal bar. The transcription direction of the Kanr and Camr cassettes is the same of that of the omcC and omcB genes and is indicated by a horizontal arrow. A partial restriction enzyme map is shown with vertical arrows indicating BamHI sites (capital B) and NcoI sites (capital N; dotted line). (B) Southern blot of genomic DNA prepared from wild-type G. sulfurreducens (DL1). Genomic DNA digested with restriction enzymes BamHI and NcoI was probed with the omcC gene amplified with primers 8901 and 8904 (primer sequences listed in Materials and Methods). Expected radiolabeled bands are 2.27, 0.5, 0.46, and 0.39 kb. However, due to the high identity between omcB and omcC, an extra band of 3.07 kb, corresponding to the omcB gene, was also detected in the blot. The blot analysis proves that both omcB and omcC genes exist in the G. sulfurreducens genome. Restriction map was based on sequence data obtained from The Institute for Genetic Research website (http://www.tigr.org).
To disrupt the omcB gene, a similar strategy was used. Using plasmid pACYC184 as the template (5, 46), three primary PCRs were carried out as follows: (i) primers 8904 (5′-CCGCAACCTGCAGTCGTGCC-3′) and 8909 (5′-GCTGTGTTGTGGGGTGAGCTC-3′) were used to amplify the sequence upstream of omcB (439 bp from −209 to +230); (ii) primers 8910 (5′-GGGTGCAGCGTTCAACGCC-3′) and 8911 (5′-GCAACTCATGAGCTAATGGG-3′) were used to amplify the downstream sequence of omcB (541 bp from +2013 to +2553); and (iii) primers 89camfor (5′-CAGAAGAGCTCACCCCACAACACGGAAGATCACTTCGC-3′) and 89camrev (5′-GGCGTTGAACGCTGCACCCAGGGCACCAATAACTGCC-3′) were used to amplify a chloramphenicol resistance (Camr) cassette (919 bp). Recombinant PCR was performed as described above by combining all three primary products with distal primers 8904 and 8911. PCR conditions were identical, except that the annealing temperature was 58°C.
Electroporation, mutant isolation, and genotype confirmation were performed as described by Coppi et al. (9) and Lloyd et al. (21). One of each of the mutants, designated DL5 (omcC::kan) and DL6 (omcB::cam), was chosen as the representative strain (Fig. 1A).
Expression of the omcB gene in trans.
The complete omcB coding sequence was amplified using the primers cyt89for (5′-GGGAATTCGGTACGTACTAATTGAGAGC-3′; the EcoRI site is italicized) and cyt89rev-2 (5′-GGGAGATCTGGAAAGCATGATCTGCTTGC-3′; the BglII site is underlined) and the following conditions: 96°C for 40 s, followed by 25 cycles of 96°C for 40s, 58°C for 1 min, and 72°C for 2 min and a final extension at 72°C for 10 min. The omcB coding sequence was digested with EcoRI and BglII and inserted into the EcoRI and BamHI sites of the expression vector pCDS (as described below) to generate pCDS-omcB. The omcB gene was then sequenced to screen for PCR artifacts.
Following electroporation of strain DL6 with pCDS-omcB, a streptomycin-resistant transformant was isolated and designated DL6/pCDS-omcB. The simultaneous presence of both the plasmid pCDS-omcB and the omcB::cam mutation in this strain was confirmed by PCR screening.
The plasmid pCDS was constructed by replacing the Kanr cassette of plasmid pCD342 (11) with a streptomycin resistance (Strr) cassette. Using primers prDL7 and prDL8 (9; BglII and BstB I sites at the 5′ end), the Strr cassette was amplified from the plasmid pJRD215 (10). Following digestion of the cassette with BglII and BstBI, it was inserted into corresponding sites of pCD342.
Detection of OmcB and OmcC in the membrane fraction.
The membrane fraction of G. sulfurreducens was isolated as described by Kaufmann and Lovley (19), except that cells were disrupted by a French pressure cell at 40,000 kPa and 25 mM MOPS (morpholinepropanesulfonic acid) (pH 6.5) was used for washing and resuspension. Membrane proteins (50 μg) were analyzed by Tris-Tricine denaturing polyacrylamide gel electrophoresis (1). c-type cytochromes were detected by staining with N,N,N′,N′-tetramethylbenzidine as previously described (13, 53). Molecular weight markers (LMW electrophoresis calibration kit) were purchased from Amersham Pharmacia Biotech Inc. and were stained separately with Coomassie blue R-250. The Tris-Tricine gel image was digitized using a Scanjet 7400C series scanner (Hewlett Packard, Palo Alto, Calif.). The levels of OmcB and OmcC proteins were determined (according to pixel density) using Labworks version 4.0 software (UVP Inc., Upland, Calif.).
Determination of Fe(III)-reducing ability of cell suspensions.
Cell suspension experiments were carried out as previously described (27), except that cells were washed and resuspended in an isosmotic basal wash medium containing the following ingredients: 0.42 g of KH2PO4/liter, 0.22 g of K2HPO4/liter, 0.38 g of KCl/liter, 4.96 g of NaCl/liter, 0.04 g of CaCl2 · 2H2O/liter, 0.1 g of MgSO4 · 7H2O/liter, 1.8 g of NaHCO3/liter, and 0.5 g of Na2CO3/liter.
Analytical techniques.
Growth of fumarate cultures was assessed by measuring turbidity at 600 nm in a Genesys 2 spectrophotometer (Spectronic Instruments, Rochester, N.Y.). Cell densities of Fe(III)-grown cultures were determined by epifluorescence microscopy using acridine orange staining (28). Fe(II) concentrations were determined with the ferrozine assay as previously described (27). Protein concentration was determined by the bicinchoninic acid method with bovine serum albumin as a standard (50).
RESULTS
Analysis of the gene cluster surrounding omcB and omcC.
The ferA gene sequence could not be found in the G. sulfurreducens genome, but two other similar genes were discovered. Both genes, designated omcB and omcC (outer membrane c-type cytochrome), respectively, encode a putative outer membrane c-type cytochrome with multiple heme-binding sites. They share 79% identity and are located within a 10-kb chromosomal duplication (DL1; Fig. 1A). Southern blot analysis confirmed that the chromosomal duplication containing the omcB and omcC genes was indeed present within the G. sulfurreducens genome and was not a sequencing artifact (Fig. 1B).
The 1.9-kb sequences preceding omcB and omcC are identical with the exception of a single base pair change. This duplicated sequence contains two consecutive open reading frames designated orf1 (1.2 kb) and orf2 (0.6 kb). The orf1 gene encodes a protein of unknown function, and the orf2 gene encodes a putative periplasmic c-type cytochrome. The 0.7 kb of sequence upstream of the two orf1 genes is divergent. There is an open reading frame, designated orfR, which is immediately upstream of the orf1/orf2/omcB cluster and which encodes a putative TetR family transcriptional regulator.
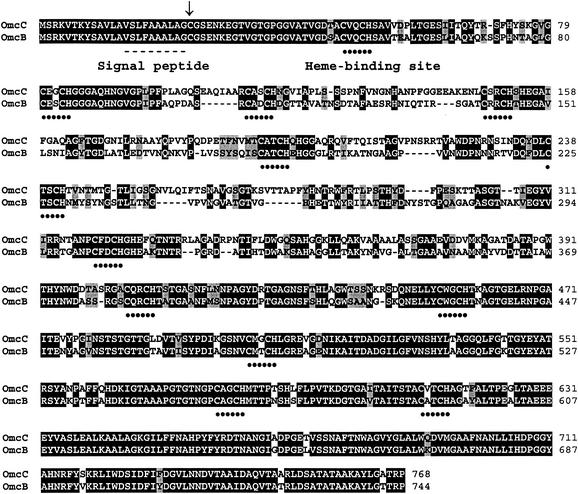
A comparison of the predicted amino acid sequences of OmcB and OmcC indicated that they share 73% identity and that the N (first 100 residues) and C termini (last ∼375 residues) share the highest degree of sequence identity, 85 and 88% for the N and C termini, respectively (Fig. 2). OmcB and OmcC have identical N-terminal signal peptides (Fig. 2). These signal peptides are homologous to those of other lipoproteins and contain a cysteine residue (Fig. 2) which is predicted to serve as a lipid attachment site (Wisconsin Package version 10.2; Genetics Computer Group, Madison, Wis.). The absence of an aspartate residue directly downstream of this cysteine suggests that OmcB and OmcC are likely to be outer membrane associated (42). Both OmcB and OmcC contain 12 heme-binding domains at similar positions in the two proteins. Both proteins are predicted to have the same isoelectric point, pH 6.57, and have very similar Kyte-Doolittle hydrophobicity plots. Hydrophobic and hydrophilic residues were distributed evenly throughout the rest of the proteins, which further suggests that OmcB and OmcC are not inner membrane associated. The predicted secondary structures of both proteins showed that both contain domains of coils intercepted with short helices and beta strands (31). The predicted molecular masses of OmcB and OmcC are 85.5 and 88.9 kDa, respectively.
FIG. 2.
Amino acid sequence alignments of OmcB and OmcC. Identical and conservatively substituted residues are highlighted in black and gray, respectively. Heme-binding domains (CXXCH) are indicated with dotted lines. The signal peptide (dashed line) and lipid attachment site (arrow) are indicated for both proteins.
There were no protein sequences significantly similar to those of OmcB or OmcC in the databases available on the National Center for Biotechnology Information web site or among the available genome sequences of Desulfovibrio vulgaris and S. oneidensis (www.tigr.org). OmcB and OmcC show very weak homology to several other multiheme c-type cytochromes; however, it is limited to the heme binding motifs.
Expression of OmcB and OmcC.
RT PCR analysis of omcB and omcC mRNA confirmed that both genes were expressed when G. sulfurreducens was grown with either fumarate or Fe(III) citrate as the electron acceptor (data not shown). However, due to the high degree of similarity in their physical properties, it was extremely difficult to distinguish the protein products of omcB and omcC on a polyacrylamide gel. To identify OmcB and OmcC and determine their physiological roles, mutants lacking OmcB (DL6 [omcB::cam]) and OmcC ([DL5 omcC::kan]) were constructed. In both of these mutants, the central portion of the gene was replaced with an antibiotic resistance cassette (Fig. 1A).
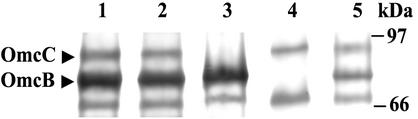
It was possible to resolve the two c-type cytochromes by running a Tris-Tricine gel until ca. 50-kDa proteins were off the gel (Fig. 3). Each mutant was missing a single membrane-bound c-type cytochrome, making it possible to identify OmcB and OmcC.
FIG. 3.
Heme-staining and Tricine-polyacrylamide gel electrophoresis of membrane fractions prepared from DL1, DL5 (omcC::kan), DL6 (omcB::cam), and the complemented strain DL6/pCDS-omcB. Membrane fractions were prepared from cultures grown with acetate-fumarate (lane 1, DL1; lane 3, DL5; lane 4, DL6) or acetate-Fe(III) citrate (lane 2, DL1; lane 5, DL6/pCDS-omcB). Membrane proteins (50 μg) were resolved on a 10% Tris-Tricine polyacrylamide gel and stained for heme. OmcB and OmcC are indicated with arrows.
Wild-type cells produced both OmcB and OmcC during growth with either fumarate or Fe(III) as the electron acceptor (Fig. 3, lanes 1 and 2). There was more (∼160%) OmcB than OmcC during growth on both electron acceptors. Levels of OmcB were comparable in fumarate- and Fe(III)-grown cells. There was ∼20% less OmcC in Fe(III)-grown cells than in fumarate-grown cells (Fig. 3).
Characterization of mutant DL5 (omcC::kan) and DL6 (omcB::cam).
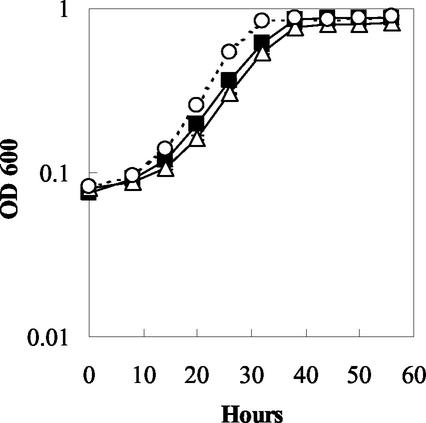
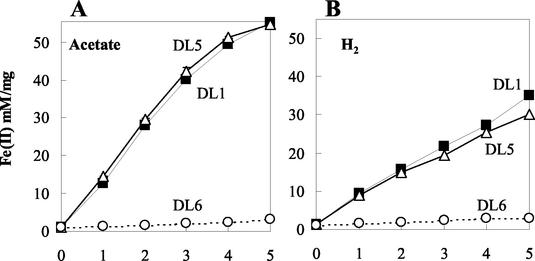
The OmcB and OmcC knockout mutants grew as well as the wild type in fumarate medium (Fig. 4). When washed cell suspensions of fumarate-grown cells were suspended in bicarbonate buffer containing either acetate or hydrogen as the electron donor and Fe(III) citrate as the electron acceptor, the mutant lacking omcC (DL5) reduced Fe(III) as well as the wild type (Fig. 5). However, Fe(III) reduction by the OmcB-deficient mutant (DL6) was greatly reduced (Fig. 5). With acetate, the rate of Fe(III) reduction in the OmcB-deficient mutant (DL6) was only 3.8% of that of the wild type. With hydrogen, Fe(III) reduction was only 6.2% of that of the wild type.
FIG. 4.
Growth study showing results for DL1 (filled square), DL5 (omcC::kan) (empty triangle), and DL6 (omcB::cam) (empty circle) in the medium containing acetate as the electron donor and fumarate as the electron acceptor. Cell growth was measured at A600 over time. Data are means ± standard deviations (SD) of triplicates.
FIG. 5.
Cell suspension assay showing the production of Fe(II) by DL1 and mutants DL5 (omcC::kan) and DL6 (omcB::cam). Each sample was assayed in triplicate, and all assays were done in parallel. Acetate (A) or hydrogen (B) was supplied as the electron donor. Symbols: DL1 (filled square); DL5 (empty triangle); DL6 (empty circle). Data are means ± SD of triplicates.
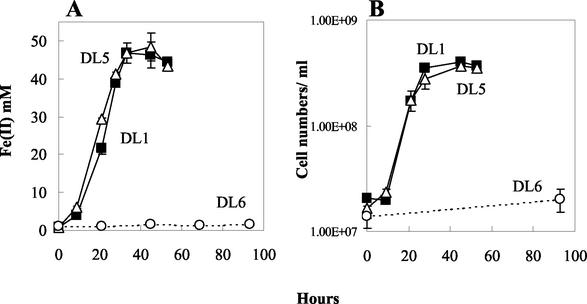
The OmcC-deficient mutant (DL5) grew as well as the wild type with acetate as the electron donor and either Fe(III) citrate (Fig. 6) or poorly crystalline Fe(III) oxide (data not shown) as the electron acceptor. In contrast, growth of the OmcB mutant (DL6) was considerably diminished with Fe(III) citrate (Fig. 6) and growth of the mutant (DL6) was not sustainable with Fe(III) oxide (data not shown).
FIG. 6.
Growth of DL1 (filled square), DL5 (empty triangle), and DL6 (empty circle) in medium containing acetate as the electron donor and Fe(III) citrate as the electron acceptor. Mid-log (A600 = ∼0.3) fumarate-grown cells were inoculated (3% inoculum) into fresh Fe(III) citrate medium. (A) Fe(II) production; (B) cell density. Data are means ± SD of triplicates.
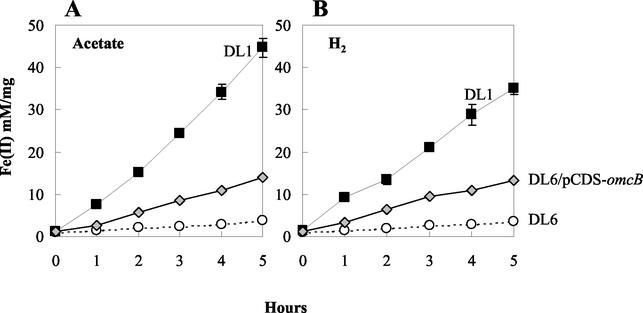
When omcB was reintroduced into the OmcB-deficient mutant (DL6) in trans, OmcB was produced at about 40% of the wild-type level (Fig. 3, lane 5). This was presumably due to two reasons. First, it is difficult to maintain selective pressure with streptomycin in liquid medium. Even though sufficient antibiotic was added, some cells lacking the omcB expression vector, pCDS-omcB, might have been able to grow. Another potential reason is that the omcB gene was not under the control of its original operator/promoter.
Washed cell suspensions of the cells in which the expression of OmcB was restored reduced Fe(III) at a rate 30 or 38% of that of the wild type in medium containing acetate or hydrogen as an electron donor, respectively (Fig. 7). This is commensurate with the level of OmcB in these cells. Reintroduction of omcB also restored the capacity for growth on Fe(III); however, the lag period prior to growth was significantly longer than that of the wild type.
FIG. 7.
Cell suspensions assay showing the production of Fe(II) by DL1 and mutants DL6 and the complemented strain DL6/pCDS-omcB. Each sample was assayed in triplicate, and all assays were done in parallel. Acetate (A) or hydrogen (B) was supplied as the electron donor. Symbols: DL1 (filled square); DL6 (empty circle); DL6/pCDS-omcB (triangle shaded in gray). Data are means ± SD of triplicates.
DISCUSSION
The results suggest that the polyheme c-type cytochrome, OmcB, is an important component in electron transport to Fe(III) in G. sulfurreducens but that the highly similar OmcC probably is not. There have been several reports of proteins purified from G. sulfurreducens that can reduce Fe(III) in vitro (19, 20, 30, 48), but OmcB is the first putative outer membrane protein shown to have an in vivo role in Fe(III) reduction in this organism. As discussed in detail below, although c-type cytochromes like OmcB have the ability to transfer electrons to Fe(III), it is not yet possible to definitively state whether OmcB is an Fe(III) reductase or an intermediate electron carrier to the terminal Fe(III) reductase in vivo.
Relationship between OmcB, OmcC, and “FerA.”
It was previously reported (30) that a membrane-bound 89-kDa c-type cytochrome from G. sulfurreducens which was purified as part of a NADH-dependent Fe(III) reductase complex and was able to reduce Fe(III) in vitro was encoded by the “ferA” gene (GenBank accession no. AY033095). However, the sequence of “ferA” (30) is not present in the genome of G. sulfurreducens and is actually a hybrid of omcB and omcC. This error likely arose from the high similarity of omcB and omcC and the presence of both of these genes in the clone that was used to obtain the “ferA” sequence. It is unlikely that the procedure for purifying the cytochrome from the Fe(III) reductase separated OmcB and OmcC. The availability of mutants that produce only OmcB or OmcC should make it possible to obtain pure preparations of these two cytochromes in the future.
Physiological role of OmcB and OmcC and implications for Fe(III) reduction.
The results clearly demonstrate a role for OmcB, but not OmcC, in Fe(III) reduction. This further suggests that outer membrane proteins play important roles in Fe(III) reduction. However, given the many similarities between OmcB and OmcC, further investigation to determine the structure and topology of both proteins and to understand their regulation mechanisms is warranted.
Neither OmcB nor OmcC appears to be required for fumarate respiration. This is consistent with the fact that in all other organisms in which fumarate reduction has been studied, the fumarate reductase is either cytoplasmic or periplasmic (7, 8, 32, 41, 49). Thus, outer membrane cytochromes should not be involved in fumarate respiration.
The finding that OmcB, a putative outer membrane cytochrome, is involved in Fe(III) reduction is consistent with the concept (6, 24, 25, 37-39) that Geobacter species reduce Fe(III) at the outer surface of the cell. The finding that many c-type cytochromes (2, 3, 12, 16, 21, 29, 30, 34-36, 48), including an 89-kDa c-type cytochrome from G. sulfurreducens (30), can reduce Fe(III) suggests that OmcB is a terminal Fe(III) reductase. Alternatively, OmcB might function as an intermediary electron carrier between electron carriers in the periplasm and a terminal Fe(III) reductase in the outer membrane. Elucidation of these possibilities requires further biochemical investigation. For example, at this point it is not known whether a substantial portion of OmcB is exposed on the extracellular surface of the outer membrane.
The fact that omcB is expressed during growth with fumarate as the electron acceptor as well as with Fe(III) is consistent with the finding that fumarate-grown cells have Fe(III) reductase levels that are comparable with those grown on Fe(III) (29). Although electron transport to fumarate might be more energetically favorable than electron transport to Fe(III), it is not surprising that G. sulfurreducens might not downregulate genes involved in electron transport to Fe(III) in the presence of fumarate. This is because fumarate is unlikely to be an abundant electron acceptor in sedimentary environments, whereas Fe(III) oxides are generally among the most abundant electron acceptors in subsurface environments and Fe(III) oxide reduction is expected to be the primary form of respiration of Geobacter species in subsurface environments (23, 24).
To date, the only other c-type cytochrome shown to have a role in Fe(III) reduction in G. sulfurreducens in vivo is a 9.6-kDa periplasmic cytochrome designated PpcA (21). When the ppcA gene was deleted from the chromosome, the rate of acetate-dependent Fe(III) reduction in cell suspensions was decreased to 60% of the wild type and growth in acetate-Fe(III) medium was also affected. Elimination of PpcA had no impact on Fe(III) reduction with hydrogen as the electron donor. These results suggest that PpcA is involved as an intermediary electron carrier in acetate oxidation coupled to Fe(III) reduction and shuttles electrons from electron carriers in the inner membrane to electron carriers in the outer membrane (21). PpcA might not be required for electron transport from hydrogen to Fe(III), due to the presence of a periplasmic hydrogenase. This contrasts with OmcB, which, as shown here, was also required for hydrogen oxidation coupled to Fe(III) reduction. This probably reflects the fact that OmcB is located in the outer membrane, accepting electrons from a variety of periplasmic electron carriers. Further investigation of potential interactions between PpcA or hydrogenases and OmcB is warranted.
Even though no orthologs of OmcB were identified in the National Center for Biotechnology Information database or in the genome of phylogenetically distinct Fe(III)-reducing organisms, orthologs of OmcB have been identified in other Geobacteraceae. Using the G. sulfurreducens OmcB coding sequence, a search of the draft databases of G. metallireducens and Desulfuromonas acetoxidans genomes (Department of Energy Joint Genome Institute) revealed that both genomes contain OmcB homologs, with 44 and 47% similarity, respectively (K. O'Neill and S. Ciufo, personal communications). When the OmcC coding sequence was used to search these genomes, the same genes were identified. These genes were more homologous to OmcB than to OmcC. Because the G. metallireducens and D. acetoxidans genomes are currently incomplete, it is possible that additional omcB and omcC homologs might be identified in the future. The identification of OmcB orthologs in other Geobacteraceae species might imply that closely related members of Geobacteraceae have similar key genes related to the crucial function of electron transport to Fe(III).
Despite the lack of either OmcB or PpcA homologs in other phylogenetically distinct Fe(III)-reducing microorganisms, these organisms contain genes for other c-type cytochromes that might function in a similar manner. These include the outer membrane decaheme c-type cytochrome MtrC (2) and the periplasmic tetraheme c-type cytochrome CymA of Shewanella oneidensis MR-1 (34, 36), as well as a periplasmic tetraheme cytochrome c3 of S. frigidimarina, CctA (16). The absence of OmcB homologs in Shewanella spp. is consistent with the suggestion that the mechanisms for Fe(III) reduction in these organisms are distinct from that of the Geobacteraceae (37-40).
In summary, the finding that OmcB plays an important role in Fe(III) reduction is consistent with the inference from biochemical studies that membrane-associated c-type cytochromes are involved in Fe(III) reduction and might even serve as the terminal Fe(III) reductase. Further investigations on the localization of OmcB and its interactions with other proteins should help elucidate whether OmcB is the terminal Fe(III) reductase or an intermediary electron carrier.
Acknowledgments
This research was supported by the Office of Biological and Environmental Research Microbial Genome Program (DE-FG02-01ER 63145) and Microbial Cell Project (DE-FG02-01ER63221) of the U.S. Department of Energy.
We are grateful for the excellent technical support from Betsy Blunt.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 2.Beliaev, A., D. Saffarini, J. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 3.Beliaev, A. S., and D. A. Saffarini. 1998. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180:6292-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childers, S. E., S. Ciufo, and D. R. Lovley. 2002. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416:767-769. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T. 1982. Nucleotide sequence coding for the flavoprotein subunit of the fumarate reductase of Escherichia coli. Eur. J. Biochem. 122:479-484. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., T. Grundstrom, B. Jaurin, J. J. Robinson, and J. H. Weiner. 1982. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur. J. Biochem. 126:211-216. [DOI] [PubMed] [Google Scholar]
- 9.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, J., M. Heusterspreute, N. Chevalier, V. Ha-Thi, and F. Brunel. 1987. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51:275-280. [DOI] [PubMed] [Google Scholar]
- 11.Dehio, M., A. Knorre, C. Lanz, and C. Dehio. 1998. Construction of versatile high-level expression vectors for Bartonella henselae and the use of green fluorescent protein as a new expression marker. Gene 215:223-229. [DOI] [PubMed] [Google Scholar]
- 12.Dobbin, P. S., J. N. Butt, A. K. Powell, G. A. Reid, and D. J. Richardson. 1999. Characterization of a flavocytochrome that is induced during the anaerobic respiration of Fe3+ by Shewanella frigidimarina NCIMB400. Biochem. J. 342:439-448. [PMC free article] [PubMed] [Google Scholar]
- 13.Francis, Jr., R. T., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509-514. [DOI] [PubMed] [Google Scholar]
- 14.Gaspard, S., F. Vazquez, and C. Holliger. 1998. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl. Environ. Microbiol. 64:3188-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorby, Y. A., and D. R. Lovley. 1991. Electron transport in the dissimilatory iron reducer, GS-15. Appl. Environ. Microbiol. 57:867-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, E. H., A. D. Pike, A. E. Hill, P. M. Cuthbertson, S. K. Chapman, and G. A. Reid. 2000. Identification and characterization of a novel cytochrome c3 from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem. J. 349:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557.. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann, F., and D. R. Lovley. 2001. Isolation and characterization of a soluble NADPH-dependent Fe(III) reductase from Geobacter sulfurreducens. J. Bacteriol. 183:4468-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd, J. R., C. Leang, A. L. Hodges-Myerson, M. V. Coppi, S. Ciufo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovley, D. R. 2000. Fe(III) and Mn(IV) reduction, p. 3-30. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 24.Lovley, D. R. 2001. Reduction of iron and humics in subsurface environments, p. 193-217. In J. K. F. Fredrickson and M. Fletcher (ed.), Subsurface microbiology and biogeochemistry. Wiley-Liss, Inc., New York, N.Y.
- 25.Lovley, D. R., and J. D. Coates. 2000. Novel forms of anaerobic respiration of environmental relevance. Curr. Opin. Microbiol. 3:252-256. [DOI] [PubMed] [Google Scholar]
- 26.Lovley, D. R., J. D. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445-448. [Google Scholar]
- 27.Lovley, D. R., and E. J. Philips. 1986. Organic matter mineralization with the reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnuson, T. S., A. L. Hodges-Myerson, and D. R. Lovley. 2000. Characterization of a membrane-bound NADH-dependent Fe3+ reductase from the dissimilatory Fe3+-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol. Lett. 185:205-211. [DOI] [PubMed] [Google Scholar]
- 30.Magnuson, T. S., N. Isoyama, A. L. Hodges-Myerson, G. Davidson, M. J. Maroney, G. G. Geesey, and D. R. Lovley. 2001. Isolation, characterization, and gene analysis of a membrane-associated 89 kDa Fe(III) reducing cytochrome c from Geobacter sulfurreducens. Biochem. J. 359:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 32.Morris, C. J., A. C. Black, S. L. Pealing, F. D. Manson, S. K. Chapman, G. A. Reid, D. M. Gibson, and F. B. Ward. 1994. Purification and properties of a novel cytochrome: flavocytochrome c from Shewanella putrefaciens. Biochem. J. 302:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 34.Myers, C. R., and J. M. Myers. 1997. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, C. R., and J. M. Myers. 1997. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim. Biophys. Acta 1326:307-318. [DOI] [PubMed] [Google Scholar]
- 36.Myers, J. M., and C. R. Myers. 2000. Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nevin, K. P., and D. R. Lovley. 2000. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl. Environ. Microbiol. 66:2248-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ. Microbiol. 68:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J. 19:141-159. [Google Scholar]
- 40.Newman, D. K., and R. Kolter. 2000. A role for excreted quinones in extracellular electron transfer. Nature 405:94-97. [DOI] [PubMed] [Google Scholar]
- 41.Pealing, S. L., A. C. Black, F. D. Manson, F. B. Ward, S. K. Chapman, and G. A. Reid. 1992. Sequence of the gene encoding flavocytochrome c from Shewanella putrefaciens: a tetraheme flavoenzyme that is a soluble fumarate reductase related to the membrane-bound enzymes from other bacteria. Biochemistry 31:12132-12140. [DOI] [PubMed] [Google Scholar]
- 42.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Res. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology (Reading) 146:551-571. [DOI] [PubMed] [Google Scholar]
- 44.Röling, W. F., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Seeliger, S., R. Cord-Ruwisch, and B. Schink. 1998. A periplasmic and extracellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J. Bacteriol. 180:3686-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, J., R. Gross, O. Klimmek, M. Ringel, and A. Kroger. 1998. A periplasmic flavoprotein in Wolinella succinogenes that resembles the fumarate reductase of Shewanella putrefaciens. Arch. Microbiol. 169:424-433. [DOI] [PubMed] [Google Scholar]
- 50.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 51.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 52.Stein, L. Y., M. T. La Duc, T. J. Grundl, and K. H. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 53.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 54.Turick, C. E., L. S. Tisa, and F. Caccavo, Jr. 2002. Melanin production and use as a soluble electron shuttle for Fe(III) oxide reduction and as a terminal electron acceptor by Shewanella algae BrY. Appl. Environ. Microbiol. 68:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Wiedner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]