Abstract
In human cells, efficient global genomic repair of DNA damage induced by ultraviolet radiation requires the p53 tumor suppressor, but the mechanism has been unclear. The p48 gene is required for expression of an ultraviolet radiation-damaged DNA binding activity and is disrupted by mutations in the subset of xeroderma pigmentosum group E cells that lack this activity. Here, we show that p48 mRNA levels strongly depend on basal p53 expression and increase further after DNA damage in a p53-dependent manner. Furthermore, like p53−/− cells, xeroderma pigmentosum group E cells are deficient in global genomic repair. These results identify p48 as the link between p53 and the nucleotide excision repair apparatus.
Global genomic repair (GGR) and transcription-coupled repair (TCR) are two pathways used by cells for nucleotide excision repair of ultraviolet radiation (UV)-damaged DNA (1). TCR removes lesions from the DNA strands transcribed by RNA polymerase II. GGR removes lesions from the nontranscribed strand as well as nontranscribed regions of the genome. Efficient GGR of cyclobutane pyrimidine dimers (CPDs) induced by UV requires activation of the p53 tumor suppressor (2, 3). The mechanism for p53-dependent GGR has not been determined but may involve transcriptional activation because p53 activates the transcription of genes involved in other cellular responses to DNA damage, such as cell cycle arrest and apoptosis (4, 5).
Defects in GGR and TCR have been documented in several inherited human diseases (6). TCR is defective in Cockayne syndrome, which is characterized by UV sensitivity, cachectic dwarfism, and mental retardation. GGR is defective in xeroderma pigmentosum (XP), which also is characterized by UV sensitivity but is associated with a severe predisposition to skin cancer not seen in Cockayne syndrome. XP consists of seven complementation groups, and cells from groups A, B, D, F, and G are defective in both GGR and TCR whereas group C cells are defective only in GGR (7).
Group E has remained uncharacterized for GGR and TCR but consists of two subsets in which a UV-damaged DNA binding activity (UV-DDB) is absent (DDB−) or present (DDB+) (8, 9). UV-DDB has a 500,000-fold preference for UV-damaged DNA over undamaged DNA (10) and can be purified as a 125-kDa polypeptide, p125, either alone or together with a 48-kDa polypeptide, p48 (10, 11, 12, 13, 14, 15). Binding activity is absent in DDB– XPE cells because of a mutation of the p48 gene (16, 17). It is notable that binding activity and p48 expression are also absent in hamster cells (16), which are deficient in GGR of CPDs (18, 19). Thus, we tested whether XPE cells are deficient in GGR. To identify the link between GGR and p53, we also tested whether p48 or p125 transcription is activated by p53.
MATERIALS AND METHODS
Cell Lines.
All cell lines were grown in DMEM supplemented with 10% fetal bovine serum at 37°C in 5% CO2. WI38 fetal lung primary human fibroblasts and XP2RO (DDB−) primary human skin fibroblasts from an XPE individual were obtained from American Type Culture Collection (Rockville, MD). XP89TO (DDB+) primary human skin fibroblasts from an XPE individual were a gift from Stuart Linn (University of California, Berkeley). The p53−/− (041 mut) human fibroblasts were isolated after spontaneous loss of the remaining wild-type copy of p53 from primary skin fibroblasts obtained from an individual with Li-Fraumeni syndrome (20) and were homozygous for a single base pair frameshift mutation at codon 184 of p53 (2). The 041 mut cells were stably transfected either with a control vector not containing p53 or with a vector containing wild-type p53 cDNA under control of a tetracycline regulated promoter, allowing for the induction of p53 expression by removal of tetracycline from the growth medium (3).
UV and Ionizing Radiation (IR) Treatment.
Cells were exposed to UV or IR as described (21). In brief, 2 × 106 cells were plated onto a 150-mm diameter dish and were grown for 24 h before treatment. Cells were rinsed with PBS and were exposed to UV (from a 15-W germicidal lamp delivering predominantly 254 nm of light at a flux of 0.7 J/m2/s) or to IR (from a 137Cs source with a flux of 11.5 Gy/min) and then were grown in fresh medium until the cells were harvested.
Immunoblot.
Whole cell extracts were prepared as described (21), were resolved by SDS/PAGE, were transferred to a nitrocellulose membrane (Schleicher & Schuell), and were probed with a 1:250 dilution of mouse anti-p53 IgG (1801, Santa Cruz Biotechnology) followed by a 1:1000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG antibody (Vector Laboratories). Antibody binding was detected by enhanced chemiluminescence (ECL plus, Amersham).
Northern Blot.
Total RNA was prepared, and Northern blots were probed first for p48, then were stripped and probed sequentially for p21, p125, actin, and 28S rRNA as described (16, 21).
Electrophoretic Mobility-Shift Assay for UV-DDB.
Whole cell extract was prepared as described (16) and was assayed for UV-DDB activity by an electrophoretic mobility-shift assay (10). In brief, the 32P-labeled 148-bp DNA probe (f148) was prepared and either was left intact or was damaged with a UV dose of 5,000 J/m2. Extracts (0.5 μg) were incubated with DNA probe (0.2 ng) and a mixture of unlabeled salmon sperm DNA (2 μg) and poly(dI-dC) (1 μg) to mask the effect of nonspecific DNA binding proteins. The mixture then was resolved by nondenaturing gel electrophoresis, and the gel was dried and exposed to x-ray film.
Global Genomic Repair.
The relative number of UV-induced photoproducts in unreplicated genomic DNA from cells collected at various times after UV was determined by using an immunoblot assay with mouse monoclonal antibodies to CPDs and 6-4 photoproducts as described (3). In brief, exponentially growing cells were labeled with 3H-thymidine, were washed with PBS, and were exposed to UV. Cells either were lysed immediately for an initial sample or were incubated in growth medium containing BrdUrd to density label newly replicated DNA and then were lysed at various times. Density labeling was performed during repair periods to allow unreplicated DNA to be isolated by cesium chloride isopycnic density gradient sedimentation. Equal amounts from each DNA sample were fixed to a Hybond N+ nylon membrane in triplicate by using a slot-blot apparatus. The membrane was incubated with anti-CPD or 6-4 photoproduct antibody and horseradish peroxidase-conjugated goat anti-mouse antibody, which permitted detection by enhanced chemiluminescence and phosphorimager analysis (Bio-Rad model GS-363).
TCR.
Repair of CPDs was examined within the transcribed or nontranscribed strand of a 20-kilobase KpnI restriction fragment of the dihydrofolate reductase gene, as described (2, 18). In brief, unreplicated DNA was isolated, as described above, from cells that were exposed to UV. The DNA was treated with KpnI, was treated or mock-treated with T4 endonuclease V (which specifically cleaves DNA at CPDs), was electrophoresed under denaturing conditions, was transferred to a membrane, and was hybridized with strand-specific RNA probes generated by in vitro transcription of the plasmid pGEM0.69EH (2). The ratio of full length restriction fragments in the T4 endonuclease V treated and untreated samples was determined by phosphorimager analysis (Bio-Rad Model GS-363), which was used to calculate the average number of CPDs per fragment by using Poisson statistics.
RESULTS
p48 mRNA Increases After DNA Damage and Is p53-Dependent.
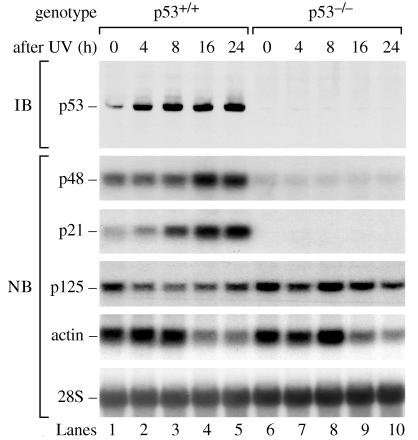
To test whether p48 transcription depends on p53, we used primary human fibroblasts, which were p53 wild-type (p53+/+, WI38) or p53 mutant (p53−/−, 041 mut). Cells were exposed to 10 J/m2 UV and were analyzed at different times afterward. As expected, p53 protein levels increased after UV irradiation in the p53+/+ cells, but p53 was absent in the p53−/− cells (Fig. 1). The basal p48 mRNA level in p53+/+ cells was 3.8-fold higher than in p53−/− cells and increased an additional 2.3-fold after UV, attaining levels 8.7-fold higher than in the p53−/− cells. The p21 protein is a cyclin-dependent kinase inhibitor that is induced by p53 (22, 23). The p21 mRNA level in p53+/+ cells was 3.2-fold higher than in p53−/− cells and, as expected, increased an additional 5.9-fold after UV. Thus, p53 was required for the induction of p48 after UV radiation. By contrast, the expression of p125 mRNA did not depend on p53. After UV, p125 mRNA failed to increase and may have decreased slightly. After UV, actin mRNA levels were initially constant but then decreased by 16 h, an effect of unknown significance reported by others (24).
Figure 1.
UV induces p48 transcription by a p53-dependent pathway. Primary human fibroblast cell lines, either p53+/+ (WI38) or p53−/− (041 mut), were treated with 10 J/m2 UV radiation and were harvested after different time intervals. The level of p53 protein was measured by immunoblot (IB) of protein extract. The levels of p48, p21, p125, actin mRNA, and 28S rRNA were measured by Northern blot (NB) of total RNA. The levels of 28S rRNA confirmed equal transfer of total RNA to the membrane.
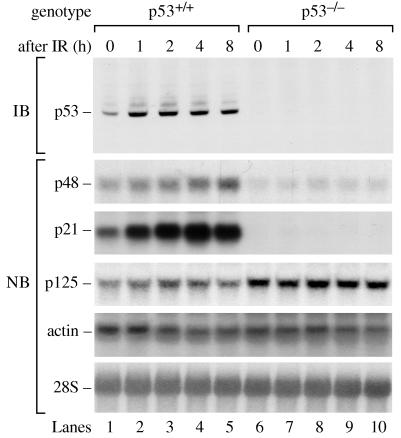
The increases in mRNA for the p48 and p21 genes occurred over 16–24 h, consistent with the gradual accumulation of p53 over this time period (Fig. 1). Although a modest increase in p21 mRNA was observed by 4 h, it was impossible to determine whether p48 mRNA also increased by this time because the higher basal levels of p48 would have obscured a modest increase. To better test whether p48 mRNA levels increase in conjunction with p53, we used a different DNA damaging agent, ionizing radiation, which induces rapid accumulation of p53. After 2 Gy of IR, p53 protein accumulated in p53+/+ cells to maximal levels by 1 h (Fig. 2) as reported (25). In this experiment, the basal p48 mRNA level in p53+/+ cells was 2.2-fold higher than in p53−/− cells and increased an additional 2.5-fold after IR. The basal p21 mRNA level in p53+/+ cells was 5-fold higher than in p53−/− cells and increased an additional 5-fold after IR. The increases in mRNA occurred within 1 h and were specific because p125 and actin mRNA levels were not significantly affected by IR.
Figure 2.
IR induces p48 transcription by a p53-dependent pathway. Cells were exposed to 2 Gy of IR and were analyzed 0, 1, 2, 4, and 8 h later for levels of p53 protein, p48, p21, p125, actin mRNA, and 28S rRNA.
To ensure that the effects on p48 mRNA were not peculiar to high doses of UV or IR, we tested the effects of a range of doses on p53 protein and p48, p21, and p125 mRNA (data not shown). In p53+/+ but not p53−/− cells, IR doses of 0.5, 1, 2, 4, and 8 Gy induced the accumulation of p53 protein and p48 and p21 mRNA. UV doses of 5 and 10 J/m2 both induced the accumulation of p53 protein and p48 and p21 mRNA. In fact, the increase in p48 mRNA was approximately the same for 5 J/m2 as for 10 J/m2.
p48 mRNA Increases After p53 Induction in the Absence of DNA Damage.
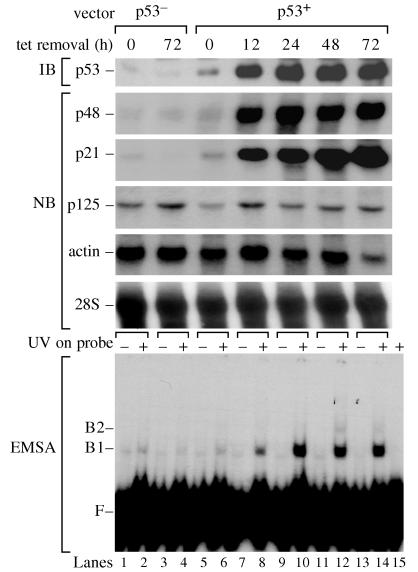
To demonstrate directly that p48 mRNA levels in the p53−/− cells could be restored by expression of p53, we analyzed p53−/− cells that had been transfected with a vector in which the expression of wild-type p53 was induced by removing tetracycline from the growth medium. In these cells, expression of p53 increases GGR of CPDs (2, 3). When we induced the expression of p53 by tetracycline withdrawal, there was a 7.6-fold increase in p48 mRNA and an 11.3-fold increase in p21 mRNA (Fig. 3). By contrast, p125 mRNA remained largely unaffected. When the p53−/− cells were transfected with a vector lacking p53, p125 mRNA was unaffected by tetracycline removal whereas p48 and p21 mRNAs remained nearly undetectable.
Figure 3.
Expression of p53 induces p48 transcription and the activity of UV-damaged DNA binding protein. We used p53−/− (041 mut) cells that had been stably transfected with a control vector not containing p53 (p53−) or with a vector containing wild-type p53 cDNA under the control of a tetracycline regulated promoter (p53+) (2). Cells were analyzed for levels of p53 protein, p48, p21, p125, actin mRNA, and 28S rRNA at different times after inducing p53 expression by removal of tetracycline. UV-DDB was measured by an electrophoretic mobility-shift assay (EMSA) with UV-damaged DNA probe.
The transfected p53−/− cells also were used to determine the effect of p53 on UV-DDB. Induction of p53 caused a large increase in UV-DDB (Fig. 3). The effect was likely to be mediated by p48 because a large increase in expression was seen for p48 but not p125 on induction of p53, and UV-DDB in human cells increases after transfection of p48 but not p125 (16). Thus, p48 transcription and UV-DDB strongly depend on the expression of p53.
XPE Cells Are Deficient in GGR.
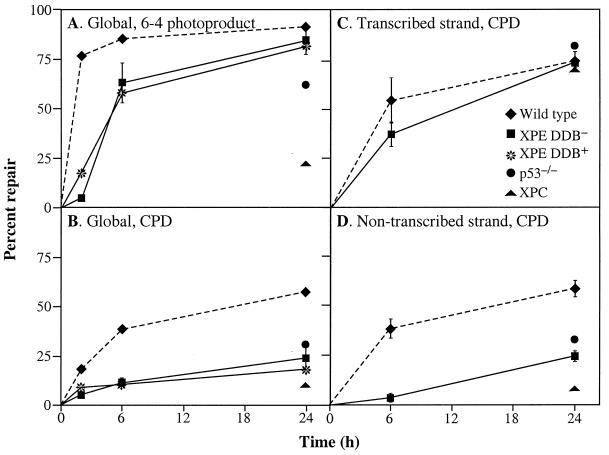
Because p53−/− cells were deficient in UV-DDB and in GGR, we wished to determine whether XPE cells, particularly those deficient in UV-DDB, are also deficient in GGR. We analyzed two primary fibroblast XPE cell lines: XP89TO, which has intact binding activity (DDB+); and XP2RO, which lacks binding activity (DDB−). Both cell lines showed significantly less GGR of CPDs than wild-type cells over the entire 24-h period (Fig. 4B). These results are consistent with the affinity of UV-DDB for DNA-containing CPDs (10, 26). Both cell lines achieved normal levels of GGR of 6-4 photoproducts by 24 h after a relatively short time delay (Fig. 4A), in contrast to the severe defect in XPC cells. Significantly, the XPE defects in GGR also were seen in p53−/− cells (2, 3).
Figure 4.
GGR and TCR in XPE cells. DDB− (XP2RO) and DDB+ (XP89TO) cells were analyzed at different times after exposure to 10 J/m2 UV. GGR of 6-4 photoproducts (A) and CPDs (B) were measured by an immunoblot assay with monoclonal antibodies specific for 6-4 photoproducts or CPDs. TCR of CPDs on the transcribed strand (C) and nontranscribed strand (D) of the dihydrofolate reductase gene were measured by restored resistance to T4 endonuclease V, which specifically cleaves CPDs. GGR and TCR also are shown for other primary fibroblast cell lines: wild type (WI38), p53−/− (041 mut), and XPC (XP10BE), as determined in previous experiments (2, 3, 34, 35). To facilitate comparison, repair in wild type (dotted lines) and XPE (solid lines) are shown over the entire time course.
To assess TCR in XPE cells, we measured the repair of CPDs in the transcribed and nontranscribed strands of the dihydrofolate reductase gene. DDB− cells showed efficient removal of CPDs from the transcribed strand, as do p53−/− cells and XPC cells, demonstrating that TCR in these cells is normal (Fig. 4C). However, DDB− cells showed a defect in repair of CPDs in the nontranscribed strand similar to that in p53−/− cells but not as severe as in XPC cells (Fig. 4D). In summary, the GGR defects in DDB− and p53−/− cells were similar in every experiment we performed, further supporting the hypothesis that p53-dependent activation of GGR is mediated by p48.
DISCUSSION
p48 Links p53 to Nucleotide Excision Repair.
This paper provides evidence that transcription of p48 depends on p53. In our experiments, exposure of the cells to UV or IR induced an increase in p48 mRNA levels that paralleled the increase in p53 protein levels. Both the basal level of p48 mRNA and its subsequent increase were not seen in p53−/− cells. Furthermore, the increase in p48 mRNA did not require UV- or IR-induced activation of factors outside the p53 pathway: when p53 was induced with a tetracycline regulated promoter in the absence of UV or IR, p48 transcription also was induced.
The p53-dependent increases in p48 mRNA and UV-DDB cannot be a secondary effect of apoptosis because induction of p53 by tetracycline withdrawal in the absence of DNA damage produces only a small number of apoptotic cells that appear after 48–72 h (3), long after the increase in p48 mRNA was observed (Fig. 3). Furthermore, p48 mRNA levels cannot be explained simply in terms of other secondary effects of p53 activation, such as cell cycle arrest, because a significant degree of p48 transcription in wild-type p53+/+ cells depends on constitutive p53 expression in the absence of DNA damage (Figs. 1 and 2). Nevertheless, further studies are required to determine whether p48 transcription is activated directly by a p53 binding site in the p48 gene.
We have presented evidence that GGR of CPDs is p48-dependent. DDB– XPE cells, which are mutant in p48, were defective in GGR of CPDs. Furthermore, it has been shown recently that p48 is required for GGR of CPDs: transfection of hamster cells with human p48 increases the GGR of CPDs to levels comparable to those found in wild-type human cells (J. Tang, J.M.F., B.J.H., P.C.H., and G.C., unpublished work). Thus, we conclude that p53-dependent activation of GGR is mediated by p48 transcription.
Genetic Heterogeneity of XPE.
Of interest, we found that DDB+ cells share the same GGR defect seen in DDB− cells. However, DDB+ cells do not appear to have mutations in either p125 or p48 (16, 17), nor are they rescued by the microinjection of preparations containing p125 and an unknown amount of p48 (27). Our finding that DDB+ and DDB− cells have a similar defect in GGR raises the possibility that DDB+ cells are mutant in a protein required for coupling p125 or p48 to other DNA repair proteins. If DDB+ and DDB− cells have defects in different proteins, why were they assigned to the same complementation group by cell fusion experiments? We speculate that the putative coupling protein forms a complex with p48 and/or p125 and that the majority of complexes remain nonfunctional. Thus, the DDB+ and DDB− defects are corrected only partially in the fused cells and are difficult to detect.
The Recognition of UV Damage.
A widely held view in the literature is that UV-DDB plays only an accessory role in the repair of UV-damaged DNA. In support of this view, Aboussekhra et al. (28) succeeded in reconstituting nucleotide excision repair of a UV-damaged DNA substrate with purified components in the absence of UV-DDB. Addition of purified UV-DDB to this system stimulated repair only 2-fold. In a DNA damage recognition-competition assay, Sugasawa et al. (29) recently found that XPC/HR23B was the earliest damage detector to initiate nucleotide excision repair in cell extracts. Specifically, XPC/HR23B acted before the damage-binding complex XPA/RPA. XPC/HR23B bound to a variety of DNA lesions, including 6-4 photoproducts. However, CPDs were not tested directly.
Our results suggest that UV-DDB plays an important role in targeting CPDs for GGR in vivo. Such a role is consistent with the 500,000-fold preference of UV-DDB for UV-damaged over undamaged nucleotides in DNA (10). By contrast XPC/HR23B has a 3,000-fold preference (29), and XPA has only a 100-fold preference (30) for UV-damaged nucleotides in DNA. UV-DDB may have failed to show a significant effect on nucleotide excision repair in vitro, either because the substrate in those experiments was naked DNA outside the context of chromatin or because a high density of lesions was required to detect repair. Sugasawa et al. irradiated their DNA substrate with an extremely high dose of 450 J/m2, producing 1 thymine dimer (CPD or 6-4 photoproduct) per 300 nucleotides (29). Under these conditions, the relatively modest affinity of XPC/HR23B for UV-damaged DNA would be sufficient to target the lesions for repair, so that the remarkable affinity of UV-DDB for UV-damaged DNA would not be required. By contrast, our in vivo measurements of GGR were done with a dose of 10 J/m2, producing only 1 thymine dimer per 13,500 nucleotides, a density low enough to require UV-DDB.
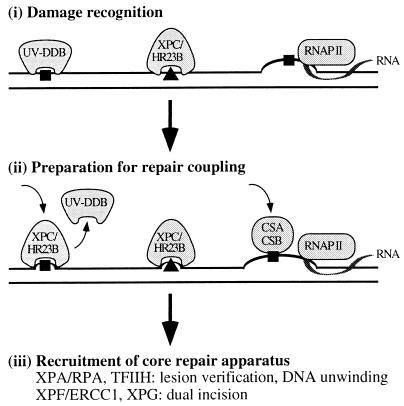
The findings in this paper add another level to the recognition of UV photoproducts for GGR. To place UV-DDB in the context of the entire nucleotide excision repair pathway, a model for photoproduct recognition in GGR and TCR is presented in Fig. 5. In particular, we propose that the GGR of CPDs is initiated by the binding of UV-DDB. XPC/HR23B then recognizes UV-DDB bound to the DNA, displaces UV-DDB, and recruits the core nucleotide excision repair apparatus to the site of the lesion. The GGR pathway also may include the gadd45 protein, which is induced by p53 (33), but the biochemical role of gadd45 remains to be fully defined.
Figure 5.
Model for photoproduct recognition in nucleotide excision repair. (i) Damage recognition. CPDs (square) are recognized by UV-DDB when p48 transcription increases in a p53-dependent manner. Although 6-4 photoproducts (triangle) can be recognized by UV-DDB (32), they may distort the DNA sufficiently to be recognized directly by XPC/HR23B. Thus, the absence of UV-DDB in XPE cells causes only a delay in the global repair of 6-4 photoproducts rather than an absolute deficiency. Either photoproduct is recognized in the transcribed strand of expressed genes by the arrest of RNA polymerase II (RNAP II) translocation (31). (ii) Preparation for repair coupling. XPC/HR23B is postulated to recognize UV-DDB bound to DNA, perhaps replacing it at the lesion site and then partially opening the DNA. A complex of the Cockayne Syndrome proteins, CSA/CSB, is postulated to recognize and displace the arrested RNA polymerase II, opening the DNA at that site. (iii) Recruitment of core repair apparatus. XPC/HR23B recruits the core nucleotide excision repair apparatus for GGR whereas CSA/CSB recruits the core apparatus for TCR. The helicases in TFIIH create a bubble of unwound DNA and, along with XPA/RPA, may participate in lesion verification before permitting the structure-specific nucleases XPF/ERCC1 and XPG to make incisions on each side of the lesion.
GGR and Cancer.
It is notable that the relatively mild UV sensitivity in XPE is nevertheless associated with an increased risk for skin cancer. A striking dissociation between UV sensitivity and skin cancer becomes evident when XPE is compared with Cockayne syndrome. Humans with Cockayne syndrome who are deficient in TCR but not GGR suffer from severe UV sensitivity without an increased risk for skin cancer. Our discovery that XPE cells are defective in GGR suggests that this pathway may be important for the prevention of skin cancer. In conclusion, UV-DDB targets CPDs for GGR, and this pathway may play an important role in minimizing UV-induced mutagenesis and maintaining genomic stability.
Acknowledgments
We thank Ann Ganesan, Virginia Goss, Ola Hammarsten, and Jean Tang for reading the manuscript and helpful discussions. This work was supported by Janet M. Shamberger Fellowship Fund (to B.J.H.), Clinical Investigator Award K08-CA64330 from the National Cancer Institute (to J.M.F.), Outstanding Investigator Grant CA44349 from the National Cancer Institute (to P.C.H), and Grant DAMD 17-94-J-4350 from the U.S. Army Medical Research and Materiel Command and gifts from Graham and Anna Nissen (to G.C.).
ABBREVIATIONS
- CPD
cyclobutane pyrimidine dimer
- GGR
global genomic repair
- IR
ionizing radiation
- TCR
transcription-coupled repair
- UV
ultraviolet radiation
- UV-DDB
UV-damaged DNA binding activity
- XP
xeroderma pigmentosum
References
- 1.Hanawalt P. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 2.Ford J M, Hanawalt P C. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford J M, Hanawalt P C. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 4.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 5.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 6.Chu G, Mayne L. Trends Genet. 1996;12:187–192. doi: 10.1016/0168-9525(96)10021-4. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen W, de Boer J, Citterio E, van Gool A J, van der Horst G T, Jaspers N G, de Laat W L, Sijbers A M, van der Spek P J, Sugasawa K, et al. Biochem Soc Trans. 1997;25:309–315. doi: 10.1042/bst0250309. [DOI] [PubMed] [Google Scholar]
- 8.Keeney S, Wein H, Linn S. Mutat Res. 1992;273:49–56. doi: 10.1016/0921-8777(92)90049-9. [DOI] [PubMed] [Google Scholar]
- 9.Chu G, Chang E. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 10.Hwang B J, Chu G. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- 11.Keeney S, Chang G J, Linn S. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 12.Abramic M, Levine A, Protic M. J Biol Chem. 1991;266:22493–22500. [PubMed] [Google Scholar]
- 13.Dualan R, Brody T, Keeney S, Nichols A, Admon A, Linn S. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 14.Hwang B J, Liao J, Chu G. Mutat Res. 1996;362:105–117. doi: 10.1016/0921-8777(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 15.Takao M, Abramic M, Moos M, Otrin V, Wootton J, McLenigan M, Levine A, Protic M. Nucleic Acids Res. 1993;21:4111–4118. doi: 10.1093/nar/21.17.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang B J, Toering S, Francke U, Chu G. Mol Cell Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols A, Ong P, Linn S. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 18.Mellon I, Spivak G, Hanawalt P. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 19.Bohr V, Smith C, Okumoto D, Hanawalt P. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y, Tainsky M, Bischoff F, Strong L, Wahl G. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 21.Rathmell W K, Kaufmann W K, Hurt J C, Byrd L L, Chu G. Cancer Res. 1997;57:68–74. [PubMed] [Google Scholar]
- 22.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 23.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 24.Guenal I, Risler Y, Mignotte B. J Cell Sci. 1997;110:489–495. doi: 10.1242/jcs.110.4.489. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Lane D. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 26.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J S, Linn S, Sancar A. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 27.Keeney S, Eker A, Brody T, Vermuelen W, Bootsma D, Hoeijmakers J, Linn S. Proc Natl Acad Sci USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aboussekhra A, Biggerstaff M, Shivji M, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 29.Sugasawa K, Ng J, Masutani C, Iwai S, van der Spek P, Eker A, Hanaoka F, Bootsma D, Hoeijmakers J. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 30.Jones C, Wood R. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 31.Donahue B A, Yin S, Taylor J S, Reines D, Hanawalt P C. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treiber D, Chen Z, Essigmann J. Nucleic Acids Res. 1992;20:5805–5810. doi: 10.1093/nar/20.21.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith M, Chen I-T, Zhan Q, Bae I, Chen C-Y, Gilmer T, Kastan M, O’Connor P, Fornace A. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 34.Ford J M, Baron E L, Hanawalt P C. Cancer Res. 1998;58:599–603. [PubMed] [Google Scholar]
- 35.Bowman K K, Smith C A, Hanawalt P C. Mutat Res. 1997;385:95–105. doi: 10.1016/s0921-8777(97)00029-3. [DOI] [PubMed] [Google Scholar]