Abstract
Inversion(16) is one of the most frequent chromosomal translocations found in acute myeloid leukemia (AML), occurring in over 8% of AML cases. This translocation results in a protein product that fuses the first 165 amino acids of core binding factor β to the coiled-coil region of a smooth muscle myosin heavy chain (CBFβ/SMMHC). CBFβ interacts with AML1 to form a heterodimer that binds DNA; this interaction increases the affinity of AML1 for DNA. The CBFβ/SMMHC fusion protein cooperates with AML1 to repress the transcription of AML1-regulated genes. We show that CBFβ/SMMHC contains a repression domain in the C-terminal 163 amino acids of the SMMHC region that is required for inv(16)-mediated transcriptional repression. This minimal repression domain is sufficient for the association of CBFβ/SMMHC with the mSin3A corepressor. In addition, the inv(16) fusion protein specifically associates with histone deacetylase 8 (HDAC8). inv(16)-mediated repression is sensitive to HDAC inhibitors. We propose a model whereby the inv(16) fusion protein associates with AML1 to convert AML1 into a constitutive transcriptional repressor.
Human acute leukemias often arise from chromosomal translocations targeting regulatory genes that affect cell growth, differentiation, and apoptosis (30). These translocations often fuse transcription factors to other proteins resulting in oncogenic chimeric proteins. inv(16) is found in approximately 8% of acute myeloid leukemia (AML) cases (35), making it one of the most frequent translocations in AML. inv(16) fuses the first 165 amino acids (aa) of core binding factor β (CBFβ) to the C-terminal coiled-coil region of a smooth muscle myosin heavy chain (SMMHC [the gene name is MYH11]) (28). CBFβ interacts with the AML1 (also known as RUNX1) transcription factor to increase the affinity of AML1 for DNA (15, 40, 47) and to stimulate the ability of AML1 to either activate or repress transcription (22, 32). Both components of the CBFβ-AML1 transcription factor complex are disrupted by chromosomal translocations. The translocations that target AML1 include t(8;21) and t(12;21) that result in the leukemogenic fusion proteins AML1/ETO and TEL/AML1 (10, 16).
The CBFβ-AML1 complex regulates genes encoding cytokines and their receptors, genes involved in differentiation such as T-cell receptors, neutrophil peptide 3, and myeloperoxidase as well as the p14ARF tumor suppressor (4, 12, 14, 27, 36, 39, 43, 46, 52). Furthermore, CBFβ/SMMHC slows the cell cycle transition from G1 to S in hematopoietic cells; thus, cell cycle regulatory genes such as cdk4 may also be targeted by this complex (5).
Animal studies indicate that CBFβ and AML1 are required for hematopoiesis. CBFβ and AML-1 knockout mice share a phenotype; these mice lack fetal liver hematopoiesis and are embryonic lethal at embryonic day 12.5 (e12.5) to e13.5 (41, 49). A similar phenotype is observed in mice expressing CBFβ/SMMHC from a “knocked-in” CBFB-MYH11 gene (7), suggesting that inv(16) creates a dominant repressor of AML1- and CBFβ-regulated genes. Chimeric mice created using Cbfβ+/cbfβ-MYH11 embryonic stem cells fail to develop leukemia at high frequency within the first year of life. However, 4- to 16-week-old chimeras that are treated with N-ethyl-N-nitrosourea develop leukemia 2 to 6 months after treatment (6). Coexpression of CBFβ/SMMHC with the human papillomavirus E7 oncogene or expression of the fusion protein in the absence of the tumor suppressors p16INK4a and p19ARF also leads to acute leukemia in mice (51). Thus, CBFβ/SMMHC predisposes mice to leukemia, and secondary mutations are necessary for the onset of leukemia in inv(16)-mediated cases.
CBFβ and CBFβ/SMMHC lack a nuclear localization signal; thus, they must complex with AML1 in the cytoplasm in order to be transported into the nucleus. Although a significant amount of CBFβ/SMMHC is retained in the cytoplasm in overexpression studies, CBFβ/SMMHC is both nuclear and cytoplasmic (1, 5, 29, 31); furthermore, the fusion protein cooperates with AML1 to repress transcription (31). AML1 binds the mSin3A and Groucho corepressors, and the inv(16) fusion protein can form a trimeric complex with AML1 and mSin3A (31). In addition, the C terminus of the CBFβ/SMMHC fusion protein is required for repression (31), suggesting that the fusion protein may cooperate with AML1 to recruit corepressors.
Other leukemogenic fusion proteins that affect AML1-dependent transcriptional control, such as AML1/ETO and TEL/AML1, repress transcription through interactions with the mSin3A, N-CoR, and SMRT corepressors and histone deacetylases (HDACs) (2, 11, 13, 18, 33, 48). We demonstrate here that the inv(16) fusion protein interacts with mSin3A and HDAC8. The C-terminal SMMHC portion of inv(16) is both necessary and sufficient for transcriptional repression and is sufficient for association with these corepressors. This region contains an assembly competence domain (ACD) that is required for multimerization of the SMMHC (24, 44). The 28-aa ACD contributes to both transcriptional repression and association with mSin3A and HDAC8. Identification of the repression domain of inv(16) and its interacting proteins may stimulate the development of novel treatments for inv(16)-mediated AML.
MATERIALS AND METHODS
Cell culture.
Cos7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (BioWhittaker, Walkersville, Md.) containing 10% fetal bovine serum (Sigma, St. Louis, Mo.), 2 mM l-glutamine (BioWhittaker), and 50 U of penicillin per ml and 50 μg of streptomycin per ml (both antibiotics from Gibco, Grand Island, N.Y.). NIH 3T3 cells were maintained in DMEM containing 10% bovine calf serum (HyClone, Logan, Utah) and antibiotics.
Deletion mutants.
inv(16) mutants were made by PCR amplification of inv(16) cDNA fragments, restriction digestion of the fragments, and ligation into the pCMV5 or pCMV5-GAL4(M1R) vector. The CBFβ portion of inv(16) (aa 1 to 165) was PCR amplified using primers at the 5′ start of the protein (5′-CGG AAT TCA TGC CGC GCG TCG TGC CCG AC-3′) and at the fusion (aa 165) to create a BamHI site (5′-CGC GGA TCC CTC CAT TTC CTC CCG GTG AGA-3′) (restriction sites are underlined). This PCR product was cloned into pCMV5-GAL4(M1R) using EcoRI and BamHI to make pCMV5-GAL4-CBFβ. Internal deletion mutants were made by amplification of the SMMHC region using a primer at the 3′ end of the cDNA (5′-GTC AAG CTT TTA TTC ACT GGC CTT GGT TCC AT-3′) with one the following primers: Δ496-720, 5′-CAG GGA TCC GAG ACG GAA CTG GAA GAC GAG-3′; Δ496-945, 5′-CAG GGA TCC GAG AT GAG AAG AAA GCC AAG AG-3′; Δ496-1170, 5′-CAG GGA TCC ATG GAG GCC ATG AGC GAC CGG-3′; Δ496-1347, 5′-CAG GGA TCC AAG TTC AAA TCC ACC ATC GCG-3′; and Δ496-1665, 5′-CAG GGA TCC ATG GGC CGC GAG GTG AAC GCA-3′. The fragments were cloned into pCMV5-GAL4-CBFβ after digestion with BamHI and HindIII. C-terminal fragments of inv(16) were PCR amplified using 5′-CGG GAT CCT TAT TCA CTG GCC TTG GTT CC-3′ with the following primers: 449-611, 5′-CGG AAT TCA AGT TCA AAT CCA CCA TCG CG-3′; 502-611, 5′-CGG AAT TCA TGG CCG AGC AGT ACA AGG AG-3′; 532-611, 5′-CGG AAT TCC AGC GCA TCA ACG CCA ACC GC-3′; and 572-611, 5′-CGG AAT TCA CCT CTT TCG TTC CTT CTA GA-3′. The fragments were cloned into pCMV5-GAL4(M1R) using EcoRI and BamHI sites. Mutants were confirmed by sequencing.
The inv(16)ΔACD mutant (24) was cloned into pCMV5-GAL4(M1R) by the following strategy. pGEM-inv(16)ΔACD was digested with XbaI, and 3′ recessed termini were filled in using Klenow fragment, followed by digestion with EcoRI. The vector was digested with HindIII, 3′ recessed termini were filled in with Klenow fragment, and the linearized vector was digested with EcoRI. These fragments were ligated in order to obtain the construct used in subsequent experiments, pCMV-GAL4-inv(16)ΔACD.
The inv(16)1-449 mutants were made by PCR amplification of these amino acids (aa 1 to 449) using the following primers: 5′-AG GGA TTC ATG CCG CGC GTC GTG CCC-3′ was used with 5′-GTC AAG CTT TTA GGA CTT GAC GGC CCC CTC-3′ or 5′-AGT GGA TCC GGA CTT GAC GGC CCC CTC-3′. These fragments were digested with EcoRI and HindIII (with stop codon) or EcoRI and BamHI (to be fused to the p53 tetramerization domain) and ligated into linearized pCMV5-M1R. The p53 tetramerization domain (aa 340 to 393) was amplified using the following primers to create restriction sites for cloning: 5′-CCC AAG CTT TCA GTC TGA GTC AGG CCC-3′ was used with 5′-CGG GAT CCA TGT TCC GAG AGC TGA AT-3′ or 5′-CGG AAT TCA TGT TCC GAG AGC TGA AT-3′. These fragments were digested with BamHI or EcoRI and HindIII and cloned into pCMV5-M1R-inv(16)1-449 and pCMV5-M1R, respectively.
Transcription assays.
GAL4 reporter assays were performed in 3T3 cells transiently transfected with the indicated amounts of pCMV5-GAL4-inv(16) or plasmid expressing mutant inv(16) proteins, 1 μg of GAL4-thymidine kinase-luciferase (GAL-TK-Luc) reporter construct, and 200 ng of pCMV5-SEAP plasmid. The GAL-TK-Luc reporter contains four GAL4 DNA-binding sites in front of the thymidine kinase promoter that drives expression of the luciferase gene. Cells were lysed in 1× reporter lysis buffer (Promega, Madison, Wis.) and centrifuged at 16,000 × g for 20 s, and the supernatant was used in luciferase assays. Luciferase activity was measured using the luciferase assay system from Promega and normalized to secreted alkaline phosphatase (SEAP) activity.
For trichostatin A (TSA) experiments, cells were transfected with 100 ng of pCMV5-GAL4-inv(16) or empty expression vector, 1 μg of GAL-TK-Luc, and 100 ng of pRL-TK plasmid (thymidine kinase promoter driving expression of Renilla luciferase). Cells were treated with TSA (Biomol, Plymouth Meeting, Pa) for 48 h posttransfection and lysed as described above. Luciferase activity of the experimental (GAL-TK-Luc) and internal control (pRL-TK) reporters was measured using the Dual Luciferase Reporter Assay System from Promega, and GAL-TK-Luc activity was normalized to that of pRL-TK.
co-IP, immunoblotting, and cell fractionation.
For mSin3A coimmunoprecipitations (co-IPs), Cos7 cells were transiently transfected with GAL4-inv(16) or the GAL4-tagged inv(16) mutants using Lipofectamine (Gibco BRL, Carlsbad, Calif.). Forty-eight hours posttransfection, cells were lysed in 300 μl of phosphate-buffered saline (PBS) containing 0.5% Triton X-100, 0.1% sodium deoxycholate (DOC), 0.1% sodium dodecyl sulfate (SDS), and protease inhibitors (0.5% Triton X-100 for co-IPs with internal deletion mutants; 0.5% Triton X-100 and 0.1% DOC for co-IPs with C-terminal fragments). ME-1 cells were lysed in 400 μl of PBS containing 0.5% Triton X-100, 0.3% DOC, and 0.2% SDS. Lysates were sonicated and precleared with pansorbin (Calbiochem, La Jolla, Calif.) and protein A (Amersham Pharmacia, Uppsala, Sweden) or protein G (Sigma) beads. Endogenous mSin3A was immunoprecipitated with anti-mSin3A antibody (K-20; Santa Cruz Biotechnology, Santa Cruz, Calif.). For ME-1 lysates, endogenous HDAC2 was precipitated with anti-HDAC2 antibody (H-54; Santa Cruz Biotechnology). The proteins were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE) (SDS-7.5% PAGE for ME-1 IPs) and transferred to nitrocellulose.
For HDAC co-IPs, Cos7 cells were cotransfected with pCMV5-GAL4-inv(16), GAL4-tagged inv(16) mutants, or AML-1B and FLAG-, hemagglutinin (HA)-, or Myc-tagged HDACs. Forty-eight hours posttransfection, cells were lysed in 300 μl of PBS containing 0.5% Triton X-100 and protease inhibitors (0.5% Triton X-100 and 0.3% DOC for co-IPs with C-terminal fragments; 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS for co-IPs with AML-1B). Lysates were sonicated and precleared with protein G. HDACs were immunoprecipitated with antibodies to their tags (anti-Flag M2 [Sigma]; anti-Myc 9E10 and anti-HA [both from Covance, Richmond, Calif.]). Proteins were separated by SDS-PAGE and transferred to nitrocellulose. Membranes were blocked in PBS containing 5% nonfat dry milk for 1 h at room temperature and immunoblotted with primary antibodies to the tagged proteins overnight at 4°C. Membranes were washed in PBS and blotted with secondary antibodies. Proteins were visualized by enhanced chemiluminescence (Pierce, Rockford, Ill.).
Cellular fractionation was performed in isotonic buffer (0.01 M Tris-HCl [pH 8.4], 0.14 M NaCl, 0.0015 M MgCl2) containing 0.5% Nonidet P-40 (NP-40) by low-speed centrifugation as described previously (31). Equal proportions of the cytoplasmic and nuclear fractions were analyzed by immunoblotting with antibodies to phospholipase Cγ1 (Robert Abraham, The Scripps Research Institute, San Diego, Calif.), HDAC2 (Santa Cruz Biotechnology), and the GAL4 DNA-binding domain (Santa Cruz Biotechnology).
RESULTS
inv(16) contains a C-terminal repression domain.
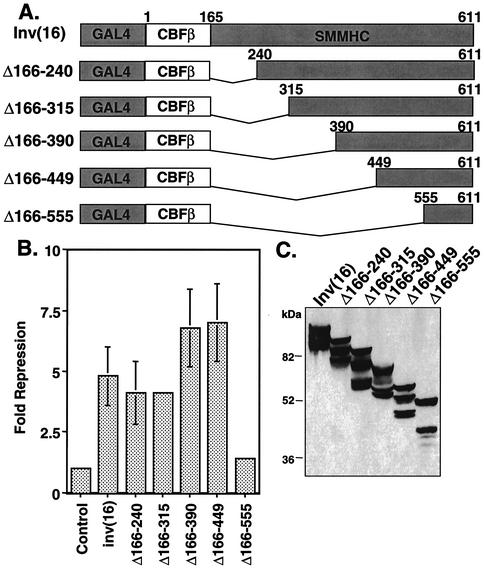
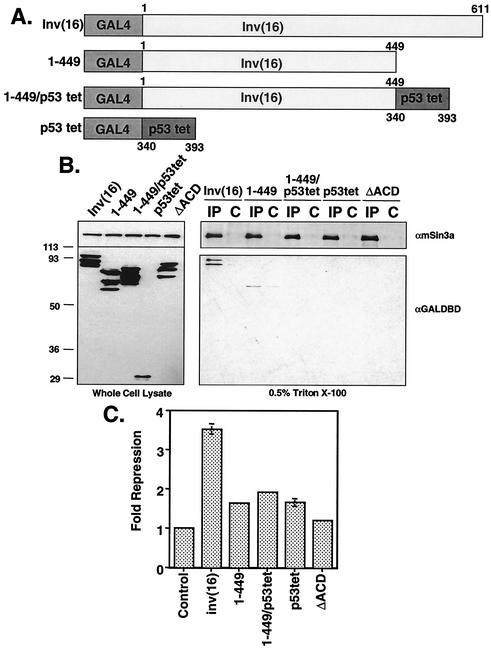
The initial analysis of deletion mutants of the inv(16) fusion protein indicated that the C-terminal 95 aa were required for active transcriptional repression (31). To define the minimal domain sufficient for transcriptional repression, we created progressive internal deletions of the SMMHC region (Fig. 1A). The inv(16) fusion protein lacks a nuclear localization signal; it is both cytoplasmic and nuclear unless it is coexpressed with AML1, which carries it into the nucleus (1, 23, 31). Therefore, to target CBFβ/SMMHC to the nucleus and measure transcriptional repression independent of AML1, we fused these deletion mutants to the GAL4 DNA-binding domain. GAL4-inv(16) repressed a GAL4-dependent reporter plasmid fivefold (using the thymidine kinase promoter linked to firefly luciferase [Fig. 1B]). Internal deletion of up to 284 aa in the SMMHC region of CBFβ/SMMHC was tolerated with little to no effect on repression (Δ166-390 [Fig. 1B]). However, further deletion past residue 449 eliminated repression. The mutants were expressed at levels similar to that of the full-length fusion protein (Fig. 1C). Note that these proteins migrated as multiple bands, possibly due to cryptic alternative splicing with 3′ vector sequences (K. L. Durst, unpublished data). Thus, the slowest migrating band is the full-length form. The transcription assay results indicate that CBFβ sequences alone are not sufficient and the C-terminal 163 aa of SMMHC are required for inv(16)-mediated repression.
FIG. 1.
The C-terminal SMMHC region of the inv(16) fusion protein contains a repression domain. (A) Schematic diagrams of internal deletion mutants of inv(16). (B) NIH 3T3 cells were transfected with 50 ng of plasmids expressing inv(16) or inv(16) mutants and 1 μg of thymidine kinase-firefly luciferase reporter plasmid. Fold repression was calculated after correcting for transfection efficiency using a plasmid expressing SEAP. Levels of repression are the averages ± standard errors of three experiments. Empty expression vector was used as a control. Note that error was too small to graph in a few samples. (C) Immunoblot analysis of internal deletion mutants of inv(16) expressed in Cos7 cells (100 μg of protein from whole-cell lysate was loaded for each mutant).
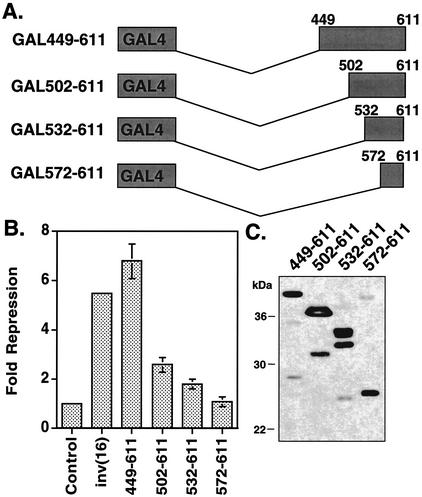
To demonstrate that the C-terminal 163 aa of SMMHC are sufficient for repression, we removed the CBFβ sequences from the fusion protein and created three additional mutations (Fig. 2A). When completely isolated, the C-terminal 163 aa of CBFβ/SMMHC were sufficient for maximal repression (449-611 [Fig. 2B]). However, residues 502 to 611 retained about 50% of maximal activity, and residues 532 to 611 retained some ability to repress transcription (twofold [Fig. 2B]). The nonhelical tail of CBFβ SMMHC, consisting of residues 572 to 611, displayed no ability to repress transcription (Fig. 2B), consistent with C-terminal deletion analysis (31). Once again, the steady-state expression levels of each mutant were similar (Fig. 2C).
FIG. 2.
The C-terminal 163 aa of the inv(16) fusion protein are sufficient for repression. (A) Schematic diagrams of inv(16) C-terminal fragments linked to the GAL4 DNA-binding domain. (B) NIH 3T3 cells were transfected with 50 ng of plasmids expressing inv(16) or the indicated mutants and 1 μg of thymidine kinase-firefly luciferase reporter plasmid. Fold repression was calculated after correcting for transfection efficiency using a plasmid expressing SEAP. Levels of repression are the averages ± standard errors of three experiments. Empty expression vector was used as a control. Note that error was too small to graph in a few samples. (C) Immunoblot analysis of internal deletion mutants of inv(16) expressed in Cos7 cells (100 μg of protein from whole-cell lysate was loaded for each mutant).
inv(16) interacts with mSin3A.
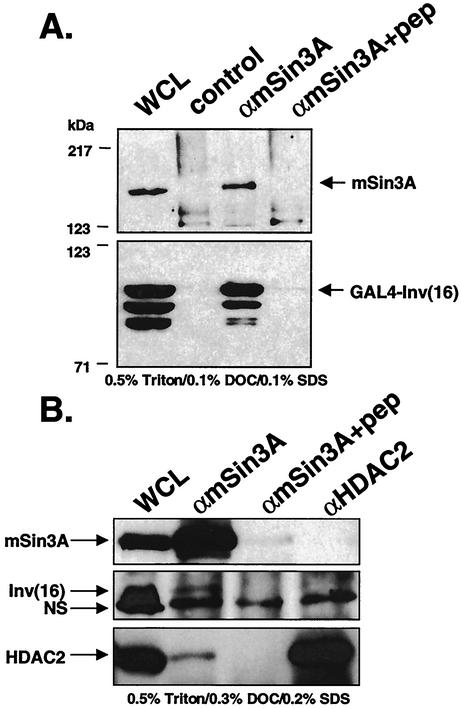
We previously reported that CBFβ/SMMHC forms a trimeric complex with AML1 and the mSin3A corepressor (31). While the inv(16) fusion protein did not contact mSin3A in the absence of AML1 in these initial assays, it is possible that it failed to localize properly in the absence of AML1 (1, 23, 31). Therefore, we used GAL4-inv(16) constructs to localize the proteins to the nucleus and to determine whether the inv(16) fusion protein could interact with mSin3A in the absence of coexpressed AML1. Cos7 cells were transiently transfected with GAL4-inv(16), and endogenous mSin3A was immunoprecipitated from lysates prepared in PBS containing 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS. GAL4-inv(16) was detected by immunoblot analysis using anti-GAL4. The GAL4-inv(16) fusion protein copurified with mSin3A, but not when the mSin3A antibody was blocked with antigenic peptide (Fig. 3A), indicating that the inv(16) fusion protein associates with mSin3A in the absence of AML1 overexpression. The association was not abrogated when the mSin3A antibody was preincubated with a nonspecific peptide (data not shown). The corepressor association was reduced when lysates were prepared in higher stringency with PBS containing 0.5% Triton X-100, 0.3% DOC, and 0.2% SDS (data not shown).
FIG. 3.
The inv(16) fusion protein interacts with mSin3A. (A) Cos7 cells were transfected with pCMV5-GAL4-inv. Cell lysates were prepared in PBS containing 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS. Endogenous mSin3A was precipitated with anti-Sin3A (αmSin3A) polyclonal antibody. Copurifying GAL4-inv(16) was detected by immunoblotting with antibodies to the GAL4 DNA-binding domain. Lanes: WCL, whole-cell lysate; control, lysate incubated with protein A-agarose (no primary immunoglobulin G [IgG]); αmSin3A, lysate incubated with mSin3A IgG; αmSin3A+pep, lysate incubated with peptide-blocked mSin3A IgG. (B) ME-1 cells were lysed in PBS containing 0.5% Triton X-100, 0.3% DOC, and 0.2% SDS. Endogenous mSin3A and HDAC2 were precipitated with anti-mSin3A (αmSin3A) and anti-HDAC2 (αHDAC2) polyclonal antibodies. Copurifying inv(16) was detected by immunoblotting with anti-CBFβ. The position of a nonspecific (NS) band is indicated by the arrow.
The association observed in Cos7 cells transiently transfected with inv(16) led us to test whether endogenous CBFβ/SMMHC associates with mSin3A. We performed the mSin3A co-immunoprecipitations in the inv(16)-expressing ME-1 cell line that was established from blood cells of a patient with AML M4E0 (50). In buffer containing 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS (data not shown) or the more stringent conditions of 0.5% Triton X-100, 0.3% DOC, and 0.2% SDS, the inv(16) fusion protein appeared to associate with mSin3, as a specific band that comigrated with CBFβ/SMMHC was detected (just above a nonspecific band labeled NS [Fig. 3B, middle panel]). The coprecipitation was blocked when the antibody was preincubated with antigen specific to the antibody. The mSin3A corepressor is known to associate with HDAC1 and HDAC2 (25). Therefore, we immunoprecipitated HDAC2 from ME-1 cells to determine if a trimeric complex containing the inv(16) fusion protein could be detected. Under the conditions used to detect the inv(16)-mSin3A association, only a small amount of HDAC2 associated with mSin3A (Fig. 3B, bottom blot) and no inv(16) fusion protein copurified with HDAC2, suggesting a direct interaction between CBFβ/SMMHC and mSin3A.
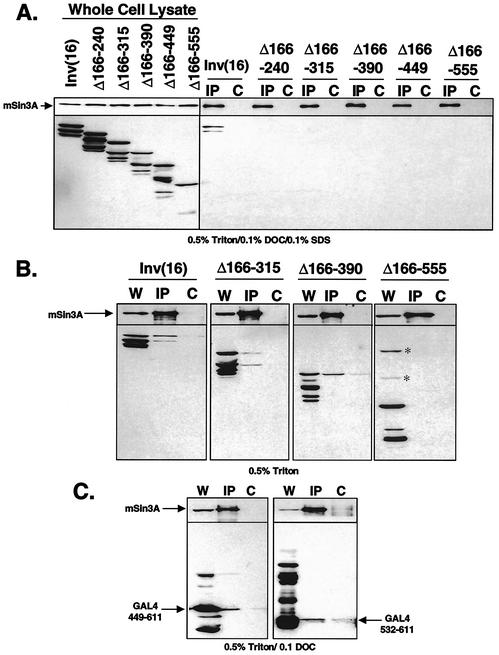
Having defined a transcriptional repression domain within the myosin heavy chain portion of the fusion protein, we asked whether mSin3A binding cosegregated with the ability to repress transcription. The GAL4-inv(16) deletion mutants (Fig. 1A and 2A) were expressed in Cos7 cells and assayed for mSin3A association by co-IP, followed by immunoblot analysis. In contrast to full-length CBFβ/SMMHC, all of the deletion mutants failed to bind mSin3A (Fig. 4A) in buffer containing 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS, even though several mutants retained the ability to repress transcription (e.g., Δ166-240 [Fig. 1B]). Therefore, we repeated the assay using only 0.5% Triton X-100 in PBS to detect weaker interactions. Under these conditions, GAL4-inv(16) as well as the GAL4-inv(16)Δ166-315 and GAL4-inv(16)Δ166-390 mutants coimmunoprecipitated with mSin3A (Fig. 4B). The association was blocked when the antigenic peptide was included in the assay (Fig. 4B, lanes labeled C). In contrast, GAL4-inv(16)Δ166-555, which fails to repress transcription (Fig. 1B), also failed to bind mSin3A (Fig. 4B). However, under this milder condition, we also observed some level of nonspecific association of GAL4-inv(16) with the protein G-Sepharose beads. By these criteria, mutants that retained the ability to repress transcription also associated with mSin3A (Fig. 1B and 4B).
FIG. 4.
The C-terminal domain of CBFβ/SMMHC is sufficient for association with mSin3A. (A) Cos7 cells were transfected with GAL4-inv(16) and the GAL4-inv(16) internal deletion mutants (Fig. 1A). Cell lysates were prepared in PBS containing 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS. Endogenous mSin3A was precipitated with anti-mSin3A antibody. Copurifying proteins were analyzed by immunoblot analysis using antibodies to mSin3A and the GAL4 DNA-binding domain. The lysates were incubated with mSin3A immunoglobulin G (IgG) (IP lanes) or with peptide-blocked mSin3A IgG (C lanes). (B) Same as panel A except that lysates were prepared in PBS containing 0.5% Triton X-100. Nonspecific bands are indicated by the asterisks. W, whole-cell lysate. (C) Cos7 cells were transfected with GAL4-inv(16)449-611 or GAL4-inv(16)532-611 (Fig. 2A). Cells were lysed in PBS with 0.5% Triton X-100 and 0.1% DOC followed by mSin3A immunoprecipitation. Immunoblot analysis was performed as in panel A.
GAL4-inv(16)Δ166-449, which contains the minimal repression domain, displayed too much background binding to protein G-Sepharose in PBS containing 0.5% Triton X-100 to make a clear determination as to whether it retained the ability to bind mSin3A (data not shown). Therefore, we surveyed a range of lysis conditions to determine whether residues 449 to 611 are sufficient to bind mSin3A. GAL4-inv(16)449-611 coimmunoprecipitated with mSin3A when cells were prepared in PBS containing 0.5% Triton X-100 and 0.1% DOC (Fig. 4C, left blot) but did not copurify in the presence of the antigenic peptide as a control (lanes labeled C). Under the same conditions, GAL4-inv(16)532-611, which retained some ability to repress transcription, also weakly associated with mSin3A (Fig. 4C, right blot). The C-terminal fragment (572-611) that had no activity in the GAL4 repression assay (Fig. 2B) failed to bind mSin3A, while the larger fragment, 502-611, showed nonspecific binding in the control immunoprecipitation (data not shown).
inv(16) interacts with HDAC8.
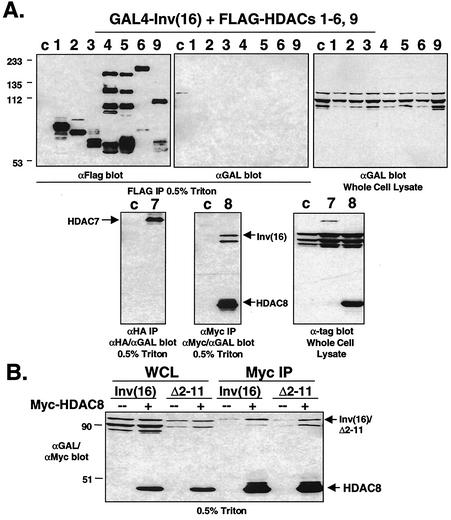
Corepressors recruit HDACs to promoters where HDACs alter chromatin structure by deacetylating histones in order to repress transcription. They are divided into two classes. The class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) are mainly composed of the catalytic domain, while the larger, class II HDACs (HDAC4 to HDAC6, HDAC7, and HDAC9) include a noncatalytic domain (3, 9). Because HDACs interact with other leukemogenic fusion proteins that disrupt AML1, we determined whether the inv(16) fusion protein could bind HDACs. GAL4-inv(16) was coexpressed with FLAG-tagged HDAC1 to HDAC6 and a large fragment of HDAC9, also known as HDAC-related protein (HDRP) (53), as well as HA-tagged HDAC7 and Myc-tagged HDAC8 (Myc-HDAC8). The inv(16) fusion protein coimmunoprecipitated with Myc-HDAC8 but failed to bind the class II HDACs and HDAC1 to HDAC3 (Fig. 5A). Unfortunately, we were unable to detect HDAC8 protein in ME-1 cells, although HDAC8 mRNA is expressed in these cells (Durst, unpublished).
FIG. 5.
The inv(16) fusion protein interacts with HDAC8. (A) Cos7 cells were transfected with pCMV5-GAL4-inv(16) and FLAG-tagged HDAC1 to HDAC6 and HDAC9 (HDRP fragment), HA-tagged HDAC7, or Myc-tagged HDAC8. Cell lysates were prepared in PBS containing 0.5% Triton X-100. Co-IPs were performed using antibodies to the tagged HDACs and were analyzed by immunoblotting with antibodies to FLAG (αFlag), HA (αHA), Myc (αMyc), and the GAL4 DNA-binding domain (αGAL). Cells transfected with GAL4-inv(16) alone are shown in c lanes. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blots. (B) GAL4-inv(16) or GAL4-inv(16)Δ2-11 expressing plasmids were cotransfected with (+) Myc-tagged HDAC8 and analyzed as in panel A. WCL, whole-cell lysate.
To confirm that the association between CBFβ/SMMHC and HDAC8 was not mediated by endogenous AML1, we repeated this assay using the GAL4-inv(16)Δ2-11 mutant that no longer binds AML1 (1). This mutant also associated with HDAC8 (Fig. 5B), suggesting that the interaction is mediated by the SMMHC sequence. Furthermore, mSin3A and HDAC8 did not associate by in Cos7 cells (data not shown), indicating that the association between the fusion protein and HDAC8 is not mediated by mSin3A.
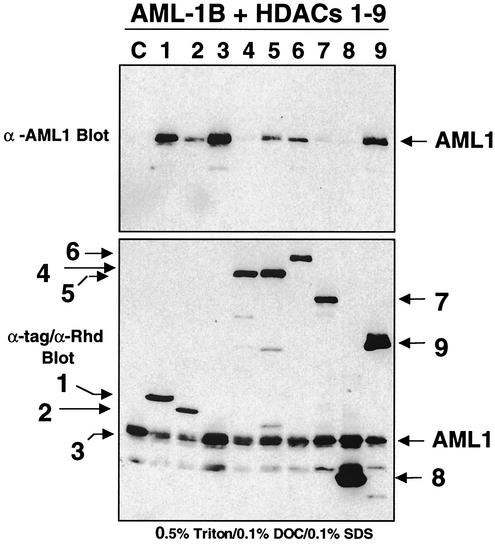
We also tested whether AML1 could associate with HDAC1 to HDAC9 in order to confirm that the inv(16)-HDAC8 interaction was not mediated by AML1. Epitope-tagged forms of each HDAC were coexpressed with AML1, and the cell lysates were immunoprecipitated with antibodies directed to the epitope tag (FLAG, HA, or Myc); coimmunoprecipitating AML1 was detected by immunoblot analysis (Fig. 6, upper blot). AML1 associated with HDAC1, HDAC3, and HDAC9 and more weakly copurified with HDAC2, HDAC5, and HDAC6. However, we did not detect an association with HDAC8.
FIG. 6.
AML-1 binds HDAC1, HDAC3, and HDAC9. AML-1 was coexpressed with HDAC1 to -8 or -9 (HDRP fragment) in Cos7 cells and coimmunoprecipitated with anti-FLAG, Myc, or HA in PBS containing 0.5% Triton X-100, 0.1% DOC, and 0.1% SDS (top blot). Copurifying AML-1 was detected by immunoblot analysis using anti-RHD (Calbiochem). The bottom blot shows HDAC and AML-1B expression in whole-cell lysate. The positions of the HDACs (shown in the figure without the HDAC prefix) are indicated by the arrows. α-AML1 and α-Rhd, anti-AML1 antibodies. Lane C, AML1 expressed in the absence of HDACs.
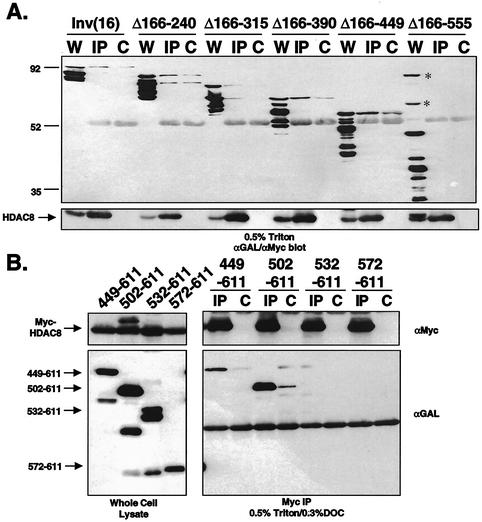
The association between CBFβ/SMMHC and HDAC8 did not appear to be mediated by AML1 or mSin3A. Therefore, we used the GAL4-inv(16) deletion mutants to define the domain within the SMMHC sequences that was required for association with HDAC8. Only GAL4-inv(16)Δ166-555, which failed to repress transcription, was impaired in binding HDAC8 (Fig. 7A). Some of the mutants showed nonspecific binding to protein G-Sepharose. Therefore, 0.3% DOC was added to the lysis buffer for copurification with the C-terminal fragments in which GAL4-inv(16)449-611 and GAL4-inv(16)502-611 coimmunoprecipitated with Myc-HDAC8 with only modest background levels (Fig. 7B). However, GAL4-inv(16)532-611, which retains some ability to repress transcription and weakly associates with mSin3A, failed to bind HDAC8 under these conditions.
FIG. 7.
Mapping the HDAC8 binding domain in CBFβ/SMMHC. (A) Cos7 cells were cotransfected with Myc-tagged HDAC8 and GAL4-inv(16) or one of the GAL4-inv(16) deletion mutants (Fig. 1A). Cells were lysed in PBS containing 0.5% Triton X-100. HDAC8 was immunoprecipitated with antibodies to the Myc tag (αMyc), and copurifying proteins were analyzed with antibodies to the GAL4 DNA-binding domain and the Myc epitope tag (αGAL/αMyc). Lanes: W, whole-cell lysate; IP, lysates immunoprecipitated with anti-Myc; C, Cos7 cells transfected with only GAL4-inv(16) or the GAL4-inv(16) deletion mutants and lysates immunoprecipitated with anti-Myc. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the blot. The positions of nonspecific bands are indicated by asterisks. (B) Cos7 cells were cotransfected with Myc-HDAC8 and GAL4-inv(16) or one of the GAL4-inv(16) deletion mutants (Fig. 2A) and analyzed as in panel A except that PBS contained 0.5% Triton X-100 and 0.3% DOC.
The multimerization domain of inv(16) contributes to repression and interactions with corepressors.
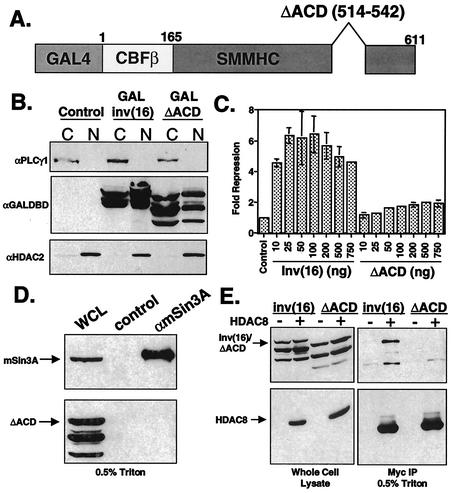
The ability of the inv(16) protein to repress transcription and bind mSin3A and HDAC8 localized to the C-terminal 163 aa. This domain contains the ACD (residues 514 to 542) that catalyzes multimerization of the SMMHC as well as the inv(16) fusion protein (24, 44). Progressive deletion of residues 502 to 532, which includes 18 aa of the ACD, significantly impaired transcriptional repression and HDAC8 binding (Fig. 2B and 7B), and complete deletion of this domain (Δ166-555 [Fig. 4B]; 572-611 [data not shown]) eliminated mSin3A binding. These data suggest that the N-terminal boundaries of the HDAC8 and mSin3A binding motifs overlap with the ACD. Therefore, we tested whether this motif was required for inv(16)-mediated repression and corepressor binding. The inv(16) cDNA with a deletion of aa 514 to 542 (ΔACD) (24) was subcloned into the GAL4-expressing plasmid (Fig. 8A). We fused this mutant to GAL4 for use in the transcription assays; however, previous data showed that it may be retained in the cytoplasm to a greater extent than the wild-type protein (24). Cell fractionation indicated that a significant amount of the GAL4-inv(16)ΔACD did reach the nucleus (Fig. 8B). In transient-transfection assays using the GAL4-TK-Luc reporter, this mutant exhibited impaired transcriptional repression compared to the full-length protein, even at high levels of input inv(16)ΔACD plasmid (Fig. 8C). Deletion of the ACD also resulted in a loss of interaction between inv(16) and mSin3A (Fig. 8D) as well as HDAC8 (Fig. 8E) in co-IP assays. Thus, the ACD contributes to transcriptional repression and corepressor binding.
FIG. 8.
ACD is required for transcriptional repression and interaction with corepressors. (A) Schematic of inv(16)ΔACD mutant. (B) Cos7 cells were transfected with GAL4-inv(16) or GAL4-inv(16)ΔACD. Cells were fractionated in isotonic buffer containing 0.5% NP-40 by low-speed centrifugation. Equal proportions of the cytoplasmic (C) and nuclear (N) fractions were analyzed by SDS-PAGE and immunoblotting with antibodies to phospholipase Cγ1 (αPLCγ1), GAL4 DNA-binding domain (αGAL4DBD), and HDAC2 (αHDAC2). Untransfected Cos7 cells were used as a control. (C) NIH 3T3 cells were transfected with increasing amounts of GAL4-inv(16)- or GAL4-inv(16)ΔACD-expressing plasmid, 1 μg of thymidine kinase-firefly luciferase reporter plasmid, and 200 ng of pCMV-SEAP. Fold repression was calculated after correcting for transfection efficiency using SEAP; each value is the average ± standard error of two experiments. Empty expression vector was used as a control. Note that error was too small to graph in several samples. (D) Cos7 cells were transfected with pCMV5-GAL4ΔACD. Cell lysates were prepared in PBS containing 0.5% Triton X-100. Co-IPs were performed using anti-mSin3A (αmSin3A) polyclonal antibody and were analyzed by immunoblotting with antibodies to mSin3A and the GAL4 DNA-binding domain. WCL, whole-cell lysate; control, immunoprecipitation using peptide-blocked anti-mSin3A. (E) Cos7 cells were transfected with pCMV5-GAL4inv(16) or pCMV5-GAL4ΔACD alone or with Myc-tagged HDAC8. Cell lysates were prepared in PBS containing 0.5% Triton X-100. Co-IPs were performed using anti-Myc and were analyzed by immunoblotting with antibodies to Myc and the GAL4 DNA-binding domain.
To determine if multimerization of the inv(16) amphipathic helices alone could induce association with mSin3A or cause transcriptional repression, we replaced the C-terminal 163 aa of inv(16) with the p53 multimerization domain (p53tet). This domain consists of residues 340 to 393, which form an α-helical basic region that is sufficient for tetramerization (45). p53tet was fused to the GAL4 DNA-binding domain and a GAL4-inv(16)1-449 mutant that lacks both the ACD and the C-terminal repression domain (Fig. 9A). Once again, the wild-type inv(16) fusion protein associated with mSin3A, and the ΔACD mutant failed to bind to mSin3A (Fig. 9B). Addition of the p53 multimerization domain to the GAL4 DNA-binding domain did not induce an mSin3A association. Although GAL-inv(16)1-449 alone did display a weak association with mSin3A, the addition of the p53 sequences did not induce binding to mSin3A (Fig. 9B). In addition, these mutants did not significantly repress transcription compared to the full-length fusion protein (Fig. 9C). Thus, the addition of a multimerization domain to the coiled-coil domain of the inv(16) fusion protein is not sufficient for transcriptional repression or corepressor binding.
FIG. 9.
Artificial multimerization of the inv(16) fusion protein does not induce corepressor binding. (A) Schematic diagrams of mutants of inv(16) and the p53 tetramerization domain. (B) Cos7 cells were transfected with pCMV5-GAL4-p53tet, pCMV5-GAL4-inv(16) or plasmids expressing mutant inv(16) proteins. Cell lysates were prepared in PBS containing 0.5% Triton X-100. Co-IPs were performed using anti-mSin3A (αmSin3A) and were analyzed by immunoblotting with antibodies to mSin3A and the GAL4 DNA-binding domain (αGAL4DBD). Lysates were incubated with mSin3A immunoglobulin G (IgG) (IP lanes) or with peptide-blocked mSin3A IgG (C lanes). (C) NIH 3T3 cells were transfected with 100 ng of plasmids expressing inv(16)- or inv(16) mutants or p53tet and 1 μg of thymidine kinase-firefly luciferase reporter plasmid. Fold repression was calculated after correcting for transfection efficiency using a plasmid expressing SEAP. Levels of repression are the averages ± standard errors of three experiments. Empty expression vector was used as a control. Note that error was too small to graph in some samples.
inv(16)-mediated repression is impaired by a HDAC inhibitor.
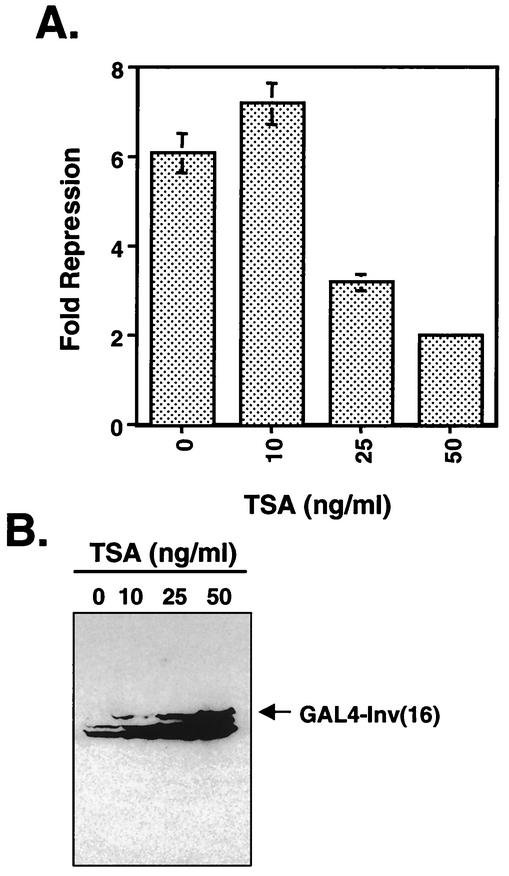
The association of CBFβ/SMMHC with mSin3A and HDAC8 led us to determine whether inhibitors of HDACs affect inv(16)-mediated repression. NIH 3T3 cells were transfected with GAL4-inv(16) and the GAL4-TK-Luc reporter plasmid, and TSA, a HDAC inhibitor, was added to the culture medium for 48 h prior to the preparation of cell lysates for luciferase assays. Even very small amounts of TSA impaired inv(16)-mediated repression (Fig. 10A) compared with treated cells transfected with empty expression vector, although the levels of CBFβ/SMMHC were greatly induced by TSA (Fig. 10B). These data suggest that HDACs play a role in inv(16)-dependent transcriptional repression.
FIG. 10.
TSA treatment impairs inv(16)-mediated transcriptional repression. (A) inv(16)-expressing plasmid (100 ng) was cotransfected with 1 μg of thymidine kinase (TK)-firefly luciferase reporter plasmid and 100 ng of pRL-TK. After transfection, TSA at the indicated concentrations was added to the culture medium for 48 h. Fold repression was calculated after normalizing to pRL-TK activity. Note that TSA affected the control (empty expression vector) to some degree (data not shown), and the values reported are relative to controls that were untreated (bar 0) or treated with TSA (bars 10, 25, and 50). Levels of repression are the averages ± standard errors of three experiments. Note that error was too small to graph in one sample. (B) Immunoblot analysis of transfected NIH 3T3 cell lysates from the experiment in panel A.
DISCUSSION
Transcriptional repression by chromosomal translocation fusion proteins is a common theme in AML. The t(15;17), t(8;21), and t(11;17) fusion proteins contact corepressors that recruit HDACs or directly bind HDACs to repress transcription (17, 19-21, 26, 34). Here we demonstrate that the inv(16) fusion protein has the ability to interact with mSin3A and HDAC8 through an unexpected repression domain within the SMMHC portion of the fusion protein. The mSin3A and HDAC8 binding domains cosegregate with the ability of the inv(16) fusion protein to repress transcription, implying that these factors contribute to inv(16)-mediated repression. In addition, inv(16)-mediated repression was sensitive to TSA, a potent, broad-spectrum HDAC inhibitor. Taken together, these data suggest that HDACs, including HDAC8 and HDACs recruited by mSin3A, contribute to active repression mediated by this chromosomal translocation fusion protein.
The contribution of a transcriptional repression domain by the myosin heavy chain portion of the inv(16) fusion protein was unexpected. This domain contains two known protein interaction motifs: the ACD, which regulates multimerization of the myosin heavy chain, and the coiled-coil domains that make extensive contacts between the chains. While the majority of the coiled-coil domains could be deleted with little or no effect on transcriptional repression (residues 166 to 449 [Fig. 1B]), deletion of these domains did impair mSin3A binding because ionic detergents could not be used to coimmunoprecipitate these deletion mutants (Fig. 4A). Nevertheless, the C-terminal 163 aa were sufficient for interaction with mSin3A and HDAC8 (Fig. 4C and 7B). Thus, the coiled-coil domain may contribute a mSin3A interaction motif, or oligomerization of these domains may yield a more stable association with corepressors binding the C-terminal domain. However, if the coiled-coil domains can contact mSin3A, this association alone is not sufficient to mediate repression (Fig. 8C).
The ability of a nucleus-localized myosin heavy chain to associate with corepressors could be a result of spurious homology to transcription factors. The Ski and Sno oncogenes that were transduced by avian leukemia viruses contain regions of homology to myosin heavy chains (8, 37, 42). Both of these oncogenes associate with mSin3A and are capable of repressing transcription and/or acting as corepressors (38). A comparison of the repression domain of the inv(16) fusion protein with the mSin3A binding domain of Ski (38) indicates 23% identity within these domains. We also considered the possibility that the inv(16) fusion protein may associate with Ski or Sno to repress transcription, given that they contain homologous protein interaction motifs. However, we did not observe an interaction between Ski or Sno and the inv(16) fusion protein (data not shown).
Our deletion mapping studies pinpointed a domain that was necessary and sufficient for active transcriptional repression. While this domain was sufficient for binding corepressors, it also contains the ACD that is required for multimerization of the inv(16) fusion protein. We were able to separate these two functions, as deletion of the first 18 aa of this domain (GAL4-532-611) impaired but did not eliminate mSin3A association (Fig. 4C). In addition, replacement of this domain with the p53 tetramerization domain failed to complement the loss of repression or corepressor binding (Fig. 9B and C). Previous studies also predict that the 502-611 and 532-611 C-terminal fragments of CBFβ/SMMHC, which coimmunoprecipitate with mSin3A, will not multimerize (24). These data argue that the ACD contributes directly to association with the corepressors, rather than causing oligomerization of a distal corepressor binding domain. However, under our assay conditions, the fusion protein most likely retains its ability to dimerize (and GAL4 forms dimers). Thus, it is possible that dimerization of the fusion protein would amplify the ability of the fusion protein to bind mSin3A and HDAC8 and repress transcription.
The inv(16) fusion protein acts as a corepressor for AML1 to repress AML1-regulated genes (31). Because AML1 can also associate with mSin3A to repress transcription and this repression is sensitive to TSA, we screened the known class I and class II HDACs for binding to AML1. Although HDAC8 failed to bind AML1, HDAC1, HDAC3, and HDAC9 associated with AML1 (Fig. 6). Given that both the inv(16) fusion protein and AML1 bind mSin3A, we speculate that association of AML1 with mSin3A is stabilized by the inv(16) fusion protein to convert AML1 from a regulated transcription factor to a constitutive repressor.
inv(16) is one of the most frequently observed chromosomal translocations associated with AML. Our data imply that those HDACs associated with mSin3, and perhaps HDAC8, contribute to the biological actions of inv(16). The observation that TSA impairs inv(16)-mediated repression provides hope that directed therapies for this leukemia may be developed using rational drug design. However, it is likely that targeted therapeutic strategies must account for the HDACs that associate with AML1, the inv(16) fusion protein, and mSin3A.
Acknowledgments
We thank K. Scott Luce and Yue Hou for expert technical assistance and the members of the Hiebert lab for helpful comments and ideas. We thank Sonia Pearson-White (University of Virginia Medical Center) for Ski and Sno cDNAs and Ed Seto (University of South Florida) for HDAC cDNAs and antibodies. We also thank the Vanderbilt Ingram Cancer Center sequencing facility for support.
This work was supported in part by National Institutes of Health (NIH) grants RO1-CA64140, RO1-CA77274, and RO1-CA87549 and a Center grant from the National Cancer Institute (CA68485) to the Vanderbilt-Ingram Cancer Center. A.D.F. is a Leukemia and Lymphoma Society Scholar.
REFERENCES
- 1.Adya, N., T. Stacy, N. A. Speck, and P. P. Liu. 1998. The leukemic protein core binding factor β (CBFβ)-smooth-muscle myosin heavy chain sequesters CBFα2 into cytoskeletal filaments and aggregates. Mol. Cell. Biol. 18:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertos, N. R., A. H. Wang, and X. J. Yang. 2001. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 79:243-252. [PubMed] [Google Scholar]
- 4.Cameron, S., D. S. Taylor, E. C. TePas, N. A. Speck, and B. Mathey-Prevot. 1994. Identification of a critical regulatory site in the human interleukin-3 promoter by in vivo footprinting. Blood 83:2851-2859. [PubMed] [Google Scholar]
- 5.Cao, W., M. Britos-Bray, D. F. Claxton, C. A. Kelley, N. A. Speck, P. P. Liu, and A. D. Friedman. 1997. CBF beta-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene 15:1315-1327. [DOI] [PubMed] [Google Scholar]
- 6.Castilla, L. H., L. Garrett, N. Adya, D. Orlic, A. Dutra, S. Anderson, J. Owens, M. Eckhaus, D. Bodine, and P. P. Liu. 1999. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat. Genet. 23:144-146. [DOI] [PubMed] [Google Scholar]
- 7.Castilla, L. H., C. Wijmenga, Q. Wang, T. Stacy, N. A. Speck, M. Eckhaus, M. Marin-Padilla, F. S. Collins, A. Wynshaw-Boris, and P. P. Liu. 1996. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell 87:687-696. [DOI] [PubMed] [Google Scholar]
- 8.Chaganti, R. S., I. Balazs, S. C. Jhanwar, V. V. Murty, P. R. Koduru, K. H. Grzeschik, and E. Stavnezer. 1986. The cellular homologue of the transforming gene of SKV avian retrovirus maps to human chromosome region 1q22—-q24. Cytogenet. Cell Genet. 43:181-186. [DOI] [PubMed] [Google Scholar]
- 9.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 10.Erickson, P., J. Gao, K. S. Chang, T. Look, E. Whisenant, S. Raimondi, R. Lasher, J. Trujillo, J. Rowley, and H. Drabkin. 1992. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 80:1825-1831. [PubMed] [Google Scholar]
- 11.Fenrick, R., J. M. Amann, B. Lutterbach, L. Wang, J. J. Westendorf, J. R. Downing, and S. W. Hiebert. 1999. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol. Cell. Biol. 19:6566-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank, R., J. Zhang, H. Uchida, S. Meyers, S. W. Hiebert, and S. D. Nimer. 1995. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene 11:2667-2674. [PubMed] [Google Scholar]
- 13.Gelmetti, V., J. Zhang, M. Fanelli, S. Minucci, P. G. Pelicci, and M. A. Lazar. 1998. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18:7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giese, K., C. Kingsley, J. R. Kirshner, and R. Grosschedl. 1995. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995-1008. [DOI] [PubMed] [Google Scholar]
- 15.Goger, M., V. Gupta, W. Y. Kim, K. Shigesada, Y. Ito, and M. H. Werner. 1999. Molecular insights into PEBP2/CBF beta-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBF beta. Nat. Struct. Biol. 6:620-623. [DOI] [PubMed] [Google Scholar]
- 16.Golub, T. R., G. F. Barker, S. K. Bohlander, S. W. Hiebert, D. C. Ward, P. Bray-Ward, E. Morgan, S. C. Raimondi, J. D. Rowley, and D. G. Gilliland. 1995. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 92:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grignani, F., S. De Matteis, C. Nervi, L. Tomassoni, V. Gelmetti, M. Cioce, M. Fanelli, M. Ruthardt, F. F. Ferrara, I. Zamir, C. Seiser, M. A. Lazar, S. Minucci, and P. G. Pelicci. 1998. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391:815-818. [DOI] [PubMed] [Google Scholar]
- 18.Guidez, F., K. Petrie, A. M. Ford, H. Lu, C. A. Bennett, A. MacGregor, J. Hannemann, Y. Ito, J. Ghysdael, M. Greaves, L. M. Wiedemann, and A. Zelent. 2000. Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood 96:2557-2561. [PubMed] [Google Scholar]
- 19.He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct interactions of PML-RARalpha and PLZF-RARalpha with corepressors determine differential responses to RA in APL. Nat. Genet. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 20.Hiebert, S. W., B. Lutterbach, and J. Amann. 2001. Role of co-repressors in transcriptional repression mediated by the t(8;21), t(16;21), t(12;21), and inv(16) fusion proteins. Curr. Opin. Hematol. 8:197-200. [DOI] [PubMed] [Google Scholar]
- 21.Hong, S. H., G. David, C. W. Wong, A. Dejean, and M. L. Privalsky. 1997. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94:9028-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno, T., Y. Kanno, L.-F. Chen, E. Ogawa, W.-Y. Kim, and Y. Ito. 1998. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol. 18:2444-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanno, Y., T. Kanno, C. Sakakura, S.-C. Bae, and Y. Ito. 1998. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor α (CBFα) subunit by the leukemia-related PEBP2/CBFβ-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol. Cell. Biol. 18:4252-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kummalue, T., J. Lou, and A. D. Friedman. 2002. Multimerization via its myosin domain facilitates nuclear localization and inhibition of core binding factor (CBF) activities by the CBFβ-smooth muscle myosin heavy chain myeloid leukemia oncoprotein. Mol. Cell. Biol. 22:8278-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 26.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 27.Linggi, B., C. Muller-Tidow, L. Van De Locht, M. Hu, J. Nip, H. Serve, W. E. Berdel, B. Van Der Reijden, D. E. Quelle, J. D. Rowley, J. Cleveland, J. H. Jansen, P. P. Pandolfi, and S. W. Hiebert. 2002. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat. Med. 8:743-750. [DOI] [PubMed] [Google Scholar]
- 28.Liu, P., S. A. Tarle, A. Hajra, D. F. Claxton, P. Marlton, M. Freedman, M. J. Siciliano, and F. S. Collins. 1993. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 261:1041-1044. [DOI] [PubMed] [Google Scholar]
- 29.Liu, P. P., C. Wijmenga, A. Hajra, T. B. Blake, C. A. Kelley, R. S. Adelstein, A. Bagg, J. Rector, J. Cotelingam, C. L. Willman, and F. S. Collins. 1996. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chromosomes Cancer 16:77-87. (Erratum, 18:71, 1997.) [DOI] [PubMed] [Google Scholar]
- 30.Look, A. T. 1997. Oncogenic transcription factors in the human acute leukemias. Science 278:1059-1064. [DOI] [PubMed] [Google Scholar]
- 31.Lutterbach, B., Y. Hou, K. L. Durst, and S. W. Hiebert. 1999. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc. Natl. Acad. Sci. USA 96:12822-12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutterbach, B., J. J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S. W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275:651-656. [DOI] [PubMed] [Google Scholar]
- 33.Lutterbach, B., J. J. Westendorf, B. Linggi, A. Patten, M. Moniwa, J. R. Davie, K. D. Huynh, V. J. Bardwell, R. M. Lavinsky, M. G. Rosenfeld, C. Glass, E. Seto, and S. W. Hiebert. 1998. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 35.Mitelman, F., and S. Heim. 1992. Quantitative acute leukemia cytogenetics. Genes Chromosomes Cancer 5:57-66. [DOI] [PubMed] [Google Scholar]
- 36.Nimer, S., J. Zhang, H. Avraham, and Y. Miyazaki. 1996. Transcriptional regulation of interleukin-3 expression in megakaryocytes. Blood 88:66-74. [PubMed] [Google Scholar]
- 37.Nomura, N., S. Sasamoto, S. Ishii, T. Date, M. Matsui, and R. Ishizaki. 1989. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res. 17:5489-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura, T., M. M. Khan, S. C. Kaul, H. D. Dong, R. Wadhwa, C. Colmenares, I. Kohno, and S. Ishii. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13:412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuchprayoon, I., S. Meyers, L. M. Scott, J. Suzow, S. Hiebert, and A. D. Friedman. 1994. PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2β/CBFβ proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol. Cell. Biol. 14:5558-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194:314-331. [DOI] [PubMed] [Google Scholar]
- 41.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 42.Pearson-White, S., D. Deacon, R. Crittenden, G. Brady, N. Iscove, and P. J. Quesenberry. 1995. The ski/sno protooncogene family in hematopoietic development. Blood 86:2146-2155. [PubMed] [Google Scholar]
- 43.Prosser, H. M., D. Wotton, A. Gegonne, J. Ghysdael, S. Wang, N. A. Speck, and M. J. Owen. 1992. A phorbol ester response element within the human T-cell receptor beta-chain enhancer. Proc. Natl. Acad. Sci. USA 89:9934-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohn, R. L., K. L. Vikstrom, M. Strauss, C. Cohen, A. G. Szent-Gyorgyi, and L. A. Leinwand. 1997. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J. Mol. Biol. 266:317-330. [DOI] [PubMed] [Google Scholar]
- 45.Sturzbecher, H. W., R. Brain, C. Addison, K. Rudge, M. Remm, M. Grimaldi, E. Keenan, and J. R. Jenkins. 1992. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene 7:1513-1523. [PubMed] [Google Scholar]
- 46.Takahashi, A., M. Satake, Y. Yamaguchi-Iwai, S. C. Bae, J. Lu, M. Maruyama, Y. W. Zhang, H. Oka, N. Arai, K. Arai, et al. 1995. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood 86:607-616. [PubMed] [Google Scholar]
- 47.Tang, Y. Y., J. Shi, L. Zhang, A. Davis, J. Bravo, A. J. Warren, N. A. Speck, and J. H. Bushweller. 2000. Energetic and functional contribution of residues in the core binding factor beta (CBFbeta) subunit to heterodimerization with CBFalpha. J. Biol. Chem. 275:39579-39588. [DOI] [PubMed] [Google Scholar]
- 48.Wang, J., T. Hoshino, R. L. Redner, S. Kajigaya, and J. M. Liu. 1998. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 95:10860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanagisawa, K., T. Horiuchi, and S. Fujita. 1991. Establishment and characterization of a new human leukemia cell line derived from M4E0. Blood 78:451-457. [PubMed] [Google Scholar]
- 51.Yang, Y., W. Wang, R. Cleaves, M. Zahurak, L. Cheng, C. I. Civin, and A. D. Friedman. 2002. Acceleration of G1 cooperates with CBFβ-SMMHC to induce acute leukemia in mice. Cancer Res. 62:2232-2235. [PubMed] [Google Scholar]
- 52.Zhang, D. E., K. Fujioka, C. J. Hetherington, L. H. Shapiro, H. M. Chen, A. T. Look, and D. G. Tenen. 1994. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol. Cell. Biol. 14:8085-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, X., V. M. Richon, R. A. Rifkind, and P. A. Marks. 2000. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. USA 97:1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]