Abstract
The expression of CD154 (CD40 ligand) by activated T lymphocytes plays a central role in humoral and cellular immunity. The fundamental importance of this protein in mounting an immune response has made it an attractive target for immunomodulation. Several studies have demonstrated that CD154 expression is regulated at the level of mRNA turnover in a manner distinct from other cytokine genes. We have purified, sequenced, and characterized the two major proteins that bind the CD154 3′ untranslated region (3′UTR) as members of the polypyrimidine tract binding protein (PTB) family. One of these proteins is a previously unreported alternatively spliced PTB isoform, which we call PTB-T. These proteins interact with a polypyrimidine-rich region within the CD154 3′UTR that lacks any known cis-acting instability elements. The polypyrimidine-rich region of the CD154 3′UTR was both necessary and sufficient to mediate changes in reporter gene expression and mRNA accumulation, indicating the presence of a novel cis-acting instability element. The presence of a cis-acting instability element in the polypyrimidine-rich region was confirmed using a tetracycline-responsive reporter gene approach. The function of this cis-acting element appears to be dependent on the relative cytoplasmic levels of PTB and PTB-T. Cotransfection of vectors encoding PTB-T consistently decreased the CD154 3′UTR-dependent luciferase expression. In contrast, transfection of plasmids encoding PTB tended to increase CD154 3′UTR-dependent luciferase expression. Thus, the CD154 3′UTR contains a novel cis-acting element whose function is determined by the binding of PTB and PTB-T. These data identify a specific pathway that regulates CD154 expression that can potentially be selectively targeted for the treatment of autoimmune disease and allograft rejection.
The expression of CD154 (CD40 ligand) by activated T lymphocytes is critical in the development of humoral and cell-mediated immunity (16, 22, 34). The interaction of CD154 with its receptor, CD40, was first shown essential for B-cell growth and differentiation and formation of germinal centers (16). With recognition that CD40 is expressed on an array of cell types came awareness that this interaction is essential to numerous elements of cell-mediated immunity. In the absence of CD154, antigen presentation by dendritic cells and macrophages is profoundly impaired, as is macrophage-mediated killing of intracellular or extracellular pathogens (22, 34). Given the breadth of importance of CD154-CD40 interaction, it is not surprising that CD154 blockade retards the development and progression of immune responses in an array of transplantation and autoimmune disease models ranging from systemic lupus erythematosus to rheumatoid arthritis to multiple sclerosis (16, 22).
The CD154 gene is located on the X chromosome and belongs to the tumor necrosis factor (TNF) gene family (25). Study of CD154 expression chiefly involves CD4+ T lymphocytes, with the earliest studies showing that resting cells express little or no CD154 (28, 35, 48). Activation of the T lymphocytes demonstrated that induction of CD154 expression was different from that of other cytokines. Signals (anti-CD3 and mitogenic lectins) that triggered resting T cells to engage in high levels of proliferation and cytokine production would elicit very little (CD4+ T cells) or no (CD8+ T cells) expression on either mouse or human T cells (28, 35, 48). Optimal expression of CD154 was found to require pharmacological stimulation provided by phorbol myristate acetate (PMA) and calcium ionophores such as ionomycin (IONO) (28, 35, 48, 49). The induction of CD154 on T lymphocytes is blocked by concurrent treatment with cyclosporine and glucocorticoids; these effects are presumed to be transcriptional (17, 48), based on the presence of NF-AT sites in the CD154 promoter (50). Since cyclosporine and glucocorticoids also inhibit cytokine production (3, 54), this pathway does not account for the differential regulation of CD154 expression by T lymphocytes.
These data prompted consideration of the role of posttranscriptional pathways in the selective regulation of CD154 expression. As stated above, CD154 is a member of the TNF gene family (25). The expression of TNF alpha (TNF-α) is primarily regulated at the level of mRNA turnover and translation, conferred by adenine-uridine rich cis-acting elements (AURE) present in its 3′ untranslated region (3′UTR) (5, 26, 51). CD154 mRNA is rapidly degraded in human peripheral blood T lymphocytes with a half-life (∼30 min) similar to that seen with interleukin 2 (IL-2) (15, 31, 44, 56). Two separate lines of evidence exist to suggest that CD154 and cytokine mRNA stability are differentially regulated in activated T lymphocytes. First, signals (CD2 engagement by LFA-3) have been shown to stabilize CD154 mRNA without altering IL-2 mRNA stability (31). Second, CD28 cross-linking has been shown to increase cytokine (TNF-α and IL-2) production at the level of mRNA stability (29), while having minimal effect on CD154 expression (15).
Given evidence that CD154 and AURE-dependent cytokine mRNA stability are independently regulated in T lymphocytes, we hypothesized the presence of a novel 3′UTR cis-acting element whose function was modulated by specific RNA binding proteins. PMA or IONO treatment rapidly increased CD154 mRNA stability in human peripheral blood lymphocytes (PBL), even in the context of transcriptional inhibition (44). In these studies, two major (p50 and p25) and two minor (p40 and p36) RNA binding proteins were shown to bind the CD154 3′UTR. The binding of the p50 and p25 mapped to a polypyrimidine-rich region (∼0.4 kb) that lacked an AURE. UV cross-linking studies demonstrated that the p50 and p25 directly contacted uridines and cytidines in this region. Signals which stabilized CD154 mRNA decreased p25 levels in both cytosolic and polysomal fractions, while a corresponding increase in p50 binding activity was observed (44). These data suggested the presence of a novel cis-acting element in this polypyrimidine-rich region that regulated CD154 mRNA turnover through the relative levels of p50 and p25 RNA binding proteins.
We addressed this hypothesis by identifying these RNA binding proteins and characterizing their functional relationship with CD154 3′UTR-dependent gene expression. The p50 binding activity was purified from calf thymus (CT) and identified as polypyrimidine tract binding protein (PTB), also known as hnRNP I (18-20, 37). The use of PTB-specific antisera confirmed the identity of the cytoplasmic p50 CD154 3′UTR binding protein as PTB. Peptide sequencing, immunoblotting, and immunoprecipitation studies additionally indicated that the p25 was a PTB-related protein. The gene encoding the p25 protein was cloned. DNA sequencing established the identity of the p25 as a previously unreported splice isoform of PTB (referred to subsequently in this work as PTB-T).
In addition, chimeric reporter gene constructs were used to demonstrate that the CD154 3′UTR inhibits expression in vivo by reducing mRNA accumulation in a 3′UTR-specific manner. The presence of the polypyrimidine-rich region of the CD154 3′UTR reduced reporter gene expression activity, mRNA accumulation, and mRNA stability. Since this region constituted the sites of p50 and p25 binding, we examined their role in regulating its function. Overexpression of PTB and PTB-T differentially regulated chimeric reporter gene expression in a CD154 3′UTR-dependent manner. These data indicate that in addition to roles in pre-mRNA splicing and internal ribosome entry site (IRES)-dependent translation (4, 10, 21, 57, 58), PTB binding to the 3′UTR can regulate mRNA stability. In this instance, the relative ratios of alternately spliced isoforms of cytoplasmic PTB in activated T lymphocytes play an important role in the regulation of CD154 mRNA stability. As such, these data provide a model of understanding of the selective regulation of CD154 expression in activated T lymphocytes.
MATERIALS AND METHODS
Preparation of cytosolic and polysomal extracts.
The Jurkat human T-cell line was maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (HyClone) and gentamicin sulfate (50 μg/ml). Human PBL cells from volunteer donors were activated with phytohemagglutinin (PHA) (Murex) at 1 μg/ml, a concentration found to give optimal proliferation and lymphokine preparation. Cytoplasmic preparations were performed using a method characterized for its lack of contamination by nuclear proteins (24). Cytoplasmic lysates were prepared by washing the cells twice in ice-cold phosphate-buffered saline. All reagents and subsequent steps were used at 4°C. The cells were lysed by gentle resuspension in 1% Triton X-100 lysis buffer (50 μl/2 × 107 cells) containing 10 mM PIPES (pH 6.8), 100 mM KCl, 2.5 mM MgCl2, 300 mM sucrose, 1 mM Pefabloc, and leupeptin and pepstatin A (2 μg/ml each) before a 3-min incubation followed by 3-min centrifugation at 500 × g. The supernatant was aliquoted and stored at −80°C as the cytoplasmic fraction. Polysomes were prepared as previously described (44). Human peripheral blood mononuclear cells from volunteer donors were homogenized in buffer A (10 mM Tris-HCl [pH 7.6], 1 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol [DTT], leupeptin and pepstatin A [each at 2 μg/ml], and 2 mM Pefabloc), and were nuclei removed by centrifugation. The supernatant was layered over a 30% sucrose cushion followed by ultracentrifugation at 36,000 rpm in a Beckman L80 ultracentrifuge with an SW41 rotor for 4 h at 4°C. The supernatant was removed as the S130 fraction, and the pellet was resuspended in buffer A and stored in aliquots at −80°C as the polysome fraction.
RNA binding assays.
Human CD154 3′UTR was generated by reverse transcriptase PCR (RT-PCR) using RNA isolated from PHA-activated (16 h) PBL, using primers that generated products encoding CD154 nucleotides (nt) 12 to 986 (referred to herein as CD154 12-986) and CD154 468-835 of the human CD154 3′UTR. Each primer set also introduced a SpeI restriction enzyme site at both ends. Following amplification the PCR products were inserted into TOPO 2.1 vector (Invitrogen) and confirmed by sequencing. CD154 12-986 and CD154 468-835 were released from TOPO 2.1 with SpeI and ligated into the XbaI site of T7/T3 α-19 (Gibco/BRL) and confirmed by sequencing. The deletion mutant T7/T3 CD154 483-814 del was generated by QuikChange (Stratagene) deletion from the T7/T3 CD154 12-986. T7/T3 CD154 12-986 was linearized with KpnI or EcoRI to generate the 12-986 and 12-292 templates, respectively. T7/T3 CD154 486-835 was linearized with EcoRI. α-32P-labeled mRNAs with specific activity of >108 cpm/μg of RNA were prepared by in vitro transcription by T7 RNA polymerase in the presence of 50 μCi of [α-32P]UTP (3,000 Ci/mmol) from Perkin-Elmer Life Sciences and 0.0125 mM UTP and 2.5 mM ATP, GTP, and CTP (Roche Biochemicals).
RNA probes (8 × 104 cpm; 3 to 14 fmol; calculated based on [α-32P]UTP incorporation) were incubated with the specified amounts of cytoplasmic extract, nucleoplasmic extract or A260 polysomes in 12 mM HEPES (pH 7.9), 15 mM KCl, 0.2 μM DTT, Saccharomyces cerevisiae tRNA (0.2 μg/ml), and 10% glycerol for 10 min at 30°C. UV cross-linking was performed at 4°C using a Stratagene UV Stratalinker 1800 (5 min, 3,000 μW/cm2) followed by RNase digestion (10 U of RNase T1 and 20 μg of RNase A) for 30 min at 37°C (44). The protein-RNA complexes were analyzed under denaturing conditions by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE), dried, and monitored by autoradiography. Immunoprecipitation of protein-RNA complexes was performed by incubation for 2 h at 4°C using the anti-PTB monoclonal antibody BB7 (generously provided by Douglas Black, Howard Hughes Medical Institute-University of California, Los Angeles) bound to protein A-Sepharose beads (Pharmacia). Parallel immunoprecipitation was performed with the anti-hnRNP A2 monoclonal antibody, EF67, as a specificity control (33). Beads were washed six times in 100 mM NaCl, boiled in SDS-PAGE loading buffer, and resolved by SDS-12% PAGE and analyzed as described above.
Protein purification and identification.
Fresh CT (1.2 kg) was obtained at a slaughterhouse, chopped into ∼1-in. cubes and snap frozen in liquid nitrogen. The tissue was thawed overnight at 4°C in buffer A (50 mM HEPES [pH 7.5], 25 mM KCl, 5 mM MgCl2, 250 mM sucrose, 10 mM 2-mercaptoethanol, and 1 mM Pefabloc). All subsequent steps were performed at 4°C, as previously described (32). The tissue was ground in a blender in 3 liters of buffer A, and then the crude homogenate was passed successively through two, four, and eight layers of cheesecloth. The suspension was centrifuged at 1,800 × g for 7 min. The supernatant was transferred to clean tubes as the cytoplasmic fraction. The nuclear pellet was resuspended in 2 liters of extraction buffer (250 mM sucrose, 400 mM NaCl, 50 mM HEPES [pH 7.5]) and then centrifuged at 8,500 rpm in a GS3 rotor for 10 min. The supernatant was saved as the nucleoplasmic extract and sequentially subjected to 25, 50, and 75% ammonium sulfate precipitation. The ammonium sulfate precipitates were resuspended in dialysis buffer (50 mM NaCl, 20 mM HEPES [pH 7.5], 2 mM EDTA, 10% [wt/vol] glycerol, 10 mM 2-mercaptoethanol, and 1 mM Pefabloc) before two 2-h rounds of dialysis at 4°C.
Fractions were analyzed for the presence of CD154 3′UTR RNA binding proteins. The nuclear 50% ammonium sulfate fraction (120 ml) was applied to a 130-ml DEAE-Sephacel (Sigma Chemicals, St Louis, Mo.) column. The flowthrough was collected, and the column was washed with 500 ml of binding buffer (20 mM HEPES [pH 7.5], 10 mM KCl, 0.2 μM DTT, 10% glycerol, 1 mM Pefabloc). Proteins were eluted with 0.1, 0.3, 0.5, and 1 M KCl and analyzed for RNA binding activity. The flowthrough and 0.1 M elution fraction were combined and passed over a carboxymethyl cellulose (CMC) column and eluted with a step gradient of 0.1, 0.3, 0.5, and 1 M KCl. The 0.3 M elution was dialyzed against poly(U) Sepharose binding buffer (12 mM HEPES [pH 7.5], 15 mM KCl, yeast tRNA [1 μg/ml], 0.2 μM DTT, 100 μM Pefabloc, 10% glycerol), applied to a 2-ml poly(U) Sepharose column, washed, and eluted. Specified elutions were resolved by SDS-12% PAGE, and the p25 and p50 Coomassie blue-stained bands were excised and sent to the HHMI Biopolymer Laboratory and the W. M. Keck foundation for identification. The p50 was identified by mass spectrometry-mass spectrometry analysis of the tryptic digest on a Q-TOF mass spectrometer. The p25 peptide was identified by internal amino acid sequencing of a tryptic digest.
Immunoblotting.
Following resolution by SDS-12% PAGE and electrotransfer to nitrocellulose, blots were blocked overnight at 25°C in TBS-T (50 mM Tris [pH 2.4], 200 mM NaCl, 3% bovine serum albumin, 0.05% Tween 20) before incubating for 1 h at 25°C with BB7 hybridoma supernatant (diluted 1:2,000) or affinity-purified rabbit antisera specific for the N-terminal 13 amino acids of PTB (diluted 1:200) in TBS-T (with 1% bovine serum albumin). The N-terminal peptide-specific antisera was generously provided by Douglas Black. Blots were then washed, incubated with a 1:10,000 dilution of goat anti-mouse horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature, washed five times with TBS-T, and visualized using chemiluminescence substrate (SuperSignal; Pierce).
Cloning of the p25/PTB-T.
The p25/PTB-T was cloned by RT-PCR amplification using upper and lower primers corresponding to the 5′UTR and 3′UTR of human PTB. For the upper primer, nt 66 to 85 (5′ CCCGCGGTCTGCTCTGTGTG 3′) were used, while the lower primer utilized nt 1816 to 1839 (5′ AATCTCTCGGCGGCTAGGTCACT 3′). RNA from two different donor PHA-activated PBL as well as the Jurkat human T-cell line was isolated, reverse transcribed (Superscript II RT from Invitrogen) using oligo(dT) (Invitrogen), and PCR amplified using Taq DNA polymerase (Roche Biochemicals). A 700-bp band was resolved by agarose gel electrophoresis, excised, cloned into TOPO 2.1 (Invitrogen), and sequenced. Identical sequences of PTB-T were seen in multiple clones derived from each RT-PCR. PTB-T was then PCR amplified from TOPO 2.1/PTB-T and TA cloned into pcDNA3.1 (Invitrogen) and sequenced to confirm that no errors were introduced during amplification. In vitro transcription and translation of [35S]methionine-labeled PTB-T was performed using pcDNA3.1 PTB-T vector and Proteinscript II (Ambion). Labeled proteins were resolved by SDS-12% PAGE and visualized by autoradiography. The pTRI-Xef1α cDNA, which encodes an ∼50-kDa protein, was supplied by the manufacturer as a positive control.
Transient transfection of cell lines.
The pRC/PTB (hnRNP I) vector was the generous gift of Stanley Lemon. PTB was released from pRC with HindIII and ligated into the HindIII site in pcDNA 3.1 to yield pcDNA3.1-PTB. Luciferase reporter constructs were generated by digesting TOPO 2.1/CD154 3′UTR 12-986 with BamHI and XhoI to release CD154 3′UTR 104-986 and ligated into the same sites in pcDNA3.1 Zeo(+) (Invitrogen) and confirmed by sequencing. The cDNA encoding firefly luciferase was released from pGL3-control vector (Promega, Madison, Wis.) by XbaI and HindIII digestions, gel purified, and ligated into the BamHI site of pcDNA3.1/CD154 104-986 to yield pcDNA3.1/LUC/CD154104-986. Digestion of pcDNA3.1/LUC/CD154 104-986 with BamHI and XhoI was used to release the CD154 3′UTR and religated to yield pcDNA3.1/LUC. The pcDNA3.1/LUC/CD154 468-835 expression plasmid was generated by digesting TOPO 2.1/CD154 468-835 with BamHI and XhoI and ligating gel-purified insert into the pcDNA3.1/LUC/CD154 104-986 that had been digested with BamHI and XhoI to remove CD154 104-986. Deletion constructs were generated by QuikChange (Stratagene) deletion from TOPO 2.1/CD154 12-986, released by BamHI/XhoI digestion, and ligated into the XbaI site of pcDNA 3.1/LUC. For generation of tetracycline-repressible luciferase expression, inserts containing the CD154 3′UTR were released by BamHI/EcoRV digestion from TOPO vectors described above and cloned into the EcoRV site downstream of the luciferase coding region in the pTRE-Luc vector (Clontech). Each vector was verified by sequencing at least twice in each direction.
Transient transfections were performed using 2 × 106 Jurkat cells or 106 HeLa cells with 0.1 μg of luciferase vectors plus 6 μl of Lipofectamine (BRL) in 0.5 ml of RPMI for 2.5 h at 37°C in 5% CO2, after which 1 ml of RPMI-20% fetal calf serum was added. After 20 h, cells were lysed and luciferase activity was determined using a luciferase reporter assay (Promega) and luminometry. In each experiment, data are shown as the CD154 3′UTR-specific effect by dividing the mean luciferase activity from triplicate transfections of pcDNA3.1/LUC/CD154 3′UTR-containing expression plasmids by that obtained from cells transfected with the pcDNA 3.1 LUC vector, which was assigned a value of 100%. In PTB and PTB-T overexpression experiments, 1 μg of pcDNA3.1 PTB-T, pcDNA3.1-PTB, or an empty vector control was used with 0.1 μg of the luciferase expression plasmids that either lacked or contained the CD154 3′UTR, and mean luciferase activity was determined. In these experiments, the percent inhibition of CD154 3′UTR-dependent luciferase expression seen with each vector was calculated and then divided by the inhibition seen with the empty control vector, which was assigned a value of 100%.
Transient transfection of primary CD4+ T cells.
Primary human CD4 T cells (>95% purity) were isolated from PBL by negative selection (StemCell Technologies, Vancouver, British Columbia, Canada) and transiently transfected as previously described (12). After being cultured overnight with an equivalent number of irradiated (3,300 rads) syngeneic whole-blood mononuclear cells in PHA (1 μg/ml), CD4 T cells were isolated and subjected to electroporation 19.5 h post-PHA stimulation. Five million CD4 T cells were transiently transfected with plasmid DNA in 250 μl of medium in 0.4-cm-gap cuvettes at 250 V and 950 μF using a Bio-Rad (Hercules, Calif.) gene pulser. Two micrograms of either pcDNA3.1/LUC/CD154 104-986 or pcDNA3.1/LUC cDNA was cotransfected with 2 μg of expression vector (pcDNA 3.1, pcDNA 3.1 PTB, or pcDNA3.1 PTB-T) along with 1 μg of a Renilla luciferase expression control vector, pRL-null (Promega). Cells were rested for 2 h, and 1 million cells per well were stimulated in vitro with PMA (25 ng/ml) and IONO (1.5 μM) or medium alone for 6 h at 37°C. Cells were washed and lysed, and luciferase activity was determined using a Dual Luciferase assay kit (Promega) and a LB9507 luminometer (EG&G Wallac, Bad Wildbad, Germany). Data were analyzed in duplicate and corrected for transfection efficiency based on Renilla luminescence (13).
Northern blotting and RT-PCR light cycler RNA analysis.
Jurkat cells were transiently transfected as described and total cellular RNA was extracted by acid guanidinium-phenol-chloroform extraction (9) modified by increasing the 2-mercaptoethanol (Sigma) from 0.1 to 0.7 M in the 5 M guanidinium thiocyanate (Fluka, Biochemika) denaturing solution. RNA was size fractionated by formaldehyde-agarose gel electrophoresis and blotted on to a Hybond-N nylon membrane (Amersham Corp., Arlington Heights, Ill.) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and baked under vacuum at 80°C × 2 h. The Northern blot was sequentially hybridized with an end-labeled luciferase primer (5′ GGTACTTCGTCCACAAACACAACTCC 3′) and oligonucleotide-labeled HLA-B7 cDNA and visualized by autoradiography, with quantification performed by phosphorimaging using the PhosphorImager 445 SI (Molecular Dynamics). A separate transfection was analyzed by real-time PCR. Total cellular RNA was extracted using an RNeasy kit (Qiagen), and poly(A)+ RNA was isolated using Oligotex beads (Qiagen). Poly(A)+ RNA was digested with DNase I (Ambion) prior to reverse transcribing with oligo(dT) and Superscript II RT (Invitrogen). Reverse transcriptions were analyzed for luciferase transcripts using 5′ GGTGGCTCCCGCTGAATTGG 3′ (upper primer) and 5′ CCGTCATCGTCTTTCCGTGC 3′ (lower primer) and SYBER Green PCR core reagents (Perkin-Elmer Biosystems) by real-time PCR using a Bio-Rad iCycler. Each RT was simultaneously examined for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcript as a control. The luciferase/GAPDH transcript ratio was calculated for each sample, based on the manufacturer's instructions. For studies of mRNA stability, Tet-Off HeLa cells (Clontech) were purchased and maintained according to manufacturer's instructions and transiently transfected as described above, allowed to recover overnight, and then treated with doxycycline (1 μg/ml) to shut off transcription for specified times. RNA extraction and analysis were performed as described above.
RESULTS
Characterization, purification and identification of PTB as CD154 3′UTR binding proteins.
Excluding an insert (nt 1 to 292) immediately distal to the translational stop site, the human and murine CD154 3′UTRs exhibit ∼70% nucleotide identity between nt 293 to 986. This level of conservation is similar to that seen in the TNF-α 3′UTR (67%), which plays a dominant role in regulating its expression (5, 26). This conserved portion of the CD154 3′UTR is distinguished by the presence of a CU-rich and polycytidine sequences, as well as a CA dinucleotide repeat (Fig. 1A). Despite the fact that CD154 mRNA stability is comparable to that of IL-2 (44), it lacks the multiple AURE seen in human TNF-α (9 AUUUA) and IL-2 (7 AUUUA) that occur within 500 nt of the translational stop codon. Rather, the conserved portion of the human CD154 3′UTR has a single distal AURE (UUAUUUAUU) at nt 951 to 959 in a context that has been shown capable of destabilizing some, but not all, mRNA (27, 60).
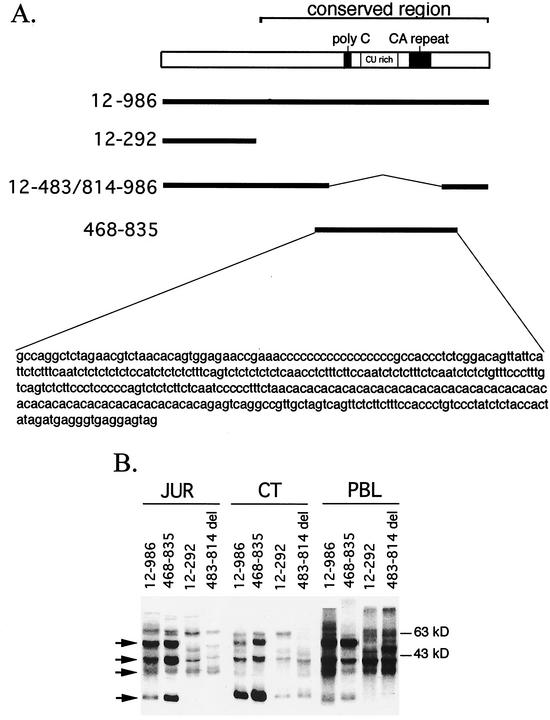
FIG. 1.
Analysis of CD154 3′UTR binding activity in CT nuclear extract, Jurkat T-cell line, and PBL. (A) Schematic diagram of human CD154 3′UTR showing poly(C), CU-rich, and CA dinucleotide repeat regions relative to radiolabeled [32P]UTP human CD154 3′UTR probes used in RNA binding assays. (B) Specified [32P]UTP-labeled CD154 3′UTR RNA probes were incubated with 50% ammonium sulfate fraction of a CT nuclear extract (5 μg) or cytoplasmic lysates (20 μg) from Jurkat (JUR) human T-cell line or PHA-activated (20 h) PBL, UV cross-linked, and RNase digested, and the digests resolved by SDS-PAGE and visualized by autoradiography. Arrows denote p50, p40, p36, and p25 binding activities.
Previous UV cross-linking studies identified RNA binding proteins with Mr of 50,000, 40,000, 36,000, and 25,000 in human PBL cytosols that directly contacted uridines and/or cytidines in the conserved region (nt 293 to 986) of the human CD154 3′UTR (44). A similar pattern of CD154 3′UTR binding was seen in ammonium sulfate (50%) precipitates of CT nuclear extracts relative to cytosols from activated human PBL and the Jurkat human T lymphocyte line (Fig. 1B). Each extract contained, to slightly varying degrees, the four major proteins previously shown to directly contact radiolabeled [32P]UTP full-length CD154 3′UTR. The p50, p40, and p25 binding activity mapped to the polypyrimidine-rich region (nt 468 to 835) defined by the BstNI-HphI restriction enzyme sites in the human CD154 3′UTR cDNA. Deletion of nt 483 to 814 resulted in loss of p50, p40, and p25 binding in Jurkat cytosol and CT extract. Activated human PBL cytosols also clearly demonstrate reduced p25 binding activity with this RNA transcript; however, the decrease in p50 and p40 are obscured by the binding of additional proteins with slightly different Mrs. These data demonstrate that CT nuclear extracts, Jurkat cells, and PBL exhibit identical patterns (p50, p40, and p25) of binding to the polypyrimidine-rich portion of the human CD154 3′UTR.
Previous work demonstrated that alterations in CD154 mRNA turnover correlated best with the binding of the p50 or p25 to its 3′UTR (44). For this reason, we utilized the 50% ammonium sulfate fraction of CT nuclear extract for purification, as it contained both p50 and p25 binding activity (Fig. 2A). Column chromatography was used to monitor binding to radiolabeled nt 468 to 835 in the CD154 3′UTR. No significant binding to DEAE was noted; the flowthrough was applied to a CMC column. The p50 and p25 binding activity eluted from the CMC column at 0.3 to 0.5 M NaCl, while the p40 binding activity was predominantly noted in the 0.1 M salt elution. The 0.3 M NaCl elution was subjected to polyuridine column chromatography, where the p25 eluted at a slightly lower salt (0.5 M) concentration relative to the p50 (1 M NaCl). The 0.5 and 1 M NaCl elutions were resolved by SDS-PAGE, with the p25 and p50 bands being excised after being visualized by Coomassie blue staining. The p50 binding protein was identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry as PTB, also known as hnRNP I (18-20, 37). In contrast, the p25 could not be identified using this method or by N-terminal sequencing, the latter suggesting blocking of its amino terminus. Internal sequencing was able to identify a nonapeptide (Asp-Tyr-Gly-Asn-Ser-Pro-Leu-His-Arg) with 100% identity to amino acids 432 to 440 present in the third RNA recognition motif (RRM)-type RNA binding domain in human PTB (18, 37).
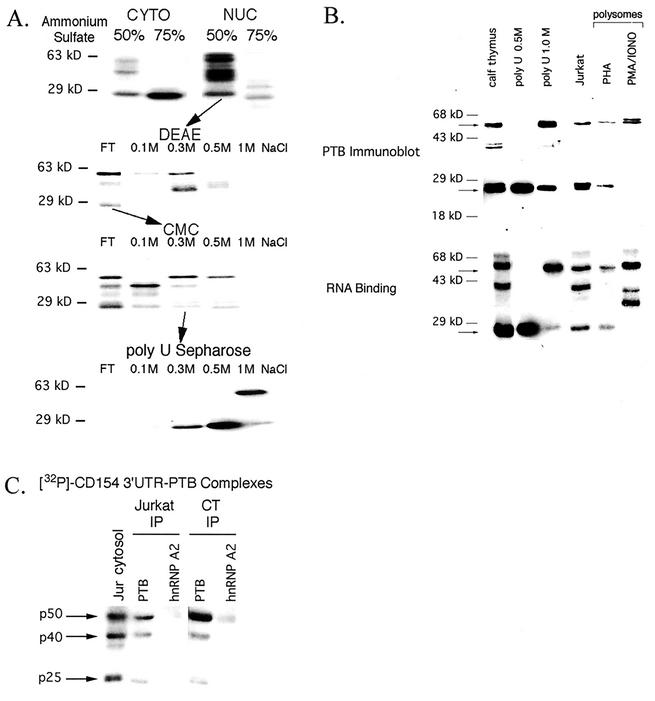
FIG. 2.
Purification and identification of p50 and p25 as PTB or PTB-related proteins. (A) Chromatographic purification of p50 and p25 through binding to [32P]UTP-labeled CD154 3′UTR 468-835 RNA probe by UV cross-linking. NUC, nuclear extract. (B) Crude CT (50% ammonium sulfate) nuclear extract, 0.5 and 1 M NaCl elutions from poly(U) Sepharose were analyzed in parallel by immunoblotting for PTB and by UV cross-linking to [32P]UTP-labeled CD154 3′UTR 468 to 835. Similar analysis was performed on Jurkat cytosols (50 μg) and polysomal fractions (0.5 A260 unit) from human PBL activated (6 h) with either PHA or PMA (20 ng/ml) and IONO (1 μM). Arrows denote p50 and p25. (C) Immunoprecipitation (IP) of PTB-CD154 3′UTR complexes from Jurkat cytosol and 50% ammonium sulfate fraction of CT nuclear extract following RNA binding. Jurkat cytosol and CT extract were incubated with [32P]UTP-labeled CD154 3′UTR 468 to 835, UV cross-linked, RNase digested, and immunoprecipitated with either BB7 (anti-PTB) or EF67 (anti-hnRNP A2) monoclonal antibodies as described in Materials and Methods. Radiolabeled proteins were detected by autoradiography. Jur cytosol, RNA binding activity from a Jurkat cytosol (50 μg).
Coordinate RNA binding assay and immunoblotting with the anti-PTB monoclonal antibody BB7 (10) demonstrated reactivity of the p50 and p25 binding proteins in the crude CT nuclear extract as well as in the purified fractions (Fig. 2B). A minor 40-kDa doublet was also detected by immunoblotting in CT, one isoform of which copurified with p50. A similar correlation of PTB-reactive proteins and CD154 3′UTR binding activity was seen with Jurkat cytosols as well as polyribosome-enriched (polysome) fractions purified by sucrose density gradients from PBL activated (6 h) with either PHA or PMA-IONO. As previously noted, PMA-IONO activation of PBL (which induces CD154 mRNA stabilization) is associated with a loss of p25 binding activity (relative to PHA activation) from the polysomes, while the p50 binding activity is increased and broadened (44). Immunoblotting demonstrated that the changes in binding activity are associated with loss of the immunoreactive p25 PTB isoform and increased levels of PTB, as well as emergence of a second, slightly larger isoform. Polysomes from PHA-activated PBL (in which CD154 mRNA is unstable) exhibit both p50 and p25 binding activity and PTB immunoreactivity.
Following incubation of Jurkat cytosol or CT extract with radiolabeled CD154 3′UTR RNA and UV-cross-linking, immunoprecipitation for PTB and an irrelevant RNA binding protein (hnRNP A2) was performed (Fig. 2C). The binding activity from a Jurkat cytosol is shown in the far left lane. Anti-PTB antibody immunoprecipitated radiolabeled RNA-protein complexes with an Mr of 50,000, 40,000, and 25,000 from both Jurkat cytosol and CT nuclear extract, indicating that each of these proteins directly contacted the RNA and was related to PTB. Anti-hnRNP A2 immunoprecipitation brought down little (CT) or no (Jurkat cytosol) binding activity, demonstrating the specificity of the observed data. We conclude that the p40 and p25 CD154 3′UTR binding proteins are related to PTB. By the same criteria, the p36 binding activity, which exhibited different binding specificity, is unrelated to PTB.
The p25 is encoded by a novel splice isoform of PTB.
The identification of the p50 and p25 as PTB or PTB-related proteins is consistent with the description of multiple PTB splice isoforms (19, 20) and related, but distinct gene products (2, 30, 42). PTB was originally defined by its regulation of alternative splicing (18, 37). Full-length PTB consists of four RRM-type RNA binding domains (Fig. 3A) and homodimerizes due to a region spanning the second RRM (39). Polypyrimidine tract binding activity is conferred by RRMs 3 and 4 (amino acids 324 to 531), particularly RRM 3 (11, 39). Based on peptide sequence, binding activity, immunoreactivity, and the inability to obtain N-terminal sequencing, we hypothesized that the p25 represented an alternatively spliced isoform of PTB. Reverse transcription with oligo(dT) or PTB 3′UTR-specific primers was followed by PCR with primers specific for the 5′- and 3′UTRs, yielding an ∼700-bp product from both human PBL or Jurkat mRNA (Fig. 3B). The predicted (1,700-bp) band corresponding to PTB was not well visualized under these conditions, suggesting preferential amplification of this smaller PCR product. This band was excised, cloned, and sequenced, which confirmed it as a novel splice variant of PTB mRNA, with exons 3 though 9 deleted (19, 46). This result has been confirmed by RT-PCR, cloning, and sequencing of three separate RNA samples—two from activated PBL from different donors and one from the Jurkat T-cell line. This splicing event results in deletion of 360 amino acids, over 50% of wild type PTB, thus corresponding in size to the p25 (Fig. 3A). In vitro translation yielded a protein of comparable size to the observed binding activity (Fig. 3C, left panel). Furthermore, immunoblotting with antisera specific for the N-terminal 13 amino acids of PTB demonstrated equivalent reactivity of the p25 to that seen with the BB7 antibody in PBL cytosol (Fig. 3C, right panel). These data establish that the p25 is an alternatively spliced isoform of PTB, clearly different from the previously described p25 form of PTB that was found to be due to proteolysis (6). We conclude that the p25 binding activity that correlated with mRNA turnover is encoded by a novel splice variant of PTB which retains most of RRM 3 and all of RRM 4, the domains that confer PTB activity (11, 39). We refer to this PTB isoform as PTB-T lymphocyte or, simply, PTB-T (Fig. 3A and 4), referring to the cell type in which it was first identified (30, 42). Preliminary studies do not indicate that expression of PTB-T mRNA or protein is restricted to lymphoid cells (data not shown).
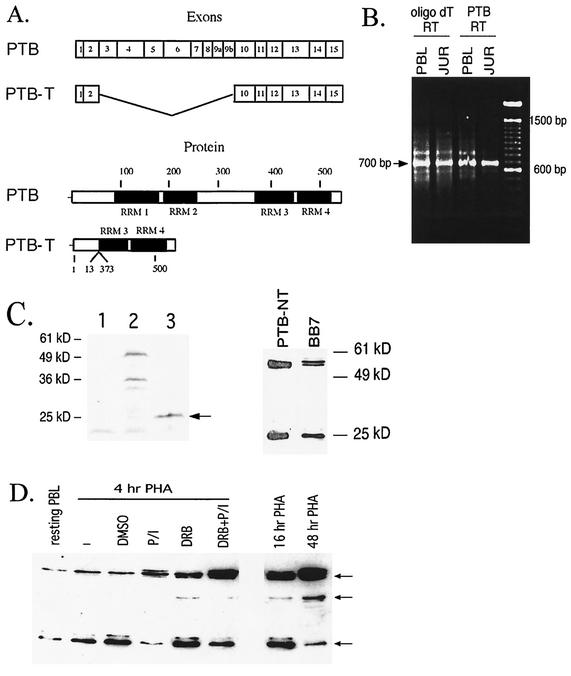
FIG. 3.
Identification of p25 as an alternately spliced isoform of PTB, PTB-T. (A) Schematic diagram of genomic (upper panel) and protein domain (lower panel) organization of PTB and PTB-T (19, 46). (B) RT-PCR of PTB was performed using RNA (2 μg) from PHA-activated (16 h) PBL (lanes 1 and 3) or Jurkat cells (lanes 2 and 4). The RT step was performed with oligo(dT) or PTB 3′UTR-specific primers and then PCR amplified with PTB primers specific for the 5′- and 3′UTRs. PCRs were analyzed by agarose gel electrophoresis and ethidium bromide staining. The ∼700-bp band was excised, cloned, and sequenced to yield PTB-T. Products corresponding to PTB (1,700 bp) were barely detectable in these experiments, probably reflecting preferential amplification of PTB-T under these conditions. (C) The left-hand panel shows in vitro translation of PTB-T. pcDNA3.1 PTB-T was transcribed and translated in vitro in the presence of [35S]methionine (lane 3) using Proteinscript II (Ambion) and visualized by autoradiography. Lane 1, no input cDNA; lane 2, input cDNA pTRI-Xef1α positive control, yielding predicted 50-kDa protein. The right-hand panel shows an immunoblot of activated (PHA-4H) PBL cytosol with affinity-purified antisera specific for the N-terminal 13 amino acids of PTB (PTB-NT) and BB7 monoclonal antibody. (D) Modulation of cytoplasmic PTB splice isoform expression in human PBL. PBL were rested overnight and PHA-activated for the specified times. After 4 h, PBL were treated for 1 h with PMA (final concentration, 10 ng/ml)-IONO (final concentration, 1 μM) (P/I) or a solvent control (dimethyl sulfoxide) in the absence or presence of 5,6-dichlorobenzimidazole 1β-d-ribofuranoside (100 μM) and cytosols analyzed by BB7 immunoblotting. Similar results were seen in three other experiments. Arrows denote p50 (PTB), p40, and p25 (PTB-T).
FIG. 4.
Primary nucleotide and amino acid sequence of PTB/hnRNP I and PTB-T. The PTB-T amino acid sequence is shown in boldface type. Underlined amino acid sequence is the nonapeptide identified by internal sequencing of the p25 binding protein. The 3′UTR sequence used for PTB-specific reverse transcription is shown in boldface type. Sequences in the 5′UTR and 3′UTR used for PCR and cloning are italicized and in boldface type.
Addition of PMA-IONO to PHA-activated human peripheral blood T cells acutely increases CD154 mRNA stability, even in the context of RNA polymerase II inhibition. This effect was accompanied by decreased p25 binding and increased p50 binding in the cytosol (44). With the identification of PTB and PTB-T as alternately spliced isoforms that bind the CD154 3′UTR, we addressed this observation by immunoblotting (Fig. 3D). Cytosolic levels of PTB and PTB-T are not significantly affected by short-term (4-h) activation by a concentration of PHA that induced maximal proliferation and IL-2 production. Addition of PMA-IONO rapidly increased PTB levels in concert with a decline in PTB-T. RNA polymerase II inhibition by 5,6-dichlorobenzimidazole 1β-d-ribofuranoside alone increased cytoplasmic PTB. This effect was further enhanced by PMA-IONO treatment, indicating that the effect of PMA-IONO was independent of de novo gene transcription. Thus, cytosolic levels of PTB-T correlate with an unstable CD154 mRNA (44). A corollary of these data are that while PHA activation induces proliferation and lymphokine production by human PBL, it does not confer stabilization of CD154 mRNA. These data account for the inability to detect CD154 expression in the absence of PMA-IONO treatment (28, 35, 44, 48). This inability of PHA activation to alter the relative cytoplasmic levels of PTB-T/PTB was maintained for at least the first 16 h. With prolonged PHA activation (48 h), a decline in cytosolic PTB-T was observed, consistent with reports that CD154 mRNA stability increases with prolonged T-cell activation (15).
Identification of a novel cis-acting element regulating mRNA turnover in the CD154 3′UTR.
Chimeric reporter gene constructs were used to analyze the role of CD154 3′UTR cis-acting elements in posttranscriptional gene regulation in vivo to avoid nonspecific toxicity and effects of transcriptional inhibitors on mRNA stability (8, 38, 41, 43, 53). Firefly luciferase reporter constructs pcDNA3.1/LUC and pcDNA3.1/LUC/CD154 104-986 were generated, in which the cytomegalovirus immediate early promoter drives transcription of a luciferase mRNA lacking or containing the conserved portion of the human CD154 3′UTR (Fig. 5A). In the Jurkat human T-cell line, the presence of the CD154 3′UTR reduced luciferase expression to 34% of that seen with cells transfected with identical reporter gene plasmids lacking this sequence (Fig. 5B). A comparable decrease in luciferase activity was conferred by the CD154 3′UTR in transient transfection of HeLa cells as well (Fig. 5B). In each cell type, the level of inhibition of luciferase activity conferred by the CD154 3′UTR was statistically significant (P < 0.001). The magnitude of this effect was equivalent to that seen with luciferase reporter constructs containing six reiterated AUUUA pentamers in their 3′UTR (data not shown). Thus, the CD154 3′UTR contains sequences that modulate luciferase reporter gene expression in a promoter-independent manner to the same extent as an AURE.
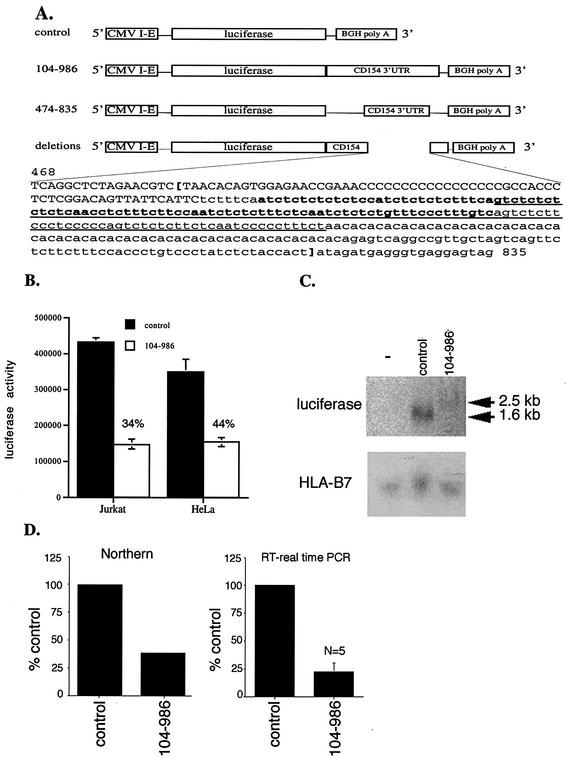
FIG. 5.
The CD154 3′UTR regulates luciferase reporter expression and mRNA accumulation. (A) Schematic diagram of pcDNA 3.1 luciferase reporter constructs lacking or containing portions of the human CD154 3′UTR with the bovine growth hormone (BGH) poly(A) signal sequence is shown. Within the nucleotide sequence (nt 468 to 835), the locations of specific deletions in the context of CD154 3′UTR 104 to 986 are shown. Capitalized nucleotides demonstrate the deletion of nt 468 to 549 (468-549 del), boldface type indicates deletion of nt 557 to 647 (557-647 del), and the underlined region indicates the deletion of nt 585 to 690 (585-690 del). Boldface brackets indicate the location of the deletion of nt 485 to 814 (485-814 del). (B) Jurkat and HeLa cells were transiently transfected with pcDNA3.1/LUC (control) or pcDNA3.1/LUC/CD154 104-986 cDNA in triplicate. Luciferase activity was measured after 24 h and is shown as the mean ± standard error of the mean (error bars). Jurkat cells received 0.1 μg of cDNA of either vector, while 0.04 μg was used for HeLa cells. Statistically significant inhibition (P < 0.0005) was seen in Jurkat and HeLa cells (n = 3). (C) Northern blot of RNA (50 μg) extracted from Jurkat cells 24 h after transfection with either the pcDNA3.1/LUC (control) or pcDNA3.1/LUC/CD154 104-986 vectors. Blots were hybridized with 32P-labeled specific cDNA probes for luciferase and HLA-B7 as a control. Arrows denote the predicted size of RNA derived from pcDNA3.1/LUC (control) and pcDNA3.1/LUC/CD154 104-986 expression plasmids of 1.6 and 2.5 kb, respectively. (D) Quantitation of inhibition of luciferase mRNA accumulation by the presence of the CD154 3′UTR 104-986 measured by phosphorimaging of the Northern blot (Northern) or real-time RT-PCR, where data are expressed as a percentage of that obtained with the pcDNA3.1/LUC (control) vector, with the mean and standard error of the mean (error bar) shown (n = 5).
The mechanism by which the CD154 3′UTR regulated luciferase reporter gene expression in transiently transfected Jurkat T lymphocytes was examined. Following transfection with the pcDNA3.1/LUC and pcDNA3.1/LUC/CD154 104-986 expression vectors, total cellular RNA was extracted and analyzed by Northern blotting (Fig. 5C). The presence of the CD154 3′UTR reduced accumulation of luciferase mRNA by 60% as measured by phosphorimaging (Fig. 5D). This effect was comparable to the magnitude (50%) of the CD154 3′UTR-dependent reduction in luciferase activity measured at the same time as the RNA extraction. These data demonstrate a direct relationship between mRNA accumulation and luciferase expression. In five separate experiments, the effect of the CD154 3′UTR on luciferase mRNA accumulation was measured by real-time quantitative RT-PCR (Fig. 5D). The level of CD154-3′UTR-dependent reduction in luciferase mRNA accumulation was comparable to that seen by Northern blotting. Thus, equivalent patterns of CD154 3′UTR-dependent changes in luciferase mRNA accumulation were shown using distinct techniques. The parallel between the magnitude of the effect of the CD154 3′UTR on luciferase mRNA and luciferase activity suggests altered mRNA stability, since the rate of transcription from each promoter is presumably equivalent. This conclusion is consistent with reports demonstrating the instability of CD154 mRNA (15, 31, 44, 56) and the recognized role of the 3′UTR in regulating mRNA turnover (47).
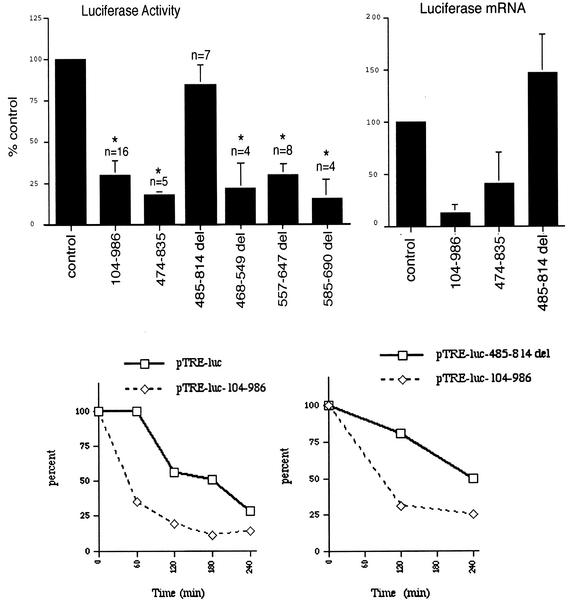
Deletion analysis was used to map the cis-acting element in the CD154 3′UTR. Removal of the entire polypyrimidine-rich region in the 485-814 del construct resulted in a loss of inhibition of luciferase activity (Fig. 6, upper-left panel). Constructs with smaller deletions of the polypyrimidine-rich region all demonstrated inhibitory activity. Deletion of the polycytidine sequence, of the CU dinucleotide repeat-rich region (nt 560 to 600), or even of >75% of the CU-rich region (nt 540 to 690) with the 468-549 del, 557-647 del, or 585-690 del reporter constructs still resulted in reduced luciferase expression in a 3′UTR-dependent manner. Finally, the polypyrimidine-rich region alone (nt 474 to 835) of human CD154 3′UTR reduced luciferase activity to a comparable degree to that seen with nt 104 to 986 in a statistically significant manner (P < 0.0005).
FIG. 6.
The polypyrimidine-rich region of CD154 3′UTR is necessary and sufficient to inhibit luciferase activity, mRNA accumulation, and mRNA stability. For the upper-left panel, Jurkat cells were transiently transfected in triplicate as described with the pcDNA3.1/LUC (control), pcDNA3.1/LUC/CD154 104-986, or pcDNA3.1/LUC/CD154 474-835, or pcDNA3.1/LUC/CD154 deletion (del) plasmids (485-514, 468-549, 557-647, 585-690). Mean luciferase activity is calculated and expressed as a function of that obtained with the pcDNA3.1/LUC (control) vector. The mean and standard error of the mean (error bar) in each experiment was then calculated, with statistically significant differences (P < 0.0005) relative to control indicated by asterisks. No inhibition of luciferase activity was seen with pcDNA3.1/LUC/CD154 485-814 deletion. For the upper-right panel, total cellular luciferase mRNA levels were measured by RT-PCR following transfection of specified plasmids into Jurkat cells. Data shown are expressed as a percentage of that obtained with the pcDNA3.1/LUC (control) vector, with the mean and standard error of the mean (error bar) from three experiments. For the lower panels, HeLa (Tet Off) cells were transiently transfected with pTRE-luc vectors that either lacked or contained various portions (nt 104 to 986 with and without deletion of nt 485 to 814) of the human CD154 3′UTR. Following transcriptional inhibition with doxycycline (1 μg/ml), luciferase mRNA levels were analyzed by RT-PCR. Data are shown as a percentage of luciferase mRNA from a specific transfection at time zero. The CD154 3′UTR increases luciferase mRNA turnover relative to the control vector pTRE-luc (left panel) or the 485-814 del (right panel). Similar results were seen in two other experiments.
When analyzed by transient transfection of Jurkat cells, both the CD154 3′UTR and the polypyrimidine-rich region alone (nt 474 to 835) were capable of reducing mRNA accumulation; deletion of this region resulted in loss of inhibition (Fig. 6, upper-right panel). Thus, the polypyrimidine-rich region of human CD154 3′UTR is both necessary and sufficient to reduce both luciferase activity and steady-state luciferase mRNA accumulation in a 3′UTR-dependent manner. This region (nt 474 to 835) lacks an AURE (Fig. 1A), indicating that this effect on reporter gene mRNA levels are mediated by a previously undefined cis-acting element.
In order to directly examine changes in mRNA stability without confounding by RNA polymerase II inhibitors, we generated a tetracycline-responsive luciferase (pTRE-luc) vector that either lacked or contained the CD154 3′UTR. Transient transfection of HeLa cells (Tet-Off) that permitted transcriptional inhibition by doxycycline treatment was followed by RNA extraction and quantitative RT-PCR analysis. The presence of the CD154 3′UTR resulted in a greater-than-twofold increase in the rate of luciferase mRNA decay (Fig. 6, lower-left panel). This effect of the CD154 3′UTR required the presence of the polypyrimidine-rich region; deletion of nt 485 to 814 exhibited increased mRNA stability relative to the CD154 3′UTR vector (Fig. 6, lower-right panel). Interestingly, the 485-814 del construct demonstrated increased mRNA stability relative to the control vector alone in two experiments (data not shown). This finding is consistent with our observation of augmented mRNA accumulation relative to the luciferase control seen with transient transfection of Jurkat T cells (Fig. 6, upper-right panel). Thus, the retained AURE (UUAUUUAUU) in the 485-814 del construct had no inhibitory effect on mRNA accumulation, suggesting it lacks activity in this context.
PTB and PTB-T differentially regulate CD154 3′UTR-dependent gene expression.
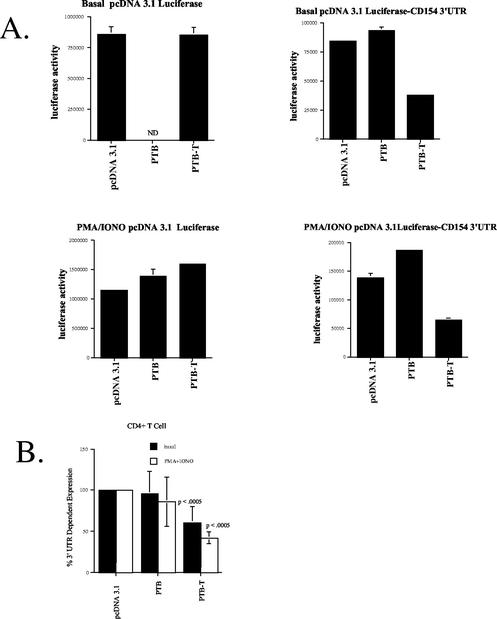
PTB and PTB-T were first identified as CD154 3′UTR RNA binding proteins whose presence in human PBL cytosols correlated with alterations in CD154 mRNA turnover (44). PTB/PTB-T were shown to bind to the CD154 3′UTR in the same polypyrimidine-rich region that contains a cis-acting instability element, suggesting their binding influenced its function. Overexpression of PTB or PTB-T in transient-transfection assay examined this possibility (Fig. 7). In the Jurkat human T-cell line, transfection of an expression vector encoding PTB increased CD154 3′UTR-dependent gene expression relative to that of empty vector controls (Fig. 7A, upper panel). In contrast, transfection of an expression vector encoding PTB-T markedly reduced luciferase activity in a CD154 3′UTR-dependent manner. These effects were statistically significant. A similar, statistically significant effect of PTB-T overexpression was seen in HeLa cells (Fig. 7A, lower panel). These data are consistent with the interpretation that levels of cytoplasmic PTB-T were limiting in both cell types. No effect of PTB overexpression was seen in HeLa cells, suggesting the possibility that PTB levels in these cells were not limiting. The presence of the polypyrimidine-rich region in the luciferase 3′UTR was both necessary and sufficient to confer the inhibitory effect of PTB-T transfection on luciferase expression (Fig. 7B).
FIG. 7.
PTB and PTB-T regulate CD154 3′UTR-dependent gene expression through the polypyrimidine-rich region. (A) Jurkat and HeLa cells were cotransfected as described with either pcDNA3.1/LUC or pcDNA3.1/LUC/CD154 104-986 expression vectors in the presence of 1 μg of pcDNA3.1 PTB-T, pcDNA3.1-PTB, or pcDNA3.1. Data are presented as a CD154 3′UTR-specific effect from each transfection as the ratio pcDNA3.1/LUC/CD154 104-986/pcDNA3.1/LUC (control). Statistically significant (P < 0.0005) differences are shown for Jurkat (n = 2) and HeLa (n = 4). (B) Jurkat cells were cotransfected as described with either pcDNA3.1/LUC, pcDNA3.1/LUC/CD154 474-835 or pcDNA3.1/LUC/CD154 485-814 del in the presence of pcDNA3.1 PTB-T or pcDNA3.1 (0.5 μg). Data are presented as a CD154 3′UTR-specific effect; PTB-T expression reduced luciferase activity by pcDNA3.1/LUC/CD154 474-835 (P < 0.001).
A similar pattern was seen in an experiment of transient transfection of purified human CD4+ T lymphocytes (Fig. 8A). Following transfection, luciferase activity in CD4+ T cells was measured after 6 h either without (basal) or with PMA-IONO stimulation. The presence of the CD154 3′UTR reduced luciferase activity, as seen in both HeLa and Jurkat cell lines under both basal and stimulated conditions. Moreover, in primary human CD4+ T cells, PTB and PTB-T overexpression differentially affected luciferase expression, in a CD154 3′UTR-dependent manner (Fig. 8). When expressed as a function of their effect on CD154 3′UTR-dependent gene expression from four experiments, the inhibitory effect of PTB-T was statistically significant (Fig. 8B). The selectivity of the effect of PTB-T transfection for the CD154 3′UTR was demonstrable through its lack of effect on reporter gene activity derived from the pcDNA3.1/LUC control. Furthermore, no effect on pRL-null-derived Renilla luciferase activity was seen, which was used to control for transfection efficiency in each experiment (data not shown). In separate experiments, we demonstrated that, as in Jurkat cells, the effect of PTB-T on luciferase expression in normal human CD4+ T lymphocytes was conferred by nt 468 to 835. In these studies, PTB-T reduced luciferase expression by pcDNA3.1/LUC/CD154 104-986 by 38% relative to controls. Transfection of PTB-T cDNA had a greater effect (58% inhibition) of luciferase expression by pcDNA3.1/LUC/CD154 468-835 (n = 4). Together, these data show that PTB-T and, in some instances, PTB can be demonstrated to regulate CD154 gene expression in both normal human T cells and cell lines solely through their interaction with the polypyrimidine-rich region found in its 3′UTR.
FIG. 8.
The effects of PTB and PTB-T on CD154 3′UTR-dependent gene expression are seen in normal human T lymphocytes. (A) Human CD4+ T lymphocytes were transfected with pcDNA3.1/LUC or pcDNA3.1/LUC/CD154 104-986 expression vectors in the presence of pcDNA3.1 PTB-T, pcDNA3.1-PTB or pcDNA3.1 control as described. Luciferase data (mean and standard error of the mean [error bar]) for pcDNA3.1/LUC (left panels) or pcDNA3.1/LUC/CD154 104-986 (right panels) are shown following transfection for 6 h under basal or stimulated (PMA/IONO) conditions. ND, not done. (B) Calculation of CD154 3′UTR-dependent effects of PTB and PTB-T (n = 4). Statistically significant effects of PTB-T expression are seen in the absence or presence of PMA-IONO treatment (P < 0.0005).
DISCUSSION
Expression of CD154 by activated T lymphocytes plays a central role in both cellular and humoral immunity (16, 22, 34). To date, the regulation of CD154 expression has been poorly understood, although several studies have demonstrated the importance of mRNA stability (15, 31, 44, 56). We describe the purification, identification, and characterization of PTB and a novel splice isoform, p25/PTB-T, as two major cytosolic proteins capable of binding to the polypyrimidine-rich region of the human CD154 3′UTR. As part of these studies, we demonstrate that the CD154 3′UTR contains a novel cis-acting element(s) which decreases reporter gene expression at the level of mRNA accumulation by increasing the rate of mRNA turnover in vivo. The activity of the CD154 3′UTR has been demonstrated in both lymphoid and nonlymphoid cell lines, but also in normal human CD4+ T cells, demonstrating its physiologic significance. The polypyrimidine-rich region (nt 474 to 835) of CD154 3′UTR was shown to be both necessary and sufficient to reduce reporter gene expression and mRNA accumulation, indicating the cis-acting element can be localized within this region. Subsequently we demonstrated that PTB and PTB-T expression differentially modulates the function of this cis-acting element. These data suggest the interpretation that CD154 expression is regulated at the level of mRNA stability in vivo through their binding to the polypyrimidine-rich portion of the human CD154 3′UTR.
This series of experiments derived from our observations that CD154 mRNA turnover rates differentially correlated with levels of specific proteins that bound to the polypyrimidine-rich region of the 3′UTR (44). These studies found that the presence of a cytoplasmic 25-kDa protein (p25) capable of binding the CD154 3′UTR correlated with CD154 mRNA instability (44). Signals (PMA-IONO) which stabilized CD154 mRNA were associated with the loss of p25 binding and increased levels of p50 binding activity (44). With identification of the p25 and p50 as PTB-T and PTB, respectively, we now demonstrate that the changes in RNA binding activity were correlated with changes in their relative protein levels in the cytoplasm as well as polysomes (Fig. 2D and 3D). These data suggested that modulation of CD154 mRNA stability is transduced by the relative cytoplasmic levels of these related proteins and their binding to the 3′UTR polypyrimidine-rich region. Using luciferase reporter constructs, we show that the 3′UTR polypyrimidine-rich region is both necessary and sufficient to reduce total cellular luciferase mRNA levels in transient transfection of Jurkat cells. Since these reporter constructs that contain identical promoters lack introns and differ only in their 3′UTR, these data are consistent with increased mRNA degradation. We have directly confirmed this interpretation using a tetracycline-repressible transcription approach in HeLa cells. The comparable effects of the CD154 3′UTR polypyrimidine-rich region on luciferase activity across cell types, including normal human CD4+ T cells, strongly supports the importance of this region and pathway in normal immune responses. Moreover, the effects of PTB-T and PTB on CD154 3′UTR-dependent luciferase expression are completely consistent with the initial observations of their differential correlation with changes in CD154 mRNA turnover.
Following the original functional characterization of the AURE type cis-acting element in mRNA stability (51), over a decade elapsed before it was shown that the fate of an mRNA can be determined by different proteins that compete for binding to this specific cis-acting element. Tristetraprolin, TIA-1, and HuR each bind the TNF-α 3′UTR AURE to mediate changes in either mRNA stability or translation (7, 14, 40). Our findings suggest a variant of this model, in which two proteins derived from the same gene compete for binding to the same cis-acting element with different functional consequences (Fig. 9). We have not yet formally tested if PTB and PTB-T compete for binding to the same sequences within the CD154 3′UTR cis-acting element; however, this interpretation seems reasonable given that PTB and PTB-T share the domains (RRMs 3 and 4) that confer polypyrimidine tract binding activity (11, 39). Moreover, we have mapped the binding of both PTB and PTB-T to the same polypyrimidine-rich region (nt 468 to 835) of the CD154 3′UTR, which contains multiple repeats [UCUUC or UUC(U/C)] of consensus binding sites for PTB (1, 55).
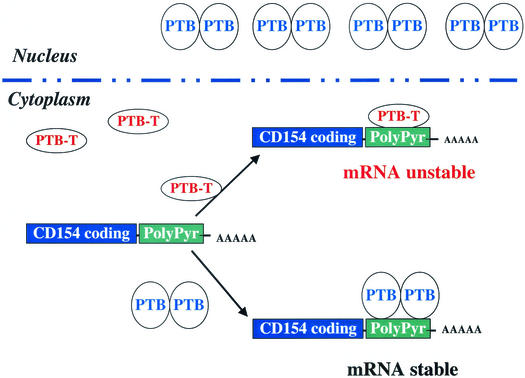
FIG. 9.
Model denoting regulation of CD154 mRNA stability by PTB and PTB-T binding to the polypyrimidine-rich region in the 3′UTR. PTB-T and PTB compete for binding to CD154 3′UTR and determine changes in mRNA stability. PTB is shown as a dimeric molecule that is primarily nuclear. PTB-T, which lacks both the nuclear localization sequence and dimerization domain of PTB, is shown to be predominantly cytoplasmic and monomeric.
The different structures of PTB and PTB-T would predict distinct consequences as a result of their binding to the CD154 3′UTR. In contrast to PTB, PTB-T lacks the homodimerization domain that encompasses RRM 2 (39). Thus, dimeric PTB would be expected to interact with the polypyrimidine-rich region of the CD154 3′UTR at multiple sites and have a distinct effect on RNA structure relative to PTB-T, potentially favoring CD154 mRNA stability. This model of multisite PTB binding to the CD154 3′UTR for mRNA stability is similar to that proposed for PTB-dependent regulation of exon silencing (10, 58).
Such a model has implications for our ability to map this cis-acting element. Deletion mutants would result in concurrent loss of both PTB-T and PTB binding sites, each having differing consequences for mRNA accumulation. This model would be consistent with our initial deletion mutant analysis, in which the inhibitory effect of this region was not attenuated by removal of the polycytidine sequence, CU dinucleotide-repeat region, or even >75% of the CU-rich region. Although it is possible we missed the site of the cis-acting element, if PTB requires multisite binding in the 3′UTR to stabilize CD154 mRNA, deletion analysis would preferentially alter the effect of PTB by reducing these interactions. In contrast, PTB-T would be predicted to mediate its effects through binding in a monomeric fashion. Only deletion of all of the PTB-T binding sites would be predicted to increase mRNA accumulation and luciferase expression, which is what was observed.
Alternatively, PTB might be able to stabilize mRNA through monomeric interactions by recruiting another protein that leads to CD154 mRNA stability. In this regard, the RRM 2 of PTB has been shown to promote interaction with hnRNP L (23, 36). The binding of hnRNP L to the vascular endothelial growth factor 3′UTR has been implicated in regulating mRNA stability (52). In each of these models, PTB, but not PTB-T, would be predicted capable of oligomerizing to favor mRNA stability. Discrimination between these two models will require identification of the minimal cis-acting element that permitted both PTB and PTB-T to modulate reporter gene expression in a 3′UTR-dependent manner.
The alternative splicing that generates PTB-T (amino acids 1 to 13 and amino acids 373 to 557) also potentially accounts for its observed cytoplasmic localization relative to PTB. This splicing event removes the nuclear localization sequence determinants in the N-terminal 55 amino acids of PTB (11, 39, 45). The relative levels of PTB and PTB-T in the cytoplasm of normal human T lymphocytes appear to be actively regulated. Treatment of PBL with PMA-IONO, which rapidly stabilizes CD154 mRNA (44), increased cytoplasmic levels of PTB in PBL as well as the appearance of a more slowly migrating isoform (Fig. 2D and 3D). In addition, RNA polymerase II inhibition alone increases cytoplasmic PTB levels. Since these effects can occur rapidly and even in the context of RNA polymerase II inhibition, it suggests the possibility that these effects are mediated by alterations in nucleocytoplasmic shuttling of PTB/PTB-T. These data are consistent with the hypothesis that signaling pathways regulate CD154 mRNA stability by increasing cytoplasmic levels of PTB. In this model, as cytoplasmic levels of PTB increase, PTB-T is displaced due to the higher avidity of dimeric PTB for binding to the CD154 3′UTR (Fig. 9).
PTB proteins have been reported to regulate internal ribosomal entry site (IRES) function (21, 59). There is no evidence to indicate that PTB is playing a role in the translational regulation of CD154 expression, as the magnitude of the CD154 3′UTR-dependent reduction in luciferase activity and total cellular mRNA accumulation are closely correlated. This finding is consistent with previous studies, which indicate that CD154 mRNA is regulated at the level of mRNA stability and not translation (15, 31, 44, 56). However, these data do not exclude the possibility that cytoplasmic levels of PTB-T and PTB may differentially affect IRES function in other mRNA.
In addition to IRES function, the PTB gene family has been shown to play a role in tissue-specific splice site selection (57, 58). It has been proposed that tissue-specific regulation may be dependent on expression of alternately spliced isoforms of PTB that include either all (PTB4) or part (PTB2) of exon 9 (19, 20). These splice isoforms differ from wild-type PTB by insertions of 19 (PTB-2) and 26 (PTB-4) amino acids and differentially regulate α-tropomyosin splice site selection and IRES function (59). Our data suggest that the presence of a more radically distinct splice isoform of PTB, PTB-T, might serve to compete with PTB and influence its function. It will be of interest to study the relative expression of PTB-T in tissues such as brain and muscle, where PTB function in alternative splicing appears to be selectively modulated (20, 37, 10).
Finally, our studies indicate the presence of another PTB-related protein, the p40, which binds to the CD154 3′UTR and may regulate turnover. We do not believe that the p40 represents a PTB splice isoform despite its reactivity with PTB-specific antisera, as it exhibited weak binding to CMC, in contrast to PTB and PTB-T. Moreover, a variety of PTB-specific primers have been unsuccessful in identifying a splice isoform corresponding to p40 by RT-PCR of T lymphocyte mRNA. We hope to resolve these issues by identifying and cloning, if necessary, p40. Thus, these studies not only prompt consideration of the roles of PTB proteins in regulating mRNA stability but also suggest a greater level of complexity of PTB gene products.
In conclusion, we have defined a novel posttranscriptional regulatory pathway that utilizes previously undescribed cis-acting elements and trans-acting factors in the regulation of expression of a critical immune response protein, CD154. As part of these studies, we have identified PTB-T, an alternatively spliced isoform of PTB. In addition to a greater consideration of the cytoplasmic roles of PTB proteins, the identification of PTB-T provides further insight into the complexity of the biologic function of PTB gene products. With regard to the immune system, this discrete regulatory pathway is consistent with previous studies demonstrating the differential regulation of cytokine and CD154 expression (28, 35, 48, 49). In activated T lymphocytes, cytokine and CD154 mRNA turnover can be independently and selectively modulated (15, 31). The demonstration that CD154 and cytokine mRNA turnover are regulated by different (polypyrimidine-rich versus AURE) cis-acting elements and trans-acting factors accounts for these findings. Moreover, these data suggest that understanding the signals that regulate cytoplasmic levels or binding of PTB/PTB-T might lead to the development of approaches that are specific in limiting CD154 expression. Given the efficacy of CD154 blockade in models of allograft rejection and autoimmune disease (16, 22), selective modulation of CD154 expression would be predicted to have significant clinical potential.
Acknowledgments
We thank Douglas Black and Stanley Lemon for the generous provision of valuable reagents used in these studies. We also express our appreciation of the thoughtful review of the manuscript by James Patton and Paul Anderson as well as the contributions and support of, in particular, Xiao-Wei Wang, Sam Rigby, and Mary Waugh.
This work was supported by grants from the Arthritis Foundation, the NIH (AI34928), and the Grimshaw-Gudewicz Foundation.
REFERENCES
- 1.Anwar, A., N. Ali, R. Tanveer, and A. Siddiqui. 2000. Hepatitis C Virus IRES-mediated translation Initiation: demonstration of functional requirement of polypyrimidine tract-binding protein (PTB) by SELEX RNA. J. Biol. Chem. 275:34231-34235. [DOI] [PubMed] [Google Scholar]
- 2.Ashiya, M., and P. J. Grabowski. 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3:996-1015. [PMC free article] [PubMed] [Google Scholar]
- 3.Ashwell, J. D., F. W. Lu, and M. S. Vacchio. 1992. Glucocorticoids in T cell development and function. Annu. Rev. Immunol. 18:309-345. [DOI] [PubMed] [Google Scholar]
- 4.Belshamm, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutler, B., and V. Kruys. 1995. Lipopolysaccharide signal transduction, regulation of tumor necrosis factor biosynthesis, and signaling by tumor necrosis factor itself. J. Cardiovasc. Pharm. 25:S1-S8. [DOI] [PubMed] [Google Scholar]
- 6.Bothwell, A. L., D. W. Ballard, W. M. Philbrick, G. Lindwall, S. E. Maher, M. M. Bridgett, S. F. Jamison, and M. A. Garcia-Blanco. 1991. Murine polypyrimidine tract binding protein. Purification, cloning, and mapping of the RNA binding domain. J. Biol. Chem. 266:24657-24663. [PubMed] [Google Scholar]
- 7.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., N. Xu, and A. B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15:5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Chou, M. Y., J. G. Underwood, J. Nikolic, M. H. Luu, and D. L. Black. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5:949-957. [DOI] [PubMed] [Google Scholar]
- 11.Conte, M. R., T. Grune, J. Ghuman, G. Kelly, A. Ladas, S. Matthews, and S. Curry. 2000. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19:3132-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cron, R. Q., L. A. Schubert, D. B. Lewis, and C. C. Hughes. 1997. Consistent transient transfection of DNA into non-transformed human and murine T-lymphocytes. J. Immunol. Methods 205:145-150. [DOI] [PubMed] [Google Scholar]
- 13.Cron, R. Q., S. R. Bartz, A. Clausell, S. J. Bort, S. J. Klebanoff, and D. B. Lewis. 2000. NFAT1 enhances HIV-gene expression in primary human CD4 T cells. Clin. Immunol. 94:179-191. [DOI] [PubMed] [Google Scholar]
- 14.Dean, J. L., R. Wait, K. R. Mahtani, G. Sully, A. R. Clark, and J. Saklatvala. 2001. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 21:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford, G. S., B. Barnhart, S. Shone, and L. R. Covey. 1999. Regulation of CD154 (CD40 ligand) mRNA stability during T cell activation. J. Immunol. 162:4037-4044. [PubMed] [Google Scholar]
- 16.Foy, T. M., et. al. 1996. Immune regulation by CD40 and its ligand gp39. Annu. Rev. Immunol. 14:591-617. [DOI] [PubMed] [Google Scholar]
- 17.Fuleihan, R., N. Ramesh, A. Horner, D. Ahern, P. J. Belshaw, D. G. Alberg, I. Stamenkovic, W. Harmon, and R. S. Geha. 1994. Cyclosporin A inhibits CD40 ligand expression in human T lymphocytes. J. Clin. Investig. 93:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Blanco, M. A., S. F. Jamison, and P. A. Sharp. 1989. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 3:1874-1886. [DOI] [PubMed] [Google Scholar]
- 19.Ghetti, A., S. Pinol-Roma, W. M. Michael, C. Morandi, and G. Dreyfuss. 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20:3671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil, A., P. A. Sharp, S. F. Jamison, and M. A. Garcia-Blanco. 1991. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 5:1224-1236. [DOI] [PubMed] [Google Scholar]
- 21.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grewal, I. S., and R. A. Flavell 1998. CD40 and CD154 in cell-mediated immunity. Annu. Rev. Immunol. 16:111-135. [DOI] [PubMed] [Google Scholar]
- 23.Hahm, B., O. H. Cho, J. E. Kim, Y. K. Kim, J. H. Kim, Y. L. Oh, and S. K. Jang. 1998. Polypyrimidine tract-binding protein interacts with hnRNP L. FEBS Lett. 425:401-406. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton, B. J., E. Nagy, J. S. Malter, B. A. Arrick, and W. F. C. Rigby. 1993. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 268:8881-8887. [PubMed] [Google Scholar]
- 25.Hollenbaugh, D., H. D. Ochs, R. J. Noelle, J. A. Ledbetter, and A. Aruffo. 1994. The role of CD40 and its ligand in the regulation of the immune response. Immunol. Rev. 138:23-37. [DOI] [PubMed] [Google Scholar]
- 26.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398. [DOI] [PubMed] [Google Scholar]
- 27.Lagnado, C. A., C. Y. Brown, and G. J. Goodall. 1994. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A). Mol. Cell. Biol. 14:7984-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane, P., A. Traunecker, S. Hubele, S. Inui, A. Lanzavecchia, and D. Gray. 1992. Activated human T cells express a ligand for the human B cell-associated antigen CD40, which participates in T cell-dependent activation of B lymphocytes. Eur. J. Immunol. 22:2573-2578. [DOI] [PubMed] [Google Scholar]
- 29.Lindsten, T., C. H. June, J. A. Ledbetter, G. Stella, and C. B. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339-343. [DOI] [PubMed] [Google Scholar]
- 30.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami, K., W. Ma, R. Fuleihan, and J. S. Pober. 1999. Human endothelial cells augment early CD40 ligand expression in activated CD4+ T cells through LFA-3-mediated stabilization of mRNA. J. Immunol. 163:2667-2673. [PubMed] [Google Scholar]
- 32.Nagy, E., and W. F. C. Rigby. 1995. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD+-binding region (Rossmann fold). J. Biol. Chem. 270:2755-2763. [DOI] [PubMed] [Google Scholar]
- 33.Nichols, R. C., X. W. Wang, J. Tang, B. J. Hamilton, F. A. High, H. R. Herschman, and W. F. C. Rigby. 2000. The RGG domain in hnRNP A2 affects sub-celluar localization. Exp. Cell Res. 256:522-532. [DOI] [PubMed] [Google Scholar]
- 34.Noelle, R. J. 1996. CD40 and its ligand in host defense. Immunity 4:415-419. [DOI] [PubMed] [Google Scholar]
- 35.Nusslein, H. G., K. H. Frosch, W. Woith, P. Lane, J. R. Kalden, and B. Manger. 1996. Increase of intracellular calcium is the essential signal for the expression of CD40 ligand. Eur. J. Immunol. 26:846-850. [DOI] [PubMed] [Google Scholar]
- 36.Oh, Y. L., B. Hahm, Y. K. Kim, H. K. Lee, J. W. Lee, O. Song, K. Tsukiyama-Kohara, M. Kohara, A. Nomoto, and S. K. Jang. 1998. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patton, J. G., S. A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237-1251. [DOI] [PubMed] [Google Scholar]
- 38.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez, I., J. G. McAfee, and J. G. Patton. 1997. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 36:11881-11890. [DOI] [PubMed] [Google Scholar]
- 40.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 42.Polydorides, A. D., H. J. Okano, Y. Y. Yang, G. Stefani, and R. B. Darnell. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 97:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajagopalan, L. E., and J. S. Malter. 1996. Turnover and translation of in vitro synthesized messenger RNAs in transfected, normal cells. J. Biol. Chem. 271:19871-19876. [DOI] [PubMed] [Google Scholar]
- 44.Rigby, W. F. C., M. G. Waugh, and B. J. Hamilton. 1999. Characterization of RNA binding proteins associated with CD40 ligand (CD154) mRNA turnover in human T lymphocytes. J. Immunol. 163:4199-4206. [PubMed] [Google Scholar]
- 45.Romanelli, M. G., F. Weighardt, G. Biamonti, S. Riva, and C. Morandi. 1997. Sequence determinants for hnRNP I protein nuclear localization. Exp. Cell Res. 235:300-304. [DOI] [PubMed] [Google Scholar]
- 46.Romanelli, M. G., P. Lorenzi, and C. Morandi. 2000. Organization of the human gene encoding heterogeneous nuclear ribonucleoprotein type I (hnRNP I) and characterization of hnRNP I related pseudogene. Gene 255:267-272. [DOI] [PubMed] [Google Scholar]
- 47.Ross, J. 1988. Messenger RNA turnover in eukaryotic cells. Mol. Biol. Med. 5:1-14. [PubMed] [Google Scholar]
- 48.Roy, M., T. Waldschmidt, A. Aruffo, J. A. Ledbetter, and R. J. Noelle. 1993. The regulation of the expression of gp39 on normal and cloned CD4+ T cells. J. Immunol. 151:2497-2510. [PubMed] [Google Scholar]
- 49.Roy, M., A. Aruffo, J. A. Ledbetter, P. Linsley, M. Kehry, and R. Noelle. 1994. Studies on the independence of gp39 and B7 expression and function during antigen-specific immune responses. Eur. J. Immunol. 25:596-603. [DOI] [PubMed] [Google Scholar]
- 50.Schubert, L. A., G. King, R. Q. Cron, D. B. Lewis, A. Aruffo, and D. Hollenbaugh. 1995. The human gp39 promoter: two distinct nuclear factors of activated T cell protein-binding elements contribute independently to transcriptional activation. J. Biol. Chem. 15:29264-29627. [DOI] [PubMed] [Google Scholar]
- 51.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-669. [DOI] [PubMed] [Google Scholar]
- 52.Shih, S. C., and K. P. Claffey. 1999. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 274:1359-1365. [DOI] [PubMed] [Google Scholar]
- 53.Shyu, A.-B., M. E. Greenberg, and J. G. Belasco. 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3:60-72. [DOI] [PubMed] [Google Scholar]
- 54.Sigal, N. H., and F. J. Dumont. 1992. Cyclosporin A, FK-506, and rapamycin: pharmacologic probes of lymphocyte signal transduction. Annu. Rev. Immunol. 10:519-560. [DOI] [PubMed] [Google Scholar]
- 55.Singh, R., J. Valcarcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 56.Suarez, A., L. Mozo, A. Gayo, J. Zamorano, and C. Gutierrez. 1997. Requirement of a second signal via protein kinase C or protein kinase A for maximal expression of CD40 ligand. Involvement of transcriptional and posttranscriptional mechanisms. Eur. J. Immunol. 27:2822-2829. [DOI] [PubMed] [Google Scholar]
- 57.Valcarcel, J., and F. Gebauer. 1997. Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 7:R705-708. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wollerton, M. C., C. Gooding, F. Robinson, E. C. Brown, R. J. Jackson, and C. W. Smith. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zubiaga, A. M., J. G. Belasco, and M. E. Greenberg. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15:2219-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]