Abstract
The chimeric fusion gene EWS/FLI-1 is detected in most cases of Ewing's sarcoma (ES), the second most common malignant bone tumor of childhood. Although 80% of ES tumors develop in skeletal sites, the remainder can arise in almost any soft tissue location. The lineage of the cell developing the EWS/FLI-1 gene fusion has not been fully characterized but is generally considered to be of either mesenchymal or neural crest origin. To study this oncogene in a conceptually relevant target cell, EWS/FLI-1 was introduced into the murine cell line C2C12, a myoblast cell line capable of differentiation into muscle, bone, or fat. In this cellular context, EWS/FLI-1 profoundly inhibited the myogenic differentiation program. The block in C2C12 myogenic differentiation required the nuclear localization and DNA-binding functions of EWS/FLI-1 and was mediated by transcriptional and posttranscriptional suppression of the myogenic transcription factors MyoD and myogenin. Interestingly, C2C12-EWS/FLI-1 cells constitutively expressed alkaline phosphatase, a bone lineage marker, and were alkaline phosphatase positive by histochemistry but showed no other evidence of bone lineage commitment. Consistent with recent findings in human ES tumor cell lines, C2C12-EWS/FLI-1 cells constitutively expressed cyclin D1 and demonstrated decreased expression of the cell cycle regulator p21cip1, even under differentiation conditions and at confluent density. This C2C12-EWS/FLI-1 cell model may assist in the identification of novel differentially expressed genes relevant to ES and provide further insight into the cell(s) of origin developing ES-associated genetic fusions.
The chimeric gene EWS/FLI-1 is identified in most tumors of the Ewing's sarcoma primitive neuroectodermal tumor (ES/PNET) family. The EWS gene encodes a ubiquitously expressed protein that has been demonstrated to interact with the RNA transcriptional machinery (32) and, in the context of EWS/FLI-1, likely acts as a transcriptional activator (2, 31). Although there are several distinct types of EWS/FLI-1 fusions, all share fusion of the EWS transcriptional activation domain with carboxy-terminal sequences from FLI-1, a member of the Ets family of DNA-binding proteins. Because the Ets DNA-binding domain is retained in the fusions, investigators have hypothesized that EWS/FLI-1 acts as a mutant transcription factor, altering normal gene regulation. Therefore, it is likely that EWS/FLI-1 plays a central role in ES/PNET, but whether this fusion protein plays a necessary and sufficient role in ES/PNET causation remains to be established. Sophisticated analyses of 3T3 cells expressing EWS/FLI-1 have identified several differentially expressed genes; however, the precise gene expression programs induced by EWS/FLI-1 to bring about transformation remain to be defined (reviewed in reference 1). At present, developing a cohesive model of EWS/FLI-1 transformation has been somewhat difficult because of limited cell line and animal model systems to study the biologic consequences of EWS/FLI-1 expression.
Patients with tumors of the ES/PNET family generally present with a bony lesion involving an extremity, but involvement of the pelvis or axial skeleton, or even an extraosseus site, is also observed. The somewhat diverse sites of origin for ES/PNET, and the observation that some tumors exhibit neural markers (13), has led to the speculation that the target cell for these tumors may involve a cell with multilineage potential, such as might be expected from a cell of mesenchymal or neural crest origin. Characterization of the cell giving rise to ES remains incomplete, largely because ES tumors provide few clues to their cellular origins (12). This is in stark contrast to other pediatric small round blue tumors, such as osteosarcoma and rhabdomyosarcoma, which arise in similar anatomical sites as ES yet offer substantial histologic evidence of their cellular lineage. These findings suggest the possibility that EWS/FLI-1 might act to suppress cellular differentiation signals as one facet of its role as an oncogene.
The mouse cell line C2C12 is best known as a murine myoblast cell line that has been extensively used to study myogenic differentiation (54). However, studies have also shown that C2C12 cells are capable of differentiating into bone (23) and fat (47), characteristics of a mesenchymal cell line. In this report, we have used the C2C12 cell line to develop a novel cellular model system to study the biologic effects of EWS/FLI-1 expression. Characterization of C2C12-EWS/FLI-1 cells revealed that EWS/FLI-1 expression was associated with an altered cellular morphology, a profound resistance to customary myogenic differentiation signals, and dysregulation of cell cycle regulatory genes.
MATERIALS AND METHODS
Cells and cell culture.
C2C12 cells were grown at 5% CO2 in Dulbecco's minimal essential (DME) (high) medium supplemented with 10% fetal calf serum, 2 mM glutamine, nonessential amino acids, and penicillin-streptomycin. Human EWS/FLI-1 cell lines were acquired and cultured as previously described (42), except for SS, which is a human cell line recently established at the Simmons Cancer Center from a patient with ES. For myogenic differentiation, the parental or C2C12-EWS/FLI-1 cell line was cultured until reaching a density of approximately 80 to 90%, washed twice in phosphate-buffered saline, and then changed to DME medium containing 2% horse serum.
To assess bone mineralization potential, parental or C2C12-EWS/FLI-1 cells were placed in bone mineralization medium, which was comprised of minimum essential medium supplemented with 10% fetal bovine serum, biotin, vitamin B12, thiotic acid, nonessential amino acids, sodium pyruvate, l-glutamine, penicillin-streptomycin, and 50 μg of ascorbic acid/ml for 8 days (29). The medium was then changed to fresh medium containing 50 μg of ascorbic acid/ml and 3 mM sodium phosphate for an additional 4 days. Calcification was assessed on day 12 by Von Kossa staining. For alkaline phosphatase induction, cultured C2C12 cells were washed in phosphate-buffered saline, placed in DME medium containing 5% fetal calf serum and 300 ng of BMP-2/ml (22), and harvested for RNA isolation at the desired time points.
Plasmids and retroviral transduction.
Wild-type or mutant forms of EWS/FLI-1 were introduced into C2C12 cells by retroviral transduction by using the vectors MINV-EWS/FLI-1 (42) and MINV-EWS/FLI-1 R2L2 (2, 42), as previously described (42). Stable C2C12-EWS/FLI-1 or control C2C12-MINV/neo (18) cell lines were generated by selecting retrovirally transduced cells in 0.8 mg of (absolute) neomycin/ml. The plasmid MINV/FLI-1(C) was constructed by digesting deltaEB/FLI-1(C) (2) with XhoI, blunting it with T4 DNA polymerase, and then digesting it with BglII to liberate the insert. The resulting cDNA was then cloned into the vector MINV/Neo by using the available BglII and SnaBI restriction sites.
To overexpress MyoD and myogenin, C2C12-EWS/FLI-1 or control cells were transduced with a replication-defective retrovirus containing murine myoD or myogenin (45) or the control MSCV/puro vector alone. Stable cell populations were obtained by selection in 2 μg of puromycin/ml. Cells were then immediately placed in medium containing 2% horse serum to assess morphological differentiation. In the case of control C2C12/MyoD cells, puromycin selection was not performed because transduced cells spontaneously differentiated into multinucleated myotubes while still in the growth medium (GM).
Antibodies, Western immunoblotting, and immunofluorescence.
Radioimmunoprecipitation assay protein lysates were prepared as previously described (42), resolved on Laemmli polyacrylamide gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose. Membranes were probed with the indicated antibody and developed by enhanced chemiluminescence. Antibodies used in this study were purchased from Santa Cruz Biotechnology, except as specifically indicated, and included anti-MyoD, anti-myogenin, anti-FLI-1, anti-EWS, anti-p21, anti-extracellular signal-regulated kinase 1 and 2 (anti-ERK1/2; Cell Signaling, Inc.), anti-Akt, and anti-p38 mitogen-activated protein kinase (MAPK) antibodies. To assess activation of signal transduction pathways important in muscle differentiation, antibodies recognizing the activated (phosphorylated) forms of ERK1/2 (pErk), p38 MAPK (pp38; Cell Signaling, Inc.), and Akt1 (pAkt; Upstate Biotechnology) were employed. For immunofluorescence analysis, C2C12 cells were grown on fibronectin coverslips or small tissue culture dishes, placed in either GM or muscle differentiation medium (DM), fixed, and probed with an anti-myosin heavy chain (anti-MHC) (Sigma) antibody. MHC-positive cells were detected with a fluorescein-conjugated secondary antibody and visualized by immunofluorescent microscopy (Nikon Instruments). Total cell nuclei were visualized by 4′,6′-diamidino-2-phenylindole (DAPI) nuclear staining.
Cell cycle analysis.
The growth arrest of C2C12-EWS/FLI-1 or C2C12-neo cells in myogenic DM was assessed by using a commercially available 5-bromo-2′-deoxyuridine (BrdU)-based assay (Roche). Briefly, cells were plated either in GM or in DM for 1 to 3 days, labeled with BrdU for 30 min, and then fixed according to the manufacturer's instructions. Cells were then incubated with a working solution containing an anti-BrdU antibody, washed, and treated with an anti-mouse immunoglobulin G-fluorescein-conjugated secondary antibody to allow detection of BrdU-positive cells. The number of BrdU-positive cells was then quantitated by fluorescence-activated cell sorting (FACS) with a FACScan cell sorter (Becton Dickinson) containing a 488-nm argon laser and analyzed with CellQuest software (Becton Dickinson).
RT-PCR and transcriptional activation analysis.
RNA was obtained from the EWS/FLI-1 or control cell line by using a Trizol-based extraction method according to the manufacturer's instructions (Invitrogen), and cDNA was generated with a commercially available reverse transcription-PCR (RT-PCR) kit (Superscript; Invitrogen). To assess muscle-related gene expression, the following primers were utilized (50) (all 5′ to 3′): L7, GGAGCTCATCTATGAGAAGGC (forward) and AAGACGAAGGAGCTGCAGAAC (reverse); myogenin, TGGAGCTGTATGAGACATCCC (forward) and TGGACAATGCTCAGGGGTCCC (reverse); myoD, GCAGGCTCTGCTGCGCGACC (forward) and TGCAGTCGATCTCTCAAAGCACC (reverse); α-skeletal actin, CAGAGCAAGCGAGGTATCC (forward) and GTCCCCAGAATCCAACACG (reverse); desmin, GTGGAGCGTGACAACCTGAT (forward) and GATGGTCTCATACTGAGCCCG (reverse). To assess bone differentiation, the primers GCCCTCTTCCAAGACACATATA (alkaline phosphatase forward), CCATGATCACGTCGATATCC (alkaline phosphatase reverse), TCTCCACTCTTCTAGTTCCT (type I collagen forward), and TTGGGTCATTTCCACATGC (type I collagen reverse) were utilized (10).
For the MyoD and myogenin transcriptional activation assay, 293T cells were plated onto 60-mm-diameter plates and transfected by a modified calcium phosphate protocol (34) by using 10 μg of MyoD or myogenin plasmid or vector alone, 250 ng of a β-galactosidase (lacZ) reporter driven by 5′ regulatory regions from the myogenin gene (myogenin-lacZ reporter) (6, 7), and 1 μg of pCMV/GFP. To assess the ability of EWS/FLI-1 to inhibit myogenin or MyoD-induced transcriptional activation, cells were cotransfected with plasmids containing either wild-type EWS/FLI-1 or the EWS/FLI-1 mutant FLI-1(C) or R2L2 (2) or vector alone (1:1 molar ratio). The number of lacZ-positive cells was determined by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining 48 to 60 h after transfection, and normalized for transfection efficiency by fluorescent microscopy (with green fluorescent protein).
Alkaline phosphatase enzyme-based histochemistry.
To assess alkaline phosphatase enzymatic activity in cultured cells, parental or C2C12-EWS/FLI-1 cells or human ES cell lines were grown on coated microscope slides until the cells reached approximately 70% confluence. Cells were then gently washed, fixed with Pen-Fix (Richard-Allan), and stained by using a commercially available 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium alkaline phosphatase substrate kit (Vector Laboratories) according to the manufacturer's instructions. After staining, slides were gently washed in 100 mM Tris-HCl (pH 9.5) for 5 min, rinsed in tap water, and air dried prior to mounting.
RESULTS
EWS/FLI-1 alters the morphology and myogenic differentiation program of murine cell line C2C12.
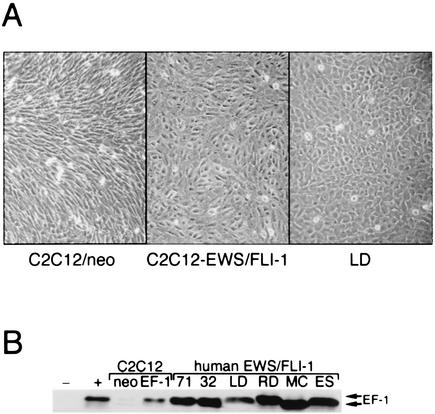
Type I EWS/FLI-1 was introduced into the cell line C2C12 by retroviral gene transfer, and a stable polyclonal population was obtained after drug selection with G418 (neomycin). Compared to cells transduced with control retrovirus, C2C12-EWS/FLI-1 cells exhibited an obvious alteration in cellular morphology, characterized by a more cuboidal appearance, even at confluent cell density (Fig. 1A). Interestingly, the appearance of the C2C12-EWS/FLI-1 cells was reminiscent of that of cell lines derived from human EWS/FLI-1 tumors, such as the cell line LD (Fig. 1A). Stable expression of EWS/FLI-1 in C2C12 cells was confirmed by anti-FLI-1 immunoblotting with an antibody that readily recognizes the EWS/FLI-1 fusion protein (Fig. 1B).
FIG. 1.
Expression of EWS/FLI-1 in the multipotential cell line C2C12. (A) A stable polyclonal population of C2C12 cells expressing either type I EWS/FLI-1 (left) or vector alone (neo) (right) is shown at confluence in standard GM. Note the cuboidal appearance of the C2C12-EWS/FLI-1 cells compared to the more-spindle-shaped control C2C12 cells. For comparison, a human EWS/FLI-1 cell line (LD) is shown at the far right. (B) C2C12-EWS/FLI-1, C2C12-neo, and the human ES cell lines TC-71 (71), TC-32 (32), LD, RD-ES (RD), SK-N-MC (MC), and SK-ES-1 (ES) were analyzed by anti-FLI-1 immunoblotting.
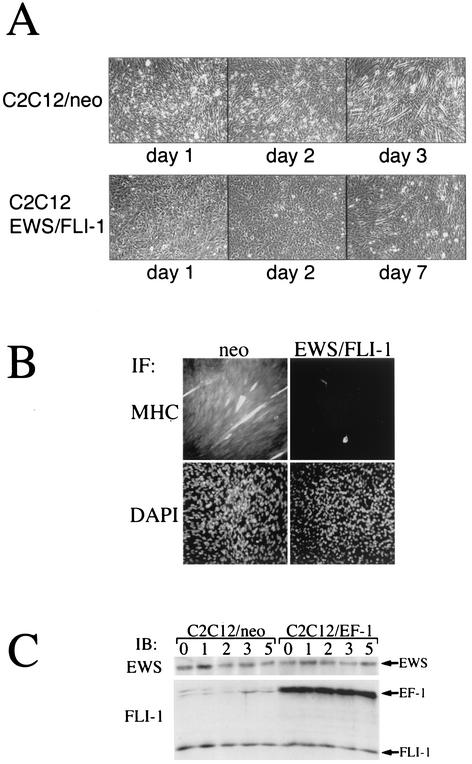
Because C2C12 cells differentiate into myotubes when subjected to low-serum conditions, this cell line has been extensively used to characterize molecules important in the muscle differentiation program. To determine the effect of EWS/FLI-1 on C2C12 differentiation, C2C12-EWS/FLI-1 cells were placed in 2% horse serum (myogenic DM) and cellular morphology was serially analyzed. Compared to vector control cells (neo), which formed readily apparent multinucleated myotubes by day 3, C2C12-EWS/FLI-1 cells showed no morphological evidence of muscle differentiation, even after 7 days in low-serum conditions (Fig. 2A). Moreover, in contrast to control C2C12 cells that became more elongated and began to line up in linear arrays as early as day 1 (Fig. 2A, left), C2C12-EWS/FLI-1 cells showed no evidence of morphological change and remained cuboidal throughout. The lack of C2C12-EWS/FLI-1 myogenic differentiation was also confirmed by immunofluorescence with an anti-MHC. Control (neo) C2C12 cells demonstrated strong anti-MHC immunoreactivity after 3 days in DM while C2C12-EWS/FLI-1 cells demonstrated only rare anti-MHC-positive cells (Fig. 2B, top panels). The levels of endogenous FLI-1 and EWS proteins were the same in C2C12-neo and C2C12-EWS/FLI-1 cells and were not affected by culturing the cells in DM (Fig. 2C). Similarly, EWS/FLI-1 protein levels (EF-1) also remained constant whether C2C12-EWS/FLI-1 cells were cultured in GM or DM.
FIG. 2.
EWS/FLI-1 inhibits the morphological differentiation of C2C12 cells to muscle. (A) C2C12-neo (top) or C2C12-EWS/FLI-1 cells were placed in DM (2% horse serum) for the indicated number of days and evaluated for morphological evidence of myogenic differentiation. Representative areas were then photographed. C2C12-neo cells differentiated to classic multinucleated myotubes by day 3 (top right), whereas C2C12-EWS/FLI-1 cells showed little evidence of morphological differentiation into myotubes, even after 7 days (bottom right) in DM. (B) C2C12-neo (neo) and C2C12-EWS/FLI-1 cells were analyzed on day 3 in DM by immunofluorescence (IF) with an antibody against MHC, a marker of myogenic differentiation, and stained with DAPI to visualize total nuclei. Note the only rare MHC-positive cell in the C2C12-EWS/FLI-1 population. (C) To confirm stable EWS/FLI-1 expression under differentiation conditions, C2C12-neo (neo) and C2C12-EWS/FLI-1 cells were analyzed by Western immunoblotting (IB) in GM (0) or after the indicated number of days in DM with an anti-FLI-1 antibody which recognizes endogenous FLI-1 and the EWS/FLI-1 fusion (EF-1) or with an antibody recognizing the endogenous EWS protein.
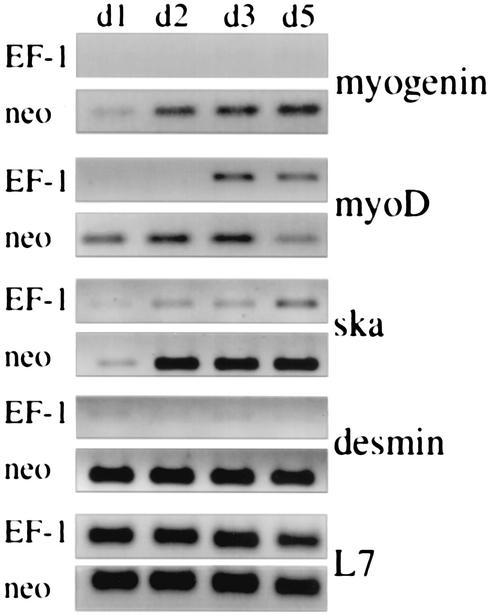
To explore possible mechanisms for the lack of myogenic differentiation, C2C12-EWS/FLI-1 and C2C12-neo cells were placed in muscle DM and analyzed for muscle-specific gene expression by semiquantitative RT-PCR (50). Consistent with previous studies, MyoD was induced in C2C12-neo cells by day 1 in DM (41) and gradually increased thereafter (Fig. 3). Myogenin and skeletal actin (ska), downstream targets of MyoD (48), were induced by day 2. Expression of desmin, another muscle-related gene, was detectable on day 0 (cells still in GM) but markedly increased further when cells were placed in DM. As expected, this orderly induction of muscle-specific genes (41) correlated with the morphological differentiation of C2C12-neo cells into classic multinucleated myotubes (Fig. 2A). This normal pattern of muscle gene expression contrasted sharply with that observed when C2C12-EWS/FLI-1 cells were placed in DM. C2C12-EWS/FLI-1 cells showed a marked delay in MyoD expression, which was not detectable until day 3 in DM. There was also evidence of impaired induction of MyoD target genes, with C2C12-EWS/FLI-1 cells exhibiting little or no expression of myogenin, desmin, and skeletal actin through day 5. Interestingly, although MyoD was expressed in C2C12-EWS/FLI-1 cells on days 3 and 5, there was no evidence of myogenin expression. This was despite the fact that myogenin was readily detected when a similar level of MyoD was expressed in parental cells (Fig. 3, compare neo-MyoD/myogenin d2 with EF-1-MyoD/myogenin d3). Moreover, C2C12-EWS/FLI-1 cells continued to show no morphological evidence of myotube formation, even after 10 days in DM (data not shown). As a control for equivalent RNA integrity and quantity among the samples, RT-PCR amplification of the mRNA encoding the ribosomal protein L7 was performed and was equivalent in all samples. The results of the muscle-specific gene RT-PCR analysis were also confirmed by Western immunoblotting. Induction of MyoD and myogenin protein expression was readily detectable in C2C12-neo cells by day 2 in DM, whereas MyoD protein was expressed at only threshold levels in C2C12-EWS/FLI-1 cells through day 5 in DM (data not shown). Consistent with the RT-PCR analysis, myogenin protein expression was not detectable by immunoblotting even when C2C12-EWS/FLI-1 cells were cultured in DM for up to 7 days (data not shown). These results demonstrate that expression of EWS/FLI-1 in the cell line C2C12 alters the gene expression program normally observed when these cells are subjected to low-serum conditions.
FIG. 3.
EWS/FLI-1 interferes with the induction of muscle-specific genes. C2C12 cells expressing EWS/FLI-1 (EF-1) or control vector (neo) were placed in DM, and the expression of myogenin, MyoD, skeletal actin (ska), and desmin was analyzed by semiquantitative RT-PCR (50) prepared on the indicated days (d1, d2, d3, and d5). To control for equivalent sample integrity and quantity, the expression of the ribosome-associated protein L7 mRNA is shown at the bottom. The data depicted are representative of three independent experiments.
Overexpression of MyoD or myogenin fails to rescue the differentiation defect of C2C12-EWS/FLI-1 cells.
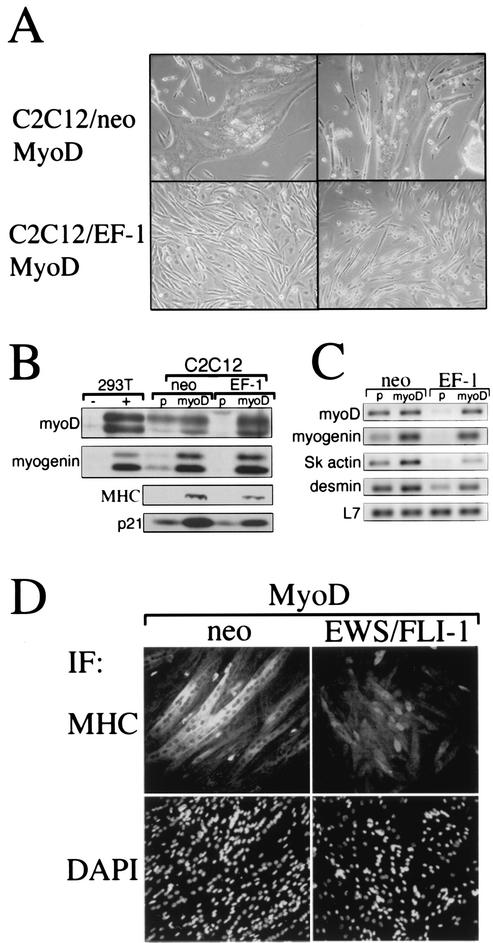
Expression of MyoD, an important myogenic transcription factor (reviewed in reference 5), was markedly delayed when C2C12-EWS/FLI-1 were subjected to low-mitogen medium (Fig. 3). To determine whether restoring MyoD expression would rescue the observed differentiation defect, C2C12-EWS/FLI-1 cells or vector control cells were transduced with a high-titer MyoD retrovirus (45) and then placed in DM. C2C12 cells transduced with MyoD formed massive multinucleated myotubes after 2 to 3 days, even while still in GM, while C2C12-EWS/FLI-1 cells showed no signs of myotube formation in GM (data not shown) or even after 7 days in DM (Fig. 4A). C2C12-EWS/FLI-1 cells transduced with MyoD retrovirus did develop an altered cellular morphology, becoming more slender and spindle-like. To confirm restoration of MyoD expression and the expression of downstream MyoD target genes, transduced cells were examined by Western immunoblotting. As anticipated, C2C12-EWS/FLI-1 and control cells transduced with MyoD retrovirus demonstrated constitutive expression of the MyoD protein (Fig. 4B). Overexpression of MyoD in EWS/FLI-1 and control C2C12 cells was sufficient to induce constitutive expression of myogenin, MHC, p21, and downstream targets of MyoD (16, 17, 48). Interestingly, despite higher expression of MyoD in C2C12-EWS/FLI-1 cells than in control cells, there was only partial rescue of MHC and p21 protein expression.
FIG. 4.
Overexpression of MyoD fails to rescue C2C12-EWS/FLI-1 myogenic differentiation despite partial rescue of downstream gene expression. (A) C2C12-neo or C2C12-EWS/FLI-1 cells were transduced with a MyoD retrovirus and examined for morphological evidence of myogenic differentiation. Note that control cells transduced with MyoD retrovirus formed large multinucleated myotubes (upper photos) while EWS/FLI-1-expressing cells showed no evidence of myotube formation (lower photos). (B) Protein lysates from C2C12-neo or C2C12-EWS/FLI-1 cells were transduced with MyoD or control (p) retrovirus and examined by anti-MyoD, anti-myogenin, anti-MHC, or anti-p21 immunoblotting. As controls, 293T cells transfected with MyoD or myogenin are shown at the left (note that both myogenin and MyoD are detected as doublet bands). (C) The same cells depicted in panel B were analyzed by semiquantitative RT-PCR for myoD, myogenin, skeletal actin (sk actin), and desmin mRNA expression. The expression of the ribosome-associated protein L7 mRNA is shown at the bottom to confirm equivalent sample integrity. (D) C2C12-neo or C2C12-EWS/FLI-1 cells overexpressing MyoD were analyzed by immunofluorescence (IF) with an anti-MHC antibody. Note the less intense MHC staining of C2C12-EWS/FLI-1 cells compared to control (neo) cells. Cells were counterstained with DAPI (bottom) to visualize total cell nuclei.
An analysis of myogenic factor mRNA expression by semiquantitative RT-PCR also revealed a similar pattern. Overexpression of MyoD in control C2C12-neo cells resulted in robust expression of myogenin, skeletal actin, and desmin mRNA (Fig. 4C). Overexpression of MyoD in C2C12-EWS/FLI-1 cells, on the other hand, restored myogenin mRNA expression but only partially rescued expression of skeletal actin and desmin. Therefore, although MyoD overexpression forced the myogenic differentiation of parental cells, restoring MyoD expression in C2C12-EWS/FLI-1 cells failed to overcome their resistance to myogenic differentiation signals and was associated with a blunted induction of muscle-specific genes.
EWS/FLI-1 inhibits MyoD- and myogenin-dependent transcriptional activation.
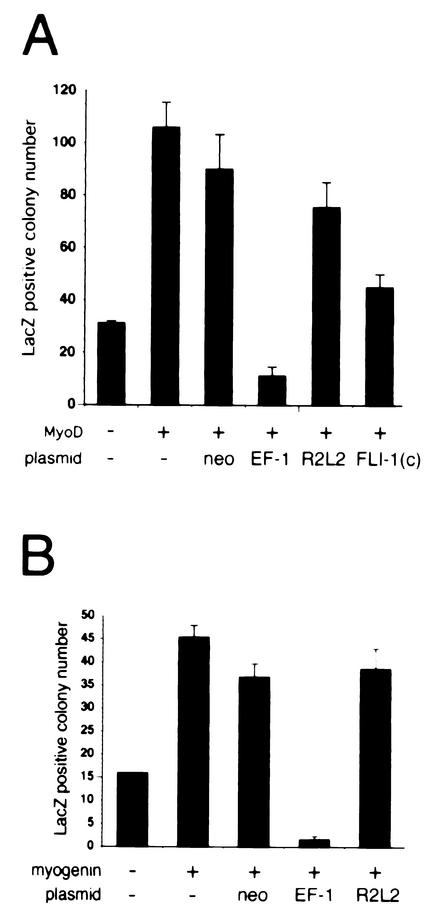
In C2C12 cells, EWS/FLI-1 led to delayed induction of MyoD and blocked myogenin expression (Fig. 3). Although overexpression of MyoD partially rescued downstream target gene expression (Fig. 4B and C), the lack of myogenic differentiation suggested that EWS/FLI-1 might interfere with MyoD and/or myogenin transcriptional activation. To explore this possibility, a MyoD transcriptional activation assay was performed by using a β-galactosidase (lacZ) reporter driven by 5′ regulatory regions from the myogenin gene (6, 7). As anticipated, 293T cells transfected with a MyoD expression plasmid exhibited a fourfold increase in the lacZ-positive colony number compared to cells transfected with control plasmid alone (Fig. 5A). Cotransfection of MyoD and EWS/FLI-1 at equimolar ratios, however, profoundly inhibited MyoD-dependent transcriptional activation, decreasing lacZ-positive colony formation 10-fold. Transfection of MyoD with vector alone (neo) or an EWS/FLI-1 point mutant (EWS/FLI-1 R2L2) defective in nuclear localization, DNA binding (2), and transformation (42) had little effect on MyoD-induced transcriptional activation. Similarly, cotransfection of MyoD with FLI-1(C), which contains only the portion of FLI-1 present in the EWS/FLI-1 fusion protein (2), had only a modest inhibitory effect on MyoD-induced transcriptional activation, decreasing lacZ colony formation by only 50%.
FIG. 5.
EWS/FLI-1 blocks MyoD-dependent and myogenin-dependent transcriptional activation of a myogenin-lacZ reporter. 293T cells were transfected with MyoD (A) or myogenin (B) plasmid and a β-galactosidase (lacZ) reporter driven by 5′ regulatory regions from the myogenin gene (6, 7) and either control vector (neo), type I EWS/FLI-1 (EF-1), the point mutant EWS/FLI-1 R2L2, or a plasmid containing only the portion of FLI-1 present in the EWS/FLI-1 fusion [Fli-1(C)]. Transcriptional activation of the reporter was scored by the lacZ-positive colony number. Data were normalized for differences in transfection efficiency by cotransfection of a green fluorescent protein plasmid. The baseline myogenin-lacZ colony number (control plasmid and myogenin-lacZ only) is shown at the far left. Error bars represent the standard errors of duplicate experiments.
Overexpression of MyoD in C2C12-EWS/FLI-1 cells did not overcome the block in myogenic differentiation, despite restored expression of myogenin. Moreover, direct rescue of myogenin expression by a myogenin retrovirus (45) also failed to rescue the inhibitory effect of EWS/FLI-1 on the C2C12 myogenic program (data not shown). These results suggested that EWS/FLI-1 might also suppress myogenin-dependent transcriptional activation. To test this hypothesis, a myogenin-induced transcriptional activation assay was performed in 293T cells with the same myogenin-lacZ reporter, taking advantage of the ability of myogenin to induce its own expression (11). Compared to vector alone, myogenin increased lacZ colony formation approximately threefold (Fig. 5B). Cotransfection of myogenin and EWS/FLI-1 completely blocked myogenin-induced transcriptional activation, suppressing the number of lacZ-positive colonies to less than background levels. Importantly, cotransfection of control plasmid or the EWS/FLI-1 R2L2 mutant had little inhibitory effect on myogenin-induced transcriptional activation in this assay. These results demonstrate that EWS/FLI-1 interferes with MyoD- and myogenin-dependent transcriptional activation and may explain why overexpression of MyoD failed to rescue C2C12-EWS/FLI-1 myogenic differentiation despite restored expression of myogenin.
Altered pattern of cell cycle gene expression in C2C12-EWS/FLI-1 cells.
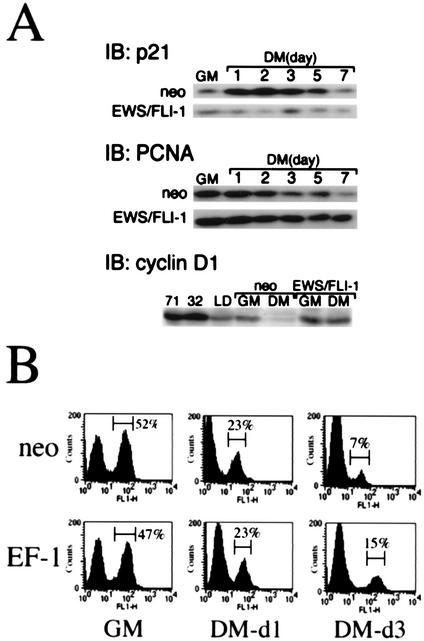
To determine whether other MyoD target genes might also be inhibited by EWS/FLI-1, the expression of the cyclin-dependent kinase inhibitor p21cip1, a transcriptional target of MyoD and a critical effector of MyoD-induced muscle differentiation (16, 17), was examined by Western immunoblotting. In C2C12-neo cells, p21 protein expression was induced within 24 h after placing the cell in DM, reaching a peak on day 2 (Fig. 6A, upper panels). As expected, expression of proliferating cell nuclear antigen (PCNA), a protein associated with cellular proliferation (46, 51), decreased upon transfer of C2C12-neo cells to DM, reflecting the growth arrest accompanying differentiation. In contrast, C2C12-EWS/FLI-1 cells showed no evidence of p21 induction in DM, consistent with the EWS/FLI-1-associated block in MyoD expression and transcriptional activation noted in earlier experiments (Fig. 3 and 5). p21 has also been recently reported to be constitutively decreased in some human ES cell lines (9, 30) and was among the most consistent member of the G1 checkpoint regulators to be dysregulated in primary ES tumor specimens (27). Interestingly, C2C12-EWS/FLI-1 cells had higher levels of constitutive PCNA expression (Fig. 6A, middle panels). Nonetheless, C2C12-EWS/FLI-1 PCNA protein levels still gradually decreased in DM, in a pattern similar to that of C2C12-neo cells. Despite suppressed p21 levels, there was no evidence of continued C2C12-EWS/FLI-1 cell proliferation in muscle DM by viable cell counts (data not shown), likely because the differentiation assays were performed at confluent density.
FIG. 6.
EWS/FLI-1 alters C2C12 cell cycle gene expression without interfering with DM-induced cell cycle arrest. Protein lysates were prepared from C2C12 cells expressing EWS/FLI-1 or control cells (neo) cultured in either GM or DM and analyzed by anti-p21 (upper panel), anti-PCNA (middle panel), or anti-cyclin D1 (lower panel) immunoblotting (IB). In the case of the cyclin D1, cells were analyzed in GM and after 3 days in DM. For comparison, protein lysates from the human ES cell lines TC-71, TC-32, and LD were probed with anti-cyclin D1 antibody and shown at the left. (B) C2C12-neo or C2C12-EWS/FLI-1 cells were cultured in GM or cultured for the indicated number of days in DM, labeled with BrdU, and then analyzed in an anti-BrdU-based immunofluorescence assay. The number of cells undergoing DNA replication (BrdU-positive cells) was quantitated by FACS, with the percentage of BrdU-positive cells shown on the right.
Although the relationship between cell cycle regulators such as p21 and cyclin D1 may depend on cellular context (40); in myogenic cells, p21 and cyclin D1 generally appear to play antagonistic roles (43, 52). To determine whether EWS/FLI-1 also altered the expression of cyclin D1, protein lysates were prepared from C2C12-EWS/FLI-1 or control cells either plated in GM or after 3 days in muscle DM. Consistent with previous findings (43, 52), cyclin D1 protein expression decreased to threshold levels when C2C12 cells were placed in DM (Fig. 6A, lower panels). Cyclin D1 expression in C2C12-EWS/FLI-1 cells, however, failed to be down-regulated in DM. Cyclin D1 was also highly constitutively expressed in the ES cell lines TC-71, TC-32, and LD, a finding also noted by others (9, 30).
The defect in p21 induction and the constitutive expression of cyclin D1 even in DM suggested the possibility that the defect in C2C12-EWS/FLI-1 myogenesis could due to the inability of these cells to undergo the growth arrest requisite for muscle differentiation. To explore this possibility, the proliferation of C2C12-EWS/FLI-1 and control cells was analyzed by a BrdU-based immunofluorescence assay. In GM, C2C12-EWS/FLI-1 and C2C12-neo cells showed similar rates of cellular proliferation, as measured by BrdU incorporation, with 47 and 52% of cells staining positive for BrdU by FACS analysis, respectively (Fig. 6B). By day 1 in DM, BrdU incorporation had decreased to 23% in both C2C12-EWS/FLI-1 and C2C12-neo cells, consistent with the onset of cell cycle arrest accompanying the transition to DM. After 3 days in DM, the vast majority of C2C12-EWS/FLI-1 and C2C12-neo cells were growth arrested, with only 15 and 7% of cells, respectively, staining positive for BrdU.
Thus, expression of EWS/FLI-1 in C2C12 cells suppressed induction of the MyoD target gene p21 and resulted in the constitutive expression of cyclin D1, a pattern shared by some human ES cell lines. Furthermore, the altered expression of p21 and cyclin D1 was not associated with a defect in DM-induced growth arrest, suggesting that EWS/FLI-1 inhibited the C2C12 myogenic differentiation program independent of its impact on the cell cycle.
Classic signal transduction pathways important in myogenesis are intact in C2C12-EWS/FLI-1 cells.
Signal transduction pathways such as MAPK (3), phosphatidylinositol 3-kinase (PI 3-kinase) (21), and p38 MAPK (8, 56) play important roles in muscle differentiation. Although the precise mechanisms are not yet fully defined, these signaling pathways likely modulate events upstream of the basic helix-loop-helix family of muscle regulatory transcription factors. Recently, the MAPK family members ERK1 and ERK2 have been demonstrated to be constitutively activated in human EWS/FLI-1 tumor cell lines (25, 42). Furthermore, activated forms of Ras have been demonstrated to inhibit skeletal muscle differentiation (33), likely through multiple mechanisms (35, 38). The PI 3-kinase signaling pathway, in contrast, has been demonstrated to be an important promoter of myogenic differentiation. Inhibitors of PI 3-kinase have been shown to block skeletal muscle differentiation (21), and constitutive activation of PI 3-kinase enhances myogenesis and the induction of muscle-specific genes (19, 20). PI 3-kinase has also been shown to play a role in the ability of EWS/FLI-1 cells to avoid chemotherapy-induced apoptotic signals (49). Like PI 3-kinase, activation of p38 MAPK, a pathway classically involved in mediating cellular responses to stress-related signals, has also been demonstrated to play an essential role in promoting skeletal muscle differentiation (8, 56).
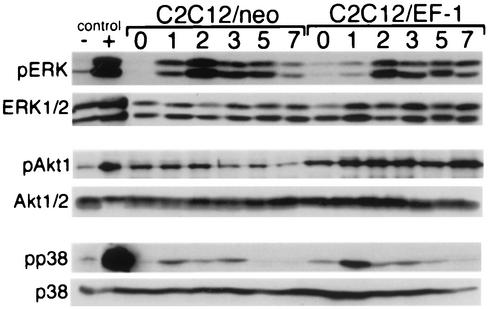
To determine whether dysregulation of any of these signal transduction pathways could account for the block in C2C12-EWS/FLI-1 muscle differentiation, C2C12 cells expressing EWS/FLI-1 were placed in DM, and the pattern of ERK (Fig. 7, top panel), Akt (an established downstream target of PI 3-kinase [14]) (Fig. 7, middle panel), and p38 MAPK (Fig. 7, lower panel) activation was compared to control C2C12 cells by phosphospecific immunoblotting. Consistent with previous results (42), C2C12-EWS/FLI-1 cells demonstrated low-level constitutive activation of ERK1/2 in GM (Fig. 7, day 0). However, once cells were placed in DM, ERK1/2 activation increased further, peaking at day 2, and gradually decreased. An almost identical pattern of ERK activation was observed in control C2C12 cells during myogenic differentiation. The pattern of Akt and p38 MAPK activation was also similar in C2C12-EWS/FLI-1 and C2C12-neo cells (Fig. 7, middle and lower panels). As expected, total ERK1/ERK2, Akt 1/2, and p38 MAPK protein levels were unchanged in GM and DM and were similar in both C2C12-EWS/FLI-1 and control cells. These results demonstrate that although ERK1/2 is constitutively activated in EWS/FLI-1 cells in GM (25, 42), the temporal regulation of ERK1/2 upon transferring the cells to DM is intact, making dysregulation of this pathway an unlikely explanation for the altered differentiation potential of C2C12-EWS/FLI-1 cells. Moreover, the lack of C2C12-EWS/FLI-1 myogenic differentiation was also not explained by insufficient activation of the promyogenic signaling pathways PI 3-kinase/Akt and p38 MAPK.
FIG. 7.
Signaling pathways important in muscle differentiation are intact in C2C12-EWS/FLI-1 cells. Protein lysates were prepared from C2C12-EWS/FLI-1 or vector control cells (neo) while in GM (day 0) or after the indicated number of days in muscle DM. Samples were analyzed by immunoblotting with an antibody recognizing the activated form of ERK1/2 (pERK) or total ERK1/2 (upper panel), an antibody recognizing the activated form of Akt1 (pAkt) or total Akt1/2 (middle panel), or an antibody recognizing the activated form of p38 MAPK (pp38) and total p38 (lower panel). Controls, consisting of NIH 3T3 cells starved (−) and stimulated (+) with serum (for ERK and Akt) and HeLa cells untreated (−) and treated (+) with anisomycin (for p38 MAPK), are shown at the left.
EWS/FLI-1 induces constitutive expression of alkaline phosphatase, a bone lineage marker, in C2C12 cells.
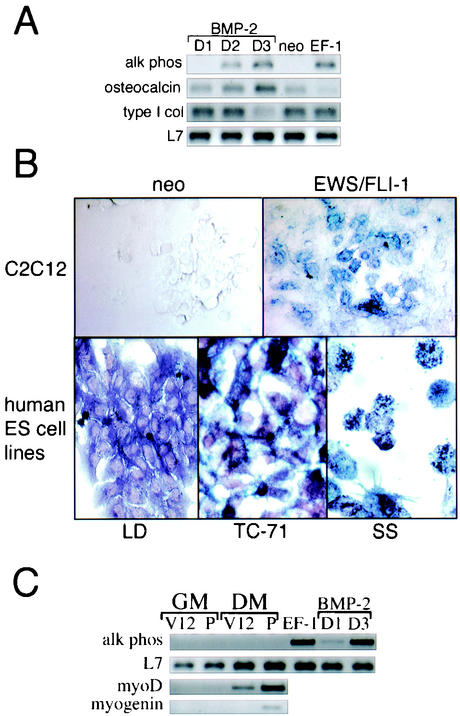
At baseline, C2C12 cells expressing EWS/FLI-1 exhibited a cuboidal shape, which persisted even when the cells were placed in culture conditions conducive to myogenic differentiation. With appropriate growth factor stimulation, C2C12 cells also have the potential to differentiate into osteoblasts (23). Moreover, induction of the bone differentiation program in C2C12 cells has been previously demonstrated to block C2C12 myogenesis, through both transcriptional and posttranslational mechanisms (22). To determine whether there was evidence for the induction of proteins associated with the bone lineage, C2C12-EWS/FLI-1 cells were examined by semiquantitative RT-PCR for expression of alkaline phosphatase, a commonly used marker to evaluate the osteoblastic differentiation of C2C12 cells (22, 23). Interestingly, C2C12-EWS/FLI-1 cells demonstrated constitutive expression of alkaline phosphatase mRNA, while parental (neo) cells showed no evidence of baseline alkaline phosphatase expression (Fig. 8A, right). C2C12-neo cells did express alkaline phosphatase mRNA, however, when treated with bone morphogenetic protein 2 (BMP-2) (Fig. 8A, left), a potent inducer of bone differentiation (22, 23, 55). In control cells, alkaline phosphatase expression peaked on day 3 of BMP-2 treatment and then decreased thereafter (data not shown), consistent with an early- to mid-bone lineage marker (28).
FIG. 8.
Constitutive alkaline phosphatase expression and activity in cells expressing EWS/FLI-1. (A) C2C12 cells expressing EWS/FLI-1 (EF-1) or vector control (neo) were analyzed for alkaline phosphatase (alk phos) expression by semiquantitative RT-PCR (top) while in GM. As a control for alkaline phosphatase expression, C2C12 cells were treated with BMP-2 and analyzed at the indicated day (D) number. To control for equivalent sample processing, the expression of the ribosome-associated protein L7 mRNA is shown at the bottom. (B) C2C12 cells expressing EWS/FLI-1 or control (neo) (top panel) and the human ES tumor cell lines LD, TC-71, and SS (bottom panel) were analyzed for alkaline phosphatase activity by alkaline phosphatase enzyme-based histochemistry. (C) C2C12 cells stably expressing V12 ras (V12) or the parental vector (P) were cultured in GM or for 3 days in DM and analyzed for the expression of alkaline phosphatase, MyoD, or myogenin by semiquantitative RT-PCR. As a control for alkaline phosphatase expression, C2C12 cells treated with BMP-2 are shown at the right. For comparison, the constitutive expression of alkaline phosphatase by C2C12-EWS/FLI-1 cells is also shown at the right. The expression of L7 is included as a control for sample integrity.
To confirm the presence of alkaline phosphatase enzymatic activity, C2C12-EWS/FLI-1 cells and control cells were assessed by alkaline phosphatase enzyme-based histochemical staining (23). Consistent with the RT-PCR analysis, C2C12-EWS/FLI-1 cells were positive for alkaline phosphatase while C2C12-neo cells were completely negative (Fig. 8B, top). To determine whether any human EWS/FLI-1 tumor cell lines showed evidence of constitutive alkaline phosphatase enzymatic activity, the ES cell lines LD, TC-71, and SS were evaluated by alkaline phosphatase-based histochemistry. LD was diffusely alkaline phosphatase positive while both TC-71 and SS demonstrated punctate cytoplasmic staining (Fig. 8B, bottom).
C2C12-EWS/FLI-1 cells did not exhibit a constitutive increase in other bone lineage markers, however, such as type I collagen or osteocalcin (28). Expression of osteocalcin, a late bone lineage marker (28), was detectable at threshold levels in C2C12-neo cells at baseline levels (neo) and steadily increased with BMP-2 treatment, whereas C2C12-EWS/FLI-1 cells showed no evidence of constitutive osteocalcin mRNA expression. Constitutive expression of type I collagen, an early bone lineage marker (28), was similar in both C2C12-EWS/FLI-1 and C2C12-neo cells. Moreover, C2C12-EWS/FLI-1 cells showed no evidence of spontaneous ossification when placed in bone mineralization medium and assessed by Von Kossa staining (data not shown), confirming that EWS/FLI-1 had not activated a definitive osteoblastic cell program. Therefore, EWS/FLI-1 expression in C2C12 cells was associated with constitutive expression of alkaline phosphatase, a marker associated with bone lineage, a characteristic also shared by some human EWS/FLI-1 cell lines.
To determine whether the constitutive expression of alkaline phosphatase by C2C12-EWS/FLI-1 cells was a default consequence of inhibiting myogenesis, C2C12 cells were transfected with a plasmid expressing the constitutively active ras mutant G12V (V12 ras), a potent inhibitor of myogenic differentiation (33). Consistent with previous results (33), C2C12 cells expressing V12 ras showed no evidence of myogenic differentiation in DM (data not shown) and, like C2C12-EWS/FLI-1 cells, showed little evidence of MyoD or myogenin gene expression when placed in DM (Fig. 8C). However, neither V12 nor parental C2C12 cells showed any evidence of constitutive alkaline phosphatase gene expression, whether cultured in GM or DM (Fig. 8C). These results suggest that the constitutive expression of alkaline phosphatase in C2C12-EWS/FLI-1 cells was not an inevitable consequence of oncogene-associated inhibition of myogenic differentiation.
DISCUSSION
The cell of origin harboring EWS/FLI-1 or one of the related EWS/Ets fusion genes implicated in ES is unknown. Approximately 80% of tumors arise from a bony site, while the remainder occur in soft tissue sites, suggesting either a multipotential target cell for ES or distinct target cells, depending on the anatomical presentation. Some, but not all, ES tumors express neural markers (13), leading some to speculate about a possible neural crest cell origin for some ES tumors. However, it is also conceivable that EWS/FLI-1 merely induces early neural markers in a nonneural, multipotential stem cell. This, together with the fact that ES tumors arise in bony or soft tissue sites, makes a mesenchymal target cell also quite a strong possibility. Although the cell line C2C12 has been a useful tool for studying muscle differentiation, its ability to differentiate into bone, fat, and muscle are characteristic of a mesenchymal stem cell (15). Therefore, expression of EWS/FLI-1 in C2C12 cells was of interest because it represented a potentially relevant target cell and provided an opportunity to determine the impact of this oncogene on cellular differentiation.
In C2C12 cells, EWS/FLI-1 profoundly inhibited the muscle differentiation program by interfering with the induction of the myogenic regulatory factors MyoD and myogenin in low-mitogen-containing medium. Because optimal MyoD and myogenin expression rely on positive auto-regulatory feedback (4, 11, 48), it is likely that the ability of EWS/FLI-1 to suppress MyoD- and myogenin-dependent transcriptional activation contributed to their suppressed expression. Moreover, cyclin D1, constitutively expressed in C2C12-EWS/FLI-1 cells even while in DM, has been demonstrated to interfere with muscle regulatory factor-induced transcriptional activation and myogenic differentiation (39, 43). Therefore, dysregulated cyclin D1 may have also played a role in the inability of MyoD and myogenin overexpression to rescue C2C12-EWS/FLI-1 myogenic differentiation. Perhaps the constitutive expression of cyclin D1 in EWS/FLI-1 cells (9, 30) suppresses differentiation signals, leading to a primitive cell lacking definitive lineage markers.
A variety of oncogenes, including activated forms of ras, have been demonstrated to inhibit muscle cell differentiation by altering either myogenic gene transcription (24) or upstream signaling (37). Muscle regulatory factors become activated by upstream signaling pathways, such as PI 3-kinase, ERK1/2 (MAPK1/2), and p38 MAPK, although the precise mechanisms still remain to be precisely defined. Activation of the ras/MAPK (25, 42) and PI 3-kinase/Akt (49) signaling pathways have been implicated in ES; however, the activation pattern of these signaling pathways in DM was similar in C2C12-EWS/FLI-1 and control cells. These results suggest a model in which EWS/FLI-1 acts as a mutant transcription factor, inhibiting the expression and activity of transcription factors important for C2C12 myogenic differentiation. Consistent with this hypothesis, the EWS/FLI-1 point mutant EWS/FLI-1 R2L2, defective in nuclear localization and transcriptional activation (2), did not interfere with either MyoD- or myogenin-dependent transcriptional activation or C2C12 muscle differentiation (data not shown).
Previous studies have generally focused on studying EWS/FLI-1 in the context of NIH 3T3 cells. Although this fibroblastic cell line is commonly used in oncogene expression analysis and transformation assays, novel cell culture models will provide additional and/or corroborative insights into the biologic consequences of EWS/FLI-1 expression. In the cell line C2C12, introduction of EWS/FLI-1 revealed some similarities with human EWS/FLI-1 tumor cell lines. C2C12-EWS/FLI-1 cells exhibited a morphology distinct from that of the parental cells, becoming more cuboidal, reminiscent of some well-characterized ES cell lines (Fig. 1A). ES tumors are well known as small round blue tumors that lack most definitive cellular lineage markers (12). Parental C2C12 cells constitutively expressed myogenic factors such as desmin and MyoD, but C2C12-EWS/FLI-1 cells lacked baseline desmin or MyoD expression by Western immunoblotting or RT-PCR (Fig. 3B and C and 4 and data not shown). This is reminiscent of primary ES tumors, which almost always lack myogenic markers (12, 44), whereas rhabdomyosarcoma, another small round blue tumor, expresses a range of myogenic factors (53). C2C12-EWS/FLI-1 cells also demonstrated a constitutive decrease in p21 expression and a constitutive increase in cyclin D1 expression, cell cycle regulators that have been previously demonstrated to be disturbed in some human EWS/FLI-1 cell lines and in samples from primary ES tumors (9, 27, 30). C2C12-EWS/FLI-1 cells also demonstrated constitutive expression of an early to mid-bone lineage marker, alkaline phosphatase. Interestingly, we found that some human EWS/FLI-1 cell lines were alkaline phosphatase positive, which have also been reported in some primary ES tumors (26, 36). Although C2C12-EWS/FLI-1 cells did develop a polygonal shape reminiscent of osteoblastic cells, increased expression of other bone lineage markers was not detected, and there was no evidence of spontaneous osteoblastic differentiation when C2C12-EWS/FLI-1 cells were placed in bone mineralization medium (data not shown). Therefore, EWS/FLI-1 expression in the cell line C2C12 was associated with some subtle bone lineage features, perhaps analogous to primary ES tumors, which generally arise in bone but lack the definitive bone morphological and histochemical features of osteosarcoma.
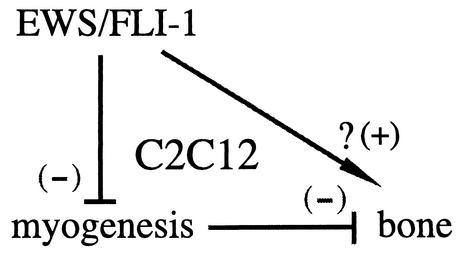
In summary, expression of EWS/FLI-1 in the cell line C2C12 induced an alteration in the pattern of myogenic and osteogenic marker expression. The resulting C2C12-EWS/FLI-1 cell line constitutively expressed an early to mid-bone lineage marker and lost the ability for myogenic differentiation. The block in myogenic differentiation required the nuclear localization and DNA-binding functions of EWS/FLI-1 and was mediated by suppression of myogenic transcription factors through transcriptional and posttranscriptional mechanisms. Differentiation of mesenchymal stem cells along a particular lineage pathway likely occurs by direct stimulation of lineage-specific signals, which may directly or indirectly inhibit differentiation towards other lineages. Our data are consistent with a model in which EWS/FLI-1 may influence C2C12 differentiation by both indirect and direct mechanisms (Fig. 9). In this model, the ability of EWS/FLI-1 to inhibit C2C12 myogenic differentiation acts to diminish the inhibitory influence of the myogenic pathway on bone differentiation. However, oncogenes like V12 ras, which also inhibit C2C12 myogenic differentiation, do not induce alkaline phosphatase gene expression. This suggests the possibility that EWS/FLI-1 may also have a direct role in modulating C2C12 lineage signals but that EWS/FLI-1 expression alone is not sufficient to direct a full-lineage commitment towards a bone lineage. Although future study will be required, our results suggest a model in which EWS/FLI-1 suppresses lineage-specific transcription factors, resulting in a primitive EWS/FLI-1-expressing cell lacking definitive signs of lineage commitment. Sophisticated gene expression analysis with C2C12-based EWS/FLI-1 cell lines will be a useful tool for exploring the molecular pathophysiology of ES and may facilitate progress in identifying potential EWS/FLI-1 targets.
FIG. 9.
A working model of the effect of EWS/FLI-1 on C2C12 mesodermal differentiation. A schematic representation of EWS/FLI-1 inhibiting C2C12 myogenic signals, which normally act to suppress bone differentiation. The possibility that EWS/FLI-1 might also directly modulate bone lineage features is also depicted; see the text for additional details.
Acknowledgments
The FLI-1(C) expression plasmid was obtained from Jacques Gysdael. We thank Orhan Oz, Charles Timmons, and Doris Brown for helpful advice, Steve Harris for providing us with BMP-2 and for assistance with the FACS analysis, and Richard Gaynor for critical review of the manuscript prior to publication.
This work was supported by the Nearburg Family Fund for Pediatric Cancer Research and NIH grant HL03310 (to R.L.I.).
REFERENCES
- 1.Arvand, A., and C. T. Denny. 2001. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 20:5747-5754. [DOI] [PubMed] [Google Scholar]
- 2.Bailly, R. A., R. Bosselut, J. Zucman, F. Cormier, O. Delattre, M. Roussel, G. Thomas, and J. Ghysdael. 1994. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 14:3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, A. M., and N. K. Tonks. 1997. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science 278:1288-1291. [DOI] [PubMed] [Google Scholar]
- 4.Braun, T., E. Bober, G. Buschhausen-Denker, S. Kohtz, K. H. Grzeschik, H. H. Arnold, and S. Kotz. 1989. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 8:3617-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckingham, M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11:440-448. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, T. C., T. A. Hanley, J. Mudd, J. P. Merlie, and E. N. Olson. 1992. Mapping of myogenin transcription during embryogenesis using transgenes linked to the myogenin control region. J. Cell Biol. 119:1649-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, T. C., M. C. Wallace, J. P. Merlie, and E. N. Olson. 1993. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 261:215-218. [DOI] [PubMed] [Google Scholar]
- 8.Cuenda, A., and P. Cohen. 1999. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J. Biol. Chem. 274:4341-4346. [DOI] [PubMed] [Google Scholar]
- 9.Dauphinot, L., C. De Oliveira, T. Melot, N. Sevenet, V. Thomas, B. E. Weissman, and O. Delattre. 2001. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene 20:3258-3265. [DOI] [PubMed] [Google Scholar]
- 10.Ducy, P., M. Starbuck, M. Priemel, J. Shen, G. Pinero, V. Geoffroy, M. Amling, and G. Karsenty. 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 13:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmondson, D. G., T. C. Cheng, P. Cserjesi, T. Chakraborty, and E. N. Olson. 1992. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 12:3665-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fechner, R. E., and S. E. Mills. 1993. Small cell sarcomas, p. 187-201. In J. Rosai (ed.), The atlas of tumor pathology, 3rd series, vol. 8. Tumors of the bones and joints. Armed Forces Institute of Pathology, Washington, D.C.
- 13.Franchi, A., G. Pasquinelli, G. Cenacchi, C. Della Rocca, C. Gambini, M. Bisceglia, G. N. Martinelli, and M. Santucci. 2001. Immunohistochemical and ultrastructural investigation of neural differentiation in Ewing sarcoma/PNET of bone and soft tissues. Ultrastruct. Pathol. 25:219-225. [PubMed] [Google Scholar]
- 14.Franke, T. F., S. I. Yang, T. O. Chan, K. Datta, A. Kazlauskas, D. K. Morrison, D. R. Kaplan, and P. N. Tsichlis. 1995. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell 81:727-736. [DOI] [PubMed] [Google Scholar]
- 15.Grigoriadis, A. E., J. N. Heersche, and J. E. Aubin. 1988. Differentiation of muscle, fat, cartilage, and bone from progenitor cells present in a bone-derived clonal cell population: effect of dexamethasone. J. Cell Biol. 106:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, K., J. Wang, V. Andres, R. C. Smith, and K. Walsh. 1995. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 15:3823-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018-1021. [DOI] [PubMed] [Google Scholar]
- 18.Hawley, R. G., F. H. Lieu, A. Z. Fong, and T. S. Hawley. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136-138. [PubMed] [Google Scholar]
- 19.Jiang, B. H., M. Aoki, J. Z. Zheng, J. Li, and P. K. Vogt. 1999. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc. Natl. Acad. Sci. USA 96:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, B. H., J. Z. Zheng, and P. K. Vogt. 1998. An essential role of phosphatidylinositol 3-kinase in myogenic differentiation. Proc. Natl. Acad. Sci. USA 95:14179-14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaliman, P., F. Vinals, X. Testar, M. Palacin, and A. Zorzano. 1996. Phosphatidylinositol 3-kinase inhibitors block differentiation of skeletal muscle cells. J. Biol. Chem. 271:19146-19151. [DOI] [PubMed] [Google Scholar]
- 22.Katagiri, T., S. Akiyama, M. Namiki, M. Komaki, A. Yamaguchi, V. Rosen, J. M. Wozney, A. Fujisawa-Sehara, and T. Suda. 1997. Bone morphogenetic protein-2 inhibits terminal differentiation of myogenic cells by suppressing the transcriptional activity of MyoD and myogenin. Exp. Cell Res. 230:342-351. [DOI] [PubMed] [Google Scholar]
- 23.Katagiri, T., A. Yamaguchi, M. Komaki, E. Abe, N. Takahashi, T. Ikeda, V. Rosen, J. M. Wozney, A. Fujisawa-Sehara, and T. Suda. 1994. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 127:1755-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lassar, A. B., M. J. Thayer, R. W. Overell, and H. Weintraub. 1989. Transformation by activated ras or fos prevents myogenesis by inhibiting expression of MyoD1. Cell 58:659-667. [DOI] [PubMed] [Google Scholar]
- 25.Lawlor, E. R., C. Scheel, J. Irving, and P. H. Sorensen. 2002. Anchorage-independent multi-cellular spheroids as an in vitro model of growth signaling in Ewing tumors. Oncogene 21:307-318. [DOI] [PubMed] [Google Scholar]
- 26.Llombart-Bosch, A., A. Peydro-Olaya, and F. Gomar. 1980. Ultrastructure of one Ewing's sarcoma of bone with endothelial character and a comparative review of the vessels in 27 cases of typical Ewing's sarcoma. Pathol. Res. Pract. 167:71-87. [DOI] [PubMed] [Google Scholar]
- 27.Maitra, A., H. Roberts, A. G. Weinberg, and J. Geradts. 2001. Aberrant expression of tumor suppressor proteins in the Ewing family of tumors. Arch. Pathol. Lab. Med. 125:1207-1212. [DOI] [PubMed] [Google Scholar]
- 28.Malaval, L., F. Liu, P. Roche, and J. E. Aubin. 1999. Kinetics of osteoprogenitor proliferation and osteoblast differentiation in vitro. J. Cell. Biochem. 74:616-627. [PubMed] [Google Scholar]
- 29.Marsh, M. E., A. M. Munne, J. J. Vogel, Y. Cui, and R. T. Franceschi. 1995. Mineralization of bone-like extracellular matrix in the absence of functional osteoblasts. J. Bone Miner. Res. 10:1635-1643. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, Y., K. Tanaka, F. Nakatani, T. Matsunobu, S. Matsuda, and Y. Iwamoto. 2001. Downregulation and forced expression of EWS-Fli1 fusion gene results in changes in the expression of G(1)regulatory genes. Br. J. Cancer 84:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.May, W. A., S. L. Lessnick, B. S. Braun, M. Klemsz, B. C. Lewis, L. B. Lunsford, R. Hromas, and C. T. Denny. 1993. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol. Cell. Biol. 13:7393-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohno, T., M. Ouchida, L. Lee, Z. Gatalica, V. N. Rao, and E. S. Reddy. 1994. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene 9:3087-3097. [PubMed] [Google Scholar]
- 33.Olson, E. N., G. Spizz, and M. A. Tainsky. 1987. The oncogenic forms of N-ras or H-ras prevent skeletal myoblast differentiation. Mol. Cell. Biol. 7:2104-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry, R. L., M. H. Parker, and M. A. Rudnicki. 2001. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol. Cell 8:291-301. [DOI] [PubMed] [Google Scholar]
- 36.Povysil, C., and Z. Matejovsky. 1977. Ultrastructure of Ewing's tumour. Virchows Arch. A 374:303-316. [DOI] [PubMed] [Google Scholar]
- 37.Puri, P. L., Z. Wu, P. Zhang, L. D. Wood, K. S. Bhakta, J. Han, J. R. Feramisco, M. Karin, and J. Y. Wang. 2000. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 14:574-584. [PMC free article] [PubMed] [Google Scholar]
- 38.Ramocki, M. B., S. E. Johnson, M. A. White, C. L. Ashendel, S. F. Konieczny, and E. J. Taparowsky. 1997. Signaling through mitogen-activated protein kinase and Rac/Rho does not duplicate the effects of activated Ras on skeletal myogenesis. Mol. Cell. Biol. 17:3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao, S. S., C. Chu, and D. S. Kohtz. 1994. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol. Cell. Biol. 14:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 41.Shimokawa, T., M. Kato, O. Ezaki, and S. Hashimoto. 1998. Transcriptional regulation of muscle-specific genes during myoblast differentiation. Biochem. Biophys. Res. Commun. 246:287-292. [DOI] [PubMed] [Google Scholar]
- 42.Silvany, R. E., S. Eliazer, N. C. Wolff, and R. L. Ilaria, Jr. 2000. Interference with the constitutive activation of ERK1 and ERK2 impairs EWS/FLI-1-dependent transformation. Oncogene 19:4523-4530. [DOI] [PubMed] [Google Scholar]
- 43.Skapek, S. X., J. Rhee, D. B. Spicer, and A. B. Lassar. 1995. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science 267:1022-1024. [DOI] [PubMed] [Google Scholar]
- 44.Sorensen, P. H., H. Shimada, X. F. Liu, J. F. Lim, G. Thomas, and T. J. Triche. 1995. Biphenotypic sarcomas with myogenic and neural differentiation express the Ewing's sarcoma EWS/FLI1 fusion gene. Cancer Res. 55:1385-1392. [PubMed] [Google Scholar]
- 45.Spencer, J. A., S. Eliazer, R. L. Ilaria, Jr., J. A. Richardson, and E. N. Olson. 2000. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J. Cell Biol. 150:771-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takasaki, Y., J. S. Deng, and E. M. Tan. 1981. A nuclear antigen associated with cell proliferation and blast transformation. J. Exp. Med. 154:1899-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teboul, L., D. Gaillard, L. Staccini, H. Inadera, E. Z. Amri, and P. A. Grimaldi. 1995. Thiazolidinediones and fatty acids convert myogenic cells into adipose-like cells. J. Biol. Chem. 270:28183-28187. [DOI] [PubMed] [Google Scholar]
- 48.Thayer, M. J., S. J. Tapscott, R. L. Davis, W. E. Wright, A. B. Lassar, and H. Weintraub. 1989. Positive autoregulation of the myogenic determination gene MyoD1. Cell 58:241-248. [DOI] [PubMed] [Google Scholar]
- 49.Toretsky, J. A., M. Thakar, A. E. Eskenazi, and C. N. Frantz. 1999. Phosphoinositide 3-hydroxide kinase blockade enhances apoptosis in the Ewing's sarcoma family of tumors. Cancer Res. 59:5745-5750. [PubMed] [Google Scholar]
- 50.Valdez, M. R., J. A. Richardson, W. H. Klein, and E. N. Olson. 2000. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 219:287-298. [DOI] [PubMed] [Google Scholar]
- 51.Waga, S., G. J. Hannon, D. Beach, and B. Stillman. 1994. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369:574-578. [DOI] [PubMed] [Google Scholar]
- 52.Wang, J., and K. Walsh. 1996. Inhibition of retinoblastoma protein phosphorylation by myogenesis-induced changes in the subunit composition of the cyclin-dependent kinase 4 complex. Cell Growth Differ. 7:1471-1478. [PubMed] [Google Scholar]
- 53.Wang, N. P., J. Marx, M. A. McNutt, J. C. Rutledge, and A. M. Gown. 1995. Expression of myogenic regulatory proteins (myogenin and MyoD1) in small blue round cell tumors of childhood. Am. J. Pathol. 147:1799-1810. [PMC free article] [PubMed] [Google Scholar]
- 54.Yaffe, D., and O. Saxel. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270:725-727. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi, A., T. Katagiri, T. Ikeda, J. M. Wozney, V. Rosen, E. A. Wang, A. J. Kahn, T. Suda, and S. Yoshiki. 1991. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J. Cell Biol. 113:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zetser, A., E. Gredinger, and E. Bengal. 1999. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 274:5193-5200. [DOI] [PubMed] [Google Scholar]