Abstract
Hypoxia, reoxygenation, and the tyrosine phosphatase inhibitor pervanadate activate the transcription factor NF-κB, involving phosphorylation of its inhibitor IκB-α on tyrosine 42. This modification does not lead to degradation of IκB by the proteasome/ubiquitin pathway, as is seen on stimulation of cells with proinflammatory cytokines. It is currently unknown how tyrosine-phosphorylated IκB is removed from NF-κB. Here we show that p85α, the regulatory subunit of PI3-kinase, specifically associates through its Src homology 2 domains with tyrosine-phosphorylated IκB-α in vitro and in vivo after stimulation of T cells with pervanadate. This association could provide a mechanism by which newly tyrosine-phosphorylated IκB is sequestered from NF-κB. Another mechanism by which PI3-kinase contributed to NF-κB activation in response to pervanadate appeared to involve its catalytic p110 subunit. This was evident from the inhibition of pervanadate-induced NF-κB activation and reporter gene induction by treatment of cells with nanomolar amounts of the PI3-kinase inhibitor wortmannin. The compound had virtually no effect on tumor necrosis factor- and interleukin-1-induced NF-κB activities. Wortmannin did not inhibit tyrosine phosphorylation of IκB-α or alter the stability of the PI3-kinase complex but inhibited Akt kinase activation in response to pervanadate. Our data suggest that both the regulatory and the catalytic subunit of PI3-kinase play a role in NF-κB activation by the tyrosine phosphorylation-dependent pathway.
A large number of stimuli including proinflammatory cytokines, antigen stimulation of T and B cells, bacterial lipopolysaccharide, UV irradiation, viral infection, phorbol esters, and reactive oxygen intermediates can activate a family of transcription factors called NF-κB/Rel [for recent reviews see (1–3)]. The prototypical NF-κB transcription factor is a heterodimer composed of p50 (NFκB1) and p65 (RelA) DNA-binding subunits (4–9). Other members of this family in mammals include p52 (NFκB2) (10, 11), RelB (12), c-Rel (13), p105 (4–7) and p100 (10, 11). These proteins share homology within an ≈300-amino acid domain termed the Rel homology domain, which mediates homo- and heterodimerization of the Rel family members as well as DNA-binding activity and nuclear localization (2, 3). Three members of the Rel family have a transcription activation domain (c-Rel, RelA, RelB) (14).
The activity of NF-κB is tightly controlled by a family of inhibitory proteins termed IκB that sequester the transcription factor in the cytosol (2, 3, 14, 15). IκB proteins are structurally characterized by a cluster of 5 to 7 ankyrin repeats, a domain mediating the interaction with the Rel homology domain of NF-κB/Rel family members. Some NF-κB stimuli trigger a cascade of events leading to the phosphorylation of IκB, its polyubiquitination, and subsequent degradation by the 26S proteasome (16). Thereafter, the liberated NF-κB is translocated to the nucleus and activates transcription of its many target genes. Among them are genes involved in the immune response [Igκ, interleukin (IL)-2 and IL-2Rα], the inflammatory response [tumor necrosis factor (TNF)α and β, IL-1, IL-6], cell adhesion [I-CAM, V-CAM, E-selectin], and cell growth (p53, Ras, c-Myc), as well as the genes coding for some Rel/NF-κB and IκB family members (14, 17- 20).
Induced phosphorylation of IκB proteins occurs at two conserved serine residues in the N-terminal domain of several IκB molecules. The residues are serines 32 and 36 in IκB-α, serines 19 and 23 in IκB-β, and serines 18 and 22 in IκB-ɛ (21- 27). Alanine or threonine substitutions at these sites greatly impair IκB degradation and consequently NF-κB activation. Last year, two cytokine-inducible kinases were identified that phosphorylate IκB-α on serines 32 and 36 (28–32). These two kinases, called IKK-α and β, are serine/threonine kinases and present common structural features such as a leucine zipper and helix–loop–helix protein interaction motifs. The leucine zippers of IKK-α and β allow the proteins to heterodimerize, which is required for kinase activity (29, 31, 32).
Recently an alternative pathway of activation of NF-κB has been identified (33–35). This pathway is triggered by hypoxia, reoxygenation, and pervanadate (pV) treatment of cells and involves phosphorylation of IκB-α on tyrosine 42. Unlike serine phosphorylation of IκB-α, tyrosine phosphorylation does not lead to degradation of the inhibitor through the proteasome pathway. Rather, the phosphorylation event triggers dissociation of IκB from NF-κB through an unknown mechanism (33). The tyrosine phosphorylation site of IκB-α is not found in other IκB proteins. The tyrosine kinase involved in this process has not been identified yet, but preliminary evidence suggests an involvement in T cells of the tyrosine kinase Lck and the tyrosine phosphatase CD45 (33). It has also been shown in an in vitro reconstitution system that tyrosine-phosphorylated IκB-α is protected from degradation induced by TNFα (35). This observation, together with the stability of tyrosine-phosphorylated IκB-α, suggested that IκB-α associates with an unknown protein. Candidates are proteins with Src homology 2 (SH2) domains, which are known to specifically bind phosphotyrosine-containing proteins and peptides (36).
We have isolated by affinity purification proteins that specifically interact with tyrosine-phosphorylated IκB-α. Protein sequencing and antibody studies identified these proteins as the regulatory (p85α) and catalytic subunits (p110) of phosphoinositide 3-kinase (PI3-kinase). Pharmacological evidence indicates that PI3-kinase activity is also necessary for the activation of NF-κB by pV. These observations would add a novel role to PI3-kinase in the activation of NF-κB by pV.
MATERIALS AND METHODS
Cell Lines and Reagents.
Jurkat T cells were grown in RPMI 1640 (Cellgro) supplemented with 10% fetal bovine serum (Cellgro). 293 cells were grown in DMEM (Cellgro) supplemented with 10% fetal bovine serum (Cellgro). Glutathione S-transferase (GST)-p85α N-SH2 and GST-p85α C-SH2 agarose conjugates and anti-p85α polyclonal antiserum were purchased from Upstate Biotechnology (Lake Placid, NY). Anti–IκB-α and anti-p110β polyclonal antisera were obtained from Santa Cruz Biotechnology. Anti-Akt and anti-Akt-pS473 polyclonal antisera were purchased from New England Biolabs.
Purification of Proteins Interacting with Tyrosine-Phosphorylated IκB-α.
Extracts from 40 liters of 293 cells were prepared in 250 ml of lysis buffer [50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (Hepes) pH 7.6/150 mM NaCl/1 mM EDTA/2 mM MgCl2/0.1% Nonidet P-40 (NP-40)/0.5 mM DTT/1 mM phenylmethysulfonyl fluoride (PMSF)/protease inhibitor mixture (Complete, Boehringer Mannheim)]. Extract (125 ml) was incubated with 250 μg of Y42 or Y42-P peptide for 30 min at 4°C. Bed volume (12.5 ml) of anti-Flag M2-agarose beads (Eastman Kodak) was added and incubated 1 h at 4°C. The beads were then washed twice with 1 liter and once with 250 ml of lysis buffer. Proteins interacting with the Y42 peptides were eluted twice in 2 ml of lysis buffer containing 0.7 mg⋅ml−1 of Flag peptide (Eastman Kodak) for 30 min at 4°C. The eluates were concentrated on Centricon 50 (Amicon) and the proteins subsequently precipitated with 80% acetone.
Spectroscopic and Microsequencing Analysis.
The proteins interacting with the tyrosine phosphorylated IκB-α peptide were separated by preparative SDS polyacrylamide gel electrophoresis (SDS/PAGE). The proteins were transferred onto a poly(vinylidene difluoride) membrane (Problot, Applied Biosystems) and visualized by Coomassie blue staining. The 85-kDa band was excised from the poly(vinylidene difluoride) membrane and wetted with 1 μl methanol. It was then reduced and alkylated with isopropylacetamide (37) followed by digestion in 20 μl of trypsin at 37°C for 12 h (38). The peptides generated were separated by capillary HPLC, as previously described (39). An aliquot (0.2 μl) of each of the isolated HPLC fractions was applied to a premade spot of matrix (0.5 μl of 20 mg/ml α-cyano-4-hydroxycinammic acid plus 5 mg/ml nitrocellulose in 50% acetone/50% 2-propanol) (40) on the target plate. Ions were formed by matrix-assisted laser desorption/ionization with a nitrogen laser at 337 nm. Spectra were acquired with a PerSeptive Biosystems (Framingham, MA) Voyager Elite time-of-flight mass spectrometer operated in linear delayed extraction mode. Subsequently, fragment ions for selected precursor masses were obtained from postsource decay experiments (41). Each peptide mass and its associated fragment ion masses were used to search a sequence database with an enhanced version of the fragfit program (42). Automated protein sequencing was performed on model 494 Applied Biosystems sequencers by using 6-mm micro cartridges; the models were equipped with on-line PTH analyzers (models 190 and 140D).
Preparation of Extracts.
Whole-cell extracts were prepared in 20 mM Hepes pH 7.9/1 mM MgCl2, 0.2 mM EDTA/10 μM ZnCl2/10% glycerol/200 mM KCl/0.2% Nonidet P-40/0.5 mM DTT/1 mM PMSF/protease inhibitor mixture (Complete, Boehringer Mannheim), 5 mM NaVO4/1 mM NaF/10 mM β-glycerophosphate/5 mM p-nitrophenyl phosphate. Nuclear and cytosolic extracts were prepared as previously described (43), except that the buffers were supplemented with 0.5 mM DTT/1 mM PMSF/protease inhibitor mixture (Complete, Boehringer Mannheim), 5 mM NaVO4/1 mM NaF/10 mM β-glycerophosphate/5 mM p-nitrophenyl phosphate.
Immunoprecipitation, Electrophoretic Mobility Shift Assay, and Pulldown Experiments.
Immunoprecipitations were performed as previously described (44) with the following modifications. All buffers were supplemented with 0.5 mM DTT/1 mM PMSF/protease inhibitor mixture (Complete, Boehringer Mannheim), 5 mM NaVO4/1 mM NaF/10 mM β-glycerophosphate/5 mM p-nitrophenyl phosphate. Preclearing of the extracts with protein A-agarose beads was not performed. Electrophoretic mobility-shift assays were performed as previously described (45), and the probe used was derived from the IL-2 gene promoter (33). Pulldown experiments were performed as described for the immunoprecipitations with the antiserum and protein A-agarose replaced by GST-protein agarose conjugates.
Preparation of GST-p85α.
The PI3-kinase p85α coding sequence was cloned in frame into pGEX-4T-2 (Amersham Pharmacia). Recombinant GST-p85α and GST were purified from cultures of the Escherichia coli strain BL21 transformed with the pGEX-p85α and pGEX-4T-2 respectively. Bacterial extracts were prepared in lysis buffer by two freeze–thaw cycles [20 mM Hepes, pH 7.9/200 mM KCl/1 mM MgCl2/0.2 mM EDTA/10 μM ZnCl2/10% glycerol/0.5 mM DTT/1 mM PMSF/protease inhibitor mixture (Complete, Boehringer Mannheim)] containing 5 mg/ml lysozyme. Extracts were incubated with glutathione-Sepharose beads (Amersham Pharmacia) for 1 h at 4°C. The beads were washed five times with lysis buffer and the purity of the preparation was analyzed by SDS/PAGE followed by Coomassie blue staining.
Transfections and Reporter Gene Assays.
Jurkat cells were transiently transfected by using the DEAE dextran method as previously described (44). Transfected cells were lysed in 100 μl of reporter lysis buffer (Promega). Ten μl of the lysate was mixed with 100 μl of luciferase assay reagent (Promega) and the luciferase activity measured in a luminometer (Turner, Palo Alto, CA). β-galactosidase activity was determined as follows. Ten μl of extract was mixed with 100 μl of Galacton reagent (Tropix, Bedford, MA) diluted 100-fold in PBS containing 1 mM MgCl2. After incubation at room temperature for 1 h, 60 μl of Emerald reagent (Tropix) that was diluted 10-fold in PBS, 0.2 n NaOH, was added, and the activity was measured in a luminometer (Turner).
RESULTS
Purification and Characterization of Proteins Interacting with a Tyrosine-Phosphorylated IκB-α Peptide.
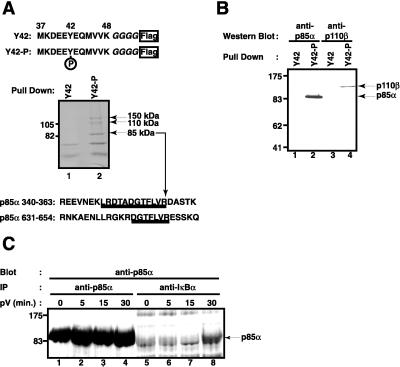
Given the stability of tyrosine-phosphorylated IκB-α in cells and its absence from pV-activated NF-κB complexes (33), we hypothesized that newly tyrosine-phosphorylated IκB-α becomes sequestered by a protein that associates with the inhibitor on stimulation of cells. In search of such a protein, we affinity-purified proteins from 293 cell extract that bind to an IκB-α peptide containing the phosphorylated tyrosine 42 (Fig. 1A Top). The peptide bait had a C-terminal Flag epitope for purification on anti-Flag IgG agarose. Proteins binding to the tyrosine-phosphorylated or nonphosphorylated control IκB-α peptide were specifically eluted from the anti-Flag resin by competition with the Flag epitope peptide and were analyzed by SDS polyacrylamide gel electrophoresis. Only three proteins with apparent molecular masses of 85, 110, and 150 kDa were found to selectively bind the tyrosine-phosphorylated peptide (Fig. 1A Middle). Microsequencing and mass spectroscopic analysis of tryptic peptides showed that the 85-kDa polypeptide contained sequences identical to the human regulatory p85α subunit of PI3-kinase (Fig. 1A Bottom) (46). No primary sequence information could be obtained from the less abundant 110- and 150-kDa polypeptides.
Figure 1.
Proteins interacting with a tyrosine-phosphorylated IκB-α peptide. (A) Affinity purification. The sequence of the peptides used for the affinity purification from 293 cell extracts is shown on top. Amino acid residues 37–48 of IκB-α were fused to four glycine residues and a Flag epitope at the C terminus. Tyrosine 42 was either phosphorylated (Y42-P) or not (Y42). Proteins interacting with the nonphosphorylated peptide (lane 1) or the phosphorylated peptide (lane 2) were analyzed by SDS polyacrylamide gel electrophoresis and Coomassie blue staining. The position of two molecular mass markers is indicated on the left; the position of three proteins interacting only with the tyrosine-phosphorylated peptide is indicated on the right. Protein sequences of two tryptic peptides obtained from the 85-kDa band are shown underlined. They are found within the indicated regions of human PI3-kinase regulatory p85α subunit. (B) Both PI3-kinase subunits bind to the IκB-α peptide. Proteins from Jurkat T cells binding to the nonphosphorylated peptide (lanes 1 and 3) or tyrosine-phosphorylated IκB-α peptide (lanes 2 and 4) were analyzed by Western blotting using an antibody directed against the regulatory p85α subunit of PI3-kinase (lanes 1 and 2) or the catalytic p110β subunit of PI3-kinase (lanes 3 and 4). The positions of p85α and p110β are indicated by arrows on the right; the positions of four molecular mass markers on the left. (C) Interaction of endogenous p85α with endogenous IκB-α on pV treatment. Jurkat T cells (1.5 × 106) pretreated with 400 nM wortmannin were induced with 400 μM pV for 0 (lanes 1 and 5), 5 (lanes 2 and 6), 15 (lanes 3 and 7), and 30 min (lanes 4 and 8). Cytosolic extracts were prepared and immunoprecipitated with anti-p85α (lanes 1–4) or anti-IκB-α (lanes 5–8). The presence of p85α in the complexes was detected by Western blotting. The position of p85α is indicated by an arrow.
PI3-kinase is a heterodimeric lipid and protein kinase composed of a 85-kDa regulatory and 110-kDa catalytic subunit and is activated in response to a number of growth factor, cytokine, and hormone stimuli (47–50). The specific binding of PI3-kinase from Jurkat T cells to the tyrosine-phosphorylated IκB-α peptide was verified by Western blot analysis. Both the regulatory p85α and the catalytic p110β subunit of PI3-kinase could be detected by Western blotting in Flag peptide eluates from the affinity resin with tyrosine 42-phosphorylated IκB-α peptide (Fig. 1B, lanes 2 and 4) but not in eluates from the resin with nonphosphorylated peptide (Fig. 1B, lanes 1 and 3). This identifies the PI3-kinase holoenzyme as a major interaction partner with the tyrosine-phosphorylated site in IκB-α.
PI3-Kinase p85α Interacts with Tyrosine-Phosphorylated IκB-α in Vivo. In an effort to establish a physiological relevance for our finding, we examined whether pV stimulation of Jurkat T cells can induce the association of endogenous tyrosine-phosphorylated IκB-α with endogenous PI3-kinase.
By using specific antibodies, IκB-α was immunoprecipitated from extracts of control and pV-treated cells and was analyzed for the presence of p85α by Western blotting. p85α was indeed found to associate with full-length endogenous IκB-α, but only after a 30-min stimulation with pV (Fig. 1C, lane 8). Association of IκB-α and p85α was preceded by tyrosine phosphorylation of IκB-α, which was first detectable after 15 min of stimulation (data not shown). A control experiment (Fig. 1C, lanes 1–4) shows an immunoprecipitation with p85α antibody from extracts of control and pV-treated cells followed by Western blot detection of p85α. The pV treatment did not alter the amounts or the mobility of the p85α band, which precisely comigrated with the band obtained when IκB-α antibodies were used for immunoprecipitation.
SH2 Domain of p85α Mediate the Interaction with Tyrosine-Phosphorylated IκB-α.
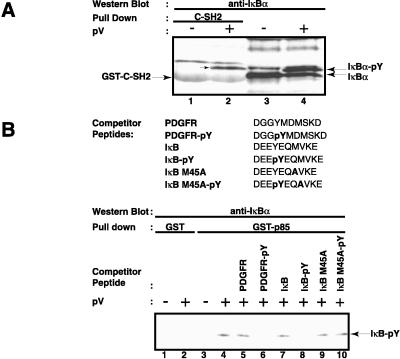
We further investigated whether a SH2 domain of p85α can interact with tyrosine-phosphorylated IκB-α and the sequence requirements of this interaction. Proteins precipitated by GST fusion proteins containing the C-terminal SH2 domain of p85α were analyzed by Western blotting using IκB-α antibodies. The C-terminal SH2 domain precipitated tyrosine-phosphorylated IκB-α from pV-treated Jurkat T cells (Fig. 2A, lane 2), but did not precipitate the nonphosphorylated IκB-α from unstimulated cells (lane 1). Some interaction between tyrosine-phosphorylated IκB-α and the N-terminal SH2 domain of p85α was also observed (data not shown).
Figure 2.
Specificity of the interaction of tyrosine-phosphorylated IκB-α with the regulatory p85α subunit of PI3-kinase. (A) Interaction of tyrosine-phosphorylated IκB-α with the C-terminal SH2 domain of p85α. Two μg of GST-C-SH2 agarose conjugate (lanes 1 and 2) were incubated with whole-cell extracts from control (−) or 200 μM pV-treated (+) Jurkat T cells. The binding of IκB-α to the GST proteins was detected by Western blotting. The analysis of total cell extract by Western blotting for IκB-α is shown in lanes 3 and 4. The positions of IκB-α and its tyrosine-phosphorylated form, as well as that of the GST fusion protein, are indicated by arrows. (B) Specificity of interaction of tyrosine-phosphorylated IκB-α with p85α. Whole-cell extracts from control (−) and pV-treated (+) Jurkat T cells were incubated with 6 μM of the indicated peptides. Two μg of GST (lanes 1 and 2) or GST-p85α (lanes 3–10) agarose conjugates were added and the presence of IκB-α in the complexes was analyzed by Western blotting. The position of tyrosine-phosphorylated IκB-α is indicated.
The specificity of the interaction between IκB-α and p85α was analyzed by a peptide competition assay using a GST-p85α fusion protein. GST alone could not pull down IκB-α, whether tyrosine-phosphorylated or not (Fig. 2B, lanes 1 and 2). However, GST-p85α efficiently precipitated IκB-α from extracts of pV-treated cells (Fig. 2B, lane 4). This interaction was inhibited by a phosphopeptide known to bind PI3-kinase (Fig. 2B, lane 6). The peptide is derived from the human platelet-derived growth factor receptor and encompasses the phosphorylated tyrosine 740 (49, 51, 52). The nonphosphorylated platelet-derived growth factor peptide did not inhibit this interaction (Fig. 2B, lane 5). As expected, the IκB-α peptide phosphorylated on tyrosine 42 also blocked the binding of GST-p85α to IκB-α but not the nonphosphorylated peptide (Fig. 2B, lanes 7 and 8).
The SH2 domains of p85α bind tyrosine-phosphorylated peptides with the consensus Y-(M/V/I/E)-X-M (53). The Y-E-Q-M sequence of IκB-α matches this consensus. The x-ray structure analysis of the p85α SH2 domain with bound peptide showed the critical importance of the methionine residue for recognition (54, 55). This residue binds into a narrow hydrophobic pocket in the SH2 domain that is able to fit only the side chain of a methionine. Consistent with these findings and with the high specificity of the interaction, we observed that a tyrosine-phosphorylated IκB-α peptide with the methionine in position +3 replaced by an alanine no longer competed for the p85α–IκB-α interaction (Fig. 2B, lane 10). The protein purification data and the peptide competition analysis suggest that tyrosine 42-phosphorylated IκB-α is very specifically recognized and bound by the SH2 domains of the p85α subunit of PI3 kinase.
Tyrosine-Phosphorylated IκB-α Is Protected from Degradation Through the TNFα Pathway in Vivo. We wanted to know whether formation of a p85α–IκB-α complex in intact cells can protect tyrosine phosphorylated IκB-α from degradation by the TNFα pathway in vivo.
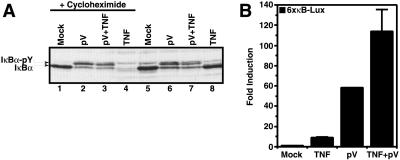
The tyrosine-phosphorylated form of IκB-α, as induced by pV treatment of T cells (Fig. 3A, lane 2), was not degraded if cells were subsequently treated with TNFα (Fig. 3A, lane 3). In contrast, treatment with TNFα alone caused a complete degradation of IκB-α (lane 4). This was however seen only in the presence of cycloheximide, which was used to prevent the rapid de novo synthesis of IκB-α in response to TNFα (compare Fig. 3A, lanes 4 and 8).
Figure 3.
The effect of TNFα on pV-induced events. (A) Protection of tyrosine-phosphorylated IκB-α from degradation through the TNFα pathway. Jurkat T cells were pretreated (lanes 1–4) or not (lanes 5–8) with 10 μg⋅ml−1 cycloheximide for 30 min, treated with 1 mM pV (lanes 2, 3, 6 and 7) and 10 min later induced with 50 ng⋅ml−1 TNFα (lanes 3, 4, 7 and 8) for 1 h. Whole-cell extracts were prepared and IκB-α was detected by Western blotting. The positions of IκB-α and its tyrosine-phosphorylated form are indicated. (B) pV does not impair TNFα signaling. Jurkat cells were transiently transfected with a plasmid containing a luciferase reporter gene driven by three repeats of the HIV type 1 (HIV-1) κB enhancer. Forty-eight hours after transfection, cells were treated with 1 mM pV and 1 h later induced with 50 ng⋅ml−1 TNFα for 5 h. Luciferase activities were measured and normalized on the basis of β-galactosidase expression from cotransfected pRSV-β-galactosidase. The values shown are averages (mean + SEM) of one representative experiment in which each transfection was performed in duplicate.
To rule out that pV inhibited TNFα signaling, we measured the effect of pV on TNFα-induced activation of NF-κB by using a luciferase reporter gene assay. The activation of a NF-κB-dependent reporter gene in response to a combination of TNFα and pV was significantly stronger than with TNFα or pV alone (Fig. 3B), suggesting that TNFα signaling was not impaired by pV. In the presence of pV, TNFα may have preferentially activated NF-κB which was associated with IκB proteins other than IκB-α.
An Inhibitor of PI3-Kinase Specifically Blocks pV-Mediated Activation of NF-κB.
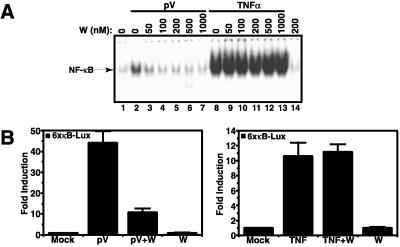
As seen in Fig. 1, the catalytic subunit of PI3-kinase (p110) is also found in the complex with IκB-α. We therefore investigated a potential role of p110 in NF-κB activation by the tyrosine phosphorylation-dependent pathway. Jurkat T cells were pretreated with increasing concentrations of the PI3-kinase inhibitor wortmannin and then induced with pV or TNFα. As measured by electrophoretic mobility shift assay, 50–100 nM wortmannin potently inhibited the NF-κB DNA-binding activity induced by pV (Fig. 4A, lanes 3 and 4). Even at 1 μM wortmannin, no such effect was seen on the TNFα-induced NF-κB activity (Fig. 4A, lane 13). Similar results were obtained with IL-1 as stimulus (data not shown). This specificity of wortmannin was also reflected at the level of nuclear gene expression. Treatment of cells with pV induced a 45-fold induction of the κB-dependent luciferase reporter gene, which was inhibited to 80% by 200 nM wortmannin (Fig. 4B Left). The same treatment had no effect on the 10-fold induction seen with TNFα (Right).
Figure 4.
PI3-kinase activity is necessary for NF-κB induction by pV. (A) Effect of wortmannin on NF-κB DNA-binding activity. Jurkat cells were pretreated for 1 h with the indicated concentrations of wortmannin (W) and were stimulated with 1 mM pV (lanes 2–7) or 50 ng⋅ml−1 TNFα (lanes 8–13) for 1 h. Nuclear extracts were prepared and electrophoretic mobility shift assays were performed by using the IL-2 gene κB site as probe. The position of the NF-κB complex is indicated by an arrow. (B) Effect of wortmannin on the activation of a NF-κB-dependent reporter gene activity. Jurkat T cells were transiently transfected with a plasmid containing the luciferase reporter gene driven by three repeats of the HIV-1 κB enhancer. Forty-eight hours after transfection, cells were pretreated with 200 nM wortmannin for 1 h and then induced with 1 mM pV or 50 ng⋅ml−1 TNFα for 6 h, as indicated. Luciferase activities were measured and normalized to the protein concentration in each extract. The values shown are averages (mean + SEM) of one representative experiment in which each transfection was performed in quadruplicate.
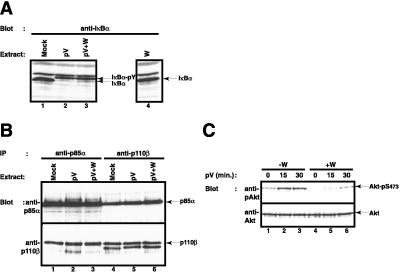
The weak induction of NF-κB DNA binding activity by pV relative to TNFα (Fig. 4A, compare lanes 2 and 8) was in obvious contrast to the much stronger induction of the reporter gene expression by pV. One explanation for this discrepancy could be that the PI3-kinase pathway may be activated by pV and is contributing to the activation of the NF-κB-dependent reporter gene. We therefore analyzed the effect of wortmannin in more detail and tested whether PI3-kinase activity was activated by pV. We could not find that wortmannin had any effect on tyrosine phosphorylation of IκB-α (Fig. 5A, compare lanes 2 and 3). Likewise, wortmannin did not affect the interaction between p85α and p110β (Fig. 5B). We observed, however, that pV treatment could induce PI3-kinase activity, as measured by increased phosphorylation of the protein kinase Akt on serine 473 (56) (Fig. 5C, lanes 1–3). The activation of Akt was partially blocked by wortmannin (Fig. 5C, lanes 4–6). These data suggest that pV induced two parallel events leading to NF-κB activation, one causing tyrosine phosphorylation of IκB and its subsequent binding to p85α, the other activating the PI3-kinase activity of p110 that significantly contributed to activation. Future studies need to address how and to what extent the two P13 kinase subunits engage in NF-κB activation.
Figure 5.
Wortmannin does not perturb tyrosine phosphorylation of IκB-α and PI3-kinase heterodimer formation. (A) Effect of wortmannin on pV-mediated tyrosine phosphorylation of IκB-α. Jurkat T cells were pretreated with 200 nM of wortmannin (lanes 3 and 4) and induced with 200 μM pV (lanes 2 and 3) for 1 h. IκB-α phosphorylation status was analyzed by Western blotting. The position of IκB-α and tyrosine phosphorylated IκB-α is indicated. (B) Effect of wortmannin on PI3-kinase heterodimer formation. Jurkat T cells were pretreated with 200 nM of wortmannin (lanes 3 and 6) and were subsequently induced with 1 mM pV (lanes 2, 3, 5, 6) for 1 h. Cytosolic extracts were prepared and immunoprecipitated with anti-p85α (lanes 1–3) or anti-p110β (lanes 4–6). The presence of p85α (Upper) and p110β (Lower) in the immunocomplexes was analyzed by Western blotting. The positions of p85α and p110β are indicated by arrows. (C) pV induces PI3-kinase activity as measured by Akt serine 473 phosphorylation. Jurkat cells were pretreated (lanes 4–6) or not (lanes 1–3) with 100 nM of wortmannin and induced with 400 μM pV for 0 (lanes 1 and 4), 15 (lanes 2 and 5), or 30 min (lanes 3 and 6). The phosphorylation status of Akt was analyzed by Western blotting with an antibody specific for phosphorylated Akt (Upper). Total levels of Akt were measured with an anti-Akt antibody (Lower). The positions of phosphoserine 473-Akt and Akt are indicated by arrows.
DISCUSSION
We have found that IκB-α, which is newly phosphorylated on tyrosine 42 in response to pV treatment of T cells, associates with the regulatory subunit of PI3-kinase. This association involves the SH2 domains of p85 and is seen in vitro as well as in vivo. This new interaction of IκB-α could explain how tyrosine phosphorylation of IκB-α can lead to NF-κB activation and how IκB-α can be inactivated without needing to be degraded. Phosphorylation of IκB-α by PI3-kinase may directly cause the release of IκB-α from the NF-κB complex. Alternatively, the p85α subunit may capture the tyrosine-phosphorylated inhibitor when it is occasionally released as a function of its natural off-rate. The rather slow activation of NF-κB in response to pV and the lag phase between tyrosine phosphorylation of IκB-α and p85α association (data not shown) may argue for the latter possibility. The sequestration of IκB-α by p85α would also explain why the inhibitor is protected from degradation when cells are stimulated with TNFα. In complex with PI3-kinase, the serine residues 32 and 36 in IκB-α, which are in close proximity to tyrosine 42 and control degradation, may not be accessible to IκB kinases. We consider it unlikely that the covalent modification of tyrosine 42 alone is sufficient to release IκB-α from NF-κB. Indeed, it is well established that phosphorylation of the closely adjacent serines 32 and 36 in IκB-α does not release the inhibitor from NF-κB (25, 45, 57). Furthermore, tyrosine 42 is within a domain of IκB-α that is dispensable for interaction with NF-κB (58). Future experiments using purified tyrosine 42-phosphorylated IκB-α, NF-κB, and PI3-kinase will allow demonstration of whether PI3-kinase can actively or passively sequester IκB-α, thereby releasing active NF-κB. Likewise it can be investigated in vitro whether both subunits of PI3-kinase are required and whether the PI3-kinase activity and ATP are required.
Our data obtained with the PI3 kinase inhibitor wortmannin suggest that binding of IκB-α to p85α may not be the sole mechanism by which PI3 kinase contributes to NF-κB activation. The inhibitory effects of nanomolar concentrations of wortmannin suggest a role for the catalytic subunit of PI3-kinase in both activation of NF-κB DNA binding and transcriptional activity. How PI3-kinase activity contributes to NF-κB activation remains unclear. It is possible that the tyrosine-phosphorylated IκB-α initially gets recruited into the PI3-kinase holoenzyme together with NF-κB. In this complex, NF-κB could be subject to direct phosphorylation by PI3-kinase. A phosphorylation event could not only release NF-κB from the PI3 kinase complex while leaving behind its inhibitor but could also account for the apparent increase in the transactivating potential of NF-κB. It has been previously reported that phosphorylation of p65 in its C terminus increases the transcriptional activity of the protein (59). Finally, the activated PI3-kinase pathway may also enhance the NF-κB pathway further downstream through the activation of synergizing transcription factors. These possibilities are currently under investigation.
NF-κB can be activated by an amazing variety of stimuli (1–3). One explanation obviously lies in the use of different IκB proteins that can couple the transcription factor to distinct upstream signaling pathways. With the use of IκB-α, the NF-κB/Rel system has the capability of specifically responding to stimuli inducing tyrosine phosphorylation, since other IκB proteins lack a site homologous to tyrosine 42 of IκB-α. Depending on the abundance of IκB species in different cell types, only subpopulations of NF-κB–IκB complexes will be activated by a particular stimulus. This allows for cell type-specific regulation and for synergistic effects if more than one kind of stimulus hits the cell.
Genetic evidence and cell-free experiments suggested an involvement of the T cell-specific proteins Lck and CD45 in the induction of tyrosine phosphorylation of IκB-α by pV (33). Future experiments need to investigate more physiological stimuli leading to changes in CD45 and Lck activity. Likewise, experiments in animal models should investigate the involvement of the tyrosine phosphorylation pathway of NF-κB activation in the biomedically important conditions of ischemia/reperfusion, hypoxia, and hyperoxia.
A still poorly understood genetic link between PI3-kinase and the NF-κB system was recently discovered in the case of Ataxia Telangiectasia (60, 61). This disorder is caused by mutations in a protein with PI3-kinase homology. The radiation sensitivity of cells from Ataxia Telangiectasia patients was found to be corrected by a transdominant-negative form of IκB-α lacking the 45 N-terminal amino acids. A study by Reddy et al. indicated that IL-1-mediated activation of NF-κB involved PI3-kinase (62). By using 293 cells expressing the IL-1 receptor, we did not see any effect of wortmannin on IL-1-induced NF-κB activation (data not shown). Moreover, IL-1 is known to induce NF-κB through phosphorylation of serines 32 and 36 in IκB-α and subsequent degradation of the inhibitor (3). It is therefore unlikely that PI3-kinase plays a similar role in IL-1 signaling as described here for pV-induced signaling.
Acknowledgments
The authors are grateful to Keith Williamson for DNA sequencing, Mike Brasseur and Linda Huang for peptide synthesis, and Wendy Schillinglaw for help with protein sequencing and mass spectrometric analysis. We also thank Charlene Liao and David V. Goeddel for critical reading of the manuscript, Lucie Cohen for helpful discussions, Dr. Masato Kasuga for the gift of the p85α cDNA, Dr. Alain Israel for the 6xκB-luciferase construct.
ABBREVIATIONS
- GST
glutathione S-transferase
- PMSF
phenylmethylsulfonyl fluoride
- TNF
tumor necrosis factor
- IL
interleukin
- pV
pervanadate
- SH2
Src homology 2
- PI3-kinase
phosphoinositide 3-kinase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 4.Bours V, Villalobos J, Burd P R, Kelly K, Siebenlist U. Nature (London) 1990;348:76–80. doi: 10.1038/348076a0. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh S, Gifford A M, Riviere L R, Tempst P, Nolan G P, Baltimore D. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- 6.Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban M B, Kourilsky P, Baeuerle P A, Israel A. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 7.Meyer R, Hatada E N, Hohmann H P, Haiker M, Bartsch C, Rothlisberger U, Lahm H W, Schlaeger E J, van Loon A P, Scheidereit C. Proc Natl Acad Sci USA. 1991;88:966–970. doi: 10.1073/pnas.88.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolan G P, Ghosh S, Liou H C, Tempst P, Baltimore D. Cell. 1991;64:961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- 9.Ruben S M, Dillon P J, Schreck R, Henkel T, Chen C H, Maher M, Baeuerle P A, Rosen C A. Science. 1991;251:1490–1493. doi: 10.1126/science.2006423. [DOI] [PubMed] [Google Scholar]
- 10.Bours V, Burd P R, Brown K, Villalobos J, Park S, Ryseck R P, Bravo R, Kelly K, Siebenlist U. Mol Cell Biol. 1992;12:685–695. doi: 10.1128/mcb.12.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercurio F, Didonato J, Rosette C, Karin M. DNA Cell Biol. 1992;11:523–537. doi: 10.1089/dna.1992.11.523. [DOI] [PubMed] [Google Scholar]
- 12.Ryseck R P, Bull P, Takamiya M, Bours V, Siebenlist U, Dobrzanski P, Bravo R. Mol Cell Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brownell E, Mittereder N, Rice N R. Oncogene. 1989;4:935–942. [PubMed] [Google Scholar]
- 14.Miyamoto S, Verma I M. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 15.Baeuerle P A, Baltimore D. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 16.Thanos D, Maniatis T. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 17.Baeuerle P A. Biochim Biophys Acta. 1991;1072:63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- 18.Collins T, Read M A, Neish A S, Whitley M Z, Thanos D, Maniatis T. FASEB J. 1995;9:899–909. [PubMed] [Google Scholar]
- 19.Grilli M, Chen-Tran A, Lenardo M J. Mol Immunol. 1993;30:1287–1294. doi: 10.1016/0161-5890(93)90045-d. [DOI] [PubMed] [Google Scholar]
- 20.Kopp E B, Ghosh S. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 21.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 23.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 25.Traenckner E B, Wilk S, Baeuerle P A. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C Y, Mayo M W, Baldwin A S., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 27.Whiteside S T, Epinat J C, Rice N R, Israel A. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 29.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, et al. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 30.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 31.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 32.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 33.Imbert V, Rupec R A, Livolsi A, Pahl H L, Traenckner E B, Mueller-Dieckmann C, Farahifar D, Rossi B, Auberger P, Baeuerle P A, et al. Cell. 1996;86:787–798. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 34.Koong A C, Chen E Y, Giaccia A J. Cancer Res. 1994;54:1425–1430. [PubMed] [Google Scholar]
- 35.Singh S, Darnay B G, Aggarwal B B. J Biol Chem. 1996;271:31049–31054. doi: 10.1074/jbc.271.49.31049. [DOI] [PubMed] [Google Scholar]
- 36.Schaffhausen B. Biochim Biophys Acta. 1995;1242:61–75. doi: 10.1016/0304-419x(95)00004-y. [DOI] [PubMed] [Google Scholar]
- 37.Krutzsch H C, Inman J K. Anal Biochem. 1993;209:109–116. doi: 10.1006/abio.1993.1089. [DOI] [PubMed] [Google Scholar]
- 38.Lui M, Tempst P, Erdjument-Bromage H. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 39.Henzel W J, Stults J T. Protocols in Protein Science. 1995;1:11.6.1–11.6.14. doi: 10.1002/0471140864.ps1106s24. [DOI] [PubMed] [Google Scholar]
- 40.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 41.Kaufmann R, Kirsch D, Spengler B. Int J Mass Spectrom Ion Processes. 1994;131:355–385. [Google Scholar]
- 42.Henzel W J, Billeci T M, Stults J T, Wong S C, Grimley C, Watanabe C. Proc Natl Acad Sci USA. 1993;90:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiber E, Matthias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beraud C, Sun S C, Ganchi P, Ballard D W, Greene W C. Mol Cell Biol. 1994;14:1374–1382. doi: 10.1128/mcb.14.2.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun S C, Elwood J, Beraud C, Greene W C. Mol Cell Biol. 1994;14:7377–7384. doi: 10.1128/mcb.14.11.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skolnik E Y, Margolis B, Mohammadi M, Lowenstein E, Fischer R, Drepps A, Ullrich A, Schlessinger J. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 47.Carpenter C L, Cantley L C. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 48.Franke T F, Kaplan D R, Cantley L C. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 49.Kapeller R, Cantley L C. BioEssays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 50.Vanhaesebroeck B, Leevers S J, Panayotou G, Waterfield M D. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 51.Escobedo J A, Kaplan D R, Kavanaugh W M, Turck C W, Williams L T. Mol Cell Biol. 1991;11:1125–1132. doi: 10.1128/mcb.11.2.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fantl W J, Escobedo J A, Martin G A, Turck C W, del Rosario M, McCormick F, Williams L T. Cell. 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- 53.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 54.Breeze A L, Kara B V, Barratt D G, Anderson M, Smith J C, Luke R W, Best J R, Cartlidge S A. EMBO J. 1996;15:3579–3589. [PMC free article] [PubMed] [Google Scholar]
- 55.Nolte R T, Eck M J, Schlessinger J, Shoelson S E, Harrison S C. Nat Struct Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 56.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 57.Miyamoto S, Maki M, Schmitt M J, Hatanaka M, Verma I M. Proc Natl Acad Sci USA. 1994;91:12740–12744. doi: 10.1073/pnas.91.26.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun S, Elwood J, Greene W C. Mol Cell Biol. 1996;16:1058–1065. doi: 10.1128/mcb.16.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitz M L, dos Santos Silva M A, Baeuerle P A. J Biol Chem. 1995;270:15576–15584. doi: 10.1074/jbc.270.26.15576. [DOI] [PubMed] [Google Scholar]
- 60.Jung M, Zhang Y, Lee S, Dritschilo A. Science. 1995;268:1619–1621. doi: 10.1126/science.7777860. [DOI] [PubMed] [Google Scholar]
- 61.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 62.Reddy S A, Huang J H, Liao W S. J Biol Chem. 1997;272:29167–29173. doi: 10.1074/jbc.272.46.29167. [DOI] [PubMed] [Google Scholar]