Abstract
In lower eukaryotes, Rad18 plays a crucial role in postreplication repair. Previously, we isolated a human homologue of RAD18 (hRAD18) and showed that human cells overexpressing hRad18 protein with a mutation in the RING finger motif are defective in postreplication repair. Here, we report the construction of RAD18-knockout mouse embryonic stem cells by gene targeting. These cells had almost the same growth rate as wild-type cells and manifested phenotypes similar to those of human cells expressing mutant Rad18 protein: hypersensitivity to multiple DNA damaging agents and a defect in postreplication repair. Mutation was not induced in the knockout cells with any higher frequencies than in wild-type cells, as shown by ouabain resistance. In the knockout cells, spontaneous sister chromatid exchange (SCE) occurred with twice the frequency observed in normal cells. After mild DNA damage, SCE was threefold higher in the knockout cells, while no increase was observed in normal cells. Stable transformation efficiencies were ∼20-fold higher in knockout cells, and gene targeting occurred with ∼40-fold-higher frequency than in wild-type cells at the Oct3/4 locus. These results indicate that dysfunction of Rad18 greatly increases both the frequency of homologous as well as illegitimate recombination, and that RAD18 contributes to maintenance of genomic stability through postreplication repair.
Exposure of cells to UV light, certain chemicals, or other mutagens can inflict lesions in the DNA. However, cells are normally equipped with repair mechanisms whereby the lesion can be removed efficiently. In the event that a lesion is not removed by base or nucleotide excision, or if the DNA replication machinery happens to meet the lesion before repair, the replication machinery stalls at the lesion, causing a gap opposite the site of damage in the newly synthesized strand. Cell death may be imminent unless the gap can be filled. Gap filling is a function of postreplication repair (PRR), a function conserved in diverse species.
In Escherichia coli, proteins involved in PRR are encoded by genes belonging to the SOS regulon and are induced in response to the appearance of single-stranded genomic DNA following DNA damage (11). DNA gaps are filled mainly by recombination in which the RecA and RecFOR complex promotes resolution of the stalled replication fork by allowing damage bypass via template-switching (8, 9; T. Kogoma, Letter, Proc. Natl. Acad. Sci. USA 94:3483-3484, 1997). However, when DNA damage is severe, another mode of PRR, named translesion synthesis is carried out by an error-prone type DNA polymerase, such as UmuD′2C (10, 42).
In the yeast Saccharomyces cerevisiae, genes belonging to the RAD6 epistasis group are responsible for the PRR pathway (8, 11, 33). RAD6, RAD18, RAD30, RAD5, REV3, and REV7 are major members of this group. DNA gaps caused by replication stalling are filled by translesion synthesis either in an error-free or an error-prone mode depending on the context of the DNA damage (19). In UV-induced PRR, error-free and error-prone types of translesion synthesis are carried out by DNA polymerase η (Rad30) and DNA polymerase ζ (a heterodimer of Rev3 and Rev7), respectively (19, 27, 29). RAD6 and RAD18 play a critical role in controlling PRR, and one of the phenotypes of rad6 and rad18 mutants is a high susceptibility to lethal effects by various DNA damaging agents (11, 23, 32). Rad18 protein binds to single-stranded DNA and forms a tight complex with Rad6 protein, a ubiquitin-conjugating enzyme (E2) (6, 7). Thus, it is proposed that Rad18 protein recruits Rad6 protein at replication-stalling sites by binding to single-stranded regions and the Rad6 protein in turn ubiquitinates certain target proteins to be degraded in the PRR process (6, 7). A homologue of RAD18 has been identified from the filamentous fungus, Neurospora crassa. This gene, uvs-2, encodes a protein which shares partial homology to Rad18 and interacts with the Rad6 homologue, MUS8 (40).
DNA gaps are also filled by recombination, but the pathway is normally down-regulated by SRS2 (8, 12, 37), which encodes a DNA helicase with 3′ to 5′ polarity (35). Hypersensitivity to DNA damaging agents in rad18 and rad6 mutants is suppressed by the srs2 mutation (1, 37). Suppression by srs2 requires RAD51, -52, -54, -55, or -57 genes which function in homologous recombination (36, 37). Thus, it is hypothesized that Srs2 channels lesions into the Rad6-dependent PRR pathway, preventing unfavorable recombinational repair (8, 37).
In mammals, two homologs of the RAD6 genes, HR6A and HR6B and a single RAD18 gene are present (17, 39, 41). The HR6B-knockout mice manifest male sterility but otherwise are normal (5, 34). Recently, it was found that human polymerase η is a homologue of yeast Rad30, and that polymerase η is mutated in patients having the variant form of the hereditary photosensitive and cancer-prone disease xeroderma pigmentosum (XP-V) (16, 26). XP-V cells exhibit a feature typical of defective PRR in that the size of newly replicated DNA is shorter than that in normal cells after UV-irradiation (24). These facts indicate the importance of error-free translesion synthesis for maintaining genomic integrity. We cloned the human RAD18 gene (hRAD18) and showed that the hRad18 protein interacts with both the hHR6A and hHR6B proteins (39). Furthermore, human cells expressing Rad18 with a mutation in the RING finger motif are defective in PRR and become sensitive to multiple DNA damaging agents (39). However, in mammalian PRR, both the role of Rad18 and recombination-mediated bypass of DNA damage remain to be determined.
In this study, in order to understand the molecular mechanism of PRR in mammals, we prepared mouse RAD18-knockout embryonic stem (ES) cells by gene targeting. These cells were sensitive to multiple DNA damaging agents and were defective in PRR. RAD18 deficiency enhanced sister chromatid exchange (SCE), stable transformation, and gene targeting, indicating that RAD18 is required for preclusion of illegitimate recombination, and is thus a key component of systems ensuring genomic stability.
MATERIALS AND METHODS
Cell culture and plasmids.
Wild-type ES cells (E14), and RAD18−/− ES cells were cultured in Glasgow modification of Eagle medium supplemented with 10% fetal calf serum. Leukemia inhibitory factor was added to the medium to prohibit differentiation of the ES cells. A plasmid containing mouse RAD18 cDNA (41) was obtained from the IMAGE Consortium (clone ID 535214). The RAD18 targeting construct was prepared using RAD18 genomic DNA and the pU-16 plasmid (3). pCAGGS-CRE was used for transient expression of the CRE recombinase in hetero-knockout ES cells (2). Stable transformation frequencies were determined by counting drug-resistant colonies using plasmids, pPGKpuro (3) and pGTIREShphpA (30). Targeting frequency was evaluated with the Oct3/4 targeting plasmid Oct3/4IREShph (30).
Generation of RAD18-deficient ES clones.
Mouse RAD18 genomic DNA was isolated from a mouse genomic library, 129/SvJ (Stratagene), by using mouse RAD18 cDNA as a probe. RAD18 targeting was performed by a two-step integration of a LacZ-neomycin (βgeo) cassette to the RAD18 genome. In the first step, one allele of the RAD18 gene was disrupted by integration of an internal ribosome entry site (IRES) βgeo-polyA cassette at the AvrII restriction enzyme site in the exon of the RAD18 genomic DNA (Fig. 1A). Then the pCAGGS-CRE plasmid was used to remove the IRES βgeo portion by transient expression of the CRE recombinase in the cells, leaving a polyA portion of the vector in the targeted exon. In the second step, the other allele was disrupted using the same targeting construct used in the first step.
FIG. 1.
Generation of mouse RAD18−/− clones. (A) Schematic representation of disruption constructs. The IRES-βgeo-polyA cassette was inserted at the AvrII site of the exon encoding amino acids Thr-90 to Lys-201. The hatched box indicates sequence used to probe Southern blots. (B) Southern blot analysis. BamHI-digested genomic DNA prepared from different ES clones (first lane, wild type; second to fourth lanes, RAD18+/−; fifth and sixth lanes, RAD18−/−) was hybridized with the probe. (C) Northern blot analysis. Total RNA from different ES clones (first lane, wild type, second lane, RAD18+/−; third and fourth lanes, RAD18−/−) was hybridized with an mRAD18 cDNA probe that corresponds to the N terminus of the gene product. Multiple mRAD18 mRNA bands, indicated by arrowheads, were detected. NADPH mRNA is shown as a control. (D) Western blot analysis. Cell lysates from different ES clones were immunoblotted with a polyclonal rabbit antibody against mRad18. Protein levels of α-tubulin in the lysates are shown as a control. (E) Wild-type (left) and RAD18−/− ES cells (right) were stained for endogenous Rad18 protein with an anti mRad18 antibody.
Immunocytochemistry.
Glutathione S-transferase (GST)-mRad18 (Met-373 to Gln-509) fusion protein was purified from E. coli extract through a glutathione-agarose column, and then PreScission protease (Amersham) was used to remove the GST portion. Polyclonal antiserum was raised by immunizing rabbits with the cleaved fragments spanning the C terminus of mRad18 (Met-373 to Gln-509). The antiserum was used at a dilution rate of 300. Cells growing on coverslips were prefixed with 3.7% formaldehyde for 2 min at room temperature, washed with phosphate-buffered saline, and then fixed with 80% methanol for 10 min at −10°C. The cells were stained with the diluted antiserum for 30 min at room temperature, washed vigorously, and then stained with fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G (Cappel) for another 30 min at room temperature.
Assessment of newly synthesized DNA.
PRR was evaluated by the method described elsewhere (24) with minor modifications. Briefly, actively growing cells (105 cells per dish) were irradiated with UV at 4 J/m2, incubated for 30 min, and then pulse labeled with 0.93 MBq/ml [methyl-3H]thymidine for 15 min. The cells were lysed with a solution containing 0.2 M NaOH and 20 mM EDTA, and the lysates were irradiated with X rays at 20 Gy and centrifuged by alkaline sucrose density as described previously (39). Control cells were treated in the same manner but without UV irradiation.
Immunoprecipitation.
Immunoprecipitation was done by the method described elsewhere (39). Briefly, COS-7 cells were transfected with 10 μg of plasmid by electroporation employing an Electro Cell Manipulator (BTX) and cultured for 48 h. Cells were lysed in 1 ml of lysis buffer and disrupted by sonication. The cleared cell lysate was prepared by centrifugation and mixed with 2 μg of an anti-T7 monoclonal antibody (Novagen), or an anti-GST polyclonal rabbit antibody (Santa Cruz) for 1 h at 4°C. After addition of 50 μl of a slurry of protein-G Sepharose beads (Pharmacia), the lysate was mixed for an additional hour at 4°C. Fifty percent of the bound fraction was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10 or 12.5% polyacrylamide). Separated proteins were identified with the enhanced chemiluminescence (ECL) System (Amersham) using either anti-mRad18 antibody or anti-MUS8 rabbit serum. An anti-N. crassa MUS8 antiserum, which cross-reacts well with mouse HR6A and HR6B proteins, was raised in rabbits by immunizing with GST-MUS8 fusion protein.
Measurement of mutation frequency.
Mutation frequency was determined by counting ouabain-resistant colonies after treatment with or without DNA damaging agents. In brief, cells (2 × 106) were cultured for 20 h and exposed to different DNA damaging agents (0.7 to 1.5 J of UV irradiation/m2 or 50 to 100 μg of methyl methanesulfonate (MMS)/ml for 1 h). The cells were incubated for 2 days. They were harvested, inoculated to new 100-mm-diameter dishes (2 × 106 cells/dish; 10 to 14 dishes). Ouabain was added to the medium at a final concentration of 2 mM and incubated for 2 weeks with medium change every 3 days. The surviving colonies were fixed with 3.7% formaldehyde and stained with 3% Giemsa solution.
SCE analysis.
SCE analysis was done by the method described elsewhere (31). In brief, cells (106) were cultured in the dark for approximately two cycles in medium containing 15 μM bromodeoxyuridine (BrdU). Colcemid was included at a concentration of 0.05 μg/ml for the final 2 h. Cells were harvested and treated with 75 mM KCl for 20 min at room temperature and then fixed with methanol-acetic acid (3:1) for 30 min. The cells were washed once with the fixative and then suspended in a small volume of the fixative. The cell suspension was dropped onto ice-cold wet glass slides and air dried. The cells on the slides were incubated with Hoechst 33258 (5 μg/ml) in H2O for 20 min, rinsed with MacIlvaine solution (164 mM Na2HPO4, 16 mM citric acid [pH 7.0]) and covered with a coverslip. Cells were exposed to black light (λ =325 nm) at a distance of 1 cm for 30 min and washed with water for 5 min. They were stained with 3% Giemsa solution at pH 6.8 for 10 min, and examined with a light microscope. To evaluate the effects of DNA damaging agents on SCE, cells were treated with these agents just before labeling in the medium containing BrdU.
Assessment of stable transformation.
ES cells were transfected with pPGKpuro, pGTIREShphpA, or Oct3/4IREShph plasmids by electroporation and then cultured in medium containing puromycin (1 μg/ml) or hygromycin B (0.2 mg/ml) (Wako Japan) for 2 weeks. The frequency of stable transformation was assessed by the frequencies of antibiotic-resistant colonies.
Evaluation of gene targeting.
The targeting efficiency of ES cells was evaluated with the construct Oct3/4IREShph. Single disruption of Oct3/4 has no effect on viability (30). After electroporation, antibiotic-resistant clones were selected in a medium containing hygromycin B (0.2 mg/ml) for 2 weeks. Surviving colonies were transferred and propagated individually. Genomic DNA from the individual clones was cleaved with EcoRI and analyzed for targeting by probing the Southern blot with Oct3/4 DNA.
RESULTS
Establishment of RAD18−/− ES cells.
RAD18 knockout ES cells were prepared in two steps by sequentially knocking out both alleles with the same targeting vector as described in Materials and Methods. The target exon encodes part of the Rad18 protein between the RING finger motif and the zinc finger (Fig. 1A). Single and double knockouts of RAD18 alleles were confirmed by Southern blotting with multiple ES clones (Fig. 1B). As expected, the amounts of RAD18 mRNA in wild-type, single-knockout, and double-knockout ES cells were roughly 2:1:0, respectively (Fig. 1C). However, the amount of Rad18 protein in RAD18+/− single-knockout ES cells was considerably less than a half of that of wild-type ES cells as revealed by Western blotting, and no Rad18 protein was detected in the RAD18−/− ES cells (Fig. 1D). As reported elsewhere (39), immunostaining showed that Rad18 was localized in the nuclei of wild-type ES cells whereas no positive signal was observed in the RAD18−/− ES cells (Fig. 1E).
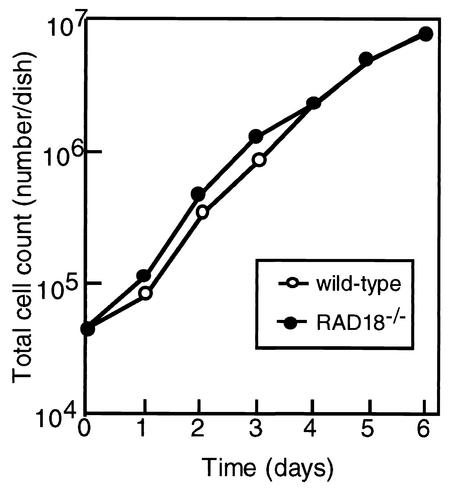
To examine the effect of RAD18 disruption on cell proliferation, we monitored the growth rates of wild-type and RAD18−/− ES cells. RAD18−/− cells proliferated at almost the same rate as the wild-type cells, having a doubling time of 16 h (Fig. 2).
FIG. 2.
Growth curves of wild-type cells (○) and RAD18−/− cells (•). Cells were counted with a hematocytometer at the indicated times. The data show typical results of three independent experiments.
Defective PRR in RAD18−/− ES cells.
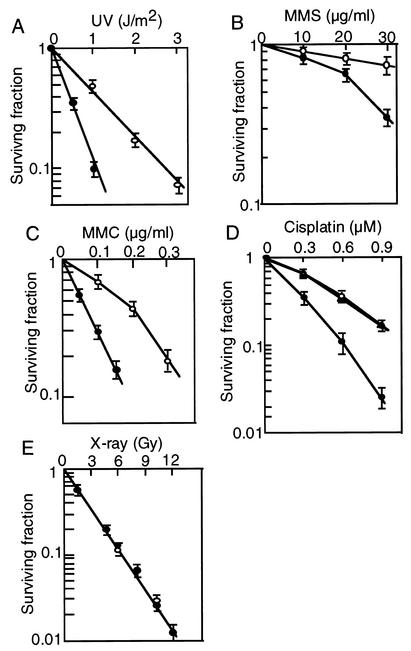
The sensitivity of RAD18−/− ES cells to a variety of DNA damaging agents was determined by the colony formation assay. The knockout ES cells showed moderate sensitivities to UV, MMS, mitomycin C (MMC), and cisplatin compared to wild-type ES cells (Fig. 3A to D). However, no difference in sensitivity to X rays was observed (Fig. 3E). Increased sensitivity to multiple types of DNA damaging agents is a characteristic of the rad18 yeast mutants defective in PRR, and is not observed in XP cells defective in nucleotide excision repair. To confirm there are defects in PRR in RAD18−/− ES cells, we measured the sizes of replicated DNA after UV-irradiation by alkaline sucrose density gradient centrifugation. There were smaller fragments of DNA in irradiated RAD18−/− ES cells in comparison to those in nonirradiated cells, while in normal ES cells the profiles more closely resembled those of nonirradiated cells (Fig. 4).
FIG. 3.
Sensitivity of RAD18−/− ES cells to DNA damaging agents. Cells were irradiated with UV (A), treated with the indicated concentrations of MMS (B) or mitomycin C (MMC) (C) for 1 h, continuously cultured in medium containing cisplatin (D), and irradiated with X-rays (E). Sensitivity was determined by the colony formation assay. Mean values of triplicate dishes are shown with standard deviations (error bars). Symbols: ○, wild-type cells; ▵, RAD18+/− cells; •, RAD18−/− cells.
FIG. 4.
Impaired PRR of RAD18−/− cells. Wild-type ES cells (A) or RAD18−/− cells (B) were irradiated with UV (4 J/m2), incubated for 30 min, and then pulse-labeled with [methyl-3H]thymidine for 15 min. Samples were sedimented on 5 to 20% alkaline sucrose gradients from right to left, and the profile of the UV-irradiated cell sample (•) was compared with that of the unirradiated control cell sample (○). The arrowheads indicate the position of size standards: bacteriophage lambda DNA (42 kb) (closed arrowhead) and bacterial artificial chromosome DNA (100 kb) (open arrowhead).
Association of mouse Rad18 protein with mouse Rad6 protein.
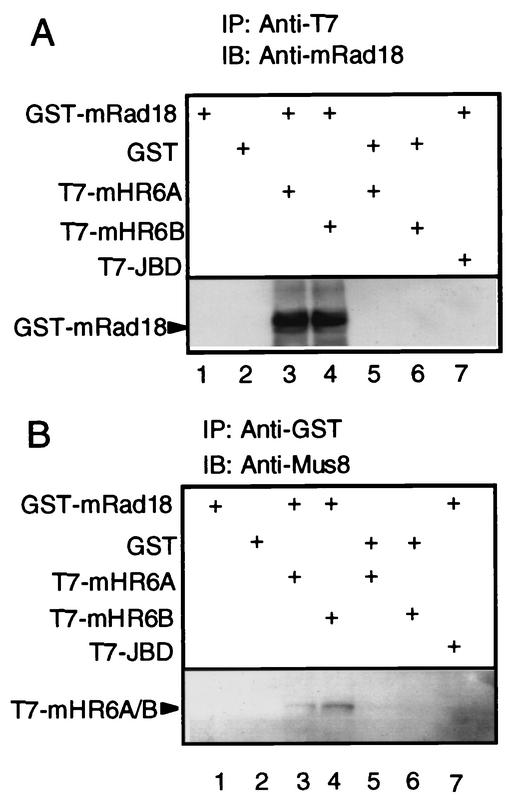
To determine whether mouse Rad18 protein (mRad18) interacts with mouse Rad6 protein (mHR6), we performed immunoprecipitation experiments. To make detection easier, GST-tagged mRad18 plasmid and T7-tagged mRad6 plasmid were transfected into COS-7 monkey cells, and tagged proteins were overproduced there. GST-mRad18 was precipitated by T7-tag-specific monoclonal antibody only when T7-mHR6A or T7-mHR6B was cotransfected with GST-mRAD18 (Fig. 5A, lanes 3 and 4). Inversely, T7-mHR6A or T7-mHR6B was precipitated by GST-tag-specific polyclonal antibody only when GST-mRAD18 was cotransfected with T7-mHR6A or T7-mHR6B (Fig. 5B, lanes 3 and 4). These results indicate that mRad18 and mHR6A/mHR6B can physically associate with each other in transfected mammalian cells.
FIG. 5.
Interaction of mouse Rad18 protein with mouse Rad6 (mHR6A or mHR6B) protein. Plasmids indicated on the left side were transfected into COS-7 cells in different combinations, and cell lysates were prepared 48 h later. Protein interaction was examined by immunoprecipitation. (A) Proteins immunoprecipitating with an anti-T7 antibody (lanes 1 to 7) were immunoblotted with an anti-mRAD18 antibody. (B) Proteins immunoprecipitating with an anti-GST antibody (lanes 1 to 7) were immunoblotted with an anti-MUS8 (N. crassa homologue of RAD6) antiserum. JBD, Jun kinase binding domain.
Mutation frequencies induced by mutagens in RAD18−/− ES cells.
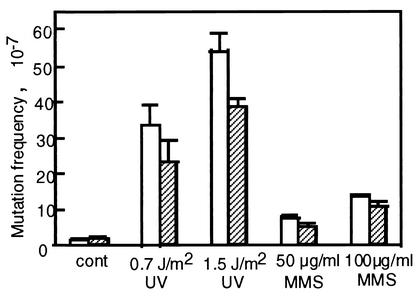
To evaluate the effects of RAD18 disruption on the frequency of induced mutation, RAD18−/− ES cells were treated with either UV or MMS, and viable colonies growing in medium containing ouabain were scored. Ouabain binds competitively at the K+ ion-binding site of the Na+-K+ pump on the plasma membrane, inhibiting ATP production (21). Under normal culture conditions, there was no significant difference in the mutation frequency of normal and RAD18−/− ES cells, with few colonies per 107 cells (Fig. 6). However, upon a low dose of UV-irradiation (0.7 J/m2), mutation frequencies in wild-type ES cells increased approximately 25-fold above the background level, while in RAD18−/− ES cells it increased approximately 13-fold. Mutation frequencies increased in both cell types with an increase in UV dose to 1.5 J/m2, but the frequency in wild-type ES cells remained higher than that of RAD18−/− ES cells. Similarly, mutation frequencies of both wild-type and RAD18−/− ES cells increased following MMS treatment, but the mutation frequency of RAD18−/− ES cells did not exceed that of wild-type cells.
FIG.6.
Damage-induced mutation frequency in RAD18−/− ES cells. ES cells were treated with either UV or MMS at the indicated doses. Mutation frequency was determined by the ratio of the numbers of ouabain-resistant colonies to the initial number of viable cells. Mean values of triplicate data are shown with standard deviations (error bars). Open columns, wild-type ES cells; hatched columns, RAD18−/− ES cells.
Enhanced sister chromatid exchange in RAD18−/− ES cells.
SCE is an event of homologous recombination between sister chromatids during DNA replication (38). SCE levels are low in normal cells, but significantly higher than in cells derived from patients with the cancer-prone hereditary disease Bloom's syndrome, which is caused by a defect in BLM encoding a DNA helicase belonging to the RecQ family. Hyper-SCE indicates a high incidence of chromosome rearrangements, which reflects genomic instability (11). To investigate whether disruption of RAD18 affects genomic stability, we measured SCE frequencies with or without genotoxic assaults. Under normal conditions, SCE frequency in RAD18−/− ES cells was more than twice that in normal cells (Fig. 7). Following mild treatments with UV, MMC, or MMS, SCE frequencies increased to nearly three times above the control level in RAD18−/− ES cells, while such treatments had little effect on wild-type ES cells. (Fig. 7B).
FIG. 7.
SCE in RAD18−/− ES cells. (A) Differential staining for SCE. Cells were cultured in the presence of BrdU through two cell cycles to enable identification of sister chromatids by differential staining. At left are shown wild-type cells; at right are shown RAD18−/− ES cells. (B) Frequencies of SCE in wild-type ES cells and RAD18−/− ES cells. Cells were irradiated with UV (0.5 J/m2) or treated with either MMC (0.05 μg/ml) or MMS (25 μg/ml) for 1 h before culture in medium containing BrdU. Control cells remained untreated before labeling with BrdU. The average number of SCE in at least 40 cells is shown with standard deviations. Open columns, wild-type ES cells; hatched columns, RAD18−/− ES cells.
Enhanced recombination in RAD18−/− ES cells.
The frequency of targeted integration and SCE is increased in BLM−/− DT40 cells (43). To investigate whether similar effects occur in RAD18−/− ES cells, transfection experiments were performed. The frequency of stable transformation was determined by counting colonies grown in selection media with three kinds of plasmids: a plasmid containing a puromycin-resistant gene and two promoterless plasmids containing a hygromycin B-resistant gene. Irrespective of the type of plasmid used, transformation frequencies of RAD18−/− ES cells were ∼20 times higher than those in wild-type ES cells (Table 1).
TABLE 1.
Frequency of stable transformationa
| Plasmid | Genotype of host cells
|
RAD18−/−/ wild type ratio | |
|---|---|---|---|
| Wild type | RAD18−/− | ||
| pPGKpuro | 20 | 400 | 20 |
| pGTIREShphpA | 30 | 590 | 20 |
| Oct3/4IREShph | 30 | 630 | 21 |
Wild-type and RAD18−/− ES cells (106) were transfected with the individual plasmids, transferred to plates of medium containing appropriate antibiotic (puromycin or hygromycin B), and incubated for 2 weeks prior to the counting of surviving colonies.
To assess targeting efficiency, the promoter-less Oct3/4IREShph plasmid was transfected into RAD18−/− ES cells and into wild-type ES cells. More than 50 hygromycin B-resistant colonies were isolated, and gene targeting at the Oct3/4 locus was determined for individual clones by genomic Southern blotting. Compared to wild-type ES cells, targeting efficiency in RAD18−/− ES cells was about twofold higher, with nearly 70% of the drug-resistant clones having targeted integration of plasmid DNA (Table 2). In consequence of the increased rate of both transformation and targeted integration, the overall targeting efficiency in RAD18−/− ES cells was about 40 times higher than that of wild-type ES cells. These results indicate that disruption of the RAD18 gene causes enhancement of both nonhomologous and homologous recombination.
TABLE 2.
Frequency of targeted integrationa
| Expt no. | No. of colonies with targeted integration/total no. tested
|
|
|---|---|---|
| Wild type | RAD18−/− | |
| 1 | 11/27 | 23/31 |
| 2 | 11/32 | 15/24 |
| Total (%) | 22/59 (37) | 38/55 (69) |
The Oct3/4 allele-targeting construct was transfected into cells. After selection, targeted integration events were determined by Southern blot analysis.
DISCUSSION
In this study, we have established mouse RAD18-knockout ES cells by gene targeting and reached the following conclusions. First, Rad18 protein is dispensable for cell viability, since RAD18−/− ES cells exhibited a similar growth rate to that of wild type cells. Second, RAD18−/− ES cells were hypersensitive to a wide variety of DNA damaging agents and defective in PRR. Third, the rate of UV-induced mutagenesis in RAD18−/− cells was reduced compared to that in wild type cells. Fourth, frequencies of spontaneous and DNA-damage induced SCE were elevated in knockout cells. Finally, efficiencies of random and targeted integration of exogenous DNA into the genome were also elevated in RAD18−/− ES cells. Although our targeting construct is designed to eliminate Rad18 protein with the exception of an N-terminal portion including the RING finger motif, we could neither detect any Rad18 mRNA with a probe corresponding to the N terminus nor any Rad18 protein by either Western blotting or immunostaining. Probably, the truncated form of mRNA is unstable and degraded rapidly. Furthermore, because nuclear import of Rad18 protein requires a nuclear localization signal located in the missing C-terminal portion (unpublished data), we conclude that the knockout ES cells do not contain any functional Rad18 protein, even if a trace amount of the truncated form is synthesized.
The conclusion that our RAD18−/− ES cells are defective in PRR is supported by the following lines of evidence, which are characteristics of rad18 mutants in lower eukaryotes. (i) Mouse Rad18 protein associated with the mouse Rad6 protein in vivo. This is a prerequisite for Rad18 protein to function in the postreplication pathway. (ii) RAD18−/− ES cells were sensitive to DNA lesions induced by various DNA damaging agents such as UV, MMS and MMC, and the lesions are repaired by different repair systems. (iii) The size range of newly synthesized DNA in UV-irradiated RAD18−/− ES cells was shorter than that of wild type cells, as determined by alkaline sucrose density gradient centrifugation.
These phenotypes of RAD18−/− ES cells are similar to those of human cells expressing Rad18 protein mutated in the RING finger motif or antisense RAD18 mRNA (39). However, sensitivity of the present cells to various DNA damaging agents was mild compared to the rad18 mutants of lower eukaryotes such as S. cerevisiae or N. crassa. Previously, we assumed that the mild sensitivity of human cells expressing mutant Rad18 or antisense mRNA was due to incomplete inhibition of Rad18 (39). However, since in the present study, RAD18−/− ES cells also showed sensitivity to DNA damage at levels comparable to those of human cells, we conclude that defects in the function of Rad18 in higher eukaryotes result in a mild phenotype compared to lower eukaryotes. Given that the PRR system operates solely on DNA replication, it is plausible that the more severe phenotype of RAD18 mutation in lower eukaryotes might reflect a higher chance for the DNA replication machinery to meet DNA lesions owing to their more rapid proliferation than higher eukaryotes. Alternatively, some back-up systems such as the Rad5-dependent PRR system or recombination systems might ameliorate the Rad18 defect in higher eukaryotes. The phenotype of cells with multiple defects in these repair systems should help resolve this issue. In lower eukaryotes, rad18 mutants show moderate sensitivity to ionizing radiation. However, we observed no differences between RAD18−/− and wild-type ES cells in their sensitivity to X ray. Currently, we do not know whether this is a general phenotype in higher eukaryotes with defective Rad18, or whether this is a specific phenotype of ES cells. Further studies, for example with other cell types derived from RAD18 knockout mice, will be required to determine whether the absence of enhanced sensitivity to ionizing radiation in RAD18−/− mutants is a peculiarity of ES cells.
In the RAD18−/− ES cells, frequency of UV- or MMS-induced mutagenesis was lower than that of wild-type ES cells (Fig. 6). In S. cerevisiae, the dramatic increase in mutation in UV-treated cells is also suppressed in rad18 mutants (4), suggesting that at least a part of DNA damage-induced mutagenesis is RAD18-dependent. In S. cerevisiae, RAD18 is required for both error-free and error-prone modes of PRR, as shown by its epistatic relationship with other members of the group, including RAD30, REV3, and REV7 (11, 15, 19, 22). While RAD30, which encodes DNA polymerase η, functions in an error-free manner in the repair of UV-induced lesions (27), DNA polymerase ζ, which consists of Rev3 and Rev7, functions in an error-prone manner (29, 32). Probably in ES cells Rad18 is required for the error-prone translesion synthesis mediated by mouse Rev3 and/or Rev7 in response to DNA damage. In contrast to the rad18 mutants of S. cerevisiae, induced mutation frequency in RAD18−/− ES cells was higher than the spontaneous level, suggesting the existence of another error-prone translesion synthesis pathway that is independent of Rad18.
In S. cerevisiae, rad18 mutants show elevated levels of spontaneous recombination, ectopic gene conversion, and recombination between direct repeats (25, 36), all of which are dependent on RAD51, RAD52, and RAD57 gene products (25). Since Rad18 is involved in repairing single-stranded gaps (13) and maintaining the integrity of single-stranded DNA (ssDNA) and linear DNA created by genotoxic assaults (28), aberrant DNA structures such as single-stranded DNA, double-strand breaks, and gaps may accumulate in rad18 mutants. These structures are known to be preferential substrates for homologous recombination (9, 14, 18, 20). Enhancement of SCE and gene targeting observed in RAD18−/− ES cells (Fig. 7; Table 2) might be caused by accumulation of the aberrant DNA structures either during normal growth or following genotoxic assaults. In addition, frequency of stable transformation was elevated in RAD18−/− ES cells (Table 1), suggesting enhancement of nonhomologous recombination. Efficient stable transformation in RAD18−/− ES cells might also be triggered by accumulation of the aberrant DNA structures. Whatever the molecular mechanism of recombination may be in mammals, RAD18−/− ES cells provide a useful tool for analysis of the Rad6/Rad18 pathway and for establishing stable transformants.
Acknowledgments
This work was supported by a Grant-in Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan; by a research grant of the Princess Takamatsu Cancer Research Fund; and by a grant from the Sagawa Foundation for Promotion of Frontier Science, Kumamoto, Japan.
We thank Kimi Araki and Ken-ichi Yamamura for providing the targeting vector pU16, pPGKpuro and pCAGGS-CRE. We thank Hideyuki Ogawa and Shunichi Takeda for valuable discussions, David Catcheside and Nell Kennedy for critical reading of the manuscript, and Chie Tateishi and Miyoko Ishizaka for technical assistance.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Cassier-Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki, K., T. Imaizumi, K. Okuyama, Y. Oike, and K. Yamamura. 1997. Efficiency of recombination by Cre transient expression in embryonic stem cells: comparison of various promoters. J. Biochem. 122:977-982. [DOI] [PubMed] [Google Scholar]
- 3.Araki, K., T. Imaizumi, T. Sekimoto, K. Yoshinobu, J. Yoshimuta, M. Akizuki, K. Miura, M. Araki, and K. Yamamura. 1999. Exchangeable gene trap using the Cre/mutated lox system. Cell Mol. Biol. 45:737-750. [PubMed] [Google Scholar]
- 4.Armstrong, J. D., D. N. Chadee, and B. A. Kunz. 1994. Roles for the yeast RAD18 and RAD52 DNA repair genes in UV mutagenesis. Mutat. Res. 315:281-293. [DOI] [PubMed] [Google Scholar]
- 5.Baarends, W. M., R. van der Laan, and J. A. Grootegoed. 2000. Specific aspects of the ubiquitin system in spermatogenesis. J. Endocrinol. Investig. 23:597-604. [DOI] [PubMed] [Google Scholar]
- 6.Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. [DOI] [PubMed] [Google Scholar]
- 7.Bailly, V., S. Prakash, and L. Prakash. 1997. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broomfield, S., T. Hryciw, and W. Xiao. 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486:167-184. [DOI] [PubMed] [Google Scholar]
- 9.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg, E. C., and V. L. Gerlach. 1999. Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell 98:413-416. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 12.Friedl, A. A., B. Liefshitz, R. Steinlauf, and M. Kupiec. 2001. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 486:137-146. [DOI] [PubMed] [Google Scholar]
- 13.Geigl, E. M., and F. Eckardt-Schupp. 1991. Repair of gamma ray-induced S1 nuclease hypersensitive sites in yeast depends on homologous mitotic recombination and a RAD18-dependent function. Curr. Genet. 20:33-37. [DOI] [PubMed] [Google Scholar]
- 14.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24:271-275. [DOI] [PubMed] [Google Scholar]
- 15.Hynes, R. H., and B. A. Kunz. 1981. DNA repair and mutagenesis, p. 371-414. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 16.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 17.Koken, M. H., P. Reynolds, I. Jaspers-Dekker, L. Prakash, S. Prakash, D. Bootsma, and J. H. Hoeijmakers. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:8865-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 19.Kunz, B. A., A. F. Straffon, and E. J. Vonarx. 2000. DNA damage-induced mutation: tolerance via translesion synthesis. Mutat. Res. 451:169-185. [DOI] [PubMed] [Google Scholar]
- 20.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb, J. F., and D. McCall. 1971. Uptake of (3H)ouabain and Na pump turnover rates in monolayer cultures of Girardi heart cells. J. Physiol. 213:57-58. [PubMed] [Google Scholar]
- 22.Lawrence, C. W. 1982. Mutagenesis in Saccharomyces cerevisiae. Adv. Genet. 21:173-254. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, C. W., and R. Christensen. 1976. UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82:207-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehman, A. R., S. Kirk-Bell, C. F. Arlett, M. C. Paterson, P. H. Lohman, E. A. de Weerd-Kastelein, and D. Bootsma. 1975. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl. Acad. Sci. USA 72:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liefshitz, B., R. Steinlauf, A. Friedl, F. Eckardt-Schupp, and M. Kupiec. 1998. Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407:135-145. [DOI] [PubMed] [Google Scholar]
- 26.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 27.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mowat, M. R., W. J. Jachymczyk, P. J. Hastings, and R. C. von Borstel. 1983. Repair of gamma-ray induced DNA strand breaks in the radiation-sensitive mutant rad18-2 of Saccharomyces cerevisiae. Mol. Gen. Genet. 189:256-262. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 30.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 31.Perry, P., and S. Wolff. 1974. New Giemsa method for the differential staining of sister chromatids. Nature 251:156-158. [DOI] [PubMed] [Google Scholar]
- 32.Prakash, L. 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 33.Prakash, S., P. Sung, and L. Prakash. 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27:33-70. [DOI] [PubMed] [Google Scholar]
- 34.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. Koken, M. Vermey, J. H. van Roijen, J. W. Hoogerbrugge, J. T. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 868:799-810. [DOI] [PubMed] [Google Scholar]
- 35.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 36.Schiestl, R. H., R. D. Gietz, P. J. Hastings, and U. Wintersberger. 1990. Interchromosomal and intrachromosomal recombination in rad 18 mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 222:25-32. [DOI] [PubMed] [Google Scholar]
- 37.Schiestl, R. H., S. Prakash, and L. Prakash. 1990. The Srs2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonoda, E., Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tateishi, S., Y. Sakuraba, S. Masuyama, H. Inoue, and M. Yamaizumi. 2000. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl. Acad. Sci. USA 97:7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomita, H., T. Soshi, and H. Inoue. 1993. The Neurospora uvs-2 gene encodes a protein which has homology to yeast RAD18, with unique zinc finger motifs. Mol. Gen. Genet. 238:225-233. (Erratum, Mol. Gen. Genet. 242:743, 1994.) [DOI] [PubMed] [Google Scholar]
- 41.van der Laan, R., H. P. Roest, J. W. Hoogerbrugge, E. M. Smit, R. Slater, W. M. Baarends, J. H. Hoeijmakers, and J. A. Grootegoed. 2000. Characterization of mRAD18Sc, a mouse homolog of the yeast postreplication repair gene RAD18. Genomics 69:86-94. [DOI] [PubMed] [Google Scholar]
- 42.Walker, G. C. 1998. Skiing the black diamond slope: Progress on the biochemistry of translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 95:10348-10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, W., M. Seki, Y. Narita, E. Sonoda, S. Takeda, K. Yamada, T. Masuko, T. Katada, and T. Enomoto. 2000. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 19:3428-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]