Abstract
Adenylate/uridylate-rich element (ARE)-mediated mRNA turnover is an important regulatory component of gene expression for innate and specific immunity, in the hematopoietic system, in cellular growth regulation, and for many other cellular processes. This diversity is reflected in the distribution of AREs in the human genome, which we have established as a database of more than 900 ARE-containing genes that may utilize AREs as a means of controlling cellular mRNA levels. The p38 mitogen-activated protein kinase (MAP kinase) pathway has been implicated in regulating the stability of nine ARE-containing transcripts. Here we explored the entire spectrum of ARE-containing genes for p38-dependent regulation of ARE-mediated mRNA turnover with a custom cDNA array containing probes for 950 ARE mRNAs. The human monocytic cell line THP-1 treated with lipopolysaccharide (LPS) was used as a reproducible cellular model system that allowed us to precisely control the conditions of mRNA induction and decay in the absence and presence of the p38 inhibitor SB203580. This approach allowed us to establish an LPS-induced ARE mRNA expression profile in human monocytes and determine the half-lives of 470 AU-rich mRNAs. Most importantly, we identified 42 AU-rich genes, previously unrecognized, that show p38-dependent mRNA stabilization. In addition to a number of cytokines, several interesting novel AU-rich transcripts likely to play a role in macrophage activation by LPS exhibited p38-dependent transcript stabilization, including macrophage-specific colony-stimulating factor 1, carbonic anhydrase 2, Bcl2, Bcl2-like 2, and nuclear factor erythroid 2-like 2. Finally, the identification of the p38-dependent upstream activator MAP kinase kinase 6 as a member of this group identifies a positive feedback loop regulating macrophage signaling via p38 MAP kinase-dependent transcript stabilization.
The regulation of mRNA stability is an important factor in modulating gene expression, in particular for transiently expressed genes that require tightly controlled mRNA levels. For different cytokines, growth factors, and proto-oncogenes with short mRNA half-lives, modulating the decay rate involves adenylate/uridylate (AU)-rich elements (AREs), often consisting of one to several copies of the sequence AUUUA located in the 3′ untranslated region (12). With a bioinformatics approach, we previously identified several hundred ARE-containing genes that were compiled in the ARE mRNA database (ARED) (2). These genes encode a wide variety of proteins, implicating ARE-mediated mRNA decay in a broader spectrum of cellular processes than was previously recognized.
The molecular mechanisms by which AREs are used to fine-tune mRNA turnover are thought to involve specific RNA-binding proteins (33, 34). trans-Acting factors from different protein families that bind AREs and influence mRNA degradation have been identified. The Hu family proteins HuR and HuB have been shown to bind many different AU-rich messages and stabilize these in several different cell systems (22, 24, 25, 30, 41, 42, 44, 46, 55, 59). AUF1/heterogeneous nuclear ribonucleoprotein D binds to and destabilizes ARE mRNAs such as c-myc, granulocyte-macrophage colony-stimulating factor and others (5, 18, 43, 70). Recently, AUF1 was implicated in apoptosis as a binding factor of the Bcl2 ARE and in tumorigenesis, causing tumor development when overexpressed in mice (26, 37). Tristetraprolin, a protein from the CCCH tandem zinc finger family, appears to regulate mRNA stability of tumor necrosis factor alpha, granulocyte-macrophage colony-stimulating factor, and interleukin-3 (7, 8, 35, 62).
The stabilizing and destabilizing activities of ARE-binding factors can in turn be regulated via a network of signal transduction, giving cells the ability to respond to extra- and intracellular signals by fine-tuning decay rates of mRNAs critical to processes such as cell growth, differentiation, and immune response. The p38 mitogen-activated protein kinase (MAP kinase) pathway has been implicated in the regulation of the mRNA half-lives of a number of AU-rich genes, including cyclooxygenase 2 (COX2), tumor necrosis factor alpha, interleukin-3, interleukin-6, interleukin-8, macrophage-inhibitory protein 1α (MIP1α), granulocyte macrophage-colony stimulating factor, vascular endothelial growth factor, and urokinase-type plasminogen activator (4, 28, 47, 48, 54, 63, 67, 70).
COX2 mRNA levels greatly increase in monocytes upon bacterial lipopolysaccharide (LPS) treatment. This induction is due to transcriptional activation and message stabilization. Inhibition of p38 with the chemical inhibitor SB203580 (SB) or by expressing dominant negative MAP kinase-activated protein kinase 2, a kinase downstream of p38, abolishes stabilization and leads to rapid degradation of COX2 mRNA (17, 39). How p38 signaling might link to ARE-binding proteins has recently been investigated for tristetraprolin. In vitro evidence shows that tristetraprolin can be directly phosphorylated by either p38 or MAP kinase-activated protein kinase 2, potentially modifying its destabilizing activity (6, 45). Alternatively, p38 may also phosphorylate ARE-stabilizing proteins that could compete with destabilizing proteins, as suggested for HuR and tristetraprolin in the case of regulating interleukin-3 decay (47).
To investigate the extent to which the p38 pathway is involved in regulating the mRNA decay of the entire spectrum of ARE-containing genes, we performed a large-scale analysis of AU-rich mRNA turnover with the AU array. This cDNA microarray with 950 ARE-containing and additional control genes was specifically constructed for this purpose. With this approach, we were able to determine the half-lives of 470 ARE mRNAs with and without p38 inhibition, allowing us to define 42 newly identified p38 MAP kinase target mRNAs.
MATERIALS AND METHODS
AU-rich gene array construction.
The AU-rich gene array used in this study contained probes for 950 AU-rich genes from the ARED (2), 18 genes potentially involved in AU-directed mRNA decay, 50 housekeeping genes, and 4 positive control sequences of bacterial origin. Sequence-verified clones for these were obtained from the 40K human clone set (Research Genetics). cDNA inserts were PCR amplified according to Research Genetics instructions. PCR products were purified with size exclusion filter plates (Millipore, MANU030 PCR) and quality controlled by gel electrophoresis. DNA in 1.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was printed on poly-l-lysine home-coated slides with the SDDC-2 microarrayer (Virtek Vision Inc.). Every gene probe was printed in duplicate. After printing, slides were UV treated (120 mJ), incubated for 10 min in blocking solution (170 mM succinic anhydride, 70 mM sodium borate in 1-methyl-2-pyrrolidinone) and for 2 min in 95°C H2O, and spun dry. These slides were used for hybridization from 2 days up to 2 months after printing with equally good results.
Cell culture, treatment, and sample preparation.
THP-1 cells were cultured in RPMI (Invitrogen) with 10% fetal calf serum at 37°C with 5% CO2 for 7 days prior to treatment. For treatments, cells were plated at 2.5 × 106 cells/ml in 25 ml of medium 1 h before adding drugs. LPS treatments (10 μg/ml, Escherichia coli serotype O111:B4; Sigma) were for 2 h. To measure RNA decay, actinomycin D (5 μg/ml, Sigma) alone or actinomycin D plus SB (1 μM, Calbiochem) was added to the LPS-treated cultures and harvested 30, 60, 90, 120, 180, and 240 min later for RNA and cell extract preparation. Cells were collected by sedimenting suspended cells and washing differentiating, adherent cells off the plate.
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer's instructions. Whole-cell extracts were prepared after two washes with ice-cold phosphate-buffered saline with a lysis buffer containing 1% Triton X-100 as the detergent, supplemented with protease and phosphatase inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, sodium orthovanadate, and pepstatin). The LPS treatments were performed 13 times independently, giving rise to the gene induction-repression data in Table 1. The RNA decay time courses were performed three times on independently cultured THP-1 cells.
TABLE 1.
AU-rich mRNAs induced or repressed in THP-1 cells after 2 h of LPS treatment
| Unigene symbol | Acca | Fold changeb | SE | Times >2-foldc |
|---|---|---|---|---|
| SCYA4 (MIP1β) | H62985 | 113.2 | 21.44 | 13/13 |
| IL1B | W47101 | 71.5 | 15.51 | 12/13 |
| SCYA3L1 | R47893 | 60.9 | 11.98 | 13/13 |
| IL8 | IL8d | 43.5 | 9.67 | 6/6 |
| TNFAIP3 | AA476272 | 42.1 | 5.53 | 13/13 |
| NR4A3 | H37761 | 37.2 | 10.71 | 13/13 |
| TNF | AA699697 | 31.3 | 6.63 | 13/13 |
| GRO2 | R50407 | 31.1 | 5.81 | 13/13 |
| SCYA3 (MIP1α) | AA677522 | 28.9 | 4.06 | 12/12 |
| GRO1 | W42723 | 27.5 | 5.86 | 13/13 |
| PTGS2 (COX2) | R80217 | 27.3 | 6.50 | 13/13 |
| CPSF6 | R18985 | 20.7 | 3.92 | 10/13 |
| GRO3 | AA935273 | 19.7 | 4.10 | 13/13 |
| IL1A | AA936768 | 14.8 | 4.58 | 13/13 |
| EGR2 | AA446027 | 14.7 | 4.26 | 11/13 |
| TNFAIP6 | W93163 | 12.9 | 1.79 | 13/13 |
| GCH1 | AA443688 | 9.9 | 1.14 | 13/13 |
| NFKBIA | W55872 | 8.7 | 0.93 | 13/13 |
| MAP2K6 | H07920 | 8.7 | 1.57 | 13/13 |
| MBL2 | T69359 | 8.7 | 0.56 | 13/13 |
| LIF | R50354 | 8.3 | 1.73 | 12/13 |
| SCYA2 | AA425102 | 7.5 | 2.37 | 5/6 |
| GLS | AI004766 | 7.1 | 2.62 | 7/7 |
| RGS16 | AA453774 | 6.9 | 1.33 | 12/12 |
| KIAA0938 | AA705735 | 6.4 | 1.17 | 10/11 |
| NFKB2 | AA952897 | 6.1 | 1.17 | 11/13 |
| DUSP2 | AA759046 | 5.9 | 0.64 | 13/13 |
| NGFB | T56316 | 5.7 | 0.88 | 13/13 |
| PLAU (uPA) | AA284668 | 5.5 | 0.78 | 13/13 |
| JUNB | N94468 | 5.2 | 0.77 | 11/11 |
| NFE2L2 | H88359 | 4.6 | 0.36 | 13/13 |
| ITGA2 | AA463610 | 4.6 | 0.85 | 6/7 |
| TWIST | AI220198 | 4.4 | 0.66 | 10/13 |
| RIPK2 | AA913804 | 4.2 | 0.26 | 13/13 |
| JUN | W96134 | 4.2 | 0.82 | 10/13 |
| PTGER2 | AI276745 | 3.8 | 0.81 | 10/13 |
| KIAA0298 | AA853966 | 3.8 | 0.40 | 11/13 |
| NAB1 | N91896 | 3.7 | 0.18 | 13/13 |
| SCYB10 | AA878880 | 3.7 | 0.78 | 8/13 |
| PDE4B | AA453293 | 3.6 | 0.39 | 13/13 |
| SLC9A6 | R45009 | 3.5 | 0.34 | 13/13 |
| PTGER3 | AA151583 | 3.4 | 0.70 | 7/12 |
| UNG2 | AA425900 | 3.4 | 0.34 | 11/13 |
| BIRC3 | H48706 | 3.3 | 0.46 | 10/13 |
| C7 | AA598478 | 3.1 | 0.28 | 11/13 |
| KIAA0105 | AA598802 | 3.1 | 0.24 | 12/13 |
| GAD1 | AA018457 | 3 | 0.35 | 11/13 |
| CBLN1 | AA495901 | 2.9 | 0.21 | 12/13 |
| SNK | AA460152 | 2.9 | 0.35 | 9/13 |
| C8orf1 | AA278836 | 2.9 | 0.24 | 11/13 |
| TIEG | AI348177 | 2.8 | 0.15 | 12/13 |
| OLR1 | AA682386 | 2.6 | 0.24 | 9/12 |
| DBY | AA447588 | 2.6 | 0.14 | 11/13 |
| BCL2 | H74208 | 2.6 | 0.34 | 9/13 |
| CHL1 | H15267 | 2.6 | 0.40 | 5/9 |
| XBP1 | W90128 | 2.6 | 0.26 | 9/13 |
| KCNA3 | AI095381 | 2.5 | 0.50 | 6/11 |
| PARG1 | AA629603 | 2.5 | 0.29 | 8/13 |
| KIAA0254 | N75979 | 2.5 | 0.49 | 7/13 |
| TFAP2C | AA399334 | 2.5 | 0.31 | 7/10 |
| CSF3 | AI074784 | 2.5 | 0.49 | 5/13 |
| ITGAV | AA029934 | 2.5 | 0.20 | 9/13 |
| PMAIP1 | AA458838 | 2.4 | 0.64 | 7/13 |
| NR4A1 | N94487 | 2.4 | 0.20 | 8/13 |
| RASGRP1 | AA884877 | 2.4 | 0.39 | 6/11 |
| TLR3 | R76099 | 2.3 | 0.10 | 10/13 |
| KIAA0852 | W80688 | 2.3 | 0.33 | 6/11 |
| SNAI1 | AA464983 | 2.3 | 0.27 | 7/13 |
| DAF | R09561 | 2.3 | 0.15 | 10/13 |
| CNK | AA489234 | 2.3 | 0.33 | 5/13 |
| PTPN1 | T57321 | 2.2 | 0.16 | 8/13 |
| DDX3 | AA626845 | 2.2 | 0.22 | 8/13 |
| KIAA0680 | AA700164 | 2.2 | 0.15 | 7/13 |
| LPL | AA633835 | 2.2 | 0.16 | 8/13 |
| REL | AI247359 | 2.2 | 0.19 | 7/13 |
| IER3 | AA480815 | 2.2 | 0.19 | 6/13 |
| CSF1 | T55558 | 2.1 | 0.26 | 5/11 |
| AKT3 | AI864214 | 2.1 | 0.31 | 5/12 |
| CTF1 | AA884403 | 2.1 | 0.38 | 5/13 |
| PIM1 | AA453663 | 2.1 | 0.17 | 6/13 |
| IRF1 | AA478043 | 2.1 | 0.15 | 8/13 |
| ZNF297B | AA465708 | 2 | 0.17 | 6/13 |
| BCL2L2 | AA456480 | 2 | 0.21 | 6/13 |
| FGF9 | AA946776 | 2 | 0.14 | 8/13 |
| TGFBR3 | H62473 | 2 | 0.16 | 5/12 |
| HIF1A | AA598526 | 1.9 | 0.12 | 7/13 |
| KAL1 | H92621 | 1.9 | 0.19 | 6/12 |
| SOX9 | AA400739 | 1.9 | 0.15 | 5/11 |
| ESMI | W46577 | 1.7 | 0.15 | 5/12 |
| DTR | R14663 | 1.2 | 0.63 | 5/13 |
| PPP1R15A | AA460168 | −1.5 | 0.23 | 5/13 |
| NET1 | R24543 | −1.6 | 0.17 | 5/13 |
| ICAM5 | R87840 | −1.7 | 0.15 | 5/13 |
| PM5 | AA629923 | −1.8 | 0.10 | 5/13 |
| GFPT1 | AA478571 | −1.9 | 0.12 | 5/13 |
| GCNT1 | AI657057 | −1.9 | 0.17 | 5/13 |
| CYP1B1 | AA448157 | −2 | 0.27 | 7/13 |
| PPP1R8 | N99208 | −2.1 | 0.08 | 7/13 |
| PPAT | AA873575 | −2.1 | 0.21 | 7/13 |
| CXCR4 | T62491 | −2.1 | 0.21 | 7/13 |
| KIAA0711 | AA702544 | −2.3 | 0.23 | 7/13 |
| PCDH8 | H29216 | −2.3 | 0.20 | 8/13 |
| SREBF1 | AA425823 | −2.4 | 0.29 | 7/13 |
| CITED2 | AA115076 | −2.4 | 0.42 | 6/13 |
| CCND3 | AI340905 | −2.6 | 0.76 | 5/13 |
| ITPKB | R94153 | −2.8 | 0.29 | 9/12 |
| CENPA | AI369629 | −3.2 | 0.35 | 11/13 |
| SERPINB2 | T49159 | −3.2 | 0.66 | 7/13 |
| B3GNT1 | H93550 | −3.5 | 0.56 | 5/6 |
| THBD | H59861 | −3.6 | 0.38 | 12/13 |
| MYC | AA464600 | −3.7 | 0.42 | 11/13 |
| GFI1 | AA418008 | −5 | 0.54 | 13/13 |
GenBank accession of the clone on the array.
Data were collected from 13 independent cell treatments. Genes shown changed twofold up or down in at least five treatments. Change is the average from all measurements for this gene centered around 1, meaning no change.
Number of times the gene showed a twofold change/number of measurements for this gene.
The IL8 probe was cloned at the Lerner Research Institute.
Western blotting.
Whole-cell extracts (30 μg) were fractionated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels, transferred to a polyvinylidene difluoride membrane, and probed with anti-phospho-p38 and anti-p38 antibodies (New England Biolabs).
RNA labeling and array hybridization.
For each array hybridization, RNAs from treated and untreated THP-1 cells were labeled with indocarbocyanine and indodicarbocyanine, respectively. The labeling reactions were standard cDNA syntheses incorporating labeled dUTP. The primer-annealing mixture contained 80 μg of total RNA, 2 μg of dT12-18 primer, 20 U of anti-RNase (Ambion), and 0.5 ng of each positive-control RNA in 33 μl. The four positive-control RNAs were obtained by in vitro transcription from poly(A) tail-modified bacterial gene constructs pGIBS-DAP, -PHE, -THR, and -TRP (American Type Culture Collection). The mixture was heated to 70°C for 10 min and cooled on ice.
Subsequent cDNA synthesis was at 42°C for 2 h in 50 μl with 400 U of Superscript II and 1× first-strand buffer (Invitrogen), 10 mM dithiothreitol, 0.5 mM each dATP, dCTP, and dGTP, 0.3 mM dTTP, and 3 nmol of carbocyanine-dUTP (New England Nuclear). The cDNA was purified with GFX columns following the manufacturer's instructions (Amersham Pharmacia Biotech), dried down, and resuspended in hybridization buffer containing 2× SSC, 0.1% SDS, 4 μg of poly(dA)40-60, and 4 μg of yeast tRNA. The indocarbocyanine- and indodicarbocyanine-labeled cDNAs were pooled and hybridized to the array slide under a coverglass in a Corning Microarray Technology hybridization chamber at 65°C for 16 h. Subsequently, slides were washed successively for 5 min each with 2× SSC-0.1% SDS, 2× SSC, and 0.2× SSC, spun dry, and scanned on a GenePix 4000A scanner (Axon).
Array data acquisition and normalization.
Raw fluorescence data were acquired with the GenePix software (Axon). Laser settings were chosen to avoid signal saturation and achieve an overall median indocarbocyanine/indodicarbocyanine ratio of 0.8 to 1.2. The raw data were imported into the GeneSpring software version 4.2 (Silicon Genetics) for further analysis. For each gene probe, the signal intensity ratio of the treated over the untreated sample was calculated with raw fluorescent intensities, with the local background subtracted. Fluorescent intensities of the untreated samples were set to a minimum value of 300 if below that. The ratios were then normalized based on the distribution of all values with locally weighted polynomial regression (LOESS). For the decay time courses, an additional normalization was necessary because the LOESS normalization localizes the overall mean of the ratios at 1, whereas the mean of ratios naturally decreases below 1 with successive decay of the treated sample. In order to recover reality, the ratios for each array were corrected so that the mean ratio of the four positive-control probes that had been spiked into every sample in a constant amount equaled 1. To be able to compare the RNA decay observed with and without the p38 inhibitor, the average ratios from the three sets of decay time courses were finally corrected for glyceraldehyde-3-phosphate dehydrogenase based on 14 glyceraldehyde-3-phosphate dehydrogenase probes on the array.
Half-life calculations.
The natural logarithms of the average normalized ratios of treated over untreated samples (y) from three independent biological repeats with their respective standard deviations (σ) were plotted against the time (x). The samples taken after 2 h of LPS treatment represent the 0-min time point from which the RNA decay measurement started. In order to exclude genes expressed at levels that are below or close to the detection limit, genes with normalized raw fluorescent intensities in the treated sample of less than the overall average background (B) plus 1 standard deviation at the 0-min time point were excluded from analysis. Lines were fit to the log-transformed data with the least-squares regression. The t1/2 was calculated as (−ln [2]/m), where m is the slope of the line fit to the data (3).
The linear regression fit was performed on from two to seven time points, and the χ2 for each fit was calculated. The fit that gave the minimum χ2 per degree of freedom was chosen for calculating the t1/2. Differences in t1/2 between decay with and without the p38 inhibitor were evaluated with a t-distributed statistic for distributions with unequal variances. The t statistic and the degree of freedom for the t statistic were processed to give a P value with SurfStat statistical tables online (K. Dear and R. Brennan, University of Newcastle [http://math.uc.edu/∼brycw/classes/148/tables.htm]) (58). To avoid distorting the half-life calculation for rapidly decaying genes by using measurements taken at later time points, when the signal detected for this message has already reached background level, we made a second expression level cut and excluded measurements from time points when the normalized raw fluorescent intensity from the treated sample reached a level below B + 2 standard deviations.
Northern blotting.
Total RNA was fractionated on 1% agarose gels containing 0.41 M formaldehyde, capillary transferred to positively charged nylon membrane with 10× SSC, and fixed by UV cross-linking. cDNA probes (25 ng) for COX2, interleukin-1β, interleukin-8, and glyceraldehyde-3-phosphate dehydrogenase were labeled with 50 μCi of [α-32P]dCTP by random priming. Hybridization overnight was done at 65°C in 0.5 M sodium phosphate buffer, pH 7.2, with 7% SDS, 1 mM EDTA, and 10% dextran sulfate. Blots were washed twice in 2× SSC-0.1% SDS at 65°C and exposed to X-OMAT AR film (Kodak). Signals were quantified by densitometry with ImageQuant software (Molecular Dynamics).
RESULTS
ARE-containing gene transcription profile in THP-1 monocytes in response to LPS.
Exposure of human monocytes, including THP-1 cells, to LPS is known to induce various genes, including those encoding AU-rich transcripts (38, 50). In order to boost the levels of AU-rich mRNAs and activate p38, THP-1 cells were treated with LPS for 2 h prior to initiating mRNA decay. As expected, the LPS treatment induced cellular differentiation from a round suspension cell to an adherent macrophage-like phenotype (1). Cell differentiation was accompanied by the transcriptional response of ARE-containing genes, which was measured by cDNA array hybridization on the AU array. Ninety genes were induced and 22 were repressed to at least twofold in LPS-treated compared to untreated cells in at least 5 of 13 independent treatments (Table 1).
The highest levels of gene induction were observed for cytokines, including interleukin-1α, tumor necrosis factor alpha, interleukin-8, and many others that are part of the inflammatory response. In addition, several genes were identified that, to the best of our knowledge, have not yet been described as LPS responsive. Examples are the cleavage- and polyadenylation-specific factor 6 (also known as CF Im), a nuclear protein implicated in mRNA 3′-end processing (61), which was induced 20.7-fold. Early growth response 2 (EGR2), a zinc finger transcription factor homologous to mouse Krox-20 that is coregulated with other immediate-early genes and plays a critical role in peripheral nerve myelination (11, 31, 49, 70), was induced 14.7-fold. The regulator of G-protein signaling 16 (RGS16), a member of the RGS gene family that has been implicated in attenuation of p38 activation via G protein-linked receptors (21, 70), was induced 6.9-fold. Some novel LPS-induced genes such as the antiapoptotic genes BCL2 and BCL2-like 2 and the transcription factor nuclear factor erythroid 2-like 2 (also abbreviated Nrf2) may play a role in self-defense mechanisms that are initiated by macrophages to protect themselves from nitric oxide.
The two most highly and consistently repressed genes included growth factor-independent 1 (GFI1) and c-Myc, decreasing in expression 5- and 3.7-fold, respectively. The latter was consistently repressed in accord with its role in maintaining proliferation and preventing differentiation and apoptosis. GFI1 has recently been suggested to limit inflammatory responses by interfering with the production of cytokines such as tumor necrosis factor, interleukin-10, and interleukin-1β (32), and hence, its repression by LPS appears logical in the context of a macrophage-initiated inflammatory response. Clearly, although our data are restricted to ARE-containing genes, the analysis of the transcriptional response of THP-1 cells to LPS can improve our understanding of macrophage defense against this bacterial toxin.
Identification of new AU-rich mRNAs stabilized by p38.
The stress-activated p38 MAP kinase has previously been implicated in regulating the expression levels of a few genes via ARE-mediated mRNA turnover. To examine the effect of p38 activity on the mRNA stability of several hundred AU-rich genes in parallel, LPS-stimulated THP-1 cells were treated with actinomycin D or a combination of actinomycin D and SB for 30 to 240 min. Total RNA from treated and untreated cells was extracted, labeled, and hybridized to the AU array. Three series of independent cell treatments, RNA isolations, and array hybridizations were performed. The average half-lives, without and with SB, of all AU-rich genes that were detectable at reliable levels were calculated from the decrease in the signal ratio of treated over untreated samples.
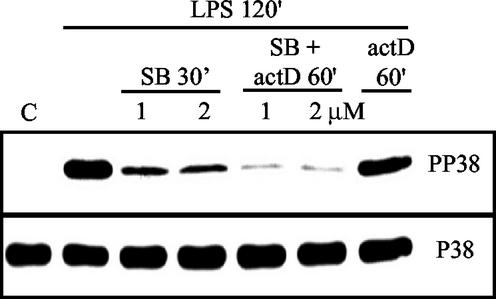
LPS treatment highly activated p38 in THP-1 cells after 2 h of exposure, and the addition of 1 or 2 μM SB with or without actinomycin D inhibited p38 to equal levels. Importantly, the addition of actinomycin D alone did not influence the p38 activity but, when combined with SB, caused nearly complete inactivation of p38 (Fig. 1). For the following decay time courses, 1 μM SB was used because the inhibitor has been shown to be largely p38 specific at this low concentration (16, 17). Minor inhibition of the c-Jun N-terminal kinase (JNK) of 10 to 15% may occur at 2 μM SB (17).
FIG. 1.
Inhibition of p38 MAP kinase by SB. Shown are phospho-p38 (PP38) and total p38 (P38) Western blots of THP-1 cell extracts. Cells were not treated (C) or treated with LPS for 2 h. After 2 h of LPS incubation, treatments were either with SB alone at 1 or 2 μM for 30 min, with a combination of SB and actinomycin D (actD) for 60 min, or with actinomycin D alone for 60 min.
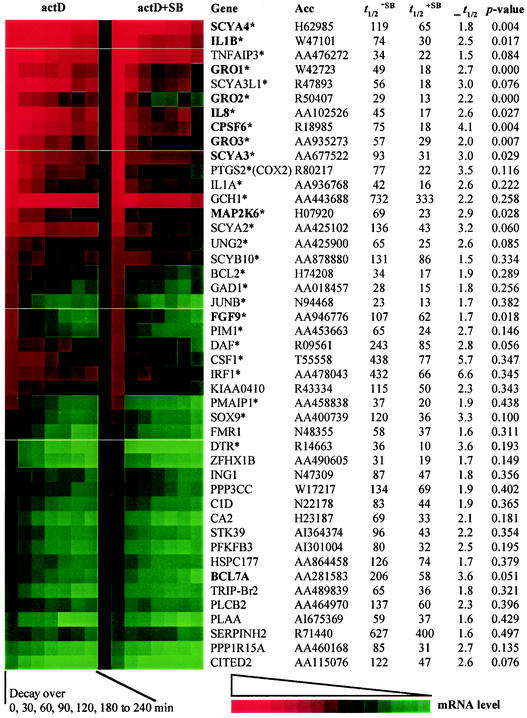
Average mRNA half-lives were calculated for 470 ARE-containing and approximately 40 housekeeping genes. The remaining 480 ARE-containing genes on the array were excluded from the analysis because their initial expression level was already below our signal intensity cutoff and consequently the decay could not be measured with any statistical certainty. We identified 45 AU-rich messages that decayed at least 1.5 times and up to 6.6 times faster in the presence of the p38 inhibitor (Fig. 2). The differences in t1/2, without and with SB, reached statistical significance (P < 0.05) for 11 of the 45 genes. However, it needs to be considered that this P value is based on the standard deviation and the degrees of freedom of t1/2 (= number of time points going into the calculation). This is important because rapidly decaying messages or genes expressed at low levels will only allow accurate calculation of t1/2 over the first two or three time points. At later time points, the mRNA will already have decayed to background levels. For example, COX2 (Unigene symbol PTGS2) mRNA decay was accelerated upon p38 inhibition 3.5-fold, from 77 to 22 min, as previously shown in other studies, yet the P value associated with this difference in our study was 0.116. Hence, we suggest that the stability of all transcripts presented in Fig. 2. is regulated via a p38-dependent pathway.
FIG. 2.
Differential mRNA decay upon p38 inhibition detected on the AU-rich gene array. mRNA decay rates were measured from three independent experiments on THP-1 cells with the AU-rich gene array. After 2 h of LPS treatment, transcription was blocked with actinomycin D (actD) with or without addition of SB. Genes are named with the Unigene database symbol. The stars (*) next to the name indicate genes that were induced after 2 h of LPS treatment (see Table 1). GenBank accession numbers (Acc) represent the clones used on the array. Half-lives (t1/2), in minutes, without and with SB, were calculated as described in the text. ρ t1/2 is the half-life ratio: t1/2−SB/t1/2+SB and the P values indicate the statistical significance of this ratio. Shown here are all genes identified in this study that decayed at least 1.5-fold faster upon p38 inhibition (ρ t1/2 > 1.5). The colored boxes (produced with TreeView by Michael Eisen, Lawrence Berkeley National Laboratory) indicate decline in mRNA levels over the decay time course from 0 to 240 min (red to green). Genes are sorted in descending order by initial expression level at 0 min. The two columns of colored boxes are separated by pale lines into five blocks. Each block has a different contrast setting, and hence the colors between blocks are not directly comparable. This was necessary to allow better horizontal comparison of the left and right column for each gene.
It should be noted that there are limitations in using actinomycin D as a tool to measure mRNA half-life, and different approaches may have to be used to arrive at consensus values. In addition to previously identified targets like COX2, interleukin-8, and MIP1α (SCYA3), we found 42 genes which we submit as newly identified targets for p38-mediated message stabilization. Though the majority of these were inflammatory response mediators, such as the members of the small inducible cytokine subfamilies A (SCYA2, -3, -3L1, and -4) and B (GRO-1, -2, and -3, interleukin-8, and SCYB10), and the acute-phase response mediators interleukin-1α and -1β, a variety of messages that do not appear to be directly involved in the immune response were also found to be stabilized by p38. Among these were mRNAs coding for apoptosis regulators (TNFAIP3, PMAIP6, and BCL2), transcription factors (JunB, IRF1, and SOX9), signaling kinases (MAP kinase kinase 6 and PIM1), phosphatases (PPP3CC), growth factors and receptors (FGF9 and DTR), various enzymes with metabolic, ion homeostasis, and DNA repair functions (GCH1, GAD1, CA2, PFKFB3, and UNG2), and nuclear mRNA processing factors (cleavage- and polyadenylation-specific factor 6). In summary, this variety of functionally diverse genes attests to the importance of the p38 pathway in the posttranscriptional regulation of gene expression for the class of ARE-containing genes.
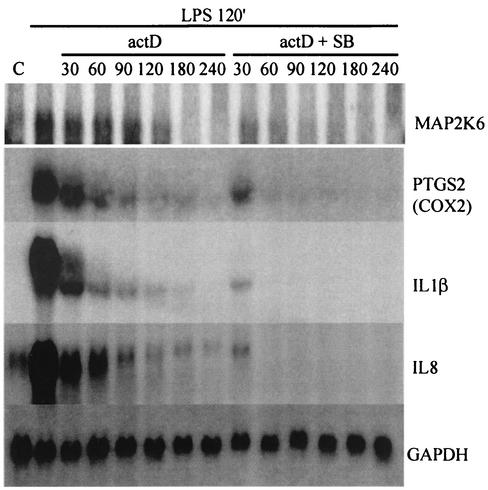
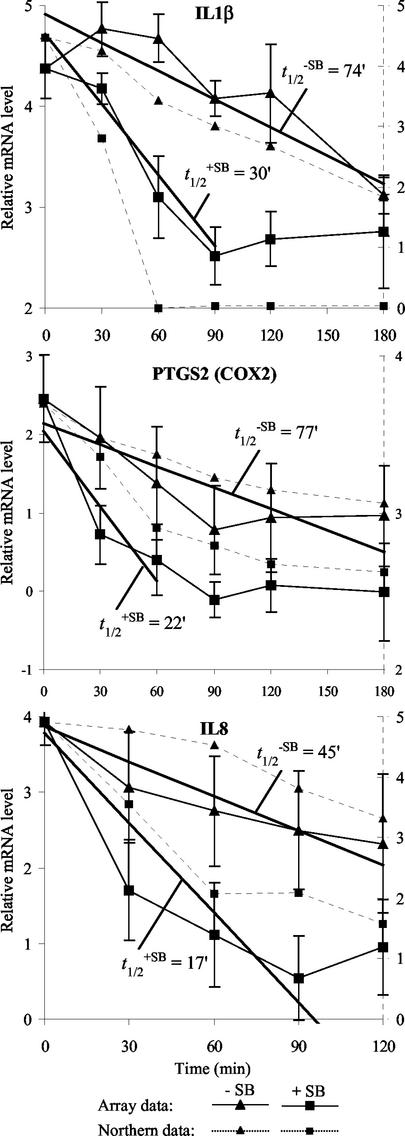
To confirm the array results for different genes with an alternative method, Northern blotting analyses were performed on RNA from one of the three time courses, probing for MAP kinase kinase 6, COX2, interleukin-8, interleukin-1β, and glyceraldehyde-3-phosphate dehydrogenase (Fig. 3). Densitometry of the Northern blots for the last four was compared with the mRNA decay measured on the AU array. The results (Fig. 4) indicate that there is a tight correlation with the results obtained by densitometry of the Northern blots compared to array analysis for the three ARE-containing genes tested.
FIG. 3.
Differential mRNA decay of MAP kinase kinase 6 (MAP2K6), COX2, interleukin-1β (IL1β), and interleukin-8 (IL8) on p38 inhibition detected by Northern blot. The samples used here are identical to one of the three used in the mRNA decay time course experiments in the array hybridizations. See the text for a description of the cell treatments. Times are in minutes. Lane C, no-treatment control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIG. 4.
p38-dependent differential mRNA decay of interleukin-1β (IL1β), COX2, and interleukin-8 (IL8): correlation of cDNA array and Northern blot data. Shown here are the graphs of array (solid lines) and Northern (dotted lines) data for interleukin-1β, COX2, and interleukin-8 mRNA decay. The array data are averages of log-transformed normalized ratios of treated over untreated samples from three repeats with standard deviations (solid y axis). The linear regression through the time points used in the half-life (t1/2) calculation is shown as a bold line with the respective t1/2 indicated (in minutes) (see text for details). The Northern data are glyceraldehyde-3-phosphate dehydrogenase-normalized, log-transformed signal intensities determined by densitometry from the Northern blots shown in Fig. 3 (dotted y axis).
Stability of 425 AU-rich mRNAs in LPS-treated THP-1 cells regulated independently of p38.
In addition to identifying genes that appeared to be stabilized by p38, we determined the half-lives of 425 AU-rich mRNAs that were not destabilized greater than 1.5-fold upon p38 inhibition in our study (Table 2). Other studies have reported that interleukin-3, interleukin-6, tumor necrosis factor alpha, and urokinase-type plasminogen activator are stabilized by p38 activation (4, 47, 48, 70). However, we did not detect any significant changes in the half-lives of these messages in our THP-1 cell experiments. For example, tumor necrosis factor alpha had the shortest half-live of all AU-rich mRNAs in all three repeat experiments in the presence and absence of the p38 inhibitor (averaged t1/2−SB = 7.6, t1/2+SB = 6.9 min). This discrepancy with other studies may be due to differences in the cell system, such as the use of fresh human blood monocytes, HeLa, or NIH 3T3 cells, or differences in experimental design or method of detection, and it seems likely that the list of genes presented in Table 2 may contain mRNAs that might be stabilized or destabilized by p38 activation under different conditions.
TABLE 2.
P38-independent half-lives of 425 AU-rich mRNAsa
| Gene | ID | t1/2 |
|---|---|---|
| AB026190 | AA479295 | 63 |
| ABCE1 | T70122 | 65 |
| ABR | W24076 | 98 |
| ACADM | N70794 | 108 |
| ACADSB | H96140 | 40 |
| ACTR1A | AI014416 | 273 |
| ADAM9 | H59231 | 75 |
| ADK | R12473 | 328 |
| AGL | AA668425 | 51 |
| AKT1 | W77811 | 49 |
| ALTE | AA630498 | 187 |
| AP3M2 | R14443 | 222 |
| APACD | AA085749 | 110 |
| APM1 | H45617 | 84 |
| ARCN1 | AA598401 | 74 |
| ARFD1 | AA706974 | 28 |
| ARHGEF7 | AA452871 | 254 |
| ARHGEF9 | AA147072 | 113 |
| ARPC5 | W55964 | 124 |
| ATP1B1 | AA598814 | 42 |
| ATP2A2 | H85355 | 98 |
| ATP6E | AI986422 | 245 |
| ATP6F | AA480826 | 941 |
| ATRX | AA410435 | 66 |
| AUH | AA448711 | 52 |
| B3GNT1 | H93550 | 29 |
| B4GALT3 | AA424578 | 63 |
| BACH | AA035455 | 199 |
| BCL2L2 | AA456480 | 47 |
| BIRC2 | R19628 | 101 |
| BIRC3 | AA002126 | 67 |
| BLZF1 | R43576 | 89 |
| BMP6 | AA424833 | 31 |
| BNIP3 | AA063521 | 57 |
| BPNT1 | AA197334 | 46 |
| BRCA2 | H48122 | 55 |
| BRD2 | AA918860 | 89 |
| BRP441 | AA459690 | 174 |
| BSN | H18306 | 68 |
| BTAF1 | AA120777 | 76 |
| BTF | H21107 | 51 |
| BTN2A1 | H68107 | 110 |
| BUB3 | H38804 | 105 |
| BYSL | AA701929 | 148 |
| C1orf16 | AA431423 | 73 |
| C1orf29 | AA410567 | 54 |
| C20orf45 | AA004210 | 63 |
| C4orf1 | AA757427 | 76 |
| C7 | AA598478 | 81 |
| C8orf1 | AA027049 | 139 |
| CALU | R78585 | 231 |
| CAP350 | AI001846 | 41 |
| CASP7 | T50828 | 274 |
| CBLN1 | AA495901 | 143 |
| CBX3 | AA682719 | 69 |
| CCT2 | N38959 | 70 |
| CD164 | AA598561 | 191 |
| CD36 | N39161 | 295 |
| CDC27 | T81764 | 122 |
| CDC42 | AA009697 | 667 |
| CDC7L1 | N62245 | 54 |
| CDK2 | AI653017 | 424 |
| CDKN2D | R77517 | 93 |
| CENTB2 | AA490493 | 104 |
| CHS1 | N74383 | 70 |
| CHSY1 | AA703453 | 61 |
| CIAS1 | AW468866 | 39 |
| CNK | AA489234 | 57 |
| COX17 | AA099855 | 76 |
| CREBBP | AA023014 | 57 |
| CREG | T71991 | 389 |
| CRLF3 | AI240562 | 37 |
| CRSP2 | AA150093 | 650 |
| CRTAP | AA486278 | 62 |
| CSF2 | AA995402 | 39 |
| CSPG2 | AA101875 | 193 |
| CTF1 | AA708512 | 39 |
| CUL3 | H98621 | 72 |
| CXCR4 | T62491 | 51 |
| CYP1B1 | AA448157 | 86 |
| CYP51 | AA477893 | 51 |
| DAZ | AA133797 | 71 |
| DBT | R89083 | 32 |
| DBY | W37634 | 101 |
| DDEF2 | N70773 | 153 |
| DDX11 | AA032090 | 88 |
| DDX16 | AA457157 | 137 |
| DDX18 | R08935 | 76 |
| DDX3 | AA626845 | 58 |
| DDX8 | AA458473 | 64 |
| DIS3 | H03208 | 45 |
| DLEU2 | N25204 | 137 |
| DMTF1 | AA129860 | 103 |
| DMXL1 | N29992 | 48 |
| DSCR3 | R97540 | 119 |
| DUSP2 | AA759046 | 17 |
| DYRK1A | AA676749 | 48 |
| E2F1 | AA424950 | 88 |
| EDN3 | T67005 | 90 |
| EDNRB | H28710 | 65 |
| EEF2 | R20379 | 187 |
| EGR2 | AA446027 | 16 |
| EHHADH | R02373 | 30 |
| EIF2AK3 | AA436178 | 139 |
| EIF3S1 | AA455070 | 54 |
| EIF4E | AA193254 | 90 |
| EIF4G3 | N92469 | 184 |
| ENC1 | H72122 | 46 |
| ENDOFIN | R26672 | 42 |
| EPB49 | N55461 | 31 |
| ESM1 | W46577 | 77 |
| EVI5 | T65001 | 53 |
| EZH2 | AA430744 | 132 |
| F2R | N20407 | 52 |
| F3 | AI313387 | 56 |
| FACL4 | AA633818 | 112 |
| FCER1G | H79353 | 697 |
| FKBP4 | AA932521 | 157 |
| FUT4 | R28447 | 67 |
| G3BP2 | AA151214 | 177 |
| GABPB2 | AW571916 | 224 |
| GALC | W85914 | 452 |
| GAS41 | T62072 | 65 |
| GC20 | AA488391 | 341 |
| GCA | R44739 | 85 |
| GCP3 | AI004751 | 253 |
| GFI1 | AA418008 | 47 |
| GLO1 | AA136710 | 103 |
| GLS | R89349 | 41 |
| GMFB | H22652 | 190 |
| GNE | T68440 | 44 |
| GNG10 | AA460286 | 121 |
| GOT1 | H22856 | 563 |
| GPC1 | AA455896 | 208 |
| GRB10 | AA136336 | 67 |
| GS2NA | AA418918 | 122 |
| GS3786 | H88599 | 79 |
| GTF2A2 | T55801 | 89 |
| GTF2B | H23978 | 75 |
| GYS2 | N72934 | 62 |
| HCS | R52654 | 172 |
| HD | T64094 | 83 |
| HERC3 | AA282253 | 57 |
| HIF1A | AA598526 | 80 |
| HIVEP2 | AA683219 | 194 |
| HK1 | AA485272 | 38 |
| HNRPA2B | W02101 | 81 |
| HSPC019 | AI017010 | 171 |
| HSU79274 | AA451900 | 91 |
| ID2 | AA482267 | 31 |
| IDH3A | AA464206 | 71 |
| IER3 | AA480815 | 71 |
| IFNA1 | M29884* | 35 |
| IFNAR1 | N59150 | 67 |
| IFNB | M28622* | 138 |
| IGF1 | N67876 | 47 |
| IL10 | M57627* | 155 |
| IL10RA | AA437226 | 81 |
| IL12B | M65272* | 93 |
| IL17 | U32659* | 101 |
| IL1RAP | AA256132 | 134 |
| IL2 | S77834* | 38 |
| IL24 | AA281635 | 247 |
| IL3 | M14743* | 124 |
| IL4 | M13982* | 37 |
| IL5 | X12705* | 89 |
| IL6 | M14584* | 204 |
| IMPA1 | H90219 | 61 |
| IQGAP1 | AA598496 | 118 |
| ITGA2 | AA463610 | 51 |
| ITGAV | AA029934 | 251 |
| ITSN1 | AA496795 | 181 |
| JUN | W96134 | 17 |
| KAL1 | H17882 | 47 |
| KDR | AA026831 | 57 |
| KIAA0010 | AA284599 | 91 |
| KIAA0022 | H60460 | 64 |
| KIAA0028 | H19822 | 193 |
| KIAA0040 | AA465479 | 62 |
| KIAA0141 | AA455516 | 103 |
| KIAA0171 | H15458 | 73 |
| KIAA0212 | AA630346 | 115 |
| KIAA0232 | AA406589 | 75 |
| KIAA0247 | N63733 | 81 |
| KIAA0254 | N75979 | 116 |
| KIAA0266 | AA598993 | 37 |
| KIAA0296 | AA890161 | 37 |
| KIAA0298 | AA853966 | 20 |
| KIAA0354 | AA062802 | 62 |
| KIAA0375 | W49494 | 75 |
| KIAA0419 | AA625653 | 58 |
| KIAA0426 | AA708279 | 45 |
| KIAA0431 | AA172053 | 44 |
| KIAA0438 | AA142966 | 63 |
| KIAA0441 | N24789 | 27 |
| KIAA0475 | AA419200 | 75 |
| KIAA0476 | AA282577 | 730 |
| KIAA0537 | AA774839 | 110 |
| KIAA0560 | AA121387 | 69 |
| KIAA0628 | N56973 | 42 |
| KIAA0669 | W60983 | 50 |
| KIAA0680 | AA700164 | 136 |
| KIAA0685 | AA490924 | 272 |
| KIAA0711 | AA702544 | 60 |
| KIAA0798 | T90374 | 32 |
| KIAA0808 | N66992 | 60 |
| KIAA0844 | AW131755 | 316 |
| KIAA0938 | AA705735 | 40 |
| KIAA0970 | R36431 | 47 |
| KIAA0982 | AA017133 | 71 |
| KIAA0997 | R28471 | 38 |
| KIAA1041 | AA629800 | 70 |
| KIAA1046 | R78541 | 66 |
| KLHL2 | AI348818 | 541 |
| KMO | R44396 | 105 |
| KNSL1 | AA504625 | 83 |
| KNTC1 | AA157787 | 79 |
| KPNA1 | AA180046 | 175 |
| KRAS2 | AA505084 | 65 |
| LEPR | AI208285 | 72 |
| LEPROTL1 | T62031 | 134 |
| LIF | R50354 | 117 |
| LIM | R92455 | 52 |
| LIV-1 | H29407 | 127 |
| LMAN1 | AA446103 | 112 |
| LOC51026 | R69622 | 87 |
| LOC51071 | N74602 | 80 |
| LPL | AA633835 | 175 |
| LRP8 | AA527256 | 168 |
| LTA | W72329 | 66 |
| M6PR | AA465223 | 189 |
| MADD | AA282445 | 154 |
| MAN1 | AA520992 | 67 |
| MATN3 | AI375563 | 41 |
| MBL2 | T69359 | 17 |
| MBTPS1 | AA447393 | 112 |
| MEF2C | AA234897 | 60 |
| METAP2 | AA283030 | 180 |
| MICB | H69835 | 59 |
| MLLT2 | AA057425 | 53 |
| MMD | AA487643 | 147 |
| MMP1 | AA143201 | 112 |
| MNAT1 | AA481759 | 247 |
| MPP2 | H39068 | 68 |
| MRPL33 | AA489478 | 81 |
| MTHFD2 | AI361330 | 192 |
| MTM1 | AA491225 | 128 |
| MTMR2 | AA436164 | 94 |
| MTR | AA233650 | 64 |
| MYC | AA464600 | 32 |
| MYO1E | AA029956 | 49 |
| NAB1 | N91896 | 118 |
| NCAM2 | AA709271 | 95 |
| NEK4 | AA496013 | 44 |
| NEURL | N30706 | 55 |
| NFE2L2 | AA629687 | 51 |
| NFKBIA | W55872 | 23 |
| NGFB | T56316 | 46 |
| NICE-3 | N76101 | 138 |
| NMT2 | AA664135 | 33 |
| NOVA1 | AI362062 | 144 |
| NR3C1 | AA664219 | 93 |
| NR4A1 | N94487 | 278 |
| NR4A3 | H37761 | 99 |
| NR6A1 | AA853954 | 226 |
| OGDH | AA856769 | 744 |
| ORC6L | N90667 | 57 |
| OSR1 | R98985 | 213 |
| P115 | AA504342 | 79 |
| PAIP1 | AA598533 | 104 |
| PARG1 | AA629603 | 26 |
| PCDHA9 | AA437139 | 56 |
| PCK1 | AA405769 | 47 |
| PDE4B | AA453293 | 23 |
| PDE7A | AA992565 | 48 |
| PDGFB | W68169 | 70 |
| PDX1 | N48320 | 125 |
| PER1 | T95053 | 34 |
| PEX1 | AA598527 | 148 |
| PGK1 | AA599187 | 163 |
| PIGN | AA033974 | 138 |
| PIK3CD | AA281784 | 100 |
| PKD1 | N27758 | 56 |
| PLAT | R38933 | 163 |
| PLAU | AA284668 | 62 |
| PLCL1 | AI222930 | 37 |
| PLOD2 | H99816 | 204 |
| PM5 | AA025160 | 895 |
| POU6F1 | N63968 | 71 |
| PPP1R8 | N99208 | 143 |
| PPP2CA | AA599092 | 79 |
| PPP3CA | AA682631 | 52 |
| PPP3CB | AA015621 | 82 |
| PRC1 | AA449336 | 69 |
| PREI3 | AA410302 | 53 |
| PREP | AA664056 | 93 |
| PRG4 | AA280514 | 127 |
| PRKX | AA778448 | 38 |
| PSMD10 | R77104 | 256 |
| PSMD12 | AA497132 | 85 |
| PTBP1 | AA677517 | 156 |
| PTK2B | R85257 | 331 |
| PTPN1 | T57321 | 365 |
| PTPRA | H82419 | 79 |
| PTPRC | H74265 | 249 |
| PWP2H | H50886 | 299 |
| RABIF | AA012984 | 53 |
| RAD21 | AA683102 | 84 |
| RB1 | AA045192 | 87 |
| RB1CC1 | AA047435 | 59 |
| RBBP8 | H23021 | 96 |
| RBL2 | N50554 | 79 |
| RCN2 | AA598676 | 81 |
| REL | AI247359 | 111 |
| RGS16 | AA453774 | 14 |
| RI58 | W24246 | 71 |
| RIPK2 | AA913804 | 72 |
| RMS1 | R77718 | 42 |
| RNF11 | W94868 | 80 |
| RNF14 | N62157 | 70 |
| RNGTT | AW137353 | 57 |
| ROR2 | AA149251 | 96 |
| RTVP1 | AA251800 | 91 |
| RYK | T77810 | 84 |
| SAC2 | R69354 | 43 |
| SBB103 | AI299601 | 166 |
| SC5DL | AA216535 | 63 |
| SCAP2 | R81177 | 49 |
| SCYA11 | W69211 | 53 |
| SCYA16 | T58775 | 59 |
| SCYD1 | R66139 | 75 |
| SDFR1 | AA130671 | 187 |
| SEMA3B | AA455145 | 82 |
| SEP15 | AA999842 | 191 |
| SERPINB2 | T49159 | 33 |
| SFRS11 | AA481054 | 239 |
| SHOX2 | AA425419 | 81 |
| SIP | W87541 | 121 |
| SIRPB1 | AI088704 | 38 |
| SIRT1 | AA460952 | 41 |
| SLC26A4 | AI139968 | 32 |
| SLC2A1 | R17667 | 42 |
| SLC2A3 | H52531 | 33 |
| SLC35A2 | H51549 | 370 |
| SLC9A6 | R45009 | 45 |
| SLK | W17289 | 143 |
| SMN1 | AA004858 | 64 |
| SNAP25 | AA663884 | 456 |
| SNAPC3 | AA043334 | 268 |
| SNK | AA460152 | 23 |
| SPG4 | AA171421 | 61 |
| SPTBN1 | H98241 | 117 |
| SRF | AA487973 | 152 |
| SRPK1 | AA630604 | 140 |
| SRPR | AA598621 | 327 |
| SRRM1 | R26536 | 65 |
| SSFA2 | AA496804 | 167 |
| ST13 | H65676 | 170 |
| STAU | AA669068 | 217 |
| STIM1 | AA157018 | 287 |
| STIP1 | AA487635 | 157 |
| STK17A | AA453754 | 78 |
| STK3 | AA136675 | 43 |
| SYBL1 | R27644 | 172 |
| TACC1 | AA664006 | 105 |
| TAF7 | AA461518 | 79 |
| TBCC | AA954188 | 43 |
| TBP | N50549 | 146 |
| TBX2 | N99243 | 209 |
| TCF12 | AA488497 | 54 |
| TCFL4 | AA134555 | 66 |
| TDE1 | AA679489 | 221 |
| TDG | T91074 | 62 |
| TEB4 | H67086 | 64 |
| TG737 | AA481585 | 63 |
| THBD | H59861 | 53 |
| THBS1 | AA464630 | 54 |
| TIEG | AI348177 | 18 |
| TIMM17A | AA708446 | 69 |
| TMEM1 | N94245 | 122 |
| TNF | AA699697 | 8 |
| TNFAIP6 | W93163 | 145 |
| TNFSF9 | AA778663 | 36 |
| TOMM20 | AA644550 | 74 |
| TOPBP1 | R97785 | 48 |
| TPD52 | AA459318 | 123 |
| TPP2 | T77959 | 63 |
| TRAF4 | AA598826 | 239 |
| TRAF5 | AA102634 | 53 |
| TRAP240 | AA434084 | 105 |
| TRPM1 | N35472 | 40 |
| TSNAX | AA477514 | 133 |
| TWIST | AI220198 | 30 |
| TXNRD1 | AA453335 | 113 |
| UAP1 | N68465 | 116 |
| UBA2 | H11320 | 69 |
| UBE2A | AA600173 | 204 |
| UBE2H | AA520978 | 150 |
| UBE2V2 | AA448676 | 80 |
| UBL3 | AA151852 | 126 |
| UGCG | N90204 | 125 |
| UGDH | AA992570 | 59 |
| UK114 | AI301696 | 207 |
| USP6 | AI203661 | 40 |
| USP8 | AI299198 | 59 |
| VBP1 | AA478108 | 162 |
| VEGF | R19956 | 106 |
| VPS26 | AA064946 | 108 |
| VPS41 | AA143559 | 137 |
| WBP4 | AA702632 | 69 |
| WDR3 | AA775806 | 97 |
| WHSC1 | AA159311 | 121 |
| WRB | AA099383 | 93 |
| WS-3 | AA451781 | 415 |
| WSB1 | AA025807 | 64 |
| WTAP | AA598802 | 200 |
| XBP1 | W90128 | 52 |
| XPR1 | AA453474 | 123 |
| YES1 | H56929 | 49 |
| ZFX | AI740859 | 97 |
| ZMPSTE24 | AA001403 | 115 |
| ZNF198 | AA251581 | 114 |
| ZNF207 | N59119 | 71 |
| ZNF238 | R79722 | 79 |
| ZNF297B | AA702698 | 47 |
Genes are named with the Unigene Database symbol and ordered alphabetically. IDs are GenBank accession numbers of the clone on the array. *, probes generated by reverse transcription-PCR. t1/2 (in minutes) was determined without p38 inhibition; calculation from three independent experiments as described in the text.
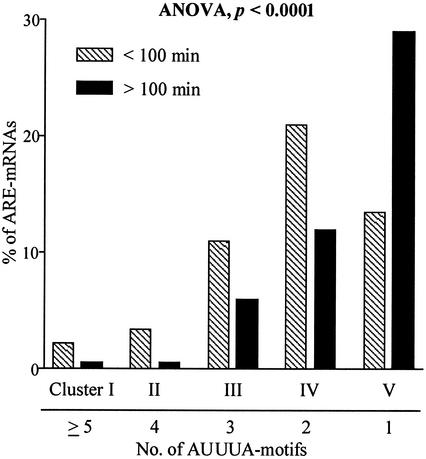
We investigated whether the number of AUUUA motifs that are present in the AREs are indicative of the half-life that one can expect for a particular message (Fig. 5). The genes in the ARED had previously been clustered on the basis of the number of ARE motifs (2). Clustering the mRNAs based on the determined half-lives into two groups, one with a t1/2 of <100 min and the other one with a t1/2 of >100 min (100 min was the mean of the ARE mRNA half-lives), we used analysis of variance (ANOVA) between groups to test the hypothesis that mRNAs with more AUUUA motifs are more likely to have a shorter half-life than those with fewer motifs. The resulting ANOVA P value of less than 0.0001 verified this hypothesis.
FIG. 5.
ARE mRNA half-lives correlate with the number of AUUUA motifs. ARE-containing mRNAs were clustered into five groups according to the number of AUUUA motifs in the 3′ untranslated regions, and their respective half-lives were clustered in two groups (<100 min and >100 min). Analysis of variance between groups (ANOVA) shows that the number of AUUUA motifs is indicative of half-life.
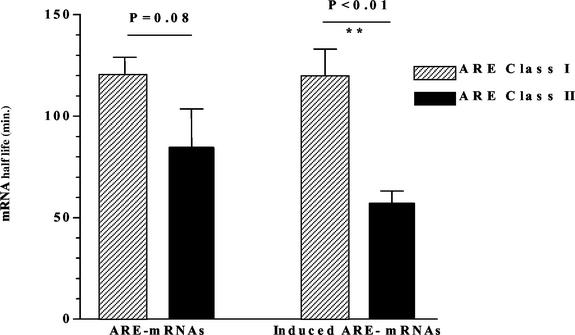
However, our analysis also showed that the presence of the AUUUA motif is not entirely predictive of a short half-life (<100 min). A recent study of mRNA turnover that used the ARE mRNA clustering provided in the our database found that AREs are present more frequently in short-lived mRNAs, yet some messages with half-lives longer than 8 h also contained the AUUUA motif (36). ARE mRNAs were also clustered into the class I and class II categories of Chen et al. (12), in which class I has discontinuous nonamers and class II has at least two overlapping copies of the nonamer. When the half-lives in each category were compared, we found that the means of half-lives in class I (121 ± 8 min) and II (84 ± 19) were marginally different but not statistically significant (Fig. 6). However, when LPS-induced ARE mRNAs were clustered, class II mRNAs had significantly (P = 0.006; unpaired t test with Welsh's variance correction) shorter half-lives (mean = 56 ± 6 min) compared to class I mRNAs (114 ± 19 min).
FIG. 6.
Class II LPS-induced ARE mRNAs have shorter half-lives than class I. ARE mRNAs were clustered into two categories, class I and class II, which have either discontinuous nonamers or at least two overlapping copies of the nonamer, respectively (12). Subsequently, the half-lives in each category were compared.
DISCUSSION
Using a computational approach, we previously identified many hundred genes, compiled in the ARE mRNA database, that have one to several copies of the AUUUA motif in the mRNA 3′ untranslated region (2). Examination of the ARE mRNA targets of the ELAV homolog protein HuB (65), which is specific to neuronal cells, showed that there were few mRNA targets in common with our array targets (notably cyclin D1, c-myc, and integrin B). In contrast, almost all of the previously reported mRNA targets for the ubiquitously expressed ARE-binding protein HuR, including tumor necrosis factor alpha, c-fos, granulocyte-macrophage colony-stimulating factor, cyclooxygenase, interleukin-3, vascular endothelial growth factor, interleukin-8, and transforming growth factor beta, are part of our array targets compiled on the basis of an ARE motif defined by bioinformatics and statistically (2).
In the present study, we used our database to make a custom cDNA array with probes representing 950 AU-rich mRNAs in order to determine the p38 dependence of mRNA stability for several hundred ARE-containing transcripts in human THP-1 monocytic cells. A role for the p38 pathway in controlling mRNA stability has so far been demonstrated for nine ARE-containing genes involved in cellular immunity, and the ARE-mediated message stabilization via p38 and the downstream MAP kinase-activated protein kinase 2 appears to be sequence specific, since the AREs of COX2 but not of c-myc or tumor necrosis factor alpha were able to confer p38-dependent stabilization on a β-globin reporter gene in HeLa-TO cells (39). Here we have identified 42 new candidates for message stability regulation by p38 by interrogating hundreds of AU-rich mRNAs in parallel. In addition, we have presented a transcription profile of AU-rich transcript induction and repression by LPS that is based on 13 independent cell treatments and array hybridizations, and furthermore we have submitted half-lives for 425 AU-rich mRNAs that did not show stabilization by p38 in this study.
When comparing the list of ARE-containing genes that were induced by LPS with those that we found to be destabilized upon p38 inhibition, one can make two important observations that differentiate regulation at the transcriptional and posttranscriptional levels. First, many ARE-containing genes that were induced by LPS were not stabilized at the mRNA level through the p38 pathway. This observation is not surprising because LPS signaling through Toll-like receptor 4 can activate the p38 as well as the nuclear factor-κB kinase pathways via TAK1, which can subsequently turn on different sets of target genes (13, 40, 57, 66). Second, we found a number of mRNAs that were destabilized by the p38 inhibitor but not upregulated upon LPS stimulation. This finding demonstrates for the first time that the p38 pathway can control the stability of some AU-rich mRNAs that are not transcriptionally induced upon p38 activation.
A number of genes identified in this study are of particular interest in terms of the cellular response of monocytes to LPS. Regarding the signaling through the p38 pathway, it should be noted that the MAP kinase kinase 6, one of the major upstream activators of p38 (27, 53), was strongly induced by LPS (8.7-fold) and destabilized upon p38 inhibition. This finding suggests that a positive feedback loop in the p38 pathway exists that may use transcriptional induction as well as message stabilization of MAP kinase kinase 6 in order to potentiate and/or prolong the signal.
Activated macrophages produce nitric oxide as an antimicrobial and antitumor effector molecule. Okada et al. established a link between self-defense against synthesized nitric oxide and induction of Bcl2-like 1 in LPS-treated macrophages, suggesting that upregulation of this antiapoptotic factor may contribute to macrophage survival (52). Similarly, we found that Bcl2 and Bcl2-like 2 were induced by LPS, suggesting that modulation of expression of several Bcl2 family members counteracts apoptosis to promote macrophage survival. A second line of defense employed by macrophages is the upregulation of autoprotective intracellular redox buffering systems (23, 56). A novel candidate potentially involved in this process could be the nuclear factor erythroid 2-like 2 gene, which was induced by LPS 4.6-fold. This basic leucine zipper transcription factor has been implicated in the regulation of expression of detoxifying genes (10, 29) and, when deficient in mice, causes increased susceptibility to oxidative stress (9).
A gene that was stabilized through p38 but not transcriptionally induced by LPS was carbonic anhydrase 2. Carbonic anhydrases are of great physiological importance in many biological processes, including respiration, calcification, acid-base balance, bone resorption, and others (20). As catalysts of the reversible hydration of carbon dioxide, they are critical in maintaining the cellular CO2/HCO3− buffering system. Recent findings suggested that carbonic anhydrase may be important in inflammatory processes, because ambient pCO2 can modulate neutrophil activity by altering intracellular pH, thereby affecting intracellular oxidant generation and interleukin-8 secretion after LPS stimulation (14). Therefore, carbonic anhydrase 2 may also play a role in maintaining intracellular pH levels in activated macrophages, and hence, it would appear logical to stabilize the carbonic anhydrase 2 mRNA in a p38-dependent manner.
The macrophage-specific colony-stimulating factor 1 was also among the p38-stabilized messages. This regulation is noteworthy because macrophage-specific colony-stimulating factor 1 may regulate host responses to pathogens by modulating Toll-like receptor expression as treatment of macrophages with macrophage-specific colony-stimulating factor 1 downregulated Toll-like receptor 1, 2, 6, and 9 expression but not the LPS receptors Toll-like receptor 4 and 5 (64).
From an evolutionary viewpoint, we find it intriguing that the five CXC chemokines interleukin-8, GRO1, GRO2, GRO3, and SCYB10, which all showed p38-dependent message stability in our study, belong to a multigene family on chromosome 4 that most likely arose by gene duplication (51). This correlation may indicate that controlling mRNA stability could be an ancestral mechanism that developed early in the evolution of immune response regulation.
When considering the practical application of this study, it should be noted that ARE-containing genes identified as posttranscriptionally regulated by p38 are involved in different diseases. For example, interleukin-1 is expressed at abnormally high levels by glial cells in Alzheimer's disease, possibly contributing to the pathophysiology of the disease (60). Interleukin-1 was also implicated in cases of rheumatoid arthritis with severe erosive disease (15) and in early-onset periodontitis (19). For ARE mRNAs implicated in disease, the knowledge of this regulation may warrant consideration of drug intervention that may influence gene expression at the level of mRNA stability. However, the degree to which the posttranscriptional regulation of the 42 novel p38 target genes contributes to their overall level of expression and the physiological importance of this regulation will first need to be assessed on a gene-by-gene basis in the appropriate cell systems.
In conclusion, this study has provided new insights into the regulatory networks that control the response of monocytes to LPS and underlined the importance of the posttranscriptional regulation of AU-rich mRNAs through the p38 MAP kinase signaling pathway. The identification of the 42 new AU-rich transcripts that are regulated via p38 at the level of mRNA stability will further aid in understanding the molecular mechanisms by which this regulation occurs.
Acknowledgments
This work was supported in part by National Institutes of Health grants RO1-AI34039 and PO1-CA62220.
REFERENCES
- 1.Adams, D. O., and T. A. Hamilton. 1984. The cell biology of macrophage activation. Annu. Rev. Immunol. 2:283-318. [DOI] [PubMed] [Google Scholar]
- 2.Bakheet, T., M. Frevel, B. R. Williams, W. Greer, and K. S. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevington, P. R., and D. K. Robinson. 1992. Method of least squares, p. 53-62. In P. R. Bevington and D. K. Robinson (ed.), Data reduction and error analysis for the physical sciences. WCB McGraw-Hill, Boston, Mass.
- 4.Brook, M., G. Sully, A. R. Clark, and J. Saklatvala. 2000. Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade. FEBS Lett. 483:57-61. [DOI] [PubMed] [Google Scholar]
- 5.Buzby, J. S., G. Brewer, and D. J. Nugent. 1999. Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J. Biol. Chem. 274:33973-33978. [DOI] [PubMed] [Google Scholar]
- 6.Carballo, E., H. Cao, W. S. Lai, E. A. Kennington, D. Campbell, and P. J. Blackshear. 2001. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 276:42580-42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 8.Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891-1899. [PubMed] [Google Scholar]
- 9.Chan, K., X. D. Han, and Y. W. Kan. 2001. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. USA 98:4611-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, K., and Y. W. Kan. 1999. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. USA 96:12731-12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavrier, P., U. Janssen-Timmen, M. G. Mattei, M. Zerial, R. Bravo, and P. Charnay. 1989. Structure, chromosome location, and expression of the mouse zinc finger gene Krox-20: multiple gene products and coregulation with the proto-oncogene c-fos. Mol. Cell. Biol. 9:787-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 13.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 14.Coakley, R. J., C. Taggart, C. Greene, N. G. McElvaney, and S. J. O'Neill. 2002. Ambient pCO2 modulates intracellular pH, intracellular oxidant generation, and interleukin-8 secretion in human neutrophils. J. Leukoc. Biol. 71:603-610. [PubMed] [Google Scholar]
- 15.Cox, A., N. J. Camp, C. Cannings, F. S. di Giovine, M. Dale, J. Worthington, S. John, W. E. Ollier, A. J. Silman, and G. W. Duff. 1999. Combined sib-TDT and TDT provide evidence for linkage of the interleukin-1 gene cluster to erosive rheumatoid arthritis. Hum. Mol. Genet. 8:1707-1713. [DOI] [PubMed] [Google Scholar]
- 16.Cuenda, A., J. Rouse, Y. N. Doza, R. Meier, P. Cohen, T. F. Gallagher, P. R. Young, and J. C. Lee. 1995. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 364:229-233. [DOI] [PubMed] [Google Scholar]
- 17.Dean, J. L., M. Brook, A. R. Clark, and J. Saklatvala. 1999. p38 mitogen-activated protein kinase regulates cyclooxygenase-2 mRNA stability and transcription in lipopolysaccharide-treated human monocytes. J. Biol. Chem. 274:264-269. [DOI] [PubMed] [Google Scholar]
- 18.DeMaria, C. T., and G. Brewer. 1996. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271:12179-12184. [DOI] [PubMed] [Google Scholar]
- 19.Diehl, S. R., Y. Wang, C. N. Brooks, J. A. Burmeister, J. V. Califano, S. Wang, and H. A. Schenkein. 1999. Linkage disequilibrium of interleukin-1 genetic polymorphisms with early-onset periodontitis. J. Periodontol. 70:418-430. [DOI] [PubMed] [Google Scholar]
- 20.Dodgson, S. J. 1991. Why are there carbonic anhydrases in the liver? Biochem. Cell Biol. 69:761-763. [DOI] [PubMed] [Google Scholar]
- 21.Druey, K. M., K. J. Blumer, V. H. Kang, and J. H. Kehrl. 1996. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature 379:742-746. [DOI] [PubMed] [Google Scholar]
- 22.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferret, P. J., E. Soum, O. Negre, E. E. Wollman, and D. Fradelizi. 2000. Protective effect of thioredoxin upon nitric oxide-mediated cell injury in THP1 monocytic human cells. Biochem. J. 346:759-765. [PMC free article] [PubMed] [Google Scholar]
- 24.Ford, L. P., J. Watson, J. D. Keene, and J. Wilusz. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, F. B., C. C. Carson, T. Levine, and J. D. Keene. 1994. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries with neuronal RNA-binding protein Hel-N1. Proc. Natl. Acad. Sci. USA 91:11207-11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouble, A., S. Grazide, F. Meggetto, P. Mercier, G. Delsol, and D. Morello. 2002. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 62:1489-1495. [PubMed] [Google Scholar]
- 27.Han, J., J. D. Lee, Y. Jiang, Z. Li, L. Feng, and R. J. Ulevitch. 1996. Characterization of the structure and function of a novel MAP kinase kinase (MKK6). J. Biol. Chem. 271:2886-2891. [DOI] [PubMed] [Google Scholar]
- 28.Holtmann, H., R. Winzen, P. Holland, S. Eickemeier, E. Hoffmann, D. Wallach, N. L. Malinin, J. A. Cooper, K. Resch, and M. Kracht. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol. 19:6742-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 30.Jain, R. G., L. G. Andrews, K. M. McGowan, P. H. Pekala, and J. D. Keene. 1997. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol. 17:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph, L. J., M. M. Le Beau, G. A. Jamieson, Jr., S. Acharya, T. B. Shows, J. D. Rowley, and V. P. Sukhatme. 1988. Molecular cloning, sequencing, and mapping of EGR2, a human early growth response gene encoding a protein with “zinc-binding finger” structure. Proc. Natl. Acad. Sci. USA 85:7164-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K. W. Schmid, U. Duhrsen, and T. Moroy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295-300. [DOI] [PubMed] [Google Scholar]
- 33.Keene, J. D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 96:5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keene, J. D. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 98:7018-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam, L. T., O. K. Pickeral, A. C. Peng, A. Rosenwald, E. M. Hurt, J. M. Giltnane, L. M. Averett, H. Zhao, R. E. Davis, M. Sathyamoorthy, L. M. Wahl, E. D. Harris, J. A. Mikovits, A. P. Monks, M. G. Hollingshead, E. A. Sausville, and L. M. Staudt. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2:0041.1-0041.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapucci, A., M. Donnini, L. Papucci, E. Witort, A. Tempestini, A. Bevilacqua, A. Nicolin, G. Brewer, N. Schiavone, and S. Capaccioli. 2002. AUF1 Is a bcl-2 A + U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J. Biol. Chem. 277:16139-16146. [DOI] [PubMed] [Google Scholar]
- 38.LaRue, K. E., and C. E. McCall. 1994. A labile transcriptional repressor modulates endotoxin tolerance. J. Exp. Med. 180:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasa, M., K. R. Mahtani, A. Finch, G. Brewer, J. Saklatvala, and A. R. Clark. 2000. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol. 20:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J., L. Mira-Arbibe, and R. J. Ulevitch. 2000. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J. Leukoc. Biol. 68:909-915. [PubMed] [Google Scholar]
- 41.Levine, T. D., F. Gao, P. H. King, L. G. Andrews, and J. D. Keene. 1993. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 13:3494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy, N. S., S. Chung, H. Furneaux, and A. P. Levy. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273:6417-6423. [DOI] [PubMed] [Google Scholar]
- 43.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 45.Mahtani, K. R., M. Brook, J. L. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurer, F., M. Tierney, and R. L. Medcalf. 1999. An AU-rich sequence in the 3′-untranslated region of plasminogen activator inhibitor type 2 (PAI-2) mRNA promotes PAI-2 mRNA decay and provides a binding site for nuclear HuR. Nucleic Acids Res. 27:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ming, X. F., G. Stoecklin, M. Lu, R. Looser, and C. Moroni. 2001. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 21:5778-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montero, L., and Y. Nagamine. 1999. Regulation by p38 mitogen-activated protein kinase of adenylate- and uridylate-rich element-mediated urokinase-type plasminogen activator (urokinase-type plasminogen activator) messenger RNA stability and urokinase-type plasminogen activator-dependent in vitro cell invasion. Cancer Res. 59:5286-5293. [PubMed] [Google Scholar]
- 49.Nagarajan, R., J. Svaren, N. Le, T. Araki, M. Watson, and J. Milbrandt. 2001. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron 30:355-368. [DOI] [PubMed] [Google Scholar]
- 50.Nau, G. J., J. F. Richmond, A. Schlesinger, E. G. Jennings, E. S. Lander, and R. A. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 99:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Donovan, N., M. Galvin, and J. G. Morgan. 1999. Physical mapping of the CXC chemokine locus on human chromosome 4. Cytogenet. Cell Genet. 84:39-42. [DOI] [PubMed] [Google Scholar]
- 52.Okada, S., H. Zhang, M. Hatano, and T. Tokuhisa. 1998. A physiologic role of Bcl-xL induced in activated macrophages. J. Immunol. 160:2590-2596. [PubMed] [Google Scholar]
- 53.Ono, K., and J. Han. 2000. The p38 signal transduction pathway: activation and function. Cell Signal. 12:1-13. [DOI] [PubMed] [Google Scholar]
- 54.Pages, G., E. Berra, J. Milanini, A. P. Levy, and J. Pouyssegur. 2000. Stress-activated protein kinases (JNK and p38/HOG) are essential for vascular endothelial growth factor mRNA stability. J. Biol. Chem. 275:26484-26491. [DOI] [PubMed] [Google Scholar]
- 55.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pierre-Jacques, P., S. Emmanuelle, N. Olivier, and D. Fradelizi. 2002. Auto-protective redox buffering systems in stimulated macrophages. BMC Immunol. 3:3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 58.Press, W. H., B. P. Flannery, S. A. Teukolsky, and W. T. Vetterling. 1986. Do two distributions have the same means or variances, p. 464-469. In W. H. Press, B. P. Flannery, S. A. Teukolsky, and W. T. Vetterling (ed.), Numerical recipes. Cambridge University Press, New York, N.Y.
- 59.Rodriguez-Pascual, F., M. Hausding, I. Ihrig-Biedert, H. Furneaux, A. P. Levy, U. Forstermann, and H. Kleinert. 2000. Complex contribution of the 3′ untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J. Biol. Chem. 275:26040-26049. [DOI] [PubMed] [Google Scholar]
- 60.Rogers, J. 2000. An interleukin-1 alpha susceptibility polymorphism in Alzheimer's disease: new fuel for the inflammation hypothesis. Neurology 55:464-465. [DOI] [PubMed] [Google Scholar]
- 61.Ruegsegger, U., D. Blank, and W. Keller. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243-253. [DOI] [PubMed] [Google Scholar]
- 62.Stoecklin, G., X. F. Ming, R. Looser, and C. Moroni. 2000. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 20:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoecklin, G., P. Stoeckle, M. Lu, O. Muehlemann, and C. Moroni. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA 7:1578-1588. [PMC free article] [PubMed] [Google Scholar]
- 64.Sweet, M. J., C. C. Campbell, D. P. Sester, D. Xu, R. C. McDonald, K. J. Stacey, D. A. Hume, and F. Y. Liew. 2002. Colony-stimulating factor-1 suppresses responses to CpG DNA and expression of toll-like receptor 9 but enhances responses to lipopolysaccharide in murine macrophages. J. Immunol. 168:392-399. [DOI] [PubMed] [Google Scholar]
- 65.Tenenbaum, S. A., C. C Carson, P. J. Lager, and J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by with cDNA arrays. Proc. Natl. Acad. Sci. USA 97:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412:346-351. [DOI] [PubMed] [Google Scholar]
- 67.Wang, S. W., J. Pawlowski, S. T. Wathen, S. D. Kinney, H. S. Lichenstein, and C. L. Manthey. 1999. Cytokine mRNA decay is accelerated by an inhibitor of p38-mitogen-activated protein kinase. Inflamm. Res. 48:533-538. [DOI] [PubMed] [Google Scholar]
- 68.Warner, L. E., P. Mancias, I. J. Butler, C. M. McDonald, L. Keppen, K. G. Koob, and J. R. Lupski. 1998. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat. Genet. 18:382-384. [DOI] [PubMed] [Google Scholar]
- 69.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, Y., S. Y. Neo, J. Han, L. P. Yaw, and S. C. Lin. 1999. RGS16 attenuates gαq-dependent p38 mitogen-activated protein kinase activation by platelet-activating factor. J. Biol. Chem. 274:2851-2857. [DOI] [PubMed] [Google Scholar]