Abstract
Phospholipase D (PLD) activity is elevated in response to the oncogenic stimulus of H-Ras but not K-Ras. H-Ras and K-Ras have been reported to localize to different membrane microdomains, with H-Ras localizing to caveolin-enriched light membrane fractions. We reported previously that PLD activity elevated in response to mitogenic stimulation is restricted to the caveolin-enriched light membrane fractions. PLD activity in H-Ras-transformed cells is dependent upon RalA, and consistent with a lack of elevated PLD activity in K-Ras-transformed cells, RalA was not activated in K-Ras-transformed cells. Although H-Ras-induced PLD activity is dependent upon RalA, an activated mutant of RalA is not sufficient to elevate PLD activity. We reported previously that RalA interacts with PLD activating ADP ribosylation factor (ARF) proteins. In cells transformed by H-Ras, we found increased coprecipitation of ARF6 with RalA. Moreover, ARF6 colocalized with RalA in light membrane fractions. Interestingly, ARF6 protein levels were elevated in H-Ras- but not K-Ras-transformed cells. A dominant-negative mutant of ARF6 inhibited PLD activity in H-Ras-transformed NIH 3T3 cells. Activated mutants of either ARF6 or RalA were not sufficient to elevate PLD activity in NIH 3T3 cells; however, expression of both activated RalA and activated ARF6 in NIH 3T3 cells led to increased PLD activity. These data suggest a model whereby H-Ras stimulates the activation of both RalA and ARF6, which together lead to the elevation of PLD activity.
Phospholipase D (PLD) is elevated in response to many oncogenic signals, including those generated by v-Src (66), v-H-Ras (9, 34, 35), v-Fps, (36), and v-Raf (23). The activation of PLD by v-Src, v-Ras, and v-Raf is dependent upon the small GTPase RalA (23, 35), which interacts directly with PLD1 (46). RalA has also been shown to be required for the transformed phenotype induced by v-Src, v-Ras, and v-Raf (1, 70). Additionally, RalA, PLD1, or PLD2 could cooperate with either c-Src or the epidermal growth factor (EGF) receptor (EGFR) to transform rat fibroblasts (38, 44). Elevated expression of either PLD1 or PLD2 was able to overcome a cell cycle block induced by high-intensity Raf signaling (39). RalA has also been implicated in the activation of PLD by phorbol esters (60) and by growth factor receptor tyrosine kinases (30, 73). These studies suggest an important role for RalA and PLD in mitogenic and oncogenic signaling. However, while RalA is necessary for activation PLD by oncoproteins and growth factors, activated RalA was not sufficient for elevation of PLD activity (35). Active RalA-PLD complexes also contain ADP ribosylation factor (ARF) GTPases (47), which activate PLD1 (7, 15, 27). The presence of ARF in active RalA-PLD1 complexes (47) also implicated ARF in mitogenic signaling. These data suggested that the activation of PLD in response to mitogenic signals requires multiple signals involving the activation of both RalA and ARF GTPases.
The activation of PLD by Ras has been somewhat controversial. While it has been reported that activated Ras leads to elevated PLD activity (9, 34, 35, 45), it has also been reported that activated Ras does not elevate PLD activity (2). Most, if not all studies showing that Ras activates PLD activity were performed with H-Ras, while the study showing that Ras did not activate PLD activity employed K-Ras. These data suggest that H-Ras activates signals not activated by K-Ras. For this report, we investigated the differential activation of RalA and PLD by H-Ras and K-Ras and describe the synergistic activation of PLD by ARF6 and RalA. A model is proposed for the activation of PLD by H-Ras whereby a RalA-PLD complex is activated, leading to the recruitment of the PLD activator ARF6 into an active RalA-PLD-ARF6 complex.
MATERIALS AND METHODS
Cells and cell culture conditions.
Parental and H-Ras-, K-Ras-, v-Src-, and v-Raf-transformed NIH 3T3 cells, NIH 3T3 cells overexpressing activated RalA (Q72L), and 3Y1 rat fibroblasts overexpressing the EGFR were characterized previously (23, 30, 34, 35) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine serum (HyClone). To reduce background PLD activity, subconfluent cell cultures were placed in fresh medium containing 0.5% bovine serum for 1 day. For transfection, cells were plated at a density of 105 cells per 100-mm dish 18 h prior to transfection. Transfections were performed with Lipofectamine Plus reagent (GIBCO) according to the vendor's instructions, in which Plus reagent was included to enhance the transfection efficiency of Lipofectamine. For transient transfection, control for efficiency was determined by transfection of pEGFP-C1 (Clonetech), which expresses green fluorescent protein (GFP). The percentage of green cells was determined microscopically and was always in excess of 80%. For stable transfections, cells were selected in the presence of G418 for 2 weeks as described previously (44), and pooled clones were used for experiments.
Materials.
[3H]myristate was obtained from New England Nuclear. Precoated silica 60A thin-layer chromatography plates were obtained from Whatman. Monoclonal antibody 1D9, which recognizes most ARF isoforms, was obtained from Richard Kahn (Emory University, Atlanta, Ga.). Antibodies raised against ARF1 and ARF6 were provided by Sylvain Bourgoin (Universite Laval, Quebec, Canada). Antibodies raised against RalA and caveolin 1 were obtained from Transduction Laboratories. The anti-Ras antibodies were obtained from Santa Cruz Biotechnology. For nonimmune controls, we used ChromPure rabbit or mouse immunoglobulin G (IgG) from Jackson ImmunoResearch.
Plasmid expression vectors.
The mammalian expression plasmids pcDNA3.1-ARF6Q67L, pcDNA3.1-ARF6T27N, pcDNA3.1-ARF1Q67L, and pcDNA3.1-ARF1T31N have been described previously (5, 18). They were constructed by PCR amplification of the corresponding cDNAs and cloned into the EcoRI site of pcDNA3.1(−) (Invitrogen). The mammalian expression plasmids for RalA (Q72L), Ki-Ras4B (G12V), and Ha-Ras (G12V) were expressed in the pZIP-NeoSV(X)1 vector and have been described previously (35).
Isolation of membranes.
The strategy for separation of light and heavy membrane fractions was based on one developed by Lisanti and colleagues (68) with modifications that excluded the use of sodium carbonate and high pH as described previously (78). Quiescent confluent cells grown in 150-mm dishes were washed twice with cold phosphate-buffered saline and scraped into 2 ml of buffer M, which consisted of 25 mM MES (morpholineethanesulfonic acid; pH 6.5), 250 mM sucrose, 1 mM EDTA, and 1× protease inhibitor cocktail. Homogenization was carried out on ice with a Wheaton Dounce homogenizer (20 to ∼25 strokes) and then by sonication (three 20-s bursts; VC 300; Sonics & Materials, Inc., Danbury, Conn.). The protein concentration was determined by the Bradford method (Bio-Rad). Five milligrams of homogenate protein was diluted to 2 ml in buffer M and adjusted to 45% (wt/vol) sucrose by adding 2 ml of 90% (wt/vol) sucrose prepared in 25 mM MES (pH 6.5). This solution was then overlaid with 4 ml of 35% (wt/vol) sucrose and 4 ml of 5% (wt/vol) sucrose in 25 mM MES (pH 6.5) to form a discontinuous gradient in an ultracentrifuge tube. The gradient was centrifuged at 39,000 rpm for 16 to 18 h in an SW41 rotor (Beckman). One-milliliter fractions were collected from top to bottom and analyzed for PLD activity and proteins as described in the text. The pellet (fraction 13) was sonicated in 1 ml of buffer M and analyzed along with the collected fractions.
Immunoprecipitation.
Quiescent confluent cells were washed twice with ice-cold phosphate-buffered saline and scraped into the modified radioimmunoprecipitation assay (RIPA) buffer: 50 mM Tris-HCl (pH 7.6), 1% Igepal CA-630, 0.25% sodium deoxycholate, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and 1× protease inhibitor cocktail, consisting of 0.5 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride], 1 μM leupeptin, 0.15 μM aprotinin, and 1 μM protease inhibitor E-64. The cells were then incubated at 4°C for 15 min by gentle rocking, sonicated for 20 s on ice, and centrifuged at 12,000 × g at 4°C for 10 min. The supernatant was precleared with protein G-Sepharose 4 Fast Flow beads (Amersham Pharmacia Biotech), and 500 μg of the precleared proteins was adjusted to 500 μl in the modified RIPA buffer and then incubated with the antibody for 1 h as described above. The immunocomplex was captured by incubation with 50 μl of protein G-Sepharose 4 Fast Flow bead slurry, collected by centrifugation at 12,000 × g for 20 s at 4°C. The beads were washed three times with the modified RIPA buffer and once with wash buffer (50 mM Tris [pH 7.6]), and subjected to Western blot analysis.
Western blot analysis.
Samples were adjusted into gel loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10% glycerol) and then heated for 3 min at 100°C prior to separation by SDS-polyacrylamide gel electrophoresis. After transfer to polyvinylidene difluoride (for caveolin) or nitrocellulose membrane (for other proteins), the membrane filters were blocked with 5% nonfat dry milk in phosphate-buffered saline with 0.05% Tween 20 (PBS-T) and then incubated with the appropriate antibody diluted in 5% milk in PBS-T. Depending upon the origin of the primary antibodies, either antimouse or antirabbit IgG conjugated with horseradish peroxidase was used, and the bands were visualized with the enhanced chemilluminescence detection system (Pierce).
Assay of PLD activity.
PLD activity was determined by the transphosphatidylation reaction in the presence of 0.8% butanol as described previously (67). Cells in 100-mm culture dishes were prelabeled with [3H]myristate for 4 to 5 h in DMEM containing 0.5% bovine serum. Lipids were extracted and characterized by thin-layer chromatography as described previously (66, 67). Relative levels of PLD activity were then determined by measuring the intensity of the corresponding phosphatidylbutanol band in the autoradiograph with a Molecular Dynamics scanning densitometer and Image-Quant software.
RalA activation assay.
The detection of activated RalA was performed essentially as described previously (44). Cells were lysed with a mixture containing 15% glycerol, 50 mM Tris-HCl (pH 7.4), 1% Igepal CA-630, 200 nM NaCl, 10 mM MgCl2, and1× protease inhibitor cocktail and precleared with glutathione-agarose beads. Lysates were then treated with glutathione S-transferase (GST)-Ral-BD fusion protein immobilized with glutathione-agarose beads (Upstate Biotechnology). Ral-BD is the Ral binding domain of Ral-BP1 that binds activated GTP-bound Ral proteins (76, 77). Activated Ral proteins were recovered by centrifugation at 14,000 × g at 4°C for 5 s, washed three times with lysis buffer, and subjected to Western blot analysis with an antibody raised against RalA (Transduction Laboratories).
RESULTS
PLD activity is elevated in H-Ras- but not K-Ras-transformed cells.
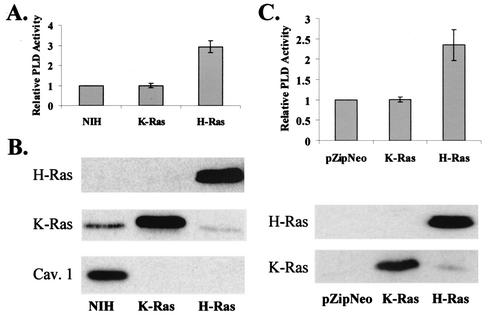
PLD activity was examined in NIH 3T3 cells transformed by either H-Ras or K-Ras. As shown in Fig. 1A, PLD activity was found to be elevated in the H-Ras- but not the K-Ras-transformed cells. To ensure that the lack of PLD activity in the K-Ras-transformed cells was not due to a lack of expression of the K-Ras protein, we examined the levels of Ras proteins in the H-Ras- and K-Ras-transformed cells. As shown in Fig. 1B, there were elevated levels of Ras proteins in both the H-Ras- and K-Ras-transformed cells. Also shown in Fig. 1B are the levels of caveolin 1, which is lost in transformed cells (42), and consistent with previous reports, caveolin 1 expression was lost in both H-Ras- and K-Ras-transformed cells. The K-Ras-transformed cells were also transformed as determined by morphology and the ability to form colonies in soft agar (data not shown). This experiment was repeated with six independent clones of H-Ras- and K-Ras-transformed cells as well as with pooled clones from H-Ras- and K-Ras-transfected NIH 3T3 cells (data not shown), indicating that the effects are not due to clonal variation in the H-Ras- and K-Ras-transformed NIH 3T3 cells. To further establish this, we examined the PLD activity in NIH 3T3 cells transiently transfected with plasmids expressing H-Ras and K-Ras. As shown in Fig. 1C, PLD activity was elevated in the H-Ras- but not the K-Ras-transfected cells. Comparable levels of H-Ras and K-Ras proteins were expressed in the transfected cells (Fig. 1C). These data reveal an apparent difference in the abilities of H-Ras and K-Ras to activate PLD activity.
FIG. 1.
PLD activity is elevated in H-Ras- but not K-Ras-transformed cells. (A) Untransformed NIH 3T3 cells and NIH 3T3 cells transformed by H-Ras and K-Ras were placed in medium containing 0.5% serum for 1 day. PLD activity was then determined by the transphosphatidylation reaction in the presence of 0.8% butanol as described in Materials and Methods. The PLD activity was normalized to the PLD activity in the parental NIH 3T3 cells, which was given a value of 1. Error bars represent the standard deviation for three independent experiments performed in duplicate. (B) Cell lysates from the parental and H-Ras- and K-Ras-transformed NIH 3T3 cells were subjected to Western blot analysis with antibodies specific for H-Ras, K-Ras, and caveolin 1 as indicated. (C) NIH 3T3 cells were transiently transfected with vectors that express H-Ras, K-Ras, and the parental pZIP-NeoSV(X)1 empty vector. Twenty-four hours after transfection, the cells were placed in fresh medium containing 0.5% serum, and after an additional 24 h, PLD activity was determined as in panel A and normalized to the PLD activity in the NIH 3T3 cells that were transfected with the empty vector pZIP-NeoSV(X)1, which was given a value of 1. Ras protein levels were determined by Western blot analysis as shown.
Differential activation of RalA in H-Ras and K-Ras.
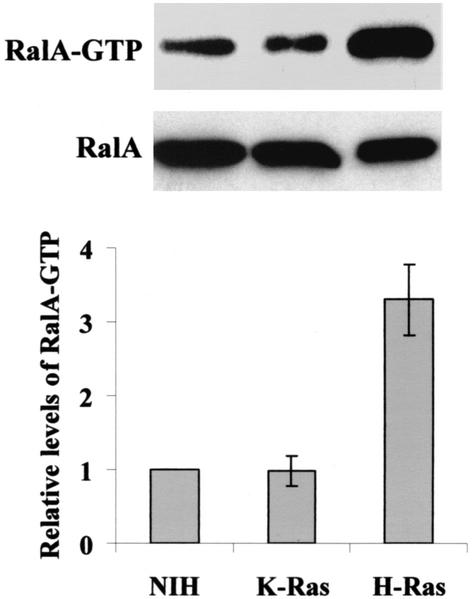
The S28N dominant-negative RalA mutant (defective for GDP/GTP exchange) inhibits PLD activation by H-Ras (35). We therefore investigated whether there was a differential ability of H-Ras and K-Ras to activate RalA. Upon activation, RalA binds GTP and associates with the downstream effector molecule Ral-BP1 (8). We took advantage of this by using the Ral-BD of Ral-BP1 fused to GST (GST-Ral-BD) to detect activated GTP-bound RalA as described previously (44). Cell lysates from parental and H-Ras- and K-Ras-transformed NIH 3T3 cells were treated with GST-Ral-BD immobilized on glutathione-agarose beads. The GST-Ral-BD was recovered by centrifugation, and the pellets were subjected to Western blot analysis with an antibody raised against RalA. As shown in Fig. 2, there was 2.5-fold more RalA detected in GST-Ral-BD precipitates from H-Ras-transformed cells than in those from the untransformed and K-Ras-transformed cells. The level of RalA detected in the Ral-BD precipitate from the K-Ras-transformed cells was equivalent to the level detected in the parental NIH 3T3 cells. These data suggest that the reduced PLD activity is likely due to the reduced ability of K-Ras to activate RalA and further confirm the different biological effects of H-Ras and K-Ras.
FIG. 2.
RalA activation in H-Ras- and K-Ras-transformed cells. Untransformed NIH 3T3 cells and NIH 3T3 cellS transformed by either H-Ras or K-Ras were lysed and then treated with immobilized GST-Ral-BD as described in Materials and Methods. GST-Ral-BD was recovered by centrifugation, and the precipitate was subjected to Western blot analysis with an antibody raised against RalA. Total RalA protein levels in the cell lysates were also examined by Western blot analysis of the untreated lysates as shown. The figure shown is representative of four independent experiments. The relative level of activated RalA was determined by densitometric analysis of the Ral-BD precipitates for four independent experiments normalized to the control NIH 3T3 cells and given a value of 1. Error bars represent the standard error for the four experiments.
Increased coprecipitation of ARF6, but not ARF1, with RalA in H-Ras-transformed cells.
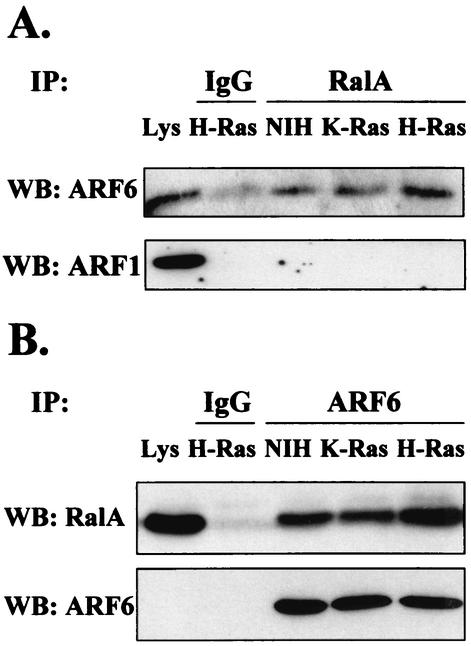
RalA coprecipitates with the PLD activating ARF GTPases (47). Both ARF1 and ARF6 have been implicated in the regulation of PLD activity (20). The ability of ARF proteins to coprecipitate with RalA from H-Ras- and K-Ras-transformed NIH 3T3 cells was therefore examined. RalA was immunoprecipitated from lysates of H-Ras- and K-Ras-transformed NIH 3T3 cells and then subjected to Western blot analysis with antibodies raised against ARF1 and ARF6. As shown in Fig. 3A, elevated levels of ARF6 were detected in the RalA immunoprecipitates from the H-Ras but not K-Ras cell lysates. ARF1 was barely detectable in RalA immunoprecipitates even though the ARF1 band in the lysate was stronger than that of ARF6 (Fig. 3A), suggesting that RalA binds a much lower percentage of ARF1 than of ARF6. After longer exposure, ARF1 was detected, but there were not elevated levels of ARF1 in RalA immunoprecipitates from either H-Ras- or K-Ras-transformed cells relative to the parental NIH 3T3 cells (data not shown). Reciprocally, an ARF6 antibody coimmunoprecipitated elevated levels of RalA from H-Ras-transformed cell lysates relative to K-Ras-transformed cell lysates, despite the equivalent levels of immunoprecipitated ARF6 (Fig. 3B). These data indicate that H-Ras stimulates the association between RalA and ARF6 and that ARF6 contributes to the elevation of PLD activity in H-Ras-transformed cells.
FIG. 3.
Increased coprecipitation of ARF6, but not ARF1, with RalA in H-Ras-transformed cells. (A) RalA was immunoprecipitated (IP) from lysates of parental and H-Ras- and K-Ras-transformed NIH 3T3 cells by using a mouse monoclonal RalA antibody. The RalA immunoprecipitates (RalA) were then subjected to Western blot (WB) analysis with antibodies raised against either ARF1 or ARF6 (rabbit IgGs). A nonimmune IgG control (IgG) is shown, as is a portion of whole-cell lysate (4%) (Lys) that was not subjected to immunoprecipitation. (B) A reciprocal experiment is shown in which lysates of parental and H-Ras- and K-Ras-transformed NIH 3T3 cells were immunoprecipitated with anti-ARF6 (rabbit IgG) in antigen excess and subjected to Western blot analysis with an antibody to RalA (mouse IgG). The membrane was stripped and reprobed with anti-ARF6 antibody to check the levels of ARF6 protein immunoprecipitated. The panels shown are representative of at least three independent experiments.
ARF6 colocalizes with RalA and is elevated in H-Ras-transformed cells.
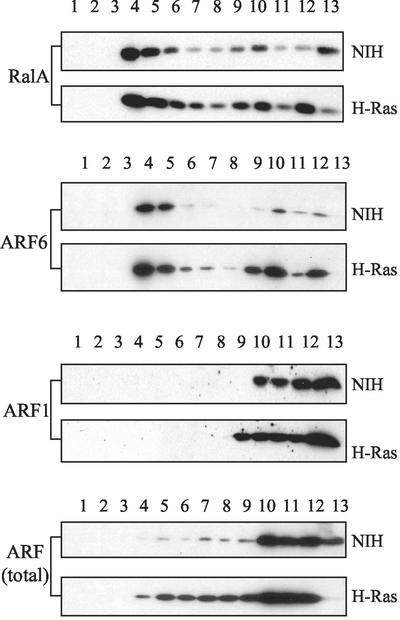
PLD activity elevated in response to mitogenic stimulation has been reported to be restricted largely to light membrane fractions (78). Therefore, the subcellular distributions of RalA, ARF6, and ARF1 were examined. As shown in Fig. 4, RalA was detected in both light and heavy membrane fractions; however, the majority of RalA was found in the light membrane fractions from H-Ras-transformed cells. ARF6 was also found predominantly in the light membrane fraction. In contrast, ARF1 localized exclusively with the heavier fractions representing heavy membranes and soluble proteins. Thus, consistent with the increased association observed between RalA and ARF6 in H-Ras-transformed cells, ARF6 and RalA colocalized with the same light membrane fraction in which the PLD activity elevated in response to mitogenic stimulation was also found (78). This is also the same membrane fraction to which both H-Ras and caveolin localize as described previously (78).
FIG. 4.
RalA colocalizes with ARF6 but not ARF1. Parental and H-Ras-transformed NIH 3T3 cells were disrupted by Dounce homogenization and then sonication as described in Materials and Methods. The membrane fragments were then run over a discontinuous gradient of 5, 35, and 45% sucrose. Twelve fractions and a pellet were recovered and subjected to Western blot analysis with antibodies to RalA, ARF1, ARF6, and total ARF (monoclonal antibody 1D9). The amount of material loaded onto the gels was normalized by volume from each of the fractions. The panels shown are representative of three independent experiments.
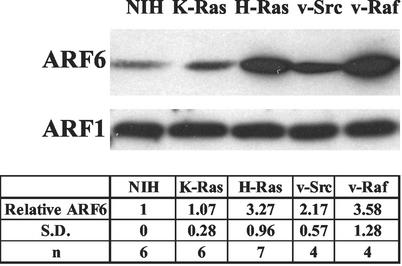
Interestingly, there appeared to be elevated levels of ARF6 in the H-Ras-transformed cells relative to the parental NIH 3T3 cells. The level of ARF proteins was therefore examined in the H-Ras- and K-Ras-transformed cells. As shown in Fig. 5, there were elevated levels of ARF6 protein found in the H-Ras-transformed cells. Elevated levels of ARF6 were also observed in cells transformed by v-Src and v-Raf (Fig. 5). We did not detect elevated levels of ARF1 in any of the cell lines examined (Fig. 5). These data suggest that Src, Raf, and H-Ras oncoproteins stimulate elevated ARF6 expression. The lack of elevated ARF6 in the K-Ras-transformed cells further distinguishes between the biological effects of H-Ras and K-Ras.
FIG. 5.
ARF6 is elevated in H-Ras- but not K-Ras-transformed cells. Homogenates from untransformed NIH 3T3 cells and NIH 3T3 cells transformed by either H-Ras, K-Ras, v-Src, or v-Raf were prepared as described in Materials and Methods and subjected to Western blot analysis with antibodies for ARF1 and ARF6. The data shown are representative of at least four independent experiments. The data from several (n) experiments were quantified by densitometry of Western blots. The level of ARF6 protein relative to that in the parental NIH 3T3 cells (Relative ARF6) was then determined with standard deviations (S.D.) as shown.
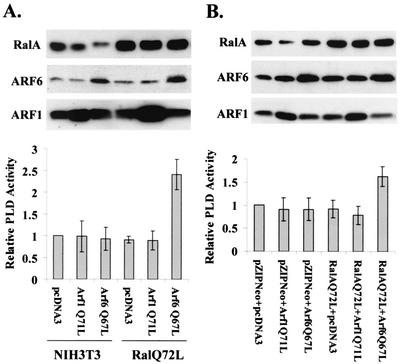
Activated ARF6 elevates PLD activity in NIH 3T3 cells expressing activated RalA.
While there is a RalA requirement for the activation of PLD by H-Ras (35), an activated form of RalA (Q72L) is not sufficient to activate PLD activity by itself (35). We therefore examined whether a combination of activated RalA and ARF could elevate PLD activity in NIH 3T3 cells. NIH 3T3 cells or NIH 3T3 cells that express activated RalA (Q72L) were transiently transfected with vectors that express activated ARF1 (Q71L), activated ARF6 (Q67L), or the empty vector pcDNA3. As shown in Fig. 6A, neither activated ARF1 nor activated ARF6 was able to increase PLD activity in the NIH 3T3 cells. However, in the NIH 3T3 cells expressing activated RalA (Q72L), activated ARF6 (Q67L) stimulated a 2.5-fold increase in PLD activity, whereas activated ARF1 (Q71L) had no effect upon PLD activity in these cells (Fig. 6A). Expression of Ral and ARF protein levels in the transfected cells was determined by Western blotting and is also shown in Fig. 6A. We also examined the effect of cotransfection of an activated RalA expression vector with activated ARF1 and ARF6 expression vectors, and as expected, only the combination of activated RalA and activated ARF6 led to increased PLD activity (Fig. 6B). These data indicate a synergistic effect of RalA and ARF6 in the activation of PLD.
FIG. 6.
Activated ARF6 elevates PLD activity in NIH 3T3 cells expressing activated RalA. (A) NIH 3T3 cells and NIH 3T3 cells overexpressing an activated RalA mutant (Q72L) (35) were transiently transfected with vectors that express activated mutants of ARF1 (Q71L) and ARF6 (Q67L), as well as the parental pcDNA3 empty vector. Twenty-four hours after transfection, the cells were placed in fresh medium containing 0.5% serum, and after an additional 24 h, PLD activity was determined as in Fig. 1A and normalized to the PLD activity in the NIH 3T3 cells that were transfected with the empty vector, which was given a value of 1. The levels of RalA, ARF1, and ARF6 were determined by Western blot analysis as shown. (B) NIH 3T3 cells were transiently transfected with either pZIP-NeoSV(X)1 empty vector or activated RalA (Q72L) mutant, along with pcDNA3 empty vector, or with vectors expressing either the activated mutant of ARF1 (Q71L) or ARF6 (Q67L) as indicated. PLD activity was determined as in panel A and normalized to the PLD activity in the NIH 3T3 cells transfected with the empty vector controls. RalA and ARF protein levels were determined as in panel A. Error bars for panels A and B represent the standard deviation for three independent experiments performed in duplicate.
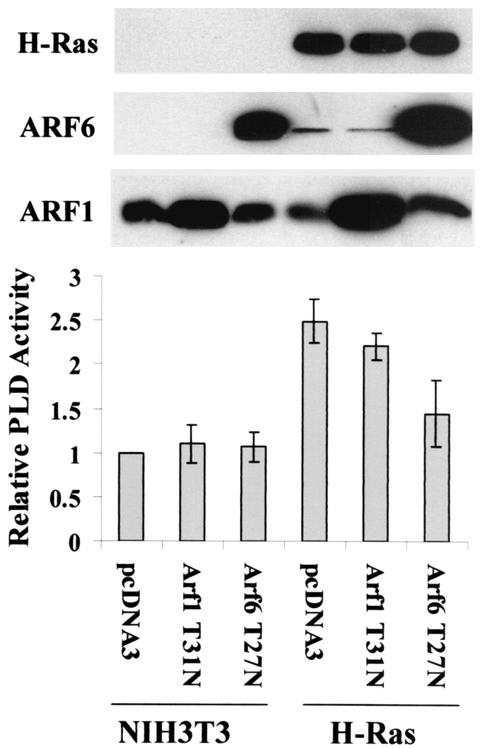
H-Ras-induced PLD activity is dependent upon ARF6.
The data in Fig. 6 indicate that the combination of activated RalA and activated ARF6 is sufficient to activate PLD activity, but do not indicate whether RalA and ARF6 are necessary for PLD activation by H-Ras. However, RalA was reported previously to be necessary for H-Ras activation of PLD activity (35). To investigate whether ARF6 is necessary for the activation of PLD activity by H-Ras, we introduced a dominant-negative mutant of ARF6 (T27N) into both parental and H-Ras-transformed NIH 3T3 cells. As shown in Fig. 7, the T27N ARF6 mutant had no effect upon the PLD activity in the NIH 3T3 cells, but reduced the elevated PLD activity in the H-Ras-transformed cells almost to the level seen in the parental NIH 3T3 cells. A dominant-negative ARF1 mutant (T31N) had no significant effect upon the PLD activity in the H-Ras-transformed cells. These data indicate that H-Ras-induced PLD activity is dependent upon ARF6.
FIG. 7.
H-Ras-induced PLD activity is dependent on ARF6. Parental and H-Ras-transformed NIH 3T3 cells were transiently transfected with vectors that express dominant-negative mutants for ARF1 (T31N) and ARF6 (T27N) as described in Materials and Methods. As a control, these cells were transfected with pcDNA3 empty vector. Forty-eight hours after transfection, PLD activity was determined as in Fig. 6. The PLD activity was normalized to the PLD activity in the empty vector control for the NIH 3T3 cells, which was given a value of 1. Ras and ARF protein levels were determined as in Fig. 6. Error bars represent the standard deviation for three independent experiments performed in duplicate.
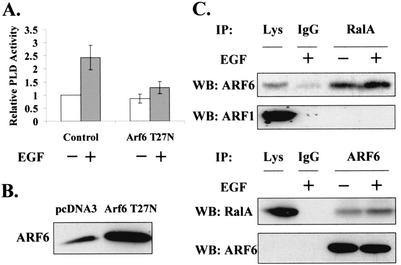
ARF6 mediates EGF-induced PLD activity.
EGF-induced PLD activity, which is dependent upon RalA (44, 61), has been shown to be restricted to light membrane fractions (78). EGF also stimulates GTP loading on ARF6 (5). Therefore, the dependence of EGF-induced PLD activity on ARF6 was examined. For these experiments, 3Y1 cells overexpressing the EGF receptor (EGFR cells) (30, 44, 61, 78) were used. These cells were stably transfected with an empty vector and the vector expressing the dominant-negative ARF6 (T27N). Several clones were pooled and then examined for the ability of EGF to induce PLD activity. Figure 8A shows that the dominant-negative mutant ARF6 inhibited EGF-induced PLD activity. We also examined whether EGF could induce the association between ARF6 and RalA seen in the H-Ras-transformed cells. As shown in Fig. 8C, EGF increased the amount of ARF6 in RalA immunoprecipitates and the amount of RalA in ARF6 immunoprecipitates. These data indicate that the role of ARF6 in regulating PLD activity is not an indirect effect of transformation. In addition, since the stimulated increase in association could be detected within 15 min, the data demonstrate that the effect of ARF6 is not due to the elevated levels of ARF6 seen in transformed cells.
FIG. 8.
ARF6 mediates EGF-induced PLD activity. (A) EGFR cells were stably transfected with pcDNA3 (empty vector) and the vector for expression of the dominant-negative mutant ARF6 (T27N). Cells were selected in the presence of G418 for 2 weeks, and clones were pooled and used for experiments. EGF (100 ng/ml) was added as indicated, and the PLD activity was determined after 15 min. The PLD activity was normalized to the PLD activity in the empty vector control cells without EGF, which was given a value of 1. Error bars represent the standard deviation for three independent experiments performed in duplicate. The level of ARF6 proteins in the transfected cells was confirmed by Western blot (WB) analysis (B). (C) Lysates from either untreated EGFR cells or EGFR cells treated with EGF (100 ng/ml) for 15 min were lysed and then immunoprecipitated (IP) with an anti-RalA antibody. Immunoprecipitates were then subjected to Western blot analysis with antibodies to either ARF1 or ARF6 as in Fig. 3. A reciprocal experiment is shown in which cell lysates were immunoprecipitated with anti-ARF6 antibody and subjected to Western blot analysis with an antibody to RalA. The membrane was stripped and reprobed with anti-ARF6. The figure shown is a representative of at least three independent experiments.
DISCUSSION
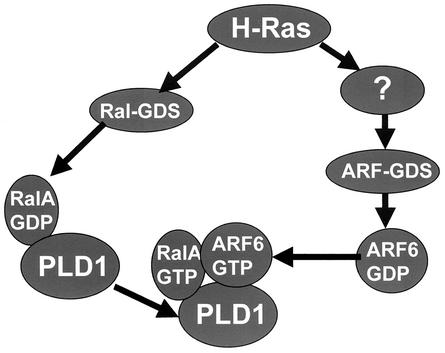
In this report, we have demonstrated that H-Ras but not K-Ras activates PLD activity. Similarly, H-Ras, but not K-Ras, led to elevated levels of ARF6 protein. H-Ras but not K-Ras activated RalA, which interacts directly with PLD1 (35, 46, 47). Neither activated RalA nor activated ARF6 was able to elevate PLD activity by itself; however, a combination of activated RalA and activated ARF6 was able to elevate PLD activity in NIH 3T3 cells. Since the activation of PLD activity in H-Ras-transformed NIH 3T3 cells was dependent on both ARF6 and RalA, the data presented here suggest a model whereby H-Ras activates PLD activity through the synergistic activation of both RalA and ARF6. In Fig. 9, we present a working model for the activation of PLD activity by H-Ras. It is proposed that H-Ras activates two parallel pathways leading to the activation of a guanine nucleotide dissociation stimulator (GDS) for RalA (70) and an as-yet-unspecified ARF-GDS. The activation of Ral-GDS by H-Ras activates RalA, which is already in a complex with PLD1 (35, 46, 47). The activation of RalA is proposed to recruit activated ARF6 into the RalA-PLD1 complex, or alternatively, activated ARF6 recruits the activated RalA-PLD1 complex. Activated ARF6, now in a RalA-PLD1-ARF6 complex, would activate PLD1.
FIG. 9.
Working model for activation of PLD1 by the synergistic action of RalA and ARF 6 in H-Ras-transformed cells. It is proposed that H-Ras activates parallel pathways leading to the activation of Ral-GDS and an as-yet-unspecified ARF-GDS. Activation of Ral-GDS activates RalA, which is already in a complex with PLD1. The activation of RalA is proposed to recruit activated ARF6 into the RalA-PLD1 complex, or alternatively activated ARF6 recruits the activated RalA-PLD1 into a RalA-PLD1-RalA complex. The activated ARF6 then activates PLD1.
Correlation between elevated PLD activity and ARF6.
Most ARF family GTPases activate PLD1 in vitro (20, 28, 48). There are three major classes of ARF family GTPases: class I (ARF1 to -3), class II (ARF4 and 5), and class III (ARF6) (50). Little is known about the functions of the class II ARF family. The class I ARFs have been implicated primarily in vesicle transport in the endoplasmic reticulum and Golgi apparatus (50). The class III ARF (ARF6) has been implicated in receptor endocytosis (18), vesicle recycling at the plasma membrane (19), and exocytosis (10, 11; reviewed by Chavrier and Goud in reference 13). PLD activity has likewise been implicated in both receptor endocytosis (61) and exocytosis (10, 21, 32, 71). PLD1 has been reported to colocalize with ARF6 in endosomes (69), and ARF6 has been implicated in the activation of PLD1 by the high-affinity IgG receptor FcγRI (49). Both ARF6 and PLD have been reported to activate phosphatidylinositol-4-phosphate-5-kinase (29, 33, 50). Thus, there is a correlation between the function of ARF6 and plasma membrane PLD activity. The data presented here provide evidence that ARF6 is a regulator of the PLD activity elevated in response to mitogenic stimulation, which like ARF6 is localized primarily in light membrane or “lipid raft” fractions (78).
RalA, which interacts directly with PLD1 (46), was also localized in the light membrane fraction and was coimmunoprecipitated with ARF6. It was previously demonstrated that ARF1 and RalA could synergistically activate PLD1 in vitro (40). In the in vivo studies presented here, ARF1 was unable to stimulate PLD activity in cells with activated RalA, and a dominant-negative ARF1 was unable to inhibit H-Ras-induced PLD activity. These data likely reflect the different distributions of ARF1 and ARF6 in cells, where ARF6 localizes to the plasma membrane and lipid rafts where the PLD activity elevated in response to mitogenic stimulation is localized (78). Whereas ARF1 is able to activate PLD1 in vitro, ARF1 would not have access to RalA-PLD1 complexes in intact cells because of the differential localization. In this regard, the recent report that antigen stimulation of RBL-2H3 cells leads to the colocalization of PLD1 and ARF6 at the plasma membrane (54) further suggests that ARF6 is a physiological regulator of PLD1.
PLD1 is likely the primary target of RalA and ARF6.
ARF proteins activate PLD1 but not PLD2 in vitro (16, 20, 28). RalA, which we have shown to coprecipitate with ARF6, interacts directly with PLD1 (47). These data implicate PLD1 as being responsible for the elevated PLD activity in cells expressing activated forms of RalA and ARF6. However, the elevated PLD activity in H-Ras-transformed cells is largely restricted to light membrane fractions where there is very little PLD1 and lots of PLD2 (78). In this regard, it is of interest that both PLD1 and PLD2 are able to cooperate with either EGFR or c-Src overexpression to transform rat fibroblasts (38, 44). In addition, both PLD1 and PLD2 were required for ligand-induced endocytosis of the EGF receptor (61). These data suggest that PLD1 and PLD2 may be working together to generate phosphatidic acid and the downstream effects of PLD signaling. A recent report by Mwanjewe et al. (51) suggested that the activation of PLD2 was dependent upon the activation of PLD1. Thus, the activation of PLD1 through RalA and ARF6 may also lead to the activation of PLD2 in the light membrane fractions where both PLD1 and PLD2 are present (17, 41, 43, 78). While it is not clear how PLD1 might lead to the activation of PLD2, the data presented here suggest that the PLD activated through the synergistic action of RalA and ARF6 is PLD1, since RalA associates with PLD1 and ARF proteins activate PLD1.
Activation of ARF6 by H-Ras.
While it has been demonstrated that H-Ras activates RalA by activating Ral-GDS through a GTP-dependent direct interaction (70), how H-Ras activates ARF6 is not clear. We reported previously that the elevated PLD activity in H-Ras-transformed cells was partially sensitive to brefeldin A (BFA) (47). However, the known GDS proteins for ARF6—ARNO, EFA6, and ARF-GEP100—have all been reported to be insensitive to BFA (12, 22, 65). Others have reported that PLD activities stimulated by the mitogenic stimulus of platelet-derived growth factor (PDGF) and insulin are also sensitive to BFA (56, 62, 63, 64). Thus, at present, we must conclude that either (i) GDS proteins for ARF6 can display differential sensitivities to BFA in different contexts or (ii) there is another ARF6-GDS that has yet to be identified that is sensitive to BFA. In support of the first hypothesis, phorbol myristate acetate (PMA)-induced PLD activity has been reported to be both BFA sensitive (62) and BFA insensitive (25). Moreover, all ARF-GDS proteins characterized to date have the s7 domain, which is critical for interaction with BFA (12, 53, 57). In this regard, the concentration of BFA used in different experimental systems could be important. A wide range of BFA concentrations have been used to block different BFA-sensitive functions. Thus, at present, it is not clear which ARF-GDS is activating ARF6 in H-Ras-transformed cells, nor is it apparent how H-Ras might activate the ARF6-GDS. This parallel signaling pathway activated by H-Ras that contributes to the activation of PLD activity might reveal another distinct signaling pathway initiated by activated Ras GTPases.
PDGF-induced PLD activity was reported to be dependent upon Ras (45) and was also blocked by dominant-negative mutants of both ARF1 and ARF6 (62). These data would appear to be in conflict with the data reported here where we showed that the elevated PLD activity in H-Ras-transformed cells was inhibited by a dominant-negative ARF6 but not a dominant-negative ARF1. This difference could indicate that the activation of PLD by PDGF involves a more elaborate mechanism or that in cells transformed by H-Ras, there is a preferential utilization of ARF6 over ARF1 that does not occur in the nontransformed cells treated with PDGF. In the cells we have used, ARF1 is not present in the light membrane fraction where the PLD activity elevated in response to mitogenic stimulation is localized (78). This apparent difference in ARF1 dependence suggests that, under some circumstances, ARF1 may localize differently and therefore regulate the PLD activity elevated in response to mitogenic signaling. The data may also reflect differences in BFA sensitivity, since activation of ARF1 is sensitive to BFA under some circumstances (50, 57). In this regard, PDGF-induced PLD activity was completely inhibited by BFA (62), whereas the elevated PLD activity in H-Ras-transformed cells was only partially inhibited by BFA (47).
Santy and Casanova (59) demonstrated that ARNO, via the activation of ARF6, can lead to elevated PLD activity in Madin-Darby canine kidney epithelial cells. These data suggested that that activation of RalA was not needed to elevate PLD activity in these cells. This observation could reflect differences between the epithelial cells used in this study and the fibroblasts we used in our studies. The data from the epithelial cell study could also indicate that ARNO can do more than activate ARF6. Although this study indicated that ARNO did not activate ARF1, Rac1 was activated in these cells, and Rac1 has been implicated as a regulator of PLD1 (6, 27). Thus, the regulation of ARF and PLD activity may be different in different cell types involving different GTPases.
Differential PLD activation by H-Ras and K-Ras.
The differential activation of PLD by H-Ras and K-Ras suggests that H-Ras and K-Ras may have different downstream targets. Differential effects of H-Ras and K-Ras have been reported previously. H-Ras activates phosphatidylinositol-3-kinase more efficiently than K-Ras (79), whereas K-Ras activates Raf1 (72, 79) and Rac1 (74) more efficiently than H-Ras. Interestingly, H-Ras induced apoptosis more efficiently than K-Ras (37, 74). H-Ras was more efficient at focus formation, and K-Ras was more efficient at inducing anchorage-independent growth (72). K-Ras was also shown to be more efficient at inducing cell migration than H-Ras (72). While several differences in the biological effects of H-Ras and K-Ras have been documented, it is not clear how the different effects of H-Ras and K-Ras are generated. It was recently reported that H-Ras and K-Ras are differentially sensitive to mutants of caveolin (58) and fractionate with different membrane microdomains (55). It has also been demonstrated that H-Ras and K-Ras take different paths to the plasma membrane (3, 14), which might explain the reported differential membrane locations of H-Ras and K-Ras (55). In the fractionation study, it was reported that H-Ras but not K-Ras associates with light membrane fractions (55). In contrast, other studies indicated that both H-Ras and K-Ras fractionate with light membrane fractions (24). This differential fractionation behavior of H-Ras and K-Ras was generated by a high-pH method of fractionation, and it has been argued that this method could lead to artifactual dissociation of K-Ras from the light membrane fraction (75). K-Ras associates with the membrane due to both prenylation and a stretch of lysines in the C terminus of K-Ras. Since the pK for lysine is about 10.5, these lysines would be largely uncharged at pH 11, and therefore the forces holding K-Ras on the membrane would be reduced. In contrast, Ha-Ras, which is both prenylated and palmitoylated, would not be similarly affected by the elevated pH. We have performed fractionation studies of membranes from both H-Ras- and K-Ras-transformed cells by using both high- and neutral-pH strategies. Our results showed that at neutral pH, both H-Ras and K-Ras fractionated with the light membrane fraction as reported by Furuchi et al. (24), and with the high-pH method, only H-Ras fractionated with the light membrane fraction as reported by Prior et al. (55; our unpublished results). However, PLD activity is elevated in H-Ras- and not in K-Ras-transformed cells, and the PLD activity in H-Ras-transformed cells is restricted to the light membrane fraction. Therefore, it would appear that H-Ras and K-Ras have some kind of differential location within light membranes or perhaps in distinct light membrane microdomains. This could reflect the different pathways taken to the plasma membrane (3, 14). Thus, while there is still much to be learned as to how H-Ras and K-Ras activate different downstream signaling machinery, the data given here provide further evidence for different signaling pathways mediated by the two GTPases.
The differential activation of PLD by H-Ras and K-Ras may also be of significance in tumor progression. Activating mutations to K-Ras are common in many human cancers, whereas activating mutations to H-Ras are relatively rare and restricted to select tumor types (4). Since the mutations that can activate either H-Ras or K-Ras are similar and since activated forms of both H-Ras and K-Ras transform cells in culture, it is likely that activating mutations to H-Ras, which in theory are just as likely to occur, are selected against. If true, then it is possible that the additional function of PLD activation by H-Ras leads to apoptosis. Consistent with this hypothesis, Walsh and Bar-Sagi (74) have shown that H-Ras induces apoptosis more efficiently than K-Ras. Elevated expression of either PLD1 or PLD2 is able to cooperate with an overexpressed tyrosine kinase to transform rat fibroblasts (38, 44), and PLD activity has been reported to be elevated in some human cancers (52, 80). These data indicate that PLD activity can play a role in mitogenic signaling. However, cells frequently respond to inappropriate mitogenic signals by undergoing apoptosis (31). In this regard, it could be hypothesized that K-Ras is tolerated in an emerging tumor because it does not stimulate the RalA-PLD pathway. Ironically, the Ral pathway appears to be the most critical pathway for H-Ras to transform human cells (26). The data presented here indicate that K-Ras does not activate the Ral pathway in murine fibroblasts. We do not know whether this is also true for human cells. However, the observation that the transformation of human cells by H-Ras is due to activation of the Ral pathway further suggests that, in an emerging tumor, the transformed phenotype induced through this pathway is selected against, preventing the appearance of tumors with activated H-Ras. We previously reported that expression of either PLD1 or PLD2 in 3Y1 rat fibroblasts leads to apoptosis (81). Thus, while elevated PLD activity can contribute to a transformed phenotype, too much PLD signaling in H-Ras-transformed cells may help sensitize these cells to apoptotic cell death and ironically make H-Ras mutations less of a problem in human cancer than mutations to K-Ras because of the ability of these cells to survive.
Acknowledgments
We are thankful to Larry Feig for communicating unpublished results and for many helpful comments on the study. We also thank Sylvain Bourgoin and Richard Kahn for antibodies used in the study.
This investigation was supported by a grant from the National Cancer Institute CA46677 and from a SCORE grant from the National Institutes of Health GM60654 to D.A.F., as well as a grant from the American Heart Association to C.D.-S. Research Centers in Minority Institutions award RR-03037 from the National Center for Research Resources of the National Institutes of Health, which supports infrastructure and instrumentation in the Biological Sciences Department at Hunter College, is also acknowledged.
REFERENCES
- 1.Aguirre Ghiso, J., P. Frankel, Z. Lu, H. Jiang, E. Farias, A. Olsen, L. A. Feig, E. B. de Kier Joffe, and D. A. Foster. 1999. RalA requirement for v-Src- and v-Ras-induced tumorigenicity and overproduction of urokinase-type plasminogen activator and metalloproteases. Oncogene 18:4718-4725. [DOI] [PubMed] [Google Scholar]
- 2.Alam, M. S., Y. Banno, S. Nakashima, and Y. Nozawa. 1995. Defective phospholipase D activation in Ki-ras-transformed NIH3T3 cells: evidence for downstream effector of PLC-γ1 in PDGF-mediated signal transduction. Biochem. Biophys. Res. Commun. 207:460-466. [DOI] [PubMed] [Google Scholar]
- 3.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos, J. L. 1989. Ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 5.Boshans, R. L., S. Szanto, L. van Aelst, and C. D'Souza-Schorey. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20:3685-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, E. P., D. J. Uhlinger, and J. D. Lambeth. 1993. Neutrophil phospholipase D is activated by a membrane-associated Rho family small molecular weight GTP-binding protein. J. Biol. Chem. 268:21509-21512. [PubMed] [Google Scholar]
- 7.Brown, H. A., S. Gutowski, C. R. Moomaw, C. Slaughter, and P. C. Sternweis. 1993. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell 75:1137-1144. [DOI] [PubMed] [Google Scholar]
- 8.Cantor, S. B., T. Urano, and L. A. Feig. 1995. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 15:4578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carnero, A., A. Cuadrado, L. del Peso, and J. C. Lacal. 1994. Activation of type D phospholipase by serum stimulation and ras-induced transformation in NIH3T3 cells. Oncogene 9:1387-1395. [PubMed] [Google Scholar]
- 10.Caumont, A. S., M. C. Galas, N. Vitale, D. Aunis, and M. F. Bader. 1998. Regulated exocytosis in chromaffin cells. Translocation of ARF6 stimulates a plasma membrane-associated phospholipase D. J. Biol. Chem. 273:1373-1379. [DOI] [PubMed] [Google Scholar]
- 11.Caumont, A. S., N. Vitale, M. Gensse, M. C. Galas, J. E. Casanova, and M. F. Bader. 2000. Identification of a plasma membrane-associated guanine nucleotide exchange factor for ARF6 in chromaffin cells. Possible role in the regulated exocytotic pathway. J. Biol. Chem. 275:15637-15644. [DOI] [PubMed] [Google Scholar]
- 12.Chardin, P., S. Paris, B. Antonny, S. Robineau, S. Beraud-Dufour, C. L. Jackson, and M. Chabre. 1996. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 384:481-484. [DOI] [PubMed] [Google Scholar]
- 13.Chavrier, P., and B. Goud. 1999. The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11:466-475. [DOI] [PubMed] [Google Scholar]
- 14.Choy, E., V. K. Chiu, J. Silletti, M. Feokitistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 15.Cockcroft, S., G. M. Thomas, A. Fensome, B. Geny, E. Cunningham, I. Gout, I. Hiles, N. F. Totty, O. Truong, and J. J. Hsuan. 1994. Phospholipase D: a downstream effector of ARF in granulocytes. Science 263:523-526. [DOI] [PubMed] [Google Scholar]
- 16.Colley, W. C., T. C. Sung, R. Roll, J. Jenco, S. M. Hammond, Y. Altshuller, D. Bar-Sagi, A. J. Morris, and M. A. Frohman. 1997. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr. Biol. 7:191-201. [DOI] [PubMed] [Google Scholar]
- 17.Czarny, M., Y. Lavie, G. Fiucci, and M. Liscovitch. 1999. Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-182-101. J. Biol. Chem. 274:2717-2724. [DOI] [PubMed] [Google Scholar]
- 18.D'Souza-Schorey, C., G. Li, M. I. Colombo, and P. D. Stahl. 1995. A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267:1175-1178. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza-Schorey, C., E. van Donselaar, V. W. Hsu, C. Yang, P. D. Stahl, and P. J. Peters. 1998. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J. Cell Biol. 140:603-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Exton, J. H. 2000. Phospholipase D. Ann. N. Y. Acad. Sci. 905:61-68. [DOI] [PubMed] [Google Scholar]
- 21.Fensome, A., E. Cunningham, S. Prosser, S. K. Tan, P. Swigart, G. Thomas, J. Hsuan, and S. Cockcroft. 1996. ARF and PITP restore GTPγS-stimulated protein secretion from cytosol-depleted HL60 cells by promoting PIP2 synthesis. Curr. Biol. 6:730-738. [DOI] [PubMed] [Google Scholar]
- 22.Franco, M., P. J. Peters, J. Boretto, E. van Donselaar, A. Neri, C. D'Souza-Schorey, and P. Chavrier. 1999. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 18:1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankel, P. A., M. Ramos, J. Flom, S. Bychenok, E. Kerkhoff, U. R. Rapp, L. A. Feig, and D. A. Foster. 1999. Ral and Rho dependent activation of phospholipase D in v-Raf transformed cells. Biochem. Biophys. Res. Commun. 255:502-507. [DOI] [PubMed] [Google Scholar]
- 24.Furuchi, T., and R. G. Anderson. 1998. Cholesterol depletion of caveolae causes hyperactivation of extracellular signal-related kinase (ERK). J. Biol. Chem. 273:21099-21104. [DOI] [PubMed] [Google Scholar]
- 25.Guillemain, I., and J. H. Exton. 1997. Effects of brefeldin A on phosphatidylcholine phospholipase D and inositolphospholipid metabolism in HL-60 cells. Eur. J. Biochem. 249:812-819. [DOI] [PubMed] [Google Scholar]
- 26.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond, S. M., Y. M. Altshuller, T. C. Sung, S. A. Rudge, K. Ross, J. Engebrecht, A. J. Morris, and M. A. Frohman. 1995. Human ARF-activated phosphatidylcholine-specific phospholipase D defines a new highly conserved gene family. J. Biol. Chem. 270:29640-29643. [DOI] [PubMed] [Google Scholar]
- 28.Hammond, S. M., J. M. Jenco, S. Nakashima, K. Cadwallader, Q. Gu, S. Cook, Y. Nozawa, G. D. Prestwich, M. A. Frohman, and A. J. Morris. 1997. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C α. J. Biol. Chem. 272:3860-3868. [DOI] [PubMed] [Google Scholar]
- 29.Honda, A., M. Nogami, T. Yokozeki, M. Yamazaki, H. Nakamura, H. Watanabe, K. Kawamoto, K. Nakayama, A. J. Morris, M. A. Frohman, and Y. Kanaho. 1999. Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99:521-532. [DOI] [PubMed] [Google Scholar]
- 30.Hornia, A., Z. Lu, T. Sukezane, M. Zhong, T. Joseph, P. Frankel, and D. A. Foster. 1999. Antagonistic effects of protein kinase C α and δ on both transformation and phospholipase D activity mediated by the epidermal growth factor receptor. Mol. Cell. Biol. 19:7672-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hueber, A. O., and G. I. Evan. 1998. Traps to catch unwary oncogenes. Trends Genet. 14:364-367. [DOI] [PubMed] [Google Scholar]
- 32.Humeau, Y., N. Vitale, S. Chasserot-Golaz, J. L. Dupont, G. Du, M. A. Frohman, M. F. Bader, and B. Poulain. 2001. A role for phospholipase D1 in neurotransmitter release. Proc. Natl. Acad. Sci. USA 98:15300-15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins, G. H., P. L. Fisette, and R. A. Anderson. 1994. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J. Biol. Chem. 269:11547-11554. [PubMed] [Google Scholar]
- 34.Jiang, H., Z. Lu, J.-Q Luo, A. Wolfman, and D. A. Foster. 1995. Ras mediates the activation of phospholipase D by v-Src. J. Biol. Chem. 270:6006-6009. [DOI] [PubMed] [Google Scholar]
- 35.Jiang, H., J.-Q. Luo, T. Urano, Z. Lu, D. A. Foster, and L. A. Feig. 1995. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature 378:409-412. [DOI] [PubMed] [Google Scholar]
- 36.Jiang, Y.-W., J. Song, Q. Zang, and D. A. Foster. 1994. Phosphatidylcholine-specific phospholipase D activity is elevated in v-Fps-transformed cells. Biochem. Biophys. Res. Commun. 203:1195-1203. [DOI] [PubMed] [Google Scholar]
- 37.Joneson, T., and D. Bar-Sagi. 1999. Suppression of Ras-induced apoptosis by the Rac GTPase. Mol. Cell. Biol. 19:5892-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph, T., R. Wooden, A. Bryant, M. Zhong, Z. Lu, and D. A. Foster. 2001. Transformation of cells overexpressing a tyrosine kinase by phospholipase D1 and D2. Biochem. Biophys. Res. Commun. 289:1019-1024. [DOI] [PubMed] [Google Scholar]
- 39.Joseph, T., A. Bryant, R. Wooden, E. Kerkhoff, U. R. Rapp, and D. A. Foster. 2002. Phospholipase D overcomes cell cycle arrest induced by high-intensity Raf signaling. Oncogene 21:3651-3658. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J. H., S. D. Lee, J. M. Han, T. G. Lee, Y. Kim, J. B. Park, J. D. Lambeth, P. G. Suh, and S. H. Ryu. 1998. Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA. FEBS Lett. 430:231-235. [DOI] [PubMed] [Google Scholar]
- 41.Kim, J. H., J. M. Han, S. Lee, Y. Kim, T. G. Lee, J. B. Park, S. D. Lee, P. G. Suh, and S. H. Ryu. 1999. Phospholipase D1 in caveolae: regulation by protein kinase Cα and caveolin-1. Biochemistry 38:3763-3769. [DOI] [PubMed] [Google Scholar]
- 42.Lisanti, M. P., Z. Tang, P. E. Scherer, E. Kubler, A. J. Koleske, and M. Sargiacomo. 1995. Caveolae, transmembrane signalling and cellular transformation. Mol. Membr. Biol. 12:121-124. [DOI] [PubMed] [Google Scholar]
- 43.Liscovitch, M., M. Czarny, G. Fiucci, Y. Lavie, and X. Tang. 1999. Localization and possible functions of phospholipase D isozymes. Biochim. Biophys. Acta 1439:245-263. [DOI] [PubMed] [Google Scholar]
- 44.Lu, Z., A. Hornia, T. Joseph, T. Sukezane, P. Frankel, M. Zhong, S. Bychenok, L. Xu, L. A. Feig, and D. A. Foster. 2000. Phospholipase D and RalA cooperate with the EGF receptor to transform 3Y1 rat fibroblasts. Mol. Cell. Biol. 20:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas, L., L. del Peso, P. Rodriguez, V. Penalva, and J. C. Lacal. 2000. Ras protein is involved in the physiological regulation of phospholipase D by platelet derived growth factor. Oncogene 19:431-437. [DOI] [PubMed] [Google Scholar]
- 46.Luo, J.-Q., X. Liu, S. M. Hammond, W. C. Colley, L. A. Feig, M. A. Frohman, A. J. Morris, and D. A. Foster. 1997. Ral interacts directly with the ARF-responsive PIP2-dependent phospholipase D1. Biochem. Biophys. Res. Commun. 235:854-859. [DOI] [PubMed] [Google Scholar]
- 47.Luo, J.-Q., X. Liu, P. Frankel, T. Rotunda, M. Ramos, J. Flom, H. Jiang, L. A. Feig, A. J. Morris, R. A. Kahn, and D. A. Foster. 1998. Functional association between RalA and ARF in active phospholipase D complexes. Proc. Natl. Acad. Sci. USA 95:3632-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massenburg, D., J. S. Han, M. Liyanage, W. A. Patton, S. G. Rhee, J. Moss, and M. Vaughan. 1994. Activation of rat brain phospholipase D by ADP-ribosylation facators 1,5, and 6: separation of ADP-ribosylation factor-dependent and oleate-dependent enzymes. Proc. Natl. Acad. Sci. USA 91:11718-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melendez, A. J., M. M. Harnett, and J. M. Allen. 2001. Crosstalk between ARF6 and protein kinase Cα in FcγRI-mediated activation of phospholipase D1. Curr. Biol. 11:869-874. [DOI] [PubMed] [Google Scholar]
- 50.Moss, J., and M. Vaughan. 1998. Molecules in the ARF orbit. J. Biol. Chem. 273:21431-21434. [DOI] [PubMed] [Google Scholar]
- 51.Mwanjewe, J., M. Spitaler, M. Ebner, M. Windegger, M. Geiger, S. Kampfer, J. Hofmann, F. Uberall, and H. H. Grunicke. 2001. Regulation of phospholipase D isoenzymes by transforming Ras and atypical protein kinase C-iota. Biochem. J. 359:211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noh, D. Y., S. J. Ahn, R. A. Lee, I. A. Park, J. H. Kim, P. G. Suh, S. H. Ryu, K. H. Lee, and J. S. Han. 2000. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 161:207-214. [DOI] [PubMed] [Google Scholar]
- 53.Peyroche, A., B. Antonny, S. Robineau, J. Acker, J. Cherfils, and C. L. Jackson. 1999. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3:275-285. [DOI] [PubMed] [Google Scholar]
- 54.Powner, D. J., M. N. Hodgkin, and M. J. Wakelam. 2002. Antigen-stimulated activation of phospholipase D1b by Rac1, ARF6, and PKCα in RBL-2H3 cells. Mol. Biol. Cell 13:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prior, I. A., A. Harding, J. Yan, J. Sluimer, R. G. Parton, and J. F. Hancock. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368-375. [DOI] [PubMed] [Google Scholar]
- 56.Rizzo, M. A., K. Shome, C. Vasudevan, D. B. Stolz, T. C. Sung, M. A. Frohman, S. C. Watkins, and G. Romero. 1999. Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway. J. Biol. Chem. 274:1131-1139. [DOI] [PubMed] [Google Scholar]
- 57.Robineau, S., M. Chabre, and B. Antonny. 2000. Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA 97:9913-9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1:98-105. [DOI] [PubMed] [Google Scholar]
- 59.Santy, L. C., and J. E. Casanova. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt, M., M. Vo, M. Thiel, B. Bauer, A. Granna, E. Tapp, R. H. Cool, J. deGunzburg, C. von Eichel-Streiber, and K. H. Jakobs. 1998. Specific inhibition of phorbol ester-stimulated phospholipase D by Clostridium sordellii lethal toxin and Clostridium difficile toxin B-1470 in HEK-293 cells. Restoration by Ral GTPases. J. Biol. Chem. 273:7413-7422. [DOI] [PubMed] [Google Scholar]
- 61.Shen, Y., L. Xu, and D. A. Foster. 2001. A role for phospholipase D in receptor-mediated endocytosis. Mol. Cell. Biol. 21:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shome, K., Y. Nie, and G. Romero. 1998. ADP-ribosylation factor proteins mediate agonist-induced activation of phospholipase D. J. Biol. Chem. 273:30836-30841. [DOI] [PubMed] [Google Scholar]
- 63.Shome, K., M. A. Rizzo, C. Vasudevan, B. Andresen, and G. Romero. 2000. The activation of phospholipase D by endothelin-1, angiotensin II, and platelet-derived growth factor in vascular smooth muscle A10 cells is mediated by small G proteins of the ADP-ribosylation factor family. Endocrinology 141:2200-2208. [DOI] [PubMed] [Google Scholar]
- 64.Shome, K., C. Vasudevon, and G. Romero. 1997. ARF proteins mediate insulin-dependent activation of phospholipase D. Curr. Biol. 7:387-396. [DOI] [PubMed] [Google Scholar]
- 65.Someya, A., M. Sata, K. Takeda, G. Pacheco-Rodriguez, V. J. Ferrans, J. Moss, and M. Vaughan. 2001. ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. USA 98:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song, J., L. M. Pfeffer, and D. A. Foster. 1991. v-Src increases diacylglycerol levels via a type D phospholipase-mediated hydrolysis of phosphatidylcholine. Mol. Cell. Biol. 11:4903-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song, J., and D. A. Foster. 1993. v-Src activates a phospholipase D activity that is distinguishable from phospholipase D activity activated by protein kinase C. Biochem. J. 294:711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song, S. K., S. Li, T. Okamoto, L. A. Quilliam, M. Sargiacomo, and M. P. Lisanti. 1996. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. J. Biol. Chem. 271:9690-9697. [DOI] [PubMed] [Google Scholar]
- 69.Toda, K., M. Nogami, K. Murakami, Y. Kanaho, and K. Nakayama. 1999. Colocalization of phospholipase D1 and GTP-binding-defective mutant of ADP-ribosylation factor 6 to endosomes and lysosomes. FEBS Lett. 442:221-225. [DOI] [PubMed] [Google Scholar]
- 70.Urano, T., R. Emkey, and L. A. Feig. 1996. Ral GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 16:810-816. [PMC free article] [PubMed] [Google Scholar]
- 71.Vitale, N., A. S. Caumont, S. Chasserot-Golaz, G. Du, S. Wu, V. A. Sciorra, A. J. Morris, M. A. Frohman, and M. F. Bader. 2001. Phospholipase D1: a key factor for the exocytotic machinery in neuroendocrine cells. EMBO J. 20:2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voice, J. K., R. L. Klemke, A. Le, and J. H. Jackson. 1999. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J. Biol. Chem. 274:17164-17170. [DOI] [PubMed] [Google Scholar]
- 73.Voss, M., P. A. Weernink, S. Haupenthal, U. Moller, R. H. Cool, B. Bauer, J. H. Camonis, K. H. Jakobs, and M. Schmidt. 1999. Phospholipase D stimulation by receptor tyrosine kinases mediated by protein kinase C and a Ras/Ral signaling cascade. J. Biol. Chem. 274:34691-34698. [DOI] [PubMed] [Google Scholar]
- 74.Walsh, A. B., and D. Bar-Sagi. 2001. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J. Biol. Chem. 276:15609-15615. [DOI] [PubMed] [Google Scholar]
- 75.White, M. A., and R. G. W. Anderson. 2001. Which Ras rides the raft? Nat. Cell Biol. 3:E172.. [DOI] [PubMed] [Google Scholar]
- 76.Wolthuis, R. M. F., B. Franke, M. van Triest, B. Bauer, R. H. Cool, J. H. Camonis, J.-W. Akkerman, and J. L. Bos. 1998. Activation of the small GTPase Ral in platelets. Mol. Cell. Biol. 18:2486-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolthuis, R. M. F., F. Zwartkruis, T. C. Moen, and J. L. Bos. 1998. Ras-dependent activation of the small GTPase Ral. Curr. Biol. 8:471-474. [DOI] [PubMed] [Google Scholar]
- 78.Xu, L., Y. Shen, T. Joseph, A. Bryant, J. Q. Luo, P. Frankel, T. Rotunda, and D. A. Foster. 2000. Mitogenic phospholipase D activity is restricted to caveolin-enriched membrane microdomains. Biochem. Biophys. Res. Commun. 273:77-83. [DOI] [PubMed] [Google Scholar]
- 79.Yan, J., S. Roy, A. Apolloni, A. Lane, and J. F. Hancock. 1998. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 273:24052-24056. [DOI] [PubMed] [Google Scholar]
- 80.Zhao, Y., H. Ehara, Y. Akao, M. Shamoto, Y. Nakagawa, Y. Banno, T. Deguchi, N. Ohishi, K. Yagi, and Y. Nozawa. 2000. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem. Biophys. Res. Commun. 278:140-143. [DOI] [PubMed] [Google Scholar]
- 81.Zhong, M., T. Joseph, D. Jackson, S. Beychenok, and D. A. Foster. 2002. Elevated phospholipase D induces apoptosis in normal rat fibroblasts. Biochem. Biophys. Res. Commun. 298:474-477. [DOI] [PubMed] [Google Scholar]