Abstract
Initiation of translation of encephalomyocarditis virus mRNA is mediated by an internal ribosome entry site (IRES) comprising structural domains H, I, J-K, and L immediately upstream of the initiation codon AUG at nucleotide 834 (AUG834). Assembly of 48S ribosomal complexes on the IRES requires eukaryotic initiation factor 2 (eIF2), eIF3, eIF4A, and the central domain of eIF4G to which eIF4A binds. Footprinting experiments confirmed that eIF4G binds a three-way helical junction in the J-K domain and showed that it interacts extensively with RNA duplexes in the J-K and L domains. Deletion of apical hairpins in the J and K domains synergistically impaired the binding of eIF4G and IRES function. Directed hydroxyl radical probing, done by using Fe(II) tethered to surface residues in eIF4G's central domain, indicated that it is oriented with its N terminus towards the base of domain J and its C terminus towards the apex. eIF4G recruits eIF4A to a defined location on the IRES, and the eIF4G/eIF4A complex caused localized ATP-independent conformational changes in the eIF4G-binding region of the IRES. This complex also induced more extensive conformational rearrangements at the 3′ border of the ribosome binding site that required ATP and active eIF4A. We propose that these conformational changes prepare the region flanking AUG834 for productive binding of the ribosome.
Translation of a subset of cellular and viral mRNAs is initiated by end-independent binding of ribosomes to an internal ribosome entry site (IRES) in the 5′ untranslated region. IRESs were first identified in different picornavirus RNA genomes, and these IRESs are now classified into two major groups, one of which includes those of the encephalomyocarditis virus (EMCV), Theiler's murine encephalitis virus (TMEV), and foot-and-mouth disease virus (FMDV) (17). The EMCV IRES is ∼450 nucleotides (nt) long and comprises H, I, J-K, and L structural domains upstream of the initiation codon AUG at nucleotide 834 (AUG834). Ribosomal initiation complexes attach directly to AUG834, and initiation does not involve scanning (19, 35). Recent analysis has begun to resolve molecular details of the mechanism of initiation on EMCV-like IRESs. Viral and cellular IRES-containing mRNAs can be translated under conditions when the cap-mediated mode of end-dependent initiation that is used by most mRNAs is inhibited (12), and studies of initiation on viral IRESs are therefore yielding insights into mechanisms that enable selective translation of mRNAs in the cell.
The stage that differentiates IRES-mediated initiation from the conventional cap-mediated mode of initiation is the mechanism by which the 43S ribosomal preinitiation complex engages mRNA to form a 48S complex at the initiation codon (16, 36). The 43S complex consists of a 40S ribosomal subunit, initiator tRNA, and eukaryotic initiation factors (eIFs), including eIF2 and eIF3. In the process of cap-mediated initiation, eIF4F is thought to create a binding site for this complex adjacent to the 5′-terminal m7G cap by inducing local conformational changes in bound mRNA (42) and by enhancing the binding of the eIF3 component of 43S complexes to mRNA (5). The 43S complex binds to the mRNA through a network of protein-protein and protein-RNA interactions and then scans to the initiation codon. Many of these interactions involve eIF4F, which comprises the cap-binding protein eIF4E, the RNA helicase eIF4A, and either the eIF4GI or eIF4GII isoform. The amino-terminal one-third of eIF4G (amino acid residues 1 to 675) binds eIF4E and the poly(A) binding protein (10, 24). The central one-third of eIF4G, which binds to eIF3 and contains one of the two eIF4A-binding sites, consists of five pairs of α-helices that are known as HEAT repeats (29).
Biochemical reconstitution of 48S complex formation from purified components on the EMCV IRES has shown that this process is ATP dependent and that it requires the same initiation factors as cap-dependent initiation, except for eIF1, eIF1A, and intact eIF4F (34, 35, 38). Similar results have been described for FMDV and TMEV IRESs (40). 48S complexes assembled in this way are competent to complete all remaining steps in initiation (T. V. Pestova, unpublished data). Initiation on the EMCV IRES has no requirement for eIF4E, and eIF4F can be replaced by eIF4A and a central domain of eIF4G, namely, eIF4GI, located from amino acid residues 737 to 989 (eIF4GI737-989) (26). The requirement for eIF4A and eIF4G is consistent with the profound inhibition of EMCV and FMDV translation by dominant-negative mutant forms of eIF4A, such as the R362Q substitution mutant, which sequester the eIF4A/eIF4G complex in an inactive form (33, 47). Initiation on the EMCV IRES does not involve scanning, and the basis for the requirement for ATP and for active eIF4A for initiation on the IRES is not known. eIF4A binds to the central α-helical domains of eIF4GI737-989 and eIF4GII745-1003 and greatly enhances their affinity for the IRES in an ATP-independent manner (26, 29).
In addition to requiring canonical initiation factors, assembly of 48S complexes on some picornavirus IRESs is enhanced by IRES trans-acting factors that bind to defined sites on them and induce conformational changes in these IRESs that facilitate binding of the eIF4G/eIF4A complex (3, 20, 35, 37, 40). IRESs are therefore not static scaffolds but undergo conformational transitions as they bind components of the translation apparatus during the initiation process. Initiation on the IRES of the EMCV strain used in the experiments described here does not require IRES trans-acting factors (35). However, the EMCV IRES becomes dependent on the pyrimidine tract-binding protein as a result of mutations either in the domain of the IRES to which eIF4G binds or in the coding region adjacent to the initiation codon (20).
EMCV-like IRESs have conserved structures that are thought to orient binding sites for components of the translation apparatus to promote recruitment of the 43S complex and consequent assembly of the 48S complex (17, 37). eIF4G binds specifically to the J-K domain of these IRESs, and this interaction is necessary for initiation (22, 26, 27, 29, 38, 40, 46). The binding site on the EMCV IRES has been localized by footprinting (22); residues are protected in the A-rich loop at the junction of the J and K domains and in the flanking J2, J3, and K1 helices (Fig. 1). Deletion of nucleotides in the A-rich loop severely reduces both binding of eIF4G/eIF4A and IRES-mediated initiation (35). Although the J-K domain is essential, it is by itself not sufficient for EMCV IRES function, and experiments to date account only for the function of the central core of this domain. The extensive sequence conservation in EMCV-like IRESs extends over much of the J-K domain (18), including regions such as the terminal loops of the J and K domains and the J3 helix that have been found to be important for initiation (13, 32, 48). The adjacent domain I also plays an essential but as yet undefined role in EMCV IRES function, and the flanking domain H, domain L, and sequences downstream of domain L all also play accessory roles (7, 9, 18, 20). The molecular basis for the importance of these regions of the IRES has not yet been determined.
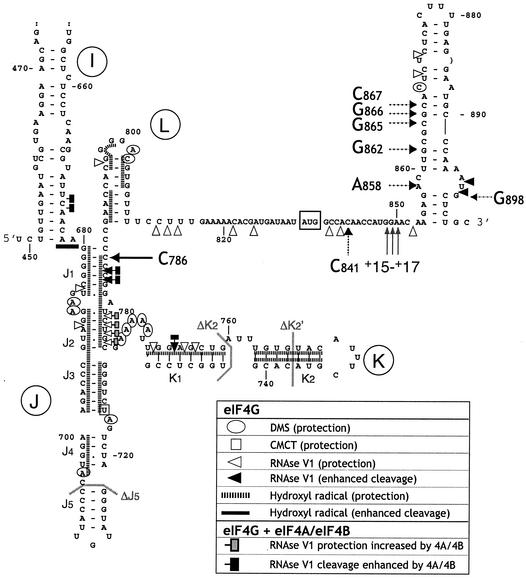
FIG. 1.
Interactions of eIF4A, eIF4B, and eIF4G with the EMCV IRES assessed by chemical and enzymatic footprinting. Sites at which chemical modification by DMS and CMCT and enzymatic cleavage by RNase V1 of the IRES were altered by eIF4G737-1600 alone and in the presence of eIFA/eIF4B are indicated by the symbols shown in the key at bottom right. Footprinting data for eIF4G alone are from previous analyses (22) except for those for protection of A704 (see Fig. 4A). These data are mapped onto a secondary structure model of nt 449 to 904 of the EMCV IRES and the adjacent coding region. Helical segments of the J-K domain in this model are designated J1 to J5 and K1 and K2 (13). The initiation codon AUG834 is boxed. A proposed RNA structure (14) in the region encoding the leader (L) protein is included. Only the lower part of domain I is shown; the structure shown is that proposed previously (39). Nucleotides deleted in the ΔJ5, ΔK2, and ΔK2′ mutants are indicated by gray lines. The positions of the toe prints due to RT arrest in the J-K domain by bound eIF4G and downstream of the initiation codon by bound eIF4G/eIF4A are indicated by black and dashed arrows, respectively. The positions of toe prints +15 to +17 nt downstream of the initiation codon caused by RT arrest by bound 48S complexes are indicated by gray arrows.
The interaction of the eIF4G/eIF4A complex with the EMCV IRES is therefore of central importance in this initiation mechanism, but important details of its function remain unresolved. We conducted a series of experiments to determine the orientation of the central HEAT-repeat domain of eIF4G on the J-K domain by using directed hydroxyl radical cleavage to characterize the nucleotide determinants in the IRES required for binding the eIF4G/eIF4A complex and to investigate the consequences on the conformation of the IRES and downstream coding sequences of binding to this complex in the presence and absence of ATP. We report that eIF4G makes much more extensive interactions with the J-K and L domains when it binds to the IRES than was previously recognized. The integrity of the terminal hairpins of J and K domains is necessary for stable binding of eIF4G and eIF4F to the EMCV IRES and for initiation on it. The bound eIF4G/eIF4A complex induced conformational changes in the coding region adjacent to the IRES that are ATP dependent and that require eIF4A to be active. We propose that these conformational changes in the IRES mediated by eIF4G/eIF4A prepare the region of the IRES flanking the initiation codon so that the 40S subunit can bind to it productively, leading to initiation of translation.
MATERIALS AND METHODS
Enzymes and reagents.
DNA restriction endonucleases and modifying enzymes were from New England BioLabs (Beverly, Mass.). Rabbit reticulocyte lysate (RRL) for in vitro translation and avian myeloblastosis virus reverse transcriptase (AMV-RT) were purchased from Promega Corp. (Madison, Wis.). RRL for purification of ribosomes and initiation factors was from Green Hectares (Oregon, Wis.). Native rabbit tRNA was from Novagen (Madison, Wis.). Unlabeled nucleoside triphosphates, RNAguard RNase inhibitor, and RNase V1 were from Amersham Biosciences (Piscataway, N.J.). Dimethyl sulfate (DMS) was from Aldrich (Milwaukee, Wis.). Radiochemicals [35S]methionine (44 TBq/mmol), 35S-dATP (37 TBq/mmol), and [32P]dATP (220 TBq/mmol) were from ICN Radiochemicals (Irvine, Calif.). Escherichia coli methionyl-tRNA synthetase from E. coli strain MRE 600, ribosomal 40S subunits, and native and recombinant initiation factors were purified as described previously (26, 29, 34, 35, 37, 38).
Plasmids.
EMCV pTE1 (9), pTE3-GUS (2), and pJK (26) transcription vectors and vectors for expression of recombinant wild-type eIF4A (35), R362Q mutant eIF4A (37), eIF4GI737-1116, eIF4GI737-1009, and eIF4GI737-1600 (26) and eIF4GII745-1003 (29) have been described previously. The eIF4GI plasmids have been renamed to take into account a recent revision to the sequence of the largest eIF4GI isoform, extending its amino terminus by 40 amino acid residues (4). Deletion mutants of the EMCV IRES were generated in pTE3-GUS and pJK plasmids by using a two-step PCR. Substitution mutations in eIF4GI737-1116 were made exactly as described previously (26). All mutations were confirmed by sequencing the complete IRES or the eIF4GI737-1116 coding sequence, as appropriate.
In vitro translation.
mRNA comprising the EMCV IRES linked to the β-glucuronidase open reading frame was transcribed by using T7 RNA polymerase and was translated in RRL in accordance with the manufacturer's instructions in the presence of [35S]methionine. Translation products were resolved by electrophoresis with 12% polyacrylamide gel. Gels were dried and exposed to X-ray film. The efficiency of translation was quantified by using a Molecular Dynamics PhosphorImager.
Assembly and analysis of ribosomal complexes.
Ribonucleoprotein and ribosomal 48S complexes were assembled on EMCV RNAs and analyzed by primer extension by using the primers 5′-GTCAATAACTCCTCTGG-3′ (complementary to EMCV nt 957 to 974) and 5′-GGGGGATGTGCTGCAACC-3′ (complementary to nt 368 to 351 in the polylinker of pTZ18R), as appropriate, and AMV-RT in the presence of [32P]dATP, as described previously (26, 35, 38).
Footprinting analysis of initiation factor-IRES complexes.
Free or initiation factor-bound RNAs in binding buffer were probed with DMS or RNase V1, and chemically modified or enzymatically cleaved RNAs were analyzed by reverse transcription, as described previously (21, 22, 40). ATP was present in all probing reactions that contained eIF4A or eIF4F. Hydroxyl radical footprinting was done essentially as described previously (15).
Preparation of Fe(II)-BABE-derivatized eIF4G.
Cysteine-containing mutants of eIF4GI737-1116 (0.2 to 0.5 μg/μl in 95 μl of buffer containing 80 mM K+-HEPES [pH 7.6], 300 mM KCl, and 10% glycerol) were conjugated with 5 μl of 20 mM Fe(II)-bromoacetyl-amidobenzyl-EDTA (BABE) (Dojindo Molecular Technologies, Inc., Gaithersburg, Md.) by incubation at 37°C for 30 min essentially as described previously (6). Unincorporated Fe-BABE was separated from derivatized eIF4G by loading reaction mixtures onto pretreated Microcon YM-30 microconcentrators and washing four times with the incubation buffer. Fe-eIF4G samples were typically recovered in 50 to 80 μl of buffer. The accessibility of each introduced cysteine residue for derivatization and the efficiency of the Fe(II)-BABE tethering reaction were assessed by using the reaction with the thiol-specific fluorescent coumarin derivative DCIA (Molecular Probes, Eugene, Oreg.) as described previously (6).
Directed hydroxyl radical probing.
Ribonucleoprotein complexes were assembled by incubating 0.2 μg of EMCV RNA (nt 280 to 974) in 40 μl of buffer containing 80 mM K+-HEPES (pH 7.6), 300 mM KCl, 2 mM MgCl2, and 10% glycerol with 0.8 to 1 μg of Fe-eIF4G (and, where indicated, 1 μg of eIF4A) at 37°C for 5 min and then placing them on ice. Fenton chemistry was initiated by the addition of 2 μl of a freshly prepared mixture of 0.1 M ascorbic acid and 0.5% H2O2 to each sample to generate hydroxyl radicals in the vicinity of the tethered Fe(II). Reactions were quenched after incubation for 10 min in ice by the addition of 20 μl of 20 mM thiourea. EMCV RNA was isolated immediately by phenol extraction followed by ethanol precipitation, and sites of hydroxyl radical cleavage were located by primer extension analysis with the primer 5′-CGGTATTGTAGAGCAGAGC-3′ (complementary to EMCV nt 854 to 836) and AMV-RT in the presence of [32P]dATP.
RESULTS
In vitro translation of mRNAs containing mutations in the EMCV IRES J-K domain.
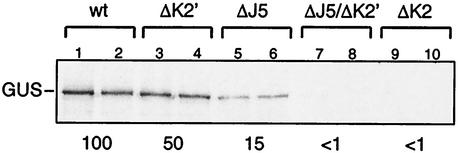
To address the basis for the importance of the terminal J5 and K2 hairpins for IRES activity, mutant IRESs lacking these hairpins were created and named IRES-ΔJ5, IRES-ΔK2′, IRES-ΔK2 and IRES-ΔJ5ΔK2′ (Fig. 1). Chemical probing and toe printing of these RNAs in the absence of initiation factors (see below) indicated that these mutations did not alter the IRES secondary structure except in the immediate vicinity of the deletions. IRES-mediated translation of a β-glucuronidase (GUS) reporter was almost completely abolished by deletion ΔK2 (Fig. 2, lanes 9 and 10), whereas the smaller ΔK2′ deletion reduced translation by half (lanes 3 and 4). Deletion ΔJ5 reduced IRES-mediated translation to 15% of the level of the wild-type IRES (lanes 5 and 6), and translation was synergistically impaired by combining deletions ΔJ5 and ΔK2′ (compare lanes 7 and 8 with lanes 3 and 4 and lanes 5 and 6). These results confirm the importance of J5 and K2 hairpins for IRES function (13, 32, 48).
FIG. 2.
The influence of the J5 and K2 subdomains on EMCV IRES function in in vitro translation. Translation in 12.5 μl of rabbit reticulocyte lysate of 0.5 (lanes 2, 3, 5, 7, and 9) or 0.75 μg (lanes 1, 4, 6, 8, and 10) of EMCV RNA (nt 315 to 844)/GUS mRNA (lanes 1 and 2) and of mutant derivatives ΔK2′ (lanes 3 and 4), ΔJ5 (lanes 5 and 6), ΔJ5/ΔK2′ (lanes 7 and 8), and ΔK2 (lanes 9 and 10) is shown. The numbers below the translation products derived from each mRNA represent the efficiencies of their translation compared to that of wild-type (wt) mRNA, which is by definition 100%. The position of the GUS translation product is indicated to the left.
Binding of eIF4G and eIF4F to IRESs containing deletions in the J-K domain.
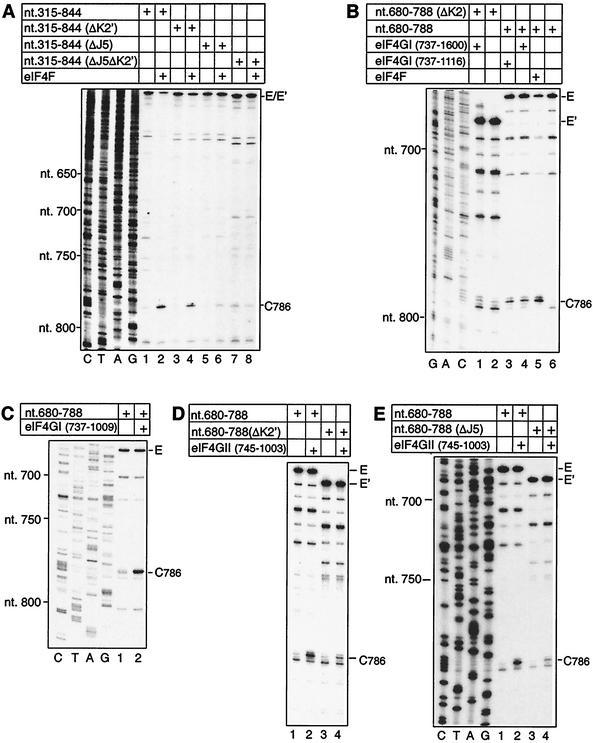
The interaction of eIF4F with the IRES was first identified by toe printing (35, 38), and this method was used to analyze the binding of eIF4F and its eIF4G subunit to these mutant IRESs. Toe printing involves cDNA synthesis by AMV-RT on RNA to which a ribosome or protein is bound. cDNA synthesis is arrested by the bound complex, yielding a stop (toe print) at its leading edge. Stops that occur in the absence of factors are due to RT arrest by stable substructures in the IRES. Binding of purified eIF4F to the IRES was not impaired by deletion of the H and I domains (data not shown) or by the additional deletion of domain L and downstream sequences (Fig. 3B, lane 5). Purified recombinant eIF4GI737-1600, eIF4GI737-1116, eIF4GI737-1009, and eIF4GII745-1003 all bound specifically to EMCV nt 680 to 788 (Fig. 3B, lanes 3 and 4; Fig. 3C, lane 2; Fig. 3D, lane 2; and Fig. 3E, lane 2). The numbering of amino acid residues in eIF4GI is based on the sequence of the longest form of this factor (4). The numbering of amino acid residues in eIF4GII is based on previous work (11). The J-K domain therefore contains determinants necessary for specific, stable binding of eIF4F and both eIF4G isoforms.
FIG. 3.
Influence of the J5 and K2 subdomains on the interaction of eIF4G and eIF4F with the J-K domain of the EMCV IRES. Shown is toe print analysis of ribonucleoprotein complex formation by binding of eIF4F (A and B), eIF4GI fragments (B and C), and eIF4GII fragments (D and E) to wild-type and mutant EMCV (nt 315 to 844) RNAs (A) and wild-type and mutant EMCV (nt 680 to 788) RNAs (B to E), as indicated. The position of the stop site due to binding of eIF4G/eIF4F is indicated at C786, and the positions of full-length cDNAs and cDNAs with internal deletions are marked as E and E′, respectively. Reference lanes C, T, A, and G depict the EMCV cDNA sequence except in panel B, which shows the sequence of the ΔK2 mutant.
The interaction of eIF4G/eIF4F with the J-K domain was reduced slightly by the ΔK2′ deletion, more by the ΔJ5 deletion, and almost completely by the ΔK2 deletion (Fig. 3A, lanes 2, 4, and 6, and Fig. 3B, lane 1). Binding of eIF4GII745-1003 to the J-K domain was similarly reduced by these deletions (e.g., Fig. 3D and E, lanes 4), consistent with previous reports that determinants for binding of eIF4GI and eIF4GII isoforms to the IRES are similar (26, 29). Combining ΔJ5 and ΔK2′ deletions had a synergistic effect, abolishing the binding of eIF4F so that no RT stops were detected on this mutant RNA other than those apparent in the absence of eIF4F (Fig. 3A, lane 8). The effects of these mutations on the binding of eIF4F/eIF4G therefore exactly paralleled their effects on IRES-mediated translation.
Footprint analysis of mutant EMCV IRES J-K domain/eIF4G complexes.
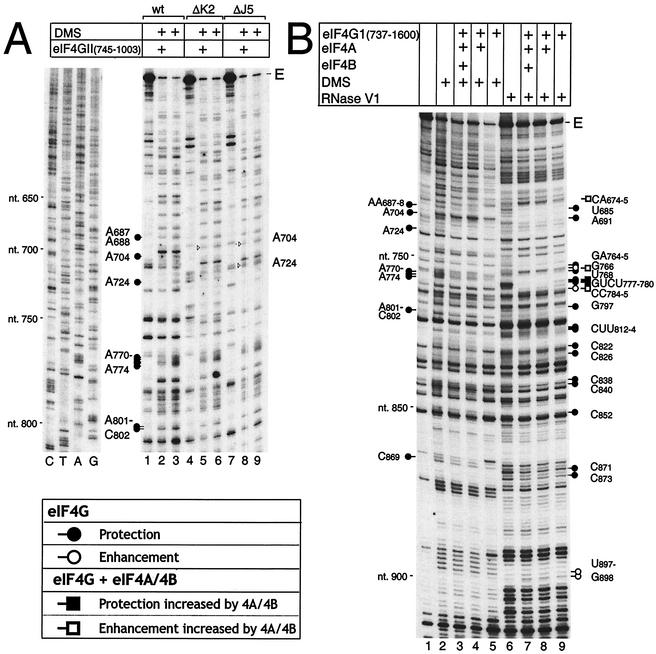
Binary complexes of eIF4GII745-1003 and wild-type, ΔJ5, or ΔK2′ mutant EMCV RNAs (nt 680 to 787) were probed with DMS, which reacts with unpaired adenine residues and to a lesser extent with unpaired cytosine residues (8). In these experiments, several residues that appear to have altered susceptibility to cleavage or modification in the presence of factors coincide with strong stops formed during primer extension by RT on this highly structured RNA in the absence of factors. Protection or enhanced modification of these nucleotides is therefore considered to be equivocal and is not discussed. In addition to the previously described protection of AA687-688, A724, and AAAAA770-774 (Fig. 1) (22), eIF4GII745-1003 and eIF4GI737-1600 also protected A704, AC801-802, and C869 (Fig. 4A, lane 2 and Fig. 4B, lane 5). Protection at these sites was weakly but reproducibly observed. The site on the IRES to which eIF4G binds is therefore more extensive than previously realized and includes residues whose conformation is directly altered in mutant ΔJ5. eIF4GII745-1003 protected AAAAA770-774 and AC801-802 in ΔJ5 and ΔK2 mutants from DMS modification as seen for wild-type RNA, but protection at AA687-688 was reduced, particularly for the ΔJ5 mutant (Fig. 4A, lanes 2, 5, and 8). Protection at A704 was abolished for the ΔJ5 mutant.
FIG. 4.
Chemical and enzymatic footprinting analyses of ribonucleoprotein complexes assembled on the EMCV IRES. (A) Chemical foot-printing of the wild-type and ΔJ5 and ΔK2′ mutant forms of EMCV (nt 315 to 1155) RNA in binary complexes formed with eIF4GII745-1003. (B) Chemical and enzymatic footprinting of wild-type EMCV (nt 315 to 1155) RNA in complexes formed with eIF4GI737-1600, eIF4A, and eIF4B, as indicated. Polyacrylamide-urea gel fractionation of cDNA products after primer extension by AMV-RT shows the sensitivity of the IRES to either DMS or RNase V1, either alone or complexed with initiation factors, as indicated. cDNA products obtained after primer extension of untreated EMCV RNAs are shown in lanes 1, 4, and 7 of panel A and in lane 1 of panel B. The positions of protected residues are indicated by the symbols shown in the key at bottom left; some sites of protection on mutant mRNAs are indicated by arrowheads on panel A. A dideoxynucleotide sequence generated with the same primer was run in parallel (lanes C, T, A, and G in panel A), and the positions of EMCV nucleotides at 50-nt intervals are indicated to the left of both panels. The full-length cDNA extension product is marked E.
Probing with RNase V1, which cleaves base-paired or stacked RNA, indicated that eIF4GI737-1600 protected the IRES from cleavage at U685, A691, GA764-765, U768, and GUCU777-780 in the J-K domain, G797 in domain L, and CUU812-814, C822, C838, C840, C852, C871, and C873 in the region flanking the initiation codon (Fig. 4B, lanes 6 and 9). The numbering of these residues indicates the nucleotide on the 3′ side of the cleaved bond. These results confirm the previously identified protection of residues in the J-K domain (22) and the interaction of eIF4G with domain L (22). Reduced cleavage of sequences flanking and downstream of the initiation codon could be due to direct binding of eIF4G to protected sites. However, sequences downstream of the J-K domain are not required for stable binding of eIF4G to the IRES, and bound eIF4G protected only one residue downstream of domain L from modification by DMS. We therefore consider it more likely that reduced RNase V1 cleavage in the region flanking and downstream of the initiation codon was due to conformational changes induced by the binding of eIF4GI737-1600.eIF4GI737-1600 also enhanced the susceptibility of the IRES to cleavage at G766 and CC784-785, as described previously (22), and slightly enhanced cleavage at UG897-898. eIF4GI737-1600 did not significantly enhance DMS modification of any part of the IRES.
Hydroxyl radical probing of EMCV IRES/eIF4G complexes.
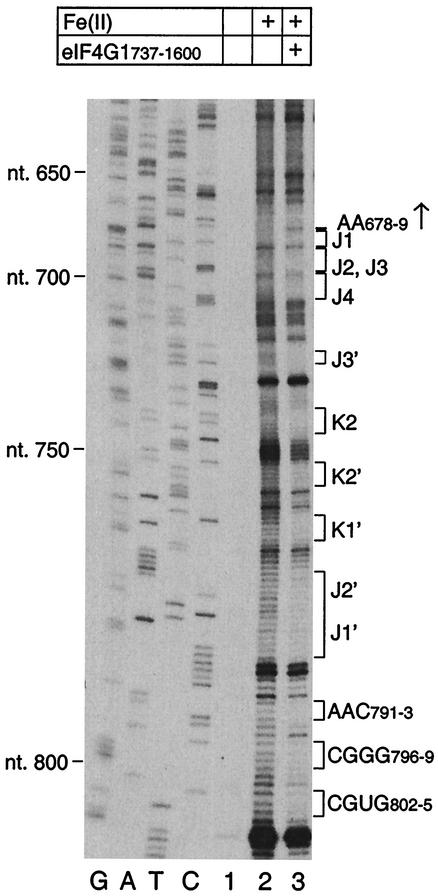
To further characterize the eIF4G binding site on the IRES, binary RNA/protein complexes were probed with hydroxyl radicals generated from free Fe(II)-EDTA. Hydroxyl radicals attack the ribose moiety in single- and double-stranded RNA in a sequence-independent manner, leading to scission of the RNA backbone (25), and this broad specificity permitted probing of the complete IRES. eIF4GI737-1600 protected extensive regions in the J, K, and L domains from cleavage (Fig. 5, lanes 2 and 3) in a manner that corresponded directly to sites of protection from RNase V1 cleavage. The protection of these helical regions of the IRES by eIF4G from cleavage by hydroxyl radicals suggests that eIF4G interacts with the minor groove of the RNA duplexes. The only site of significantly enhanced cleavage due to binding of eIF4GI737-1600 was at AA678-679 at the junction of the I and J-K domains. These results are summarized in Fig. 1.
FIG. 5.
Hydroxyl radical probing of ribonucleoprotein complexes comprising EMCV RNA (nt 280 to 1155) and eIF4GI737-1600. Polyacrylamide-urea gel fractionation of cDNA products after primer extension by AMV-RT shows the sensitivity of the IRES to cleavage by hydroxyl radicals, either alone or with initiation factors, as indicated. cDNAs obtained after primer extension of untreated EMCV RNAs are shown in lane 1. Residues or subdomains with altered sensitivity to cleavage in the presence of initiation factors are indicated to the right of the panel. A site of increased sensitivity to cleavage is indicated by an arrow. A dideoxynucleotide sequence generated with the same primer was run in parallel (lanes C, T, A, and G), and the positions of EMCV nucleotides at 50-nt intervals are indicated to the left.
Together, these findings confirm and extend previous observations concerning the interaction of eIF4G with the IRES (22) and indicate that the binding site is more extensive than previously recognized. These findings are consistent with the effects, described above, of deletions in apical regions of the J-K domain on binding of eIF4G and consequently on IRES function.
Mapping the orientation of eIF4G on the IRES by directed hydroxyl radical probing.
The core domain of eIF4G that is necessary and sufficient to support initiation on the IRES consists of five stacked pairs of α-helical repeats, numbered 1a-1b, 2a-2b, etc. (29) (Fig. 6D). We used directed hydroxyl radical probing to orient this core domain of eIF4GI on the IRES. In this approach, based on methods used to map the binding sites for factors and ribosomal proteins on prokaryotic rRNA (6), Fe(II) tethered to a specific site on the surface of eIF4G is used to generate hydroxyl radicals to cleave the IRES.
FIG. 6.
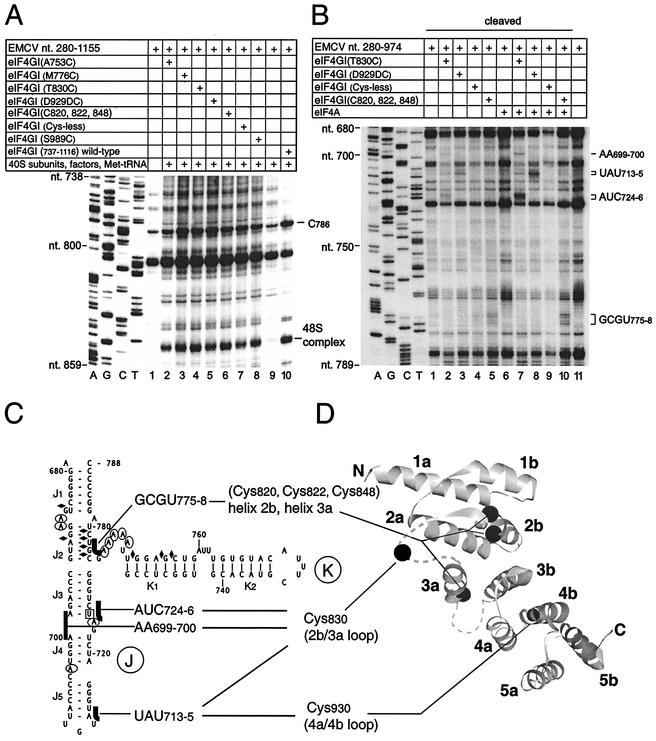
Defining the orientation of eIF4G on the J-K domain of the EMCV IRES by directed hydroxyl radical cleavage. (A) Binding and activity of Fe(II)-BABE-derivatized eIF4GI737-1116. Binding to the IRES (yielding a characteristic toe print at C786) and activity in supporting 48S complex formation on the IRES (yielding toe prints +15 to +17 nt downstream of the initiation codon AUG834 [48S complex]) of unmodified wild-type and derivatized mutant eIF4G polypeptides, as indicated, were assayed in reconstituted initiation reactions containing EMCV RNA (nt 280 to 1155), 40S subunits, eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, ATP, GTP, and amino-acylated initiator tRNA as indicated, at concentrations described previously (34). Reference lanes A, G, C, and T depict the negative-strand EMCV sequence derived by using the same primer and pTE1 DNA. For clarity, only the lower part of the gel is shown. (B) Directed hydroxyl radical probing. Probing reactions contained EMCV RNA (nt 280 to 974), eIF4GI737-1116 derivatives with Fe(II) tethered to residues as indicated, and eIF4A (lanes 6 to 10). Specific hydroxyl radical cleavages are seen as additional bands, as indicated to the right of the panel. Lanes A, G, C, and T are dideoxynucleotide sequencing lanes. (C) Summary of directed hydroxyl radical cleavages in the J-K domain of the EMCV IRES from Fe(II) tethered to specific positions on eIF4G. For comparison, residues in this domain that are protected from chemical modification or enzymatic cleavage by eIF4G are indicated exactly as in Fig. 1. (D) Positions of cysteines used as tethering sites for Fe(II)-BABE are indicated as black filled circles on a ribbon drawing of the central region of eIF4G (29), prepared by using WebLab Viewer Pro version 4.0 (Molecular Simulations, Inc.), viewed along the central axis of the α-helices, which are labeled 1a, 1b, etc. Crystallographically disordered loops are indicated by dashes.
eIF4GI737-1116 contains six cysteine residues: C820, C822, C848, C920, C935, and C937. Site-directed mutagenesis was used to create mutants of this eIF4GI domain lacking C920, C935, and C937 (mutant 1), lacking C820, C822, C920, C935, and C937 (mutant 2) or lacking all cysteine residues (mutant 3). All cysteine residues were replaced by alanine residues except for substitution of C848 by threonine. The cysteineless mutant 3 was used as the basis for the introduction of single cysteine residues at five different positions on the surface of eIF4GI (based on the known structure of the homologous eIF4GII [29]). In addition to A753C, M776C, T830C, and S989C substitution mutants, a D929DC insertion mutant was also obtained. The activity of these mutant polypeptides in binding to the EMCV IRES and in supporting 48S complex formation on the IRES in in vitro assembly reactions was comparable to that of wild-type eIF4GI737-1116 (data not shown).
Fe(II) was then tethered to the protein via BABE. The activity of each Fe(II)-conjugated mutant polypeptide was comparable to the activity of unmodified wild-type eIF4GI737-1116 in binding to the IRES and in supporting 48S complex formation on the IRES in vitro (Fig. 6A and data not shown). To identify the nucleotides in the IRES that are in close proximity to bound eIF4GI, Fe(II)-conjugated polypeptides were bound to the IRES in the presence and absence of eIF4A (which enhances eIF4G's interaction with the IRES [26]). Hydroxyl radicals were then generated to cleave the IRES in the vicinity of the tethered Fe(II). Sites of cleavage mapped by primer extension (Fig. 6B) are indicated on the secondary structure of the J-K domain (Fig. 6C). Three of the eight mutants reproducibly caused specific and characteristic cleavages in the IRES that were all restricted to the J-K domain. The T830C mutation is located in the long, crystallographically disordered loop between helices 2b and 3a. The eIF4GI737-1116 T830C mutant induced weak cleavage at AA699-700 and AUC724-6 on opposing sides of the J3/J4 helical junction, and at UAU713-715; cleavage at AA699-700 and particularly at AUC724-726 was strongly enhanced by eIF4A (Fig. 6B, lanes 2 and 7). The D929DC insertion is located immediately after helix 4a, and it induced cleavage at UAU713-715 that was strongly enhanced by eIF4A (Fig. 6B, lanes 3 and 8). The eIF4GI737-1116 (C920A, C935A, C937A) substitution mutant retains C820 and C822 residues in helix 2b and C848 in helix 3a. The cleavage that this mutant induced at GCGU775-778 was enhanced by eIF4A (Fig. 6B, lanes 5 and 10). Mutant 3 retains only a single cysteine residue (C848) and did not induce cleavage at this site (data not shown). These results suggest a model for the eIF4GI737-1116/IRES complex (Fig. 6C and D) in which the N terminus of this domain of eIF4G is orientated towards helix J1 of the J-K domain and the C terminus of this domain is oriented towards helix J5.
Footprinting complexes of eIF4G, eIF4A, and eIF4B on the EMCV IRES.
eIF4A, eIF4B, and eIF4G interact synergistically to form a ribonucleoprotein complex on the IRES of EMCV and related viruses (26, 38, 40). We used RNA footprinting to compare the interactions of this complex and of eIF4G alone with the EMCV IRES. eIF4A enhanced eIF4GI737-1600's protection of the IRES from RNase V1 cleavage at GUCU777-780, reduced protection at CUU812-814, and enhanced cleavage at CA674-675 at the base of domain I, at G766 and CC784-785 in the J-K domain, and weakly at UG897-898 downstream of the initiation codon AUG834 (Fig. 4B, lanes 7 to 9). Some of these changes, for example at CA674-675 and G766, were slightly enhanced by eIF4B. These changes were also observed when the R362Q trans-dominant mutant form of eIF4A replaced the wild-type factor (data not shown). eIF4A and eIF4B did not significantly alter the pattern of DMS modification of RNA in the eIF4GI737-1600/IRES complex (Fig. 4B, lanes 3 to 5). These results are summarized in Fig. 1.
ATP-dependent conformational changes in EMCV RNA downstream of the initiation codon induced by eIF4F.
The ATP dependence of initiation on the IRES and the requirement for active eIF4A (33, 35) suggest that ribosomal loading onto the initiation codon AUG834 may involve restructuring of the IRES by the eIF4G/eIF4A complex. Our probing analyses (22) (Fig. 4B) indicated that sequences immediately downstream of the J-K domain are not extensively unwound by eIFs 4A, 4B, and 4G but that some conformational changes are induced in structures flanking and downstream of the initiation codon.
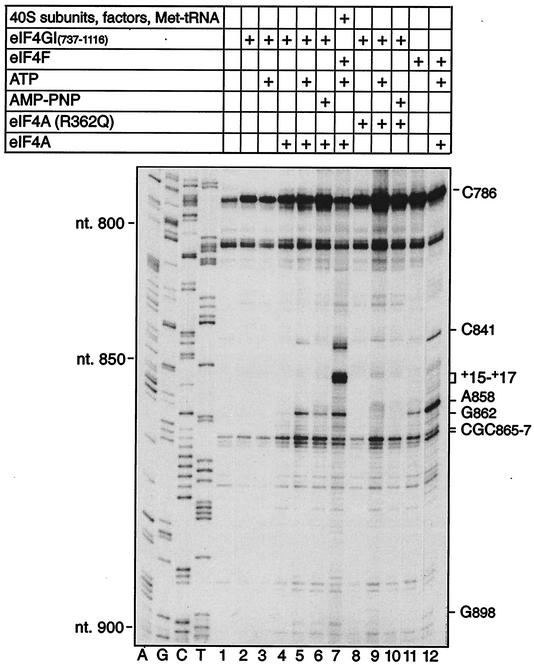
ATP-dependent conformational changes downstream of the initiation codon AUG834 induced by eIF4G/eIF4A were also identified by toe printing done by using EMCV RNA (nt 378 to 1155, comprising the entire IRES and 322 nt of the adjacent coding region), eIF4A, and eIF4GI737-1116 (which contains only one of the two eIF4A-binding sites in eIF4G [Fig. 7]). Binding of eIF4GI737-1116 in the presence of eIF4A enhanced stops at CGC865-867 in a manner that was further enhanced by but did not depend on the presence of ATP or on ATP hydrolysis (lanes 3 and 6). Additional new toe prints appeared at C841, A858 and (more weakly) G862, and G898 only in the presence of eIF4F, wild-type eIF4A, and ATP (lanes 7 and 12) or eIF4GI737-1116, wild-type eIF4A, and ATP (lane 5). All but one of these stops occur in a proposed irregular stem-loop structure (14) encompassing nt 854 to 902 (Fig. 1). These stops were not detected in reactions lacking eIF4A (lanes 2 and 3). They were only weakly apparent in the absence of ATP (lanes 4 and 11) or on inclusion of AMP-PNP (a nonhydrolyzable ATP analog) in place of ATP (lane 6). However, this result must be treated cautiously because these reactions also contained dATP (an essential constituent of primer extension reactions), which can partially substitute for ATP in eIF4A-dependent initiation on the EMCV IRES [38]). Significantly, these additional stops were not detected in reactions in which R362Q trans-dominant mutant eIF4A replaced wild-type eIF4A in the presence or absence of ATP (lanes 8 and 9). This eIF4A mutant is unable to utilize ATP or dATP to mediate conformational changes in RNA. Inclusion of R362Q mutant eIF4A in toe printing reactions with eIF4GI737-1116 enhanced the toe prints at C786 and CGC865-867 to a similar extent as when wild-type eIF4A was present (compare lanes 2, 4, and 8 and lanes 5 and 9). These results indicate that mutant eIF4A was incorporated into the eIF4G/eIF4A complex on the J-K domain but that the resulting complex was unable to induce ATP-dependent conformational changes in the IRES/factor complex downstream of the initiation codon.
FIG. 7.
ATP-dependent conformational changes induced eIF4G/4A in EMCV RNA downstream of the initiation codon. Primer extension analysis was done on EMCV RNA (nt 378 to 1155) in the presence of ATP or AMP-PNP and translation initiation components, as indicated. The reaction mixture in lane 7 that yielded a 48S complex on the EMCV IRES contained eIF1, eIF1A, eIF2, eIF3, 40S subunits, GTP, and amino-acylated initiator tRNA as well as other translation initiation components as indicated, at concentrations described previously (34). The cDNA products marked C786, C841, A858, G862, CGC865-867, and G898 terminated at these nucleotides. The cDNA products marked +15-+17 terminated at stop sites 15, 16, and 17 nucleotides from the initiation codon AUG834. Reference lanes A, G, C, and T depict the negative strand EMCV sequence derived by using the same primer and pTE1 DNA. For clarity, only the lower part of the gel downstream of EMCV nt 774 is shown.
DISCUSSION
We have extended our previous characterization of the binding site for eIF4G on the EMCV IRES (22) by using footprinting and mutagenesis and have determined the orientation of the essential core domain of eIF4G on this IRES by using directed hydroxyl radical cleavage. eIF4G recruits eIF4A to the IRES, and we found that the bound eIF4G/eIF4A complex induces limited local conformational changes in the IRES in an ATP-independent manner as well as more extensive ATP-dependent changes immediately downstream of the ribosome binding site. eIF4B, which also binds to the IRES, slightly enhanced binding of the eIF4G/eIF4A complex. These observations extend our understanding of the mechanism of IRES-mediated translation initiation by suggesting how binding of initiation factors to the IRES can actively prepare it for ribosomal attachment.
eIF4G recognizes a large and structurally complex binding site on the EMCV IRES that encompasses sequences in the J, K, and L domains. This conclusion is based on the results of hydroxyl radical and chemical and enzymatic probing (Fig. 4A, 4B, and 5A). We have previously reported that interaction of eIF4G, either alone or as part of eIF4F, with the IRES and consequent initiation of translation require the oligo(A) loop at the junction of the J and K domains (35). We have now found that binding eIF4G and consequent initiation also depend on the apical J5 and K2 hairpins, which act synergistically in these processes (Fig. 2 and 3). Taken together with observations that eIF4G/eIF4F binds specifically to the J-K domains of human parechovirus type 1 (our unpublished data), FMDV, and TMEV IRESs (27, 28, 40, 46), these findings suggest that the sequence conservation in the central region of the J-K domain and the extensive but covariant sequence differences in these peripheral helices of the J-K domain (17) are indicative of a need to maintain its structural integrity to enable eIF4G to bind. This requirement would account for the deleterious effects of mutations in these apical helices on EMCV IRES function noted here and elsewhere (13, 14, 32, 48).
The central domain of eIF4G that binds specifically to the J-K domain consists of five α-helical HEAT repeats: eIF4G is the first HEAT-repeat polypeptide that is known to bind specifically to nucleic acid (29), and the results reported here therefore define aspects of a new mode of RNA-protein interaction. The HEAT-repeat domain of eIF4G binds directly to a small number of bases in the J-K domain, most significantly in the A-rich bulge at the junction of the J2, J3, and K1 helices (Fig. 1), but hydroxyl radical probing indicated that eIF4G bound more extensively to RNA duplexes in the J, K, and L domains (Fig. 1 and 5A). Hydroxyl radicals cleave the sugar-phosphate backbone of RNA at ribose C1′ and C4′ positions (25), so the observed protection indicates that eIF4G binds in the minor groove of these RNA duplexes. Other polypeptides that bind double-stranded RNA, such as the double-stranded RNA (dsRNA)-binding domain of proteins such as Staufen, Xlrbpa, and the dsRNA-dependent protein kinase PKR (1, 41, 45), all do so through its wide minor groove. The functional 2′-hydroxyl and phosphate groups in the RNA minor groove do not allow sequence-specific recognition, and these proteins show no sequence specificity in their interactions with RNA in vitro. A similar ability of eIF4G to bind dsRNA in a sequence-independent manner could bear on its activity as a cofactor that enhances the ATP-dependent RNA helicase activity of eIF4A (43, 44), for example by reducing dissociation of eIF4A from duplex RNA during cycles of step-wise unwinding of RNA.
eIF4B also binds directly to EMCV-like IRESs (28, 31, 38), and footprinting data (Fig. 4B) indicate that it weakly enhances binding of the eIF4G/eIF4A complex to the EMCV IRES. This conclusion is consistent with previous results obtained by using EMCV and FMDV IRESs in UV cross-linking and toe printing assays (38, 40). However, this enhancement is weak, so it may be a less important contributing factor in eIF4B's activity in enhancing ribosomal binding to this group of IRESs (35) than the interaction of eIF4B with the eIF3 component of the 43S complex (30).
eIF4GI and eIF4GII both bind specifically to the J-K domain and recruit eIF4A, which by itself does not bind specifically or stably to the IRES. eIF4A interacts strongly with the IRES in the resulting heterotrimeric complex and also significantly enhances eIF4G's binding to the IRES (22, 26, 38). The eIF4G/eIF4A complex induced limited local conformational changes in the immediate vicinity of its binding site on the IRES (Fig. 4B), suggesting that binding may involve a degree of induced fit. It also induced structural rearrangements in the IRES that reduced RNase V1 cleavage at several sites flanking the initiation codon, in the region between domain L and a proposed irregular stem-loop structure (14) encompassing nt 854 to 902 (Fig. 1). These changes are probably due to a reduction in secondary structure in this region. Toe printing of 48S complexes indicated that the leading edge of a bound 40S subunit is at G849, in the immediate vicinity of the induced conformational changes (Fig. 7, lane 7). We therefore suggest that the eIF4G/eIF4A complex prepares a binding site for the ribosome on the IRES by inducing concerted conformational changes that include a reduction in the secondary structure of sequences centered on the initiation codon so that this region becomes accessible to an incoming 43S ribosomal complex.
The eIF4G/eIF4A complex also enhanced existing weak stops and induced new toe prints in an ATP-dependent manner downstream of the initiation codon, primarily at and in the vicinity of nt 858. These new toe prints indicate that the stability of this region had increased, which could be due to induced and possibly long-range conformational rearrangements involving sequences in this region and upstream sequences that had become unpaired by the helicase activity of the eIF4A/eIF4G complex. The possibility that sequences in the vicinity of nt 858 switch between two alternative conformations that involve similar degrees of base pairing could account for the minor changes in sensitivity of this region to chemical and enzymatic probes. Alternatively, the increased stability of this region could be due to binding of the eIF4A and/or the eIF4GI737-1116 fragment (which contains a binding site for one molecule of eIF4A). Such interactions could involve additional molecules of one or both of these factors, or could involve a single eIF4G/eIF4A complex anchored on the J-K domain that makes additional interactions with downstream sequences, which would be possible if the active center of this complex that is involved in its helicase activity is distinct from the RNA-binding surface that interacts with the J-K domain.
We are currently unable to distinguish between these two possible explanations for the enhanced stability of sequences in the vicinity of nt 858, which, we note, are not mutually exclusive. Either possibility would imply that the region between ∼nt 850 to 900 plays a specific role in IRES function; there is evidence for this and, more specifically, for both genetic and functional interactions between this domain and the J-K domain (14, 20). A substitution in this domain suppressed the defect caused by a substitution in the A-rich loop between the J and K domains (14), and the translation defect caused by a 1-nt insertion in this loop became apparent only if sequences including the domain from ∼nt 850 to 900 were deleted (20). The position of this stabilized domain at the 3′ border of the sequence covered by a 40S subunit bound to the initiation codon AUG834 could constrain the site of ribosomal attachment and thus play a role in enhancing the accuracy of initiation codon selection.
In summary, we suggest that the eIF4G/eIF4A complex induces concerted conformational changes in the IRESs that prepare a site on the IRES to which the ribosome can bind efficiently and accurately. A requirement for these conformational changes for initiation on the IRES can account for the dependence of EMCV translation initiation on ATP (35) and on an enzymatically active eIF4G/eIF4A complex (33, 47). The basis for these requirements has until now been obscure. Thus, in addition to enhancing the affinity of eIF4G for the EMCV IRES (26) and for eIF3, a major component of the 43S complex (23), eIF4A also induces local changes in IRES conformation that prepare the region flanking and downstream of the initiation codon AUG834 so that the 43S complexes can bind to it productively, leading to initiation of translation.
Acknowledgments
This material is based upon work supported by the National Science Foundation under grant no. 0110834.
V.G.K. and I.B.L. contributed equally to this work.
REFERENCES
- 1.Bevilacqua, P. C., and T. R. Cech. 1996. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35:9983-9994. [DOI] [PubMed] [Google Scholar]
- 2.Borovjagin, A. V., M. V. Ezrokhi, V. M. Rostapshov, T. Y. Ugarova, T. F. Bystrova, and I. N. Shatsky. 1991. RNA-protein interactions within the internal translation initiation region of encephalomyocarditis virus RNA. Nucleic Acids Res. 19:4999-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borovjagin, A., T. Pestova, and I. Shatsky. 1994. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 351:299-302. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, C. A., J. C. Padovan, T. L. Thompson, C. A. Benoit, B. T. Chait, and R. E. Rhoads. 2002. Mass spectrometric analysis of the N terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J. Biol. Chem. 277:12559-12571. [DOI] [PubMed] [Google Scholar]
- 5.Carberry, S. E., and D. J. Goss. 1991. Interaction of wheat germ protein synthesis initiation factors eIF-3, eIF-(iso)4F, and eIF4F with mRNA analogues. Biochemistry 30:6977-6982. [DOI] [PubMed] [Google Scholar]
- 6.Culver, G. M., and H. F. Noller. 2000. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 318:461-475. [DOI] [PubMed] [Google Scholar]
- 7.Duke, G. M., M. A. Hoffman, and A. C. Palmenberg. 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis viral RNA translation. J. Virol. 66:1602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J. P. Ebel, and B. Ehresmann. 1987. Probing the structure of RNA in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evstafieva, A. G., T. Y. Ugarova, B. K. Chernov, and I. N. Shatsky. 1991. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 19:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 11.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, M. A., and A. C. Palmenberg. 1995. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J. Virol. 69:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman, M. A., and A. C. Palmenberg. 1996. Revertant analysis of J-K mutations in the encephalomyocarditis virus internal ribosomal entry site detects an altered leader protein. J. Virol. 70:6425-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hüttenhofer, A., and H. F. Noller. 1994. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 13:3892-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, R. J. 2000. A comparative view of initiation site selection mechanisms, p.127-183 In N. Sonenberg, M. B. Mathews and J. W. B. Hershey (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 18.Jang, S. K., and E. Wimmer. 1990. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 4:1560-1572. [DOI] [PubMed] [Google Scholar]
- 19.Kaminski, A., M. T. Howell, and R. J. Jackson. 1990. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 9:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminski, A., and R. J. Jackson. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolupaeva, V. G., C. U. T. Hellen, and I. N. Shatsky. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199-1212. [PMC free article] [PubMed] [Google Scholar]
- 22.Kolupaeva, V. G., T. V. Pestova, C. U. T. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 23.Korneeva, N. L., B. J. Lamphear, F. L. Hennigan, and R. E. Rhoads. 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 275:41369-41376. [DOI] [PubMed] [Google Scholar]
- 24.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 25.Latham, J. A., and T. R. Cech. 1989. Defining the inside and outside of a catalytic RNA molecule. Science 245:276-282. [DOI] [PubMed] [Google Scholar]
- 26.Lomakin, I. B., C. U. T. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez de Quinto, S., and E. Martinez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez de Quinto, S., E. Lafuente, and E. Martinez-Salas. 2001. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B and eIF4GII. RNA 7:1213-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcotrigiano, J., I. B. Lomakin, N. Sonenberg, T. V. Pestova, C. U. T. Hellen, and S. K. Burley. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7:193-203. [DOI] [PubMed] [Google Scholar]
- 30.Méthot, N., M. S. Song, and N. Sonenberg. 1996. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 16:5328-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer, K., A. Petersen, M. Niepmann, and E. Beck. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J. Virol. 69:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oudshoorn, P., A. Thomas, G. Scheper, and H. O. Voorma. 1990. An initiation signal in the 5′ untranslated leader sequence of encephalomyocarditis virus RNA. Biochim. Biophys. Acta 1050:124-128. [DOI] [PubMed] [Google Scholar]
- 33.Pause, A., N. Methot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestova, T. V., S. I. Borukhov, and C. U. T. Hellen. 1998. Eukaryotic ribosomes require eIFs 1 and 1A to locate initiation codons. Nature 394:854-859. [DOI] [PubMed] [Google Scholar]
- 35.Pestova, T. V., C. U. T. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. T. Hellen. 2001. Molecular events in initiation of translation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of hepatitis C virus and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestova, T. V., I. N. Shatsky, and C. U. T. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilipenko, E. V., V. M. Blinov, B. K. Chernov, T. M. Dmitrieva, and V. I. Agol. 1989. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 17:5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. T. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos, A., S. Grünert, J. Adams, D. R. Micklem, M. R. Proctor, S. Freund, M. Bycroft, D. St. Johnston, and G. Varani. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19:997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray, B. K., T. G. Lawson, J. C. Kramer, M. H. Cladaras, J. A. Grifo, R. D. Abramson, W. C. Merrick, and R. E. Thach. 1985. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 260:7651-7658. [PubMed] [Google Scholar]
- 43.Rogers, G. W., N. R. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 44.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryter, J. M., and S. C. Schultz. 1998. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17:7505-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saleh, L., R. C. Rust, R. Fullkrug, E. Beck, G. Bassili, K. Ochs, and M. Niepmann. 2001. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J. Gen. Virol. 82:757-763. [DOI] [PubMed] [Google Scholar]
- 47.Svitkin, Y. V., A. Pause, A. Haghighat, S. Pyronnet, G. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witherell, G. W., C. S. Schultz-Witherell, and E. Wimmer. 1995. Cis-acting elements of the encephalomyocarditis virus internal ribosomal entry site. Virology 214:660-663. [DOI] [PubMed] [Google Scholar]