Abstract
The c-myc proto-oncogene plays a key role in the proliferation, differentiation, apoptosis, and regulation of the cell cycle. Recently, it was demonstrated that the 5′ nontranslated region (5′ NTR) of human c-myc mRNA contains an internal ribosomal entry site (IRES). In this study, we investigated cellular proteins interacting with the IRES element of c-myc mRNA. Heterogeneous nuclear ribonucleoprotein C (hnRNP C) was identified as a cellular protein that interacts specifically with a heptameric U sequence in the c-myc IRES located between two alternative translation initiation codons CUG and AUG. Moreover, the addition of hnRNP C1 in an in vitro translation system enhanced translation of c-myc mRNA. Interestingly, hnRNP C was partially relocalized from the nucleus, where most of the hnRNP C resides at interphase, to the cytoplasm at the G2/M phase of the cell cycle. Coincidently, translation mediated through the c-myc IRES was increased at the G2/M phase when cap-dependent translation was partially inhibited. On the other hand, a mutant c-myc mRNA lacking the hnRNP C-binding site, showed a decreased level of translation at the G2/M phase compared to that of the wild-type message. Taken together, these findings suggest that hnRNP C, via IRES binding, modulates translation of c-myc mRNA in a cell cycle phase-dependent manner.
The c-myc proto-oncogene participates in various cellular events including proliferation, differentiation, apoptosis, cell cycle, and regulation of mammalian body size (14, 27, 50, 60). The c-myc mRNA encodes a helix-loop-helix-leucine zipper class transcription factor that binds to E-box (CAGGTG) promoter sequences in conjunction with its dimerization partner Max. Myc-Max heterodimers function as transcriptional activators of critical target genes (3, 4, 13). Regulation of c-myc expression occurs at multiple levels, including transcription (36), stability of both the mRNA and protein (49), and translation (41, 56).
The human c-myc transcripts are generated from four alternative promoters (P0, P1, P2, and P3). In normal cells, transcription of most of the c-myc mRNA (75 to 90%) is directed by the P2 promoter (5, 6). P2 mRNA directs translation of two distinct proteins of 67- and 64-kDa, designated c-Myc1 and c-Myc2, respectively, by an alternative initiation mechanism involving two in-frame start codons, CUG and AUG, residing at nt 364 and 409, respectively (20). Recently, there have been reports suggesting that the 5′ nontranslated region (5′ NTR) of c-myc contains an internal ribosomal entry site (IRES) (41, 56). In the human c-myc mRNA, the IRES lies in the 5′ NTR upstream of the AUG start codon of the major c-Myc protein (c-Myc2) (Fig. 1A and see below). Moreover, translation of c-myc mRNA was shown to occur via a cap-dependent mechanism as well as an IRES-dependent mechanism (57).
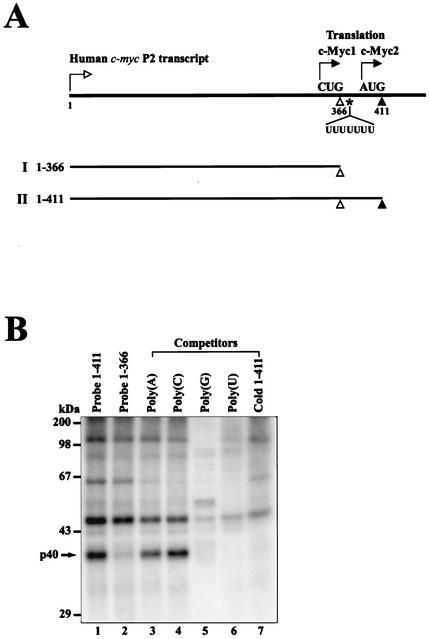
FIG. 1.
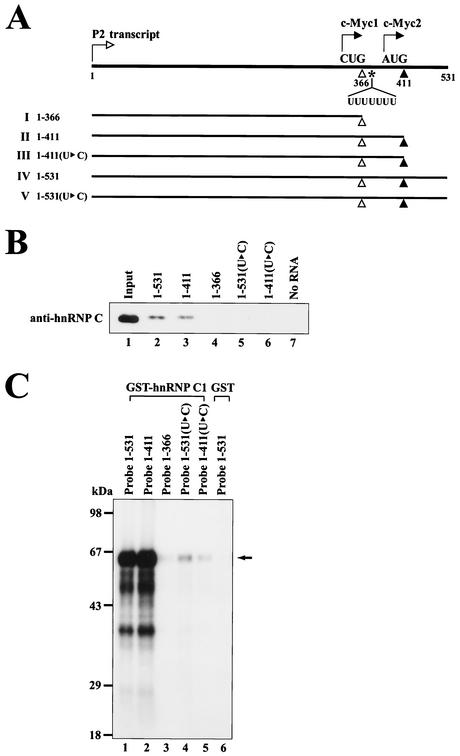
Detection of cellular proteins interacting with the human c-myc IRES. (A) Schematic diagram of the 5′ leader of human c-myc P2 transcript used in the UV cross-linking assay. Transcription and translation start sites are depicted by the arrows with open and solid heads, respectively. Transcript I (nt 1 to 366) spans from the transcription start site to the minor translation initiation codon CUG364, indicated by an open triangle, used in the translation of c-Myc1. Transcript II (nt 1 to 411) spans from the transcription start site to the major translation initiation codon AUG409, indicated by a solid triangle, used in the translation of c-Myc2. The position of the heptameric U sequence is indicated by UUUUUUU. (B) UV cross-linking experiments with 32P-labeled transcripts I and II. HeLa S3 cell cytoplasmic extracts were incubated with 32P-labeled RNA probes I and II without competitor RNAs (lanes 1 and 2). Competition experiments were carried out with RNA probe II in the presence of competitor RNAs. For RNA competition, 50 ng of four homopolymeric RNAs [poly(A), poly(C), poly(G), and poly(U)] (lanes 3 to 6) and a 50-fold molar excess of unlabeled transcript II (lane 7) were included in the binding reaction mixtures. After UV-irradiation, samples were treated with RNase cocktail before analysis by SDS-12% PAGE.
The IRES is a specialized RNA structure that recruits the ribosome to the mRNA in a cap-independent manner. After the discovery of IRESs from picornavirus mRNAs (31, 44), several viral and cellular mRNAs have been shown to contain IRESs (22, 61). Studies of factors required for IRES-dependent translation reveal that canonical translation initiation factors and RNA-binding proteins such as poly(rC)-binding protein, polypyrimidine tract-binding protein (PTB), upstream of N-ras (unr), and the autoantigen La are required for translation of certain IRES-containing mRNAs (8, 23, 29, 37). The demand for the canonical and mRNA-specific initiation factors varies among IRES-containing mRNAs (32).
Much of what we have learned about mRNA-specific translation factors has been derived from virus expression studies. The La autoantigen, also known as the transcription termination factor of RNA polymerase III (17), stimulates translation of poliovirus RNA and reduces aberrant translation (37). La also binds to the initiation codon of the hepatitis C virus IRES to enhance translation (2). PTB, also known as heterogeneous nuclear ribonucleoprotein I (hnRNP I), is a negative regulator of pre-RNA splicing (15, 43) and was suggested to augment IRES-dependent translation of picornavirus mRNAs. Poly(rC)-binding protein, also known as hnRNP E, is required for IRES-dependent translation of poliovirus and hepatitis A virus RNAs (8, 18). unr, another cellular factor, is required for efficient translation of human rhinovirus mRNA (29).
Several cellular mRNAs also contain IRESs. The list of IRES-containing mRNAs includes multifunctional c-myc (41, 56), proto-oncogene platelet-derived growth factor 2 (PDGF2)/c-sis (7), cell survival- and death-related X-linked inhibitor of apoptosis (25), apoptotic protease-activating factor 1 (11), the growth regulators fibroblast growth factor 2 (62) and vascular endothelial growth factor (28), ornithine decarboxylase (47), and p58PISTLRE (12), etc. For further information, an IRES database is available at http://www.rangueil.inserm.fr/iresdatabase.
Several RNA-binding proteins have been shown to augment cellular IRES-dependent translation. For instance, the La autoantigen regulates the translation of X-linked inhibitor of apoptotis mRNA (24). Apoptotic protease-activating factor 1 IRES activity is enhanced by overexpression of PTB and unr in cell lines deficient in these molecules (40). RNA-binding proteins also may have inhibitory effects on IRES-dependent translation. For example, PTB inhibits translation of BiP mRNA (34). Despite the many examples of IRES-dependent translation, the detailed mechanism of modulation of IRES-dependent translation by RNA-binding proteins remains to be elucidated.
We therefore sought to identify the cellular protein(s) required for translation of c-myc mRNA via the IRES element. We found that hnRNP C could interact with a heptameric U sequence within the IRES of c-myc mRNA. Moreover, coexpression of hnRNP C1 in a rabbit reticulocyte lysate (RRL) enhanced translation of c-myc mRNA in a heptameric U sequence-dependent manner. Interestingly, hnRNP C was partially redistributed between the nucleus and cytoplasm in a cell cycle phase-dependent manner. At the interphase, most of hnRNP C resided in the nucleus, whereas a portion of hnRNP C migrated to the cytoplasm at the G2/M phase. Concomitantly, translation of c-myc mRNA was enhanced relative to translation of a mutant c-myc mRNA lacking the hnRNP C-binding site. These data indicate that the c-myc IRES-dependent translation is regulated by hnRNP C in a cell cycle phase-dependent manner.
MATERIALS AND METHODS
Plasmid construction.
The 5′ NTR and a portion of the c-Myc protein-encoding region of the human c-myc P2 transcript fragment (nucleotides [nt] 1 to 531 of the P2 transcript) were obtained by PCR of a human fetal liver cDNA library (Clontech) with the primers 5′-CCATCGATAACTCGCTGTAGTAATTCCAGCG-3′ and 5′-AACTGCAGCTCGCTCTGCTGCTG-3. The resulting clone contains 41 amino acids of the c-Myc2 N-terminal coding region. The cDNA corresponding to the c-myc P2 transcript was cloned into the reporter plasmid pSK(+)/CATΔ-ICS-CAT (34) digested with ClaI and PstI in order to generate pCATΔ-M531CAT. Plasmid pM531CAT was generated by treating pCATΔ-M531CAT with ClaI, Asp718, Klenow, and T4 DNA ligase consecutively. In order to generate a mutant clone with changes in the c-myc IRES from UUUUUUU to CUUCUUC, a PCR was performed with plasmid pCATΔ-M531CAT as a template and oligonucleotides 5′-GGGGTACCAGCTGCTTAGACGCTGGACTTCTTCCGGGTAGTG-3′ and 5′-GCTCTAGATTATCACTTATTCAGG-3′ as primers. The amplified DNA fragment was digested with KpnI and XbaI and then cloned into plasmid pSK(−) (Stratagene) to generate pMPvu(U▸C)CAT. Plasmid pCATΔ-M531(U▸C)CAT, which has the U-to-C changes in the heptameric U sequence, was generated by an insertion of the PvuII-XbaI fragment of pMPvu(U▸C)CAT and the PvuII-ClaI fragment of pCATΔ-M511CAT into pSK(+)/CATΔ-ICS-CAT digested with ClaI and XbaI. The dicistronic constructs pCATΔ-M411CAT, pCATΔ-M366CAT, and pCATΔ-M411(U▸C)CAT were generated from PCR products. A common primer 5′-CCATCGATAACTCGCTGTAGTAATTCCAGCG-3′ was used in each of the PCRs together with the following specific primers and templates to generate the three different constructs. The specific primer 5′-CGGGATCCCATCGTCGCGGGAGGCTGCTG-3′ and template pCATΔ-M531CAT were used to generate pCATΔ-M411CAT, the specific primer 5′-CGGGATCCCAGCGTCTAAGCAGCTGCAAG-3′ and template pCATΔ-M531CAT were used to generate pCATΔ-M366CAT, and the specific primer 5′-CGGGATCCCATCGTCGCGGGAGGCTGCTG-3′ and template pCATΔ-M531(U▸C)CAT were used to generate pCATΔ-M411(U▸C)CAT. For the generation of monocistronic constructs, plasmids pCATΔ-M411CAT, pCATΔ-M366CAT, and pCATΔ-M411(U▸C)CAT were treated with ClaI-Asp718-Klenow and self-ligated. For in vivo analyses, dicistronic constructs under the control of the cytomegalovirus (CMV) promoter were generated by inserting the c-myc IRES in the intercistronic region between the Renilla luciferase and firefly luciferase reporter genes. Plasmids pCATΔ-M531CAT and pCATΔ-M531(U▸C)CAT were treated with XhoI-BamHI-Klenow, and the DNA fragments containing the c-myc IRES were isolated and ligated into the pGL3 control vector (Promega) treated with NcoI-Klenow to generate pGL3-M531 and pGL3-M511(U▸C). pGL3-M511 and pGL3-M531(U▸C) were treated with HindIII-XbaI-Klenow, and then the DNA fragments containing the c-myc IRES and the firefly luciferase reporter were isolated. These DNA fragments were ligated with pRL-CMV (Promega) treated with XbaI-Klenow to generate the dicistronic constructs pRM531F and pRM531(U▸C)F. The construction procedure for plasmid pGEX-KG/hnRNP C1 (1-290) was described previously (33). All mutations in the various constructs were verified by DNA sequencing.
Cell culture, synchronization, fluorescence-activated cell sorter analysis, and transient transfection.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal bovine serum (complete medium; Clontech). For cell cycle synchronization, G1/S-phase HeLa cells were obtained by a double-thymidine block (59). The cells were synchronized at the G2/M boundary by a double-thymidine block, released for 6 h, and then exposed to nocodazole (0.2 μg/ml; Sigma) (59). Synchronized HeLa cells were harvested and fixed with cold 100% ethanol. Fixed cells were stained with propidium iodide (40 μg/ml; Sigma) and treated with RNase A (50 μg/ml; Sigma) for 30 min at room temperature. Samples of 104 cells were analyzed for DNA content on a FACScan (Becton-Dickinson). Transient transfections were performed by electroporation 12 h prior to the thymidine block.
Purification of recombinant GST-hnRNP C1.
Recombinant glutathione S-transferase (GST)-fused hnRNP C1 (nt 1 to 290) (GST-hnRNP C1) was expressed in the Escherichiacoli strain BL21(DE3)/pLysS by using plasmid pGEX-KG/hnRNP C1 (1-290). After induction of GST-hnRNP C1 by the addition of isopropyl-β-d-thiogalactopyranoside, cells were harvested and resuspended in lysis buffer (20 mM Na-phosphate [pH 7.6], 300 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM β-mercaptoethanol, 10% [vol/vol] glycerol). GST fusion proteins were allowed to bind to glutathione-Sepharose 4B resin (Amersham-Pharmacia Biotech) in lysis buffer for 2 h at 4°C. GST-hnRNP C1 protein was eluted by 10 mM reduced glutathione (Roche). The fractions containing GST-hnRNP C1 were loaded onto a poly(U)-Sepharose 4B column (Amersham-Pharmacia Biotech). After the column was washed with 0.1 M NaCl binding buffer (20 mM Na-phosphate [pH 7.6], 100 mM NaCl, 0.5 mM PMSF, 10 mM β-mercaptoethanol, 10% [vol/vol] glycerol), the GST-hnRNP C1 was eluted with 1.0 M NaCl.
Preparation of HeLa cell lysates.
Cytoplasmic extract (S10) of HeLa S3 cells for UV cross-linking, immunoprecipitation, and pull-down assay was prepared as described by Oh et al. (42). To obtain asynchronous and synchronized HeLa cell extracts, asynchronous or synchronized HeLa cells from a 100-mm-diameter dish were trypsinized, rinsed with phosphate-buffered saline (PBS), incubated in 200 μl of hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2) supplemented with a 0.5 mM protease inhibitor PMSF on ice, and lysed by the addition of 25 μl of hypotonic buffer containing 2.5% Nonidet P-40 (NP-40) and PMSF. The samples were freeze-thawed five times and centrifuged at 850 × g for 10 min. Cytosolic fractions were prepared with an additional high-speed centrifugation of cytoplasmic fractions at 12,000 × g for 30 min, and the supernatants were kept for analysis. For preparing nuclear fractions, nuclear pellets were incubated in extraction buffer (20 mM HEPES [pH 7.9], 0.45 M NaCl, 1 mM EDTA) with protease inhibitors and centrifuged at 12,000 × g for 10 min, and the supernatants were kept for analysis.
Generation of biotinylated RNAs and RNA pull-down assay.
For the generation of biotinylated RNA probes, 5 μg of the monocistronic constructs pM366CAT, pM411CAT, pM531CAT, pM411(U▸C)CAT, and pM531(U▸C)CAT were linearized by using the following restriction enzymes: BamHI for pM366CAT, pM411CAT, and pM411(U▸C)CAT and SmaI for pM531CAT and pM531(U▸C)CAT. RNA transcripts were produced by T7 RNA polymerase (Stratagene) with nucleoside triphosphates in biotinylation composition buffer (0.5 mM [each] ATP, CTP, GTP, and UTP and 0.05 mM Bio-21-UTP [Stratagene]). After a 2-h incubation at 37°C, 5 U of RNase-free DNase (Promega) was added to remove template plasmids. RNA pull-down experiments were performed by using cytoplasmic extracts of HeLa S3 cells and biotinylated RNAs corresponding to wild-type c-myc RNAs (nt 1 to 531, 1 to 411, and 1 to 366) and to mutant c-myc RNAs [nt 1 to 531(U▸C) and 1 to 411(U▸C)]. After incubation of the biotinylated RNAs (4 μg) and the HeLa S3 cell cytoplasmic extracts (50 μg) in 1 ml of incubation buffer (10 mM HEPES [pH 7.4], 1.5 mM magnesium acetate, 90 mM potassium acetate, 2.5 mM dithiothreitol [DDT], 0.05% NP-40) for 30 min at 4°C, the samples were subjected to streptavidin-agarose resin (Pierce) adsorption and further incubated for 2 h. As nonspecific competitors, 8 μg of yeast tRNAs (Roche) was added in the binding mixtures. After incubation, the resin was washed four times with incubation buffer, and then the resin-bound proteins were resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). Immunoblot analysis was performed by using an hnRNP C-specific monoclonal antibody (4F4; a generous gift from Gideon Dreyfuss, University of Pennsylvania, Philadelphia).
In vitro mRNA synthesis.
The dicistronic constructs pRM531F and pRM531(U▸C)F were linearized with the restriction enzyme HpaI. Transcripts were generated in vitro from the linearized plasmids with T7 RNA polymerase in the presence of m7G(5′)pppG(5′) (Amersham-Phamacia Biotech) according to the manufacturer's protocol. Capped mRNA was extracted with phenol-chloroform and then precipitated with ethanol. The plasmids pHC12, which contains hnRNP C1 cDNA, and pT7-7/PTB1, which contains PTB1 cDNA, were linearized with StyI and SalI, respectively. In vitro transcription was carried out with the respective linearized plasmids with SP6 RNA polymerase (Roche) and T7 RNA polymerase. The amount of mRNA was measured by optical absorbance at 260 nm, and the integrity of mRNA was verified by agarose gel electrophoresis.
UV cross-linking.
32P-labeled RNA probes were synthesized by in vitro transcription and isolated by push column chromatography (Stratagene). 32P-labeled RNAs (3 × 105 cpm) were incubated with 20 μg of HeLa cell extracts or 100 ng of purified GST-hnRNP C1 dialyzed in the translation buffer (16 mM HEPES [pH 7.5], 36 mM KCl, 169 mM potassium acetate, 1.2 mM magnesium acetate, 1.6 mM DTT, 2.8 mM β-mercaptoethanol) with or without unlabeled c-myc RNAs or homopolymeric ribonucleotides [poly(A), poly(C), poly(G), and poly(U); Amersham-Pharmacia Biotech]. The RNA-protein mixture binding reaction was carried out in a 30-μl reaction mixture containing 0.5 mM DTT, 5 mM HEPES (pH 7.6), 75 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 20 U of RNasin (Promega), and 3 μg of yeast tRNA. After 20 min of incubation at 30°C, the samples were irradiated with UV on ice for 15 min with a UV-Stratalinker (Stratagene). Unbound RNAs were digested with 5 μl of RNase cocktail (2 μl of RNase A [10 mg/ml], 2 μl of RNase T1 [100 U/ml; Roche], 1 μl of RNase V1 [700 U/ml; Amersham-Pharmacia Biotech]) at 37°C for 45 min and then analyzed by SDS-10 to 12% PAGE.
Immunoprecipitation of UV cross-linked proteins.
For UV cross-linking and immunoprecipitation assays, the samples were precleared by centrifugation in a microcentrifuge for 5 min at 12,000 × g following RNase cocktail treatment. Then 1.5 μl of c-Myc protein-specific monoclonal antibody (9E10; Santa Cruz) or anti-hnRNP C monoclonal antibody (4F4) was added to the preloaded sample diluted in 1 ml of immunoprecipitation buffer (20 mM HEPES [pH 7.4], 125 mM KCl, 0.05% NP-40, 0.5 mM DTT, 0.5 mM PMSF, and 0.5 mM EDTA). After a 2-h incubation at 4°C in a rotary mixer, 30 μl of protein G-agarose slurry (Roche) was added and incubated further for 2 h. After washing four times with 1 ml of immunoprecipitation buffer, resin-bound proteins were detached from the beads by adding 30 μl of Laemmli sample buffer and heating the mixture to 100°C for 5 min and then the proteins were resolved on an SDS-10% PAGE gel.
In vitro translation with RRL.
In vitro translation assays were carried out as described by Jackson and Hunt (30) with minor modifications. One milliliter of RRL (Green Hectares) was treated with micrococcal nuclease (final concentration, 150 U/ml; Roche) at 20°C for 15 min in the presence of 20 μM hemin (Sigma), 50 μg of creatine phosphokinase (Sigma)/ml, and 1 mM CaCl2. The nuclease reaction was terminated with the addition of 10 μl of 0.2 M EGTA. After EGTA treatment, 12 μl of calf liver tRNA solution (5 mg/ml; Novagen) was added to the reaction mixture and mixed well. The resulting nuclease-treated RRL was dispensed into 200-μl aliquots and stored frozen in a liquid nitrogen tank until use. For the synthesis of hnRNP C1 and PTB1 (a negative-control RNA-binding protein), 14 μl of nuclease-treated RRL was mixed with 1 μl of an amino acid mixture containing 1 mM concentrations of each amino acid, 1 μl of 2 M KCl, 0.5 μl of 20 mM MgCl2, 1 μl of 0.2 M creatine phosphate, and 0.4 μg of mRNA encoding either hnRNP C1 or PTB1. The reaction volume was adjusted to 20 μl by adding diethyl pyrocarbonate-treated H2O. Translation reactions were carried out for 100 min at 30°C. For a negative control, the translation reaction was performed in the absence of mRNA (mock RRL). To investigate the effect of hnRNP C1 on c-myc IRES-dependent translation, the reporter dicistronic mRNAs RM531F and RM531(U▸C)F (see Fig. 5A) were incubated in fresh RRL with the addition of the RRL containing hnRNP C1 or PTB1 synthesized in vitro as described above. For the pretreatment of RNA-binding proteins, 0.2 μg of each reporter mRNA [RM531F and RM531(U▸C)F] was incubated for 10 min at 30°C in 10 μl of RNA-protein interaction mixture, which was composed of 4 μl of diethyl pyrocarbonate-treated H2O, 4 μl of RRL containing the newly synthesized hnRNP C1 or PTB1, and 2 μl of RNA-binding buffer (75 mM HEPES [pH 8.0], 50 mM KCl, 1 mM DTT, 50% glycerol). Translation reactions of dicistronic mRNAs [RM531F and RM531(U▸C)F] were initiated by the addition of 12.5 μl of fresh nuclease-treated RRL-amino acid mixture, 1 μl of 2 M KCl, 0.5 μl of 20 mM MgCl2, and 1 μl of 0.2 M creatine phosphate to 5 μl of the RNA-protein interaction mixture. Translation reactions were carried out at 30°C for 45 min. Translation reactions through cap- and IRES-dependent mechanisms were monitored by Renilla and firefly luciferase activities, respectively. All translation reactions were performed at least three times to obtain standard deviations and mean values.
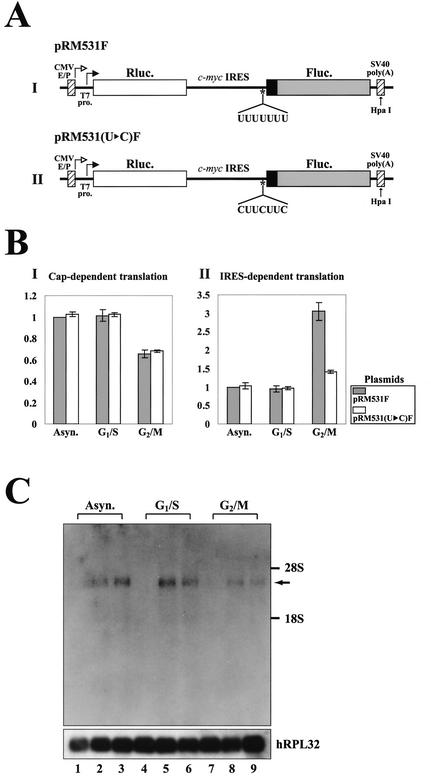
FIG. 5.
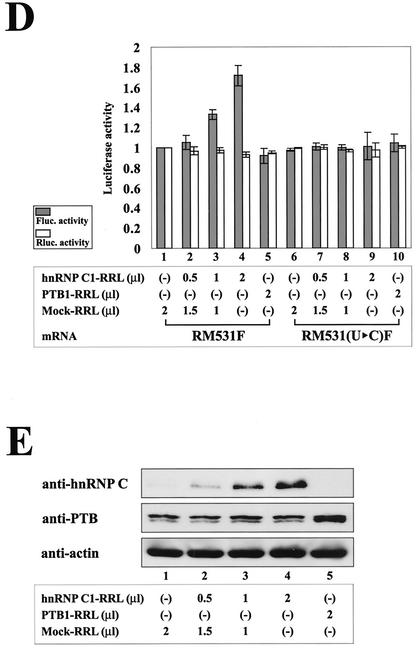
Effects of hnRNP C on IRES-dependent translation of c-myc mRNA at the G2/M phase. (A) Schematic diagrams of dicistronic mRNA used in monitoring efficiency of cap- and c-myc IRES-dependent translation in vivo and in vitro. The vectors contain the CMV immediate-early enhancer/promoter (CMV E/P) directing transcription in vivo and a T7 promoter (T7 pro.) directing transcription in vitro. The transcription start sites through the CMV E/P and T7 promoters are indicated by open and solid arrows, respectively. The dicistronic mRNA is composed of five consecutive elements: the 5′ NTR directing cap-dependent translation, Renilla luciferase gene (Rluc. open box), c-myc IRES directing IRES-dependent translation, firefly luciferase gene (Fluc. shaded box), and the 3′ NTR. The mRNA RM531F contains the complete c-myc IRES (nt 1 to 411) and the N-terminal part of the c-Myc2 protein coding sequence up to nt 531 (solid box), which is fused to the firefly luciferase open reading frame. The mRNA RM531(U▸C)F is a derivative of RM531F with the U-to-C change in the heptameric U sequence of the c-myc IRES. The restriction enzyme site HpaI used for linearization of the plasmids is also designated by an arrow. (B) Relative translation activities of dicistronic mRNAs at different cell cycle phases. The dicistronic constructs [pRM531F or pRM531(U▸C)F] and transfection control plasmid pCMV•SPORT-βgal cotransfected into HeLa cells 12 h prior to chemical treatments for synchronization. After synchronization, cells were harvested and luciferase activity was measured. Panels BI and BII represent relative Renilla and firefly luciferase activities at different cell cycle phases, respectively. Shaded and open bars represent luciferase activities of cells transfected with pRM531F and pRM531(U▸C)F, respectively. For cap-dependent translation, Renilla luciferase activities were measured, and the relative value of each condition was normalized to the Renilla luciferase activity of asynchronized (Asyn.)cells transfected with pRM531F and that value was set to 1. For IRES-dependent translation, firefly luciferase activities were measured, and the relative value of each condition was normalized to the firefly luciferase activity of asynchronized cells transfected with pRM531F and that value was set to 1. Transfection efficiency was also determined for each luciferase value relative to that of the transfection control β-galactosidase activity. The lines represent standard deviation values. Experiments were repeated at least three times to ensure reproducibility. (C) Northern blot analysis of dicistronic mRNAs produced from cells transfected with pRM531F and pRM531(U▸C)F. After harvesting synchronized cells, poly(A)+ RNAs were isolated and then subjected to Northern blotting with a 32P-labeled probe corresponding to the firefly luciferase gene. The positions of 28S and 18S ribosomal RNAs are depicted as 28S and 18S in panel C. The human ribosomal protein large subunit 32 (hRPL32) blot was used as an internal control of poly(A)+ mRNAs. The arrow indicates the position of dicistronic mRNAs. RNAs from asynchronized cells, G1/S-arrested cells, and G2/M-arrested cells are shown on lanes 1 to 3, 4 to 6, and 7 to 9, respectively. RNAs from cells transfected with no plasmid, pRM531F, and pRM531(U▸C)F were loaded on lanes 1, 4, and 7; 2, 5, and 8; and 3, 6, and 9, respectively. (D) The effect of hnRNP C on translation through the c-myc IRES element. Translation reactions were carried out as described in Materials and Methods. Variable amounts of mock-translated RRL (lanes 1, 2, 3, 6, 7, and 8) and of RRL containing presynthesized hnRNP C1 (lanes 2, 3, 4, 7, 8, and 9) and PTB1 (lanes 5 and 10) were preincubated with the mRNA RM531F (lanes 1 to 5) and RM531(U▸C)F (lanes 6 to 10) for 10 min at 30°C. The amounts of RRL used in the RNA-protein interaction reactions are indicated at the bottom. hnRNP C1-RRL, PTB1-RRL, and Mock-RRL represent RRL containing proteins synthesized by in vitro translation with hnRNP C1 mRNA, PTB1 mRNA and no mRNA, respectively. After translation of the dicistronic reporter mRNAs [RM531F and RM531(U▸C)F], firefly and Renilla luciferase activities were measured by using a dual-luciferase reporter assay system. Shaded and open bars represent firefly and Renilla luciferase activities of each dicistronic reporter mRNA [RM531F and RM531(U▸C)F], respectively. For IRES-dependent translation, firefly luciferase activity, which was directed by mRNA RM531F, measured in the translation mixture with the addition of mock-translated RRL was arbitrarily set to 1. For cap-dependent translation, Renilla luciferase activity, which was directed by mRNA RM531F, measured in the translation mixture with the addition of mock-translated RRL was arbitrarily set to 1. Lines represent standard deviation values. (E) Protein levels of hnRNP C, PTB, and actin in each in vitro translation mixture. Immunoblot analysis was used to monitor the protein levels of hnRNP C, PTB, and actin in translation mixtures that contained both endogenously and exogenously synthesized proteins. About 50 μg of total proteins was used in each immunoblot analysis. The level of actin was monitored to confirm that the amount of total protein was the same in each immunoblot analysis. RRL mixtures were analyzed by using monoclonal antibodies against hnRNP C (anti-hnRNP C), PTB (anti-PTB), and actin (anti-actin), respectively.
Cellular RNA purification and Northern blot analysis.
Expression of dicistronic mRNA was analyzed by the Northern blot method with a probe specific for firefly luciferase. A 32P-labeled probe was generated by using a random primer labeling system (rediprime II; Amersham-Pharmacia Biotech) according to the manufacturer's instructions. Unincorporated 32P was removed by passing the sample through a push column. Plasmids containing the wild-type and mutant forms of c-myc IRESs were transfected into semiconfluent HeLa cells by using Lipofectamine (Gibco-BRL) 18 h prior to synchronization. After transfection and synchronization, total RNA was harvested from HeLa cells by using the TRI reagent (Molecular Research Center) according to manufacturer's directions. After polyadenosine-containing [poly(A)+] mRNAs were purified by using oligo(dT) cellulose (Amersham-Pharmacia Biotech), 2 μg of poly(A)+ mRNAs were resolved on 1.3% agarose gels in the presence of formaldehyde and 3-(N-morpholino)propanesulfonic acid buffer and then blotted onto a nylon membrane (Qiagen). Hybridization was carried out in hybridization buffer (10% polyethylene glycol, 7% SDS, 1 mM EDTA, 250 mM NaCl, 80 mM Na2HPO4 [pH 7.3], salmon sperm single-stranded DNA [Sigma]) with 2 × 104 cpm of 32P-labeled probe/ml. After an 18-h incubation at 67°C, the nylon membrane was washed and then subjected to autoradiography.
Reporter gene analysis.
The dicistronic constructs, which contain wild-type or mutant c-myc IRES were cotransfected with pCMV•SPORT-βgal (Invitrogen) to HeLa cells by using electroporation. After synchronization, cells were harvested and freeze-thawed for three cycles. To obtain the cytoplasmic extract, centrifugation was carried out at 10,000 × g for 10 min. Aliquots of in vitro-translated RRL were diluted with PBS. The cytoplasmic extracts and PBS-diluted RRLs were analyzed with a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. The transfection efficiency was also determined for each luciferase value relative to that of the transfection control β-galactosidase activity.
Immunoblot analysis.
Immunoblot analysis was performed with monoclonal antibodies against hnRNP C, PTB, actin (ICN Biomedicals), and poly(ADP-ribose) polymerase (PARP; Santa Cruz) as primary antibodies and a horseradish peroxidase-linked anti-mouse immunoglobulin G as a secondary antibody. Membrane-bound antibodies were detected by enhanced chemiluminescence (Amersham-Pharmacia Biotech).
RESULTS
Identification of a 40-kDa protein that interacts with the human c-myc IRES.
To identify cellular factor(s) interacting with the c-myc IRES, a UV cross-linking experiment was performed with HeLa S3 cell cytoplasmic extracts and 32P-labeled c-myc RNAs I (nt 1 to 366) and II (nt 1 to 411) shown in Fig. 1A. Four cellular proteins with apparent molecular masses of 40, 52, 65, and 110 kDa were found to bind to c-myc RNA II (nt 1 to 411) (Fig. 1B, lane 1). In contrast, the binding to RNA I (nt 1 to 366) was dramatically and specifically reduced for the 40-kDa protein while identical binding was observed for the other three proteins of 52, 65, and 110-kDa (Fig. 1B, compare lanes 1 and 2). These binding data indicate that the c-myc RNA region between nt 367 and 411 is required for the strong interaction with the 40-kDa protein.
The specificity of RNA-binding activity was investigated by adding unlabeled competitor RNA II (nt 1 to 411) to the UV cross-linking reaction mixture. Binding of the 40-kDa protein was the most sensitive to competition, essentially abolishing binding activity to the RNA II probe (Fig. 1B, lane 7). When homopolymeric ribonucleotides were used as competitors in the UV cross-linking reaction mixtures, poly(U) and poly(G) but not poly(A) and poly(C) strongly inhibited binding of the 40-kDa protein to the c-myc IRES (Fig. 1B, lanes 3 to 6). The 40-kDa protein was further characterized because it binds specifically to the c-myc IRES.
hnRNP C interacts with the heptameric U sequence in the c-myc IRES.
Our binding results indicate that the region between the two alternative translation initiation codons (nt 367 to 411) of c-myc RNA is required for the binding of the 40-kDa protein (Fig. 1B, lanes 1 and 2). Moreover, poly(U) and poly(G) RNAs can compete for this binding to the c-myc IRES RNA (Fig. 1B, lanes 5 to 6). Both the molecular mass and RNA-binding specificity of the 40-kDa protein are similar to those of hnRNP C, a member of hnRNPs (58). Moreover, hnRNP C was shown to bind strongly to U- and G-rich sequences by a systematic evolution of ligands by exponential enrichment and competition binding assays (16, 54). These analyses identified a U- and G-rich sequence (AGUAUUUUUGUGGA) that is similar to a sequence present within the high-affinity binding region of the 40-kDa protein in the c-myc IRES (nt 365 to 381; UGGAUUUUUUUCGGGUA) (hereafter, we refer to this UUUUUUU sequence as the heptameric U sequence). Therefore, we next examined whether the 40-kDa protein is immunologically related to hnRNP C by immunoprecipitation of UV cross-linked proteins with 32P-labeled c-myc RNA II (nt 1 to 411). The 40-kDa protein was detected after immunoprecipitation of UV cross-linked proteins by a specific monoclonal antibody against hnRNP C (4F4) (Fig. 2A, lane 2). On the other hand, no band was detected when a monoclonal antibody against c-Myc protein (9E10) was used as a negative control (Fig. 2A, lane 1). These data identify hnRNP C as the 40-kDa protein that binds to the c-myc IRES.
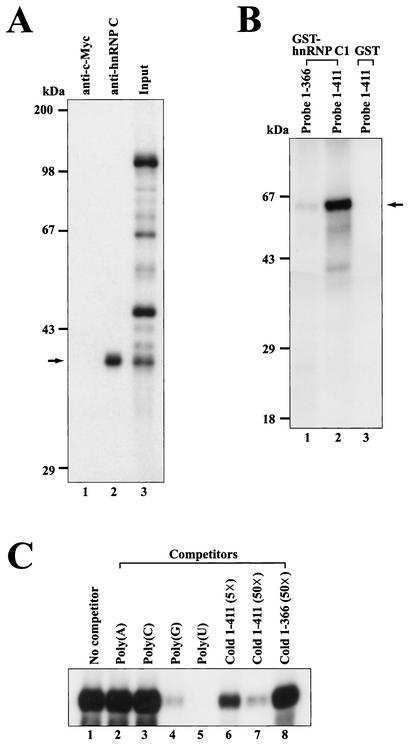
FIG. 2.
hnRNP C interacts with the c-myc IRES. (A) Immunoprecipitation of proteins labeled by UV cross-linking with 32P-labeled RNA probe II (nt 1 to 411). After RNase cocktail treatment, samples were precleared with streptavidin-agarose resin and then a monoclonal antibody against hnRNP C (4F4) (lane 2) or c-Myc protein (9E10) (lane 1) was added to the immunoprecipitation buffer. After immunoprecipitation, bound proteins were resolved on an SDS-10% PAGE gel. Lane 3 shows the labeled proteins after UV cross-linking. (B) Direct binding of purified hnRNP C1 to the c-myc IRES. One hundred nanograms of purified GST-hnRNP C1 and GST were UV cross-linked with RNA probes I and II (3 × 105 cpm), respectively. After UV-irradiation, samples were treated with RNase cocktail before being analyzed by SDS-12% PAGE. The arrow represents the position of cross-linked GST-hnRNP C1 (65 kDa). GST-hnRNP C1 protein and GST protein were used in lanes 1 and 2 and lane 3 as protein sources, respectively. Transcripts I and II were used in lane 1 and lanes 2 and 3 as probes, respectively. (C) Effects of competitor RNAs on binding of purified hnRNP C1 to probe II. One hundred nanograms of recombinant GST-hnRNP C1 was incubated with RNA probe II in the UV cross-linking experiments (lanes 1 to 8). As competitor RNAs, 50 ng of four homopolymeric RNAs (lanes 2 to 5), 5 (lane 6)- and 50 (lane 7)-fold molar excesses of unlabeled transcript II, and a 50-fold molar excess of transcript I (lane 8) were added in the competition assays.
The interaction between the c-myc IRES and hnRNP C was confirmed by the UV cross-linking method with a purified recombinant hnRNP C protein. We purified a recombinant GST-hnRNP C1 from E. coli. As shown in Fig. 2B, GST-hnRNP C1 strongly bound to probe II (nt 1 to 411) containing the heptameric U sequence. In contrast, GST-hnRNP C1 bound poorly to probe I (nt 1 to 366) lacking the heptameric U sequence (Fig. 2B, lane 1). The GST protein itself did not bind to probe II (nt 1 to 411) (Fig. 2B, lane 3).
The interaction between hnRNP C and c-myc RNA was further confirmed by competition assays. UV cross-linking experiments with purified GST-hnRNP C1 and 32P-labeled c-myc RNA were performed in the presence or absence of competitor RNAs (Fig. 2C). The interaction between GST-hnRNP C1 and probe II was strongly inhibited by poly(U), poly(G), and unlabeled c-myc RNA (nt 1 to 411) but not by poly(A), poly(C), or unlabeled c-myc RNA (nt 1 to 366) lacking the heptameric U sequence (Fig. 2C). These competition patterns with purified GST-hnRNP C1 are well matched with the binding of the 40-kDa protein to c-myc RNA obtained by UV cross-linking with a HeLa S3 cell cytoplasmic extract (compare Fig. 1B with 2C). Taken together, these data indicate that hnRNP C directly interacts with c-myc RNA and that a specific RNA-protein interaction can occur in cells.
The heptameric U sequence resides at nt 369 to 375, which is downstream of the minor translational initiation codon CUG364 but upstream of the major translational initiation codon AUG409. We next determined whether the heptameric U sequence participates in the binding of hnRNP C since this protein has a high affinity to oligo(U) (16). The heptameric U sequence was changed to CUUCUUC while maintaining the amino acid sequences of the c-Myc1 translated from CUG364 (Fig. 3A). The effect of the mutation on hnRNP C binding to c-myc RNA was assessed by RNA pull-down assays (Fig. 3B) or UV cross-linking experiments (Fig. 3C). In vitro pull-down experiments were performed by using HeLa S3 cell cytoplasmic extracts and biotinylated c-myc RNAs (Fig. 3B). HnRNP C was coprecipitated with wild-type c-myc RNAs containing the intact heptameric U sequence such as c-myc RNA (nt 1 to 531) and c-myc RNA (nt 1 to 411) (Fig. 3B, lanes 2 and 3), whereas c-myc RNAs lacking the heptameric U sequence were not associated with hnRNP C (Fig. 3B, lanes 4 to 6). We also found that a biotinylated RNA of 45 nt corresponding to nt 367 to 411 was sufficient for binding to hnRNP C (data not shown).
FIG. 3.
The heptameric U sequence plays a key role in the interaction with hnRNP C. (A) Schematic diagram of RNA transcripts used in RNA pull-down experiments and UV cross-linking experiments. The elements are represented as described in the legend to Fig. 1A. In the constructs of nt 1 to 411(U▸C) and nt 1 to 531(U▸C), there are nucleotide changes in the heptameric U sequence from UUUUUUU to CUUCUUC. The amino acid sequence of the c-Myc1 protein remains the same even after the U-to-C mutation. (B) In order to investigate whether hnRNP C can make a complex with the c-myc IRES, RNA pull-down experiments were performed by using cytoplasmic extracts of HeLa S3 cells and biotinylated RNAs corresponding to wild-type c-myc RNAs (nt 1 to 531, 1 to 411, and 1 to 366 are shown on lanes 2 to 4, respectively) and to mutant c-myc RNAs [nt 1 to 531(U▸C) and 1 to 411(U▸C) are shown on lanes 5 to 6]. After incubation of the biotinylated RNAs and the cytoplasmic extracts in incubation buffer, the samples were subjected to streptavidin-agarose resin adsorption. After washing the resins, the resin-bound proteins were resolved by an SDS-12% PAGE gel. Immunoblot analysis was performed by using an hnRNP C-specific monoclonal antibody (4F4). (C) UV cross-linking experiments were performed with 100 ng of purified proteins (GST-hnRNP C1 [lanes 1 to 5] and GST [lane 6]) and 32P-labeled RNA probes (3 × 105 cpm) corresponding to c-myc wild type (nt 1 to 531, 1 to 411, and 1 to 366 [lanes 1 to 3]) and mutant RNAs [nt 1 to 531(U▸C) and 1 to 411(U▸C) (lanes 4 and 5)]. After UV-irradiation, samples were treated with RNase cocktail and then resolved by SDS-12% PAGE. The arrow represents the position of cross-linked GST-hnRNP C1.
Direct binding of hnRNP C to the heptameric U sequence was assessed by using GST-hnRNP C1 and 32P-labled c-myc RNAs (Fig. 3C). Purified GST-hnRNP C1 interacted strongly with c-myc RNA containing the intact heptameric U sequence (Fig. 3C, lanes 1 and 2) but weakly with mutant c-myc RNAs with U-to-C changes (Fig. 3C, lanes 4 and 5). Together, these data indicate that the heptameric U sequence is critical for the binding of hnRNP C to the c-myc IRES.
hnRNP C specifically interacts with c-myc IRES in a cell cycle phase-dependent manner.
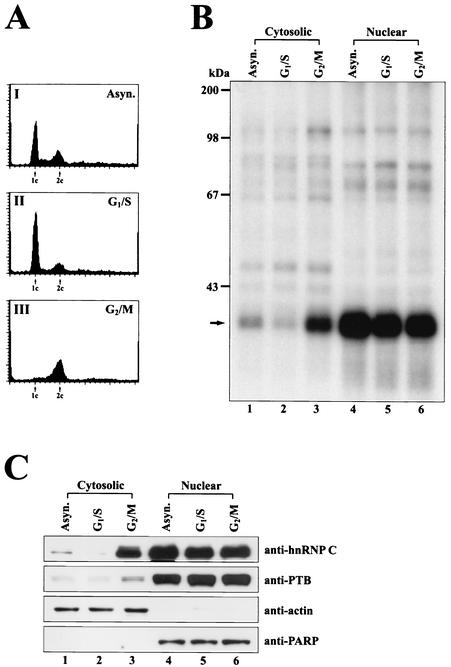
In normal cells, hnRNP C is mainly confined to the nucleus (10), and therefore, its function in translation is enigmatic, although several cytoplasmic roles of hnRNP C have been reported (38, 48, 52, 63). Recently, enrichment of hnRNP C in the cytoplasm has been reported at a specific cell cycle phase and during differentiation (38, 52). To investigate the role of hnRNP C in c-myc translational regulation, we determined the level of hnRNP C at specific cell cycle phases with synchronized HeLa cells. The proliferation of HeLa cells was arrested at the specific cell cycle phases G1/S and G2/M. Cell cycle progression was monitored by flow cytometry. As shown in Fig. 4A, cells were predominantly arrested at the G1/S phase (Fig. 4AII) and G2/M phase (Fig. 4AIII).
FIG. 4.
The level of hnRNP C protein in the cytoplasm fluctuates in a cell cycle phase-dependent manner. (A) Synchronization of cells by the chemical treatments was monitored by flow cytometry analysis of cells stained with propidium iodide. The G1 and G2 peaks are depicted as 1c and 2c, respectively. (B) For UV cross-linking experiments, 25 μg of HeLa cell extract was incubated with a wild-type c-myc RNA probe II (nt 1 to 411) (Fig. 1A) labeled with 32P (3 × 105 cpm). Cytosolic (lanes 1 to 3) and nuclear (lanes 4 to 6) extracts were prepared from asynchronous exponentially growing HeLa cells (Asyn.), G1/S phase-arrested cells (G1/S), and G2/M phase-arrested cells (G2/M). After UV-irradiation, samples were treated with RNase cocktail and then resolved by SDS-12% PAGE. The arrow depicts the position of cross-linked hnRNP C. (C) Fluctuation of hnRNP C level in the cytoplasm was monitored by immunoblot analysis. The cytosolic and nuclear fractions of HeLa cell extracts were prepared as described for panel B. Fractionated HeLa cell extracts (20 μg each) were analyzed by immunoblotting with specific monoclonal antibodies against hnRNP C, PTB, actin, and PARP as shown in the rows anti-hnRNP C, -PTB, -actin, and -PARP, respectively.
After synchronization, cells were harvested, and the cytosolic and nuclear extracts were subjected to UV cross-linking assays by using an RNA probe II (nt 1 to 411) (Fig. 1A) containing the heptameric U sequence (Fig. 4B). The level of hnRNP C in the cytosolic extracts was dramatically increased in cells arrested at the G2/M phase (Fig. 4B, lane 3) compared with cytosolic extracts of the asynchronized cells (lane 1) or cells arrested at the G1/S phase (lane 2). The level of hnRNP C in the nucleus remains at similar levels throughout the cell cycle (lanes 4 to 6 in Fig. 4B).
The level of hnRNP C in the cytoplasm was also monitored by immunoblot analysis (Fig. 4C). hnRNP C was dramatically enriched in the cytoplasm at the G2/M phase compared to the G1/S phase (Fig. 4C, compare lanes 3 and 2 of anti-hnRNP C). Similar levels of hnRNP C were detected in the nucleus of the asynchronized and synchronized cells (Fig. 4C, lanes 4 to 6 of anti-hnRNP C). To confirm the total amounts of proteins and the quality of fractionation of the cell extracts, levels of actin, which is confined in the cytoplasm, and PARP, which is confined in the nucleus, were monitored with anti-actin and anti-PARP antibodies. As shown in Fig. 4C, actin and PARP were detected mostly in the cytosolic (Fig. 4C, lanes 1 to 3) and nuclear fractions (Fig. 4C, lanes 4 to 6), respectively, and levels of these proteins remain the same throughout the cell cycle. These data indicate that the fractionations of cell extracts were accomplished properly and that the fluctuation of the level of hnRNP C in the cell cycle is not a general phenomenon observed for all proteins. Interestingly, PTB, which shuttles between the nucleus and the cytoplasm, was increased in the cytoplasm at the G2/M phase even though the degree of fluctuation in protein levels is much lower than that of hnRNP C (Fig. 4C). The fluctuation of hnRNP C levels during the cell cycle phases was also shown by Millard et al. (38).
hnRNP C augments translation of c-myc mRNA at the G2/M phase.
In order to investigate the role of the hnRNP C binding to c-myc IRES function, we generated dicistronic mRNA constructs containing the c-myc IRES or its variant at the intercistronic regions of two consecutive genes, Renilla luciferase and firefly luciferase (Fig. 5A). In these constructs, the firefly luciferase and Renilla luciferase are produced by c-myc IRES-dependent translation and cap-dependent translation, respectively. HeLa cells were transfected with plasmid pRM531F, which produces a dicistronic mRNA containing wild-type c-myc IRES, or pRM531(U▸C)F, which produces a dicistronic mRNA with the mutations in the heptameric U sequence in the c-myc IRES. After transfection, the cells were synchronized at the G1/S or G2/M phase and assessed for luciferase activity. As shown in Fig. 5BI, cap-dependent translation measured by Renilla luciferase activity was inhibited at the G2/M phase relative to the G1/S phase. This partial inhibition of cap-dependent translation at the G2/M phase occurred independently of the c-myc IRES and has been previously documented (reference 46 and references therein).
In direct contrast, translation of c-myc IRES-dependent translation was increased at the G2/M phase relative to the G1/S phase (Fig. 5BII). Interestingly, translation through the wild-type c-myc IRES was increased at the G2/M phase by about threefold, whereas translation through the mutant c-myc IRES lacking the hnRNP C-binding site was increased by only about 1.5-fold at the G2/M phase (Fig. 5BII). This marginal activation of the mutant c-myc IRES at the G2/M phase may be attributed to the general enhancement of IRES-dependent translation at mitosis (see below). Alternatively, it could be attributed to the residual activity of the mutated heptameric U sequence. These data strongly suggest that the heptameric U sequence plays an important role in the translation of c-myc mRNA at the G2/M but not at the G1/S phase. The augmentation of c-myc IRES activity by the heptameric U sequence at the G2/M phase is most likely to occur through interaction with hnRNP C, which becomes enriched in the cytoplasm at that phase (see below).
To assess the level and integrity of the c-myc mRNA directly, the dicistronic mRNAs were monitored by Northern blot analysis with a 32P-labeled DNA corresponding to the firefly luciferase gene (Fig. 5C). Dicistronic mRNAs of expected sizes were detected from all of the transfected cells (Fig. 5C, lanes 2, 3, 5, 6, 8, and 9). Similar intensities of mRNA bands were detected from the wild-type and U-to-C mutant c-myc IRESs at specific cell cycle phases (Fig. 5C, compare lanes 5 and 8 with lanes 6 and 9, respectively). The intensities of mRNA bands at the G1/S phase were slightly higher than those at the G2/M phase. From the Northern blot analysis, it appears that the translation efficiency of the c-myc mRNA at the G2/M phase compared with that at the G1/S phase may be higher than the threefold measured by the firefly luciferase assay as described above.
To demonstrate that hnRNP C augments translation of c-myc IRES-dependent translation, the effect of the hnRNP C1 protein on the in vitro translation system (RRL) was investigated by using the dicistronic mRNAs RM531F and RM531(U▸C)F as reporters. Capped mRNA was generated by in vitro transcription with T7 RNA polymerase. The proteins hnRNP C1 and PTB1 (a negative-control RNA-binding protein) were synthesized in RRL by in vitro translation of mRNAs encoding hnRNP C1 and PTB1 and then added to in vitro translation mixtures. Only a barely detectable amount of hnRNP C protein exists in the RRL (Fig. 5E, lanes 1 and 5 of anti-hnRNP C). Increased amounts of hnRNP C1 proteins were detected by immunoblot analysis in RRL mixtures where newly synthesized hnRNP C1 proteins were added (Fig. 5E, lanes 2, 3, and 4, anti-hnRNP C). A large amount of PTB protein exists in RRL (Fig. 5E, lanes 1 to 4, anti-PTB), and an increased amount of PTB1 protein was detected by the addition of newly synthesized PTB1 protein (Fig. 5E, lane 5, anti-PTB). The translation of firefly luciferase through the wild-type c-myc IRES, which contains an hnRNP C-binding site, was increased in a dose-dependent manner by the addition of exogenous hnRNP C1 protein (Fig. 5D, lanes 1 to 4). In contrast, the translation of firefly luciferase through a mutant c-myc IRES, which lacks an hnRNP C-binding site, was not affected by the addition of exogenous hnRNP C1 protein (Fig. 5D, lanes 6 to 9). Moreover, the addition of exogenous PTB1 protein did not affect translation either through the wild-type c-myc IRES (Fig. 5D, lane 5) or through the mutant c-myc IRES (Fig. 5D, lane 10). The translation of cap-dependent mRNA, which was monitored by Renilla luciferase activity, was not influenced by the addition of either hnRNPC1 or PTB1 (Fig. 5D, lanes 1 to 10). These results strongly suggest that hnRNP C augments the translation of c-myc mRNA via interaction with the heptameric U sequence of the c-myc IRES.
DISCUSSION
Cap-dependent translation is inhibited during the mitotic phase of the cell cycle by hypophosphorylation of 4E-BP1, a suppressor of eukaryotic initiation factor 4E function, and by hyperphosphorylation of eukaryotic initiation factor 4GII (reference 46 and references therein). Therefore, an alternative cap-independent mechanism of translation for mRNA encoding unstable proteins is required at mitosis. Several viral and cellular mRNAs has been shown to be functional at mitosis through IRES-dependent mechanisms. For instance, poliovirus (9), hepatitis C virus (26), ornithine decarboxylase (47), and p58PITSLRE (12) IRESs are functional at the G2/M phase where the switch from cap-dependent translation to IRES-dependent translation may occur. This switch may be mediated either indirectly through the increased availability of translation factors that are utilized in both cap-dependent and IRES-dependent translations due to the inhibition of cap-dependent translation at this phase or through the enrichment in the cytoplasm of the specific factor(s) required for IRES function.
The c-Myc protein is involved in critical cellular processes, including proliferation, transformation, and apoptosis (1, 45). The c-Myc protein is required at different phases of the cell cycle. For instance, the c-Myc protein plays a critical role in G1/S progression (reference 45 and references therein) and is necessary for DNA damage-induced apoptosis in the G2 phase of the cell cycle (1). c-Myc is a very labile protein with a half-life of 20 to 40 min (19) irrespective of the phase of the cell cycle (21) and is degraded via the ubiquitin/26S proteasome pathway (51). Therefore, c-myc mRNA should be translated under the conditions when most cellular mRNAs are not functional. Indeed, c-myc mRNA contains an IRES element at the 5′ NTR that is functional during apoptosis when cap-dependent translation is impaired (55).
Here we show that the c-Myc protein is synthesized through an IRES element and that the IRES-dependent translation of c-myc mRNA is augmented by hnRNP C, which is enriched in the cytoplasm during the G2/M phase of the cell cycle (Fig. 5BII). The augmentation of c-myc IRES-dependent translation is likely to be a way to maintain a constant level of c-Myc protein throughout the cell cycle since cap-dependent translation (which is also used by c-myc mRNA) (57) of most of the cellular mRNA decreases at the mitotic phase of the cell cycle (Fig. 5BI) (reference 46 and references therein).
The boundary of the c-myc IRES was originally defined between nt −363 and −94 upstream from the CUG364 start codon (41). However, the authors did not show that the RNA sequence between −363 and −94 upstream from CUG364 start codon was sufficient for IRES function, even though it was required for IRES activity (41). Stoneley and colleagues (56) showed that the RNA sequence between nt 353 and 408 was not essential for c-myc IRES activity by using dicistronic reporter mRNAs.
From our results, we suggest that the heptameric U sequence residing downstream of the CUG364 initiation codon plays an important role in IRES-dependent translation at the G2/M phase (Fig. 5BII). The requirement of the heptameric U sequence in the c-myc IRES-dependent translation seems to be limited to the G2/M phase, since the wild-type c-myc IRES and a mutant c-myc IRES lacking the heptameric U sequence showed similar translational efficiency in cells arrested at the G1/S phase as well as in asynchronized cells where interphase cells predominate (Fig. 5BII). It should be noted that asynchronized cells were used in the previous studies determining the boundary of the c-myc IRES (41, 56). Therefore, the differences in the results are most likely due to differences in the systems used in analyzing the IRES function of c-myc mRNA.
Here we show that the amounts of hnRNP C in the cytoplasm increase dramatically at the G2/M phase even though most of the hnRNP C still remains in the nucleus (Fig. 4B and C) (38). We also showed that hnRNP C interacts specifically with the heptameric U sequence, and this interaction is required for the augmentation of IRES-dependent translation at the G2/M phase. Moreover, we showed that the addition of hnRNP C1 protein to an in vitro translation system (RRL) significantly enhanced the function of the wild-type c-myc IRES, which contains an hnRNP C-binding site, but not that of a mutant c-myc IRES lacking a functional hnRNP C-binding site. From these data we speculate that hnRNP C augments IRES-dependent translation occurring at the G2/M phase through an interaction with the heptameric U sequence of the c-myc IRES.
The enrichment of hnRNP C in the cytoplasm at the G2/M phase was shown by Millard et al. (38) through investigations into RNA-binding proteins interacting with p27KIP1 mRNA, which plays a key role in cell cycle regulation through the inhibition of cyclin E-cdk2 (53). Cell cycle phase-dependent fluctuation of hnRNP C in the cytoplasm may be attributed to the redistribution of preexisting hnRNP C from the nucleus to the cytoplasm and/or to the blockage of transportation of newly synthesized hnRNP C to the nucleus. This study also showed that the region including a U-rich element residing about 40 nt upstream of the translation initiation codon is required for efficient translation of p27KIP1 mRNA and that hnRNP C and HuR specifically interact with the U-rich element (38).
Intriguingly, there is a heptameric U sequence in the U-rich element of p27KIP1 mRNA. However, the effect of changes in the heptameric U sequence on hnRNP C binding has not yet been investigated. Recently, Miskimins and colleagues showed that p27KIP1 mRNA contains an IRES element (39), but the effect of hnRNP C on the p27KIP1 IRES-dependent translation was not investigated. It is possible that the functions of the IRESs in both c-myc and p27KIP1, which play critical roles in G1/S phase progression (35), are regulated by the same protein, namely hnRNP C. Moreover, Sella and colleagues suggested that hnRNP C is also involved in PDGF2/c-sis IRES-dependent translation (52). The authors showed that hnRNP C interacts with a part of the PDGF2/c-sis 5′ NTR which contains a differentiation-linked IRES element. In order to understand the molecular mechanism of hnRNP C function, detailed roles of hnRNP C in the cell cycle and differentiation need to be further investigated.
Acknowledgments
We thank Gideon Dreyfuss for providing monoclonal antibody against hnRNP C (4F4) and plasmid pHC12 and Eckard Wimmer for providing monoclonal antibody against PTB. We thank Yon Woo Jung and Chang Woo Lee for helpful discussions and technical assistance in cell cycle analysis, Kwan Hyuck Baek for fluorescence-activated cell sorter analysis, and Ok-Kyu Song for critical reading of the manuscript. We are also grateful to members of our laboratory for helpful discussions and technical assistance.
The present study was supported in part by grants from the NRL (M10204000018-02J0000-01610), the MMRG (M10106000056-02B1700-01210), and the G7 Program of MOST and by KOSEF through PNRC.
REFERENCES
- 1.Adachi, S., A. J. Obaya, Z. Han, N. Ramos-Desimone, J. H. Wyche, and J. M. Sedivy. 2001. c-Myc is necessary for DNA damage-induced apoptosis in the G2 phase of the cell cycle. Mol. Cell. Biol. 21:4929-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, N., and A. Siddiqui. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 94:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amati, B., M. W. Brooks, N. Levy, T. D. Littlewood, G. I. Evan, and H. Land. 1993. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72:233-245. [DOI] [PubMed] [Google Scholar]
- 4.Amati, B., T. D. Littlewood, G. I. Evan, and H. Land. 1993. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 12:5083-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ar-Rushdi, A., K. Nishikura, J. Erikson, R. Watt, G. Rovera, and C. M. Croce. 1983. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science 222:390-393. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, D. L., and M. Groudine. 1986. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol. Cell. Biol. 6:3481-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, J., O. Sella, S. Y. Le, and O. Elroy-Stein. 1997. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES). J. Biol. Chem. 272:9356-9362. [DOI] [PubMed] [Google Scholar]
- 8.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonneau, A. M., and N. Sonenberg. 1987. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem. 262:11134-11139. [PubMed] [Google Scholar]
- 10.Choi, Y. D., and G. Dreyfuss. 1984. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 99:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coldwell, M. J., S. A. Mitchell, M. Stoneley, M. MacFarlane, and A. E. Willis. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19:899-905. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis, S., Y. Bruynooghe, G. Denecker, S. Van Huffel, S. Tinton, and R. Beyaert. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5:597-605. [DOI] [PubMed] [Google Scholar]
- 13.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evan, G., and T. Littlewood. 1998. A matter of life and cell death. Science 281:1317-1322. [DOI] [PubMed] [Google Scholar]
- 15.Gil, A., P. A. Sharp, S. F. Jamison, and M. A. Garcia-Blanco. 1991. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 5:1224-1236. [DOI] [PubMed] [Google Scholar]
- 16.Gorlach, M., C. G. Burd, and G. Dreyfuss. 1994. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J. Biol. Chem. 269:23074-23078. [PubMed] [Google Scholar]
- 17.Gottlieb, E., and J. A. Steitz. 1989. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 8:851-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graff, J., J. Cha, L. B. Blyn, and E. Ehrenfeld. 1998. Interaction of poly(rC) binding protein 2 with the 5′ noncoding region of hepatitis A virus RNA and its effects on translation. J. Virol. 72:9668-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hann, S. R., and R. N. Eisenman. 1984. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol. Cell. Biol. 4:2486-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hann, S. R., M. W. King, D. L. Bentley, C. W. Anderson, and R. N. Eisenman. 1988. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell 52:185-195. [DOI] [PubMed] [Google Scholar]
- 21.Hann, S. R., C. B. Thompson, and R. N. Eisenman. 1985. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature 314:366-369. [DOI] [PubMed] [Google Scholar]
- 22.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 23.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holcik, M., and R. G. Korneluk. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holcik, M., C. Lefebvre, C. Yeh, T. Chow, and R. G. Korneluk. 1999. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1:190-192. [DOI] [PubMed] [Google Scholar]
- 26.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. A. Abell, and S. M. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 27.Hueber, A. O., and G. I. Evan. 1998. Traps to catch unwary oncogenes. Trends Genet. 14:364-367. [DOI] [PubMed] [Google Scholar]
- 28.Huez, I., L. Creancier, S. Audigier, M. C. Gensac, A. C. Prats, and H. Prats. 1998. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 18:6178-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt, S. L., J. J. Hsuan, N. Totty, and R. J. Jackson. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson, R. J., and T. Hunt. 1983. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 96:50-74. [DOI] [PubMed] [Google Scholar]
- 31.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang, S. K., and E. Wimmer. 2001. The role of RNA-binding proteins in IRES-dependent translation, p. 1-33. In K. Sandberg and S. E. Mulroney (ed.), RNA binding proteins: new concepts in gene expression. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 33.Kim, J. H., B. Hahm, Y. K. Kim, M. Choi, and S. K. Jang. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298:395-405. [DOI] [PubMed] [Google Scholar]
- 34.Kim, Y. K., B. Hahm, and S. K. Jang. 2000. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J. Mol. Biol. 304:119-133. [DOI] [PubMed] [Google Scholar]
- 35.Luscher, B. 2001. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 277:1-14. [DOI] [PubMed] [Google Scholar]
- 36.Marcu, K. B., S. A. Bossone, and A. J. Patel. 1992. myc function and regulation. Annu. Rev. Biochem. 61:809-860. [DOI] [PubMed] [Google Scholar]
- 37.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millard, S. S., A. Vidal, M. Markus, and A. Koff. 2000. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 20:5947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miskimins, W. K., G. Wang, M. Hawkinson, and R. Miskimins. 2001. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 21:4960-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell, S. A., E. C. Brown, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2001. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 21:3364-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nanbru, C., I. Lafon, S. Audigier, M. C. Gensac, S. Vagner, G. Huez, and A. C. Prats. 1997. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 272:32061-32063. [DOI] [PubMed] [Google Scholar]
- 42.Oh, Y. L., B. Hahm, Y. K. Kim, H. K. Lee, J. W. Lee, O. Song, K. Tsukiyama-Kohara, M. Kohara, A. Nomoto, and S. K. Jang. 1998. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patton, J. G., S. A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237-1251. [DOI] [PubMed] [Google Scholar]
- 44.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 45.Prober, D. A., and B. A. Edgar. 2001. Growth regulation by oncogenes-new insights from model organisms. Curr. Opin. Genet. Dev. 11:19-26. [DOI] [PubMed] [Google Scholar]
- 46.Pyronnet, S., J. Dostie, and N. Sonenberg. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev. 15:2083-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pyronnet, S., L. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 48.Rajagopalan, L. E., C. J. Westmark, J. A. Jarzembowski, and J. S. Malter. 1998. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 26:3418-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan, K. M., and G. D. Birnie. 1996. Myc oncogenes: the enigmatic family. Biochem. J. 314:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sears, R., F. Nuckolls, E. Haura, Y. Taya, K. Tamai, and J. R. Nevins. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sella, O., G. Gerlitz, S. Y. Le, and O. Elroy-Stein. 1999. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell. Biol. 19:5429-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 54.Soltaninassab, S. R., J. G. McAfee, L. Shahied-Milam, and W. M. LeStourgeon. 1998. Oligonucleotide binding specificities of the hnRNP C protein tetramer. Nucleic Acids Res. 26:3410-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoneley, M., S. A. Chappell, C. L. Jopling, M. Dickens, M. MacFarlane, and A. E. Willis. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20:1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoneley, M., F. E. Paulin, J. P. Le Quesne, S. A. Chappell, and A. E. Willis. 1998. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16:423-428. [DOI] [PubMed] [Google Scholar]
- 57.Stoneley, M., T. Subkhankulova, J. P. Le Quesne, M. J. Coldwell, C. L. Jopling, G. J. Belsham, and A. E. Willis. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson, M. S., T. Y. Nakagawa, K. LeVan, and G. Dreyfuss. 1987. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol. Cell. Biol. 7:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, S. S., and F. McKeon. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89:727-735. [DOI] [PubMed] [Google Scholar]
- 60.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 61.Vagner, S., B. Galy, and S. Pyronnet. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO J. 2:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vagner, S., M. C. Gensac, A. Maret, F. Bayard, F. Amalric, H. Prats, and A. C. Prats. 1995. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 15:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaidi, S. H., and J. S. Malter. 1995. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J. Biol. Chem. 270:17292-17298. [DOI] [PubMed] [Google Scholar]