Abstract
The interferon (IFN)-induced signal transduction and transcription activation complex, ISGF3, is assembled from three proteins, STAT1, STAT2, and IRF9. Of these components, STAT2 provides a fundamental and essential transcriptional activation function for ISGF3. In the present study, we show that ISGF3-mediated transcription is dependent on STAT2 interactions with DRIP150, a subunit of the multimeric Mediator coactivator complex. Other Mediator subunits, DRIP77 and DRIP130, were found either to bind STAT2 without augmenting ISGF3 transcriptional activity or to enhance ISGF3 transcription without binding STAT2, but only DRIP150 both enhanced IFN-dependent transcription and coimmunoprecipitated with STAT2. Endogenous DRIP150 and STAT2 were able to interact in solution, and DNA affinity chromatography and chromatin immunoprecipitation assays demonstrated that DRIP150 binds to the mature, activated ISGF3-DNA complex and is recruited to target gene promoters in an IFN-dependent fashion. IFN-dependent recruitment of DRIP130 to an ISGF3 target promoter and SRB10-STAT2 coprecipitation suggest indirect association with a multisubunit Mediator complex. The site of STAT2 interaction was mapped to DRIP150 residues 188 to 566, which are necessary and sufficient for interaction with STAT2. Expression of this DRIP150 fragment, but not DRIP150 fragments outside the STAT2 interaction region, suppressed ISGF3-mediated transcriptional activity in a dominant-negative fashion, suggesting a direct functional role of this domain in mediating STAT2-DRIP150 interactions. These findings indicate that the IFN-activated ISGF3 transcription factor regulates transcription through contact with DRIP150 and implicate the Mediator coactivator complex in IFN-activated gene regulation.
Alpha interferon (IFN-α) and IFN-β, collectively referred to as IFN in this paper, trigger a cascade of signal transduction events that leads to the rapid activation of a gene transcription program that regulates the cellular innate antiviral immune response and influences adaptive immunity (5, 53). Binding of IFN to cell surface receptors leads to the tyrosine phosphorylation of the cytoplasmic signal transducer and activator of transcription (STAT) proteins STAT1 and STAT2. In combination with a third protein, a DNA binding subunit called interferon regulatory factor 9 (IRF9), the activated STAT proteins assemble into a heterotrimeric complex, the IFN-stimulated gene factor 3 (ISGF3) (for reviews, see references 1, 10, 17, 25, and 28). The ISGF3 complex rapidly enters the nucleus, where it binds to conserved IFN-stimulated response element (ISRE) sequences in the promoters of target IFN-stimulated gene (ISG) loci, increasing their transcription rates. While the cell-signaling events downstream of the IFN receptor have been well characterized, the mechanisms by which nuclear ISGF3 communicates with and galvanizes the activation of the RNA polymerase II holoenzyme complex remain unclear.
Numerous studies examining transcriptional mechanisms in Saccharomyces cerevisiae and metazoan systems have revealed that enhancer-binding transcription activators recruit multiprotein nuclear coactivator complexes possessing diverse catalytic activities to the promoter initiation region. A single activator may simultaneously or sequentially recruit an assortment of coactivators (31, 37, 42) that can direct specific local chromatin modifications that alter promoter accessibility and/or directly interact with RNA polymerase and its associated proteins at the transcriptional initiation region (40, 58). Recruited nucleosome modification enzymes such as histone acetyltransferases (HATs) are frequently associated with chromatin-remodeling coactivator complexes. For ISGF3, IRF9 contributes DNA binding specificity but is transcriptionally inert in the absence of STAT proteins (60; J. F. Lau, T. A. Kraus, J.-P. Parisien, and C. M. Horvath, unpublished data), and the C-terminal STAT1 transcriptional activation domain is dispensable for ISGF3 transcriptional activity (39). Instead, the C terminus of STAT2 provides the transcriptional activity of ISGF3, and current evidence suggests that it provides essential transcriptional activation domain contact surfaces for the recruitment of transcriptional coactivators (47). The STAT2 C terminus is known to interact with the CBP/p300 HAT proteins (4, 43, 44), and more recent evidence indicates that interactions between STAT2 and a GCN5/TAFII130-containing TATA-binding protein-free transcription complex are also used for mediation of ISGF3-dependent transcription (45).
Additional conduits for transmission of regulatory signals from the activator at distal enhancer sites to the RNA polymerase preinitiation complex are also required downstream of chromatin reorganization. Biochemical and genetic elucidation of the structure and function of yeast and metazoan Mediator complexes provides an additional paradigm for transcriptional activation through activator-RNA polymerase communication (22, 36, 40). Mediator is a transcriptional regulatory complex that contains more than 25 distinct polypeptide subunits. Mammalian homologues of yeast Mediator subunits, which were originally characterized in S. cerevisiae as essential components of RNA polymerase II holoenzyme preparations, have been identified in association with a variety of transcription activators such as thyroid hormone receptor (TR) and vitamin D3 receptor (VDR) (for reviews, see references 22 and 48). Originally copurified with TR as the TRAP complex (12), many related Mediator complexes have been identified in association with various activators and have been variously named DRIP (51), ARC (41), NAT (59), CRSP (52), and PC2 (35); in this report, the individual Mediator polypeptides are referred to as DRIP subunits. In mammalian cells, Mediator complexes have been identified as being essential for most nuclear hormone receptor (NR) signaling pathways, including those mediated by the estrogen, peroxisome proliferator-activated, and glucocorticoid receptors (GRs) (8, 9, 13, 16, 24). The available evidence suggests that Mediator complexes are modular in organization and function as a bridge between the distal transcription activator and the RNA polymerase II complex that is positioned at the transcription initiation site (reviewed in references 6, 36, and 48). Transcription factor activation domains recruit the Mediator through direct contact with one or more subunit protein surfaces, and modular subunits of mammalian Mediators have been proposed (21, 35). Consistent with this notion, the mammalian Mediator has been characterized as providing a unifying mechanism by which diverse transcriptional activators can gain access to and regulate the RNA polymerase II preinitiation complexes (3, 6, 27, 36).
A wealth of biochemical and genetic evidence has demonstrated the essential role of Mediator in gene regulation. As an essential component of the RNA polymerase holoenzyme, yeast Mediator is required for almost all transcriptional events. It possesses acetyltransferase activity (34) and the ability to enhance the phosphorylation of the RNA polymerase II C-terminal domain (CTD) that connects transcription initiation with elongation (26). The magnitude and diversity of Mediator complex involvement in mammalian transcription are not yet completely understood (40). Nevertheless, a select number of transcription regulators in addition to NRs have been found to signal through the Mediator for gene regulation. For instance, TR and VDR use DRIP205 to access Mediator while DRIP77 is the contact subunit for p53 (23, 49). DRIP77 is just one of several Mediator subunits used by herpes simplex virus VP16 (20, 23, 41). GR recruits Mediator through DRIP150 interactions (16), while another Mediator protein, DRIP130, has been implicated in adenovirus E1A and RAS-dependent signaling pathways (7, 36). The Mediator complex is also essential for the regulation of SREBP and NF-κB (41) as well as for SP1-dependent transcriptional events (52).
Given the strong implication of the importance of Mediator proteins in the functions of many transcription activators, a potential role for this coactivator complex in STAT-mediated transcription was examined in the IFN model system. The results indicate that increased expression of some Mediator subunits potentiates IFN-inducible, ISGF3-dependent transcription. One of these ISGF3-coactivating subunits, DRIP150, is capable of contacting STAT2, but neither STAT1 nor STAT3 coimmunoprecipitated with the tested DRIP proteins. DNA affinity chromatography experiments revealed DRIP150 recruitment by the ISGF3 complex in an IFN-dependent fashion, and chromatin immunoprecipitation assays demonstrated that IFN-dependent DRIP150 recruitment occurs at physiological target loci. Furthermore, coprecipitation of STAT2 with the distinct Mediator subunit, SRB10, and IFN-dependent recruitment of DRIP130 to ISGF3 targets support a role for the Mediator complex in IFN signaling. Mapping the site of DRIP150-STAT2 interaction revealed an essential and discrete domain of DRIP150 between amino acid residues 188 and 566 that binds to STAT2. Furthermore, expression of this STAT2-binding DRIP150 fragment specifically inhibits IFN-induced transcription in a dominant-negative fashion, emphasizing the importance of this DRIP150 region in the mediation of functional contacts with ISGF3 via STAT2. Together, these results demonstrate the specific STAT2-DRIP150 interactions that are required for ISGF3-dependent transcriptional responses, providing a mechanism for direct communication between ISGF3 and the RNA polymerase II holoenzyme.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney 293T and HeLa (S3) cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% Cosmic calf serum (HyClone). Transfection of cells with cDNAs was carried out by standard CaPO4 procedures as described previously (2, 29). To establish stable transformants, parental HeLa cells were transfected with 5 μg of FLAG-DRIP150 cDNA and cultured in G418 for selection of drug-resistant transformants as described previously (19). Treatment of cells with IFN was carried out as indicated by using 1,000 U of IFN-α/ml as described previously (18). HeLa S3 cells stably expressing FLAG-SRB10 were the generous gift of Wei Gu (Columbia University) and were used as described previously (14).
Plasmids.
The construction of FLAG epitope-tagged DRIP77, DRIP130, and DRIP150 cDNAs has been described elsewhere (50). FLAG-DRIP150 carboxyl-terminal truncation mutants were constructed from full-length pcDNA3 FLAG-DRIP150 by using the following unique restriction sites: XhoI for pcDNA3 FLAG-DRIP150 Δ1, EcoRI for pcDNA3 FLAG-DRIP150 Δ2, ScaI for pcDNA3 FLAG-DRIP150 Δ3, BglII for pcDNA3 FLAG-DRIP150 Δ4, and NotI for pcDNA3 FLAG-DRIP150 Δ5. The FLAG-DRIP150 fragments were constructed by using a PCR-based method and cloned into pcDNA3 by using BamHI and NotI restriction sites. FLAG-DRIP150 fragments containing the following amino acids were constructed with the indicated primers: 188 to 566 (top, 5′-GGGGGATCCGACCCAATTACCAAAATTG-3′; bottom, 5′-GGGCGGCCGCACTAGTACTTGTACGACAGTTG-3′), 566 to 1000 (top, 5′-GGGGGATCCTACAAGTACTACTTTATGTC-3′; bottom, 5′-GGGCGGCCGCACTAGAGCTGCGATATTAAAGAA-3′), and 1000 to 1457 (top, 5′-GGGGGATCCATATCGCAGCTCCAGCC-3′; bottom, 5′-GGGCGGCCGCACTATGGACGCCCAACAG-3′).
Reporter gene assays.
Luciferase assays were carried out according to the manufacturer's instructions (Promega) and were normalized to total protein concentrations measured from lysis extracts. For detection of ISRE-dependent luciferase activity, 293T cells were transiently cotransfected with vectors expressing various DRIP subunits or FLAG-DRIP fragments and a reporter gene plasmid containing 5 copies of the ISG54 ISRE element upstream of a TATA box and firefly luciferase open reading frame.
Immunoprecipitations, DNA affinity purification, and protein assays.
Polyclonal antisera to STAT2 (C-20), RNA polymerase II (CTD), and DRIP130 (CRSP130) antibodies were obtained from Santa Cruz Biotechnology, and FLAG epitope antibodies were obtained from Sigma; these were all used according to the manufacturers' instructions. Polyclonal DRIP150 antibodies were prepared against the DRIP150 peptide sequences KTGKQTRTNAKRKLSD and SNQDARRRSVNEDDNPP (D. Burakov and L. P. Freedman, unpublished data). To immunoprecipitate endogenous or FLAG epitope-tagged proteins, total extracts were prepared with whole-cell extract buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% IGEPAL, 0.2 nM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol) supplemented with a protease inhibitor cocktail (Complete; Roche) and incubated either with specific antibodies overnight or with M2-FLAG affinity agarose bead slurry for 4 h at 4°C. Specific antibody-protein complexes were purified by adding salmon sperm DNA-protein A agarose slurry (Upstate Biotechnology) for 1 h at 4°C. Beads were then washed with whole-cell extract buffer, incubated with protein loading buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose filters. For ISRE DNA affinity purification assays, 1 μg of double-stranded biotinylated oligodeoxynucleotides encompassing a high-affinity ISRE from the human ISG15 promoter (upper, 5′-GATCAGTTTCGGTTTCCGATC-3′; lower, 5′-GATCGGAAACCGAAACTGATC-3′; MWG Biotech) were coincubated with 500 μg of HeLa nuclear extracts in modified whole-cell extract buffer (with 100 mM NaCl) at 4°C for 4 h. DNA-binding protein complexes were washed three times with whole-cell extract buffer, eluted in SDS sample buffer, fractionated by SDS-PAGE, and transferred to nitrocellulose filters. All Western immunoblotting analyses were prepared for chemiluminescent detection according to the manufacturer's protocol (NEN Renaissance).
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed essentially as described by the protocol accompanying the ChIP assay kit (Upstate Biotechnology), with slight modifications. Briefly, following treatment with IFN, 106 human cervical carcinoma HeLa cells or HeLa cells stably expressing FLAG-DRIP150 were incubated for 10 min at 37°C in culture medium containing 1% formaldehyde. Cells were then washed two times with phosphate-buffered saline and collected by scraping with 5 ml of buffer (100 mM Tris-HCl [pH 9.4], 10 mM dithiothreitol). To prepare extracts, cells were lysed in lysis buffer (0.25% Triton X-100, 0.5% NP-40, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris [pH 8.0]) supplemented with protease inhibitor cocktail (Complete; Roche). Cell lysates were then sonicated to yield chromatin fragments of approximately 500 bp as determined by agarose gel electrophoresis. Cellular debris was removed by centrifugation for 10 min at 20,800 × g. The sonicated protein extracts were diluted 1:10 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 150 mM NaCl) and then precleared with 80 μl of protein A Sepharose beads-sheared salmon sperm slurry (Upstate Biotechnology) for 1 h at 4°C. Immunoprecipitation was then performed with the appropriate antibodies overnight at 4°C. Immune complexes were isolated by the addition of 60 μl of protein A Sepharose beads-sheared salmon sperm slurry for 1 h. The beads were washed extensively, and cross-links were reversed by incubation in elution buffer (1% SDS, 0.1 M NaHCO3) for 6 h at 65°C. DNA was extracted with phenol-chloroform, ethanol precipitated, and dissolved in 50 μl of H2O. The input and precipitated DNA were analyzed by radioactive PCR using primers encompassing two ISRE binding sites on the human ISG54 promoter (ISG54 promoter: top, 5′-GAGGAAAAAGAGTCCTCTA-3′; bottom, 5′-AGCTGCACTCTTCAGAAT-3′). PCR products were separated by 5% polyacrylamide gel electrophoresis and detected by autoradiography.
RESULTS
Enhancement of IFN-induced transcription by Mediator subunit expression.
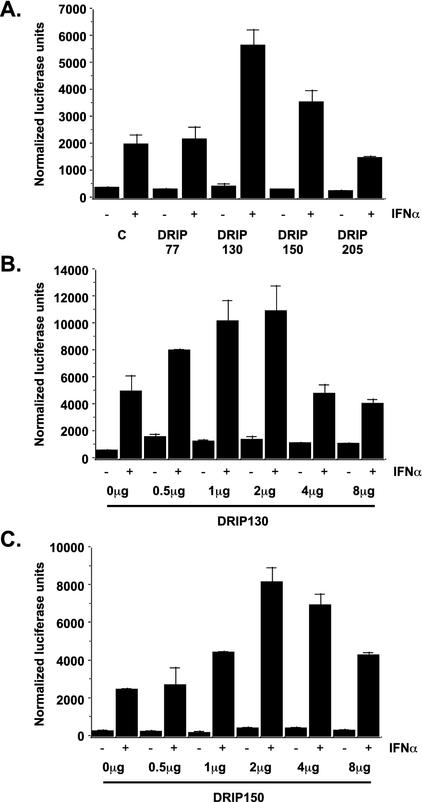
The expression of transcriptional coactivating proteins by plasmid transfection can often enhance specific activator-dependent transcription responses as a consequence of increased coactivator concentration. To specifically test whether Mediator proteins play a role in IFN-activated transcriptional responses, several DRIP protein subunits (DRIP77, DRIP130, DRIP150, and DRIP205) were expressed by cDNA transfection in 293T cells in the context of an ISGF3-dependent, IFN-responsive reporter gene assay (Fig. 1A). Two of the expressed proteins, DRIP130 and DRIP150, elicited modest increases in IFN-dependent ISRE-luciferase activity. DRIP130 provided an approximately 2.5-fold stimulation in activity compared with that of the control, whereas DRIP150 coexpression provided a twofold induction. By contrast, expression of DRIP77 or DRIP205 did not potentiate transcriptional responses upon cytokine treatment. This result suggested a functional interaction between ISGF3 and Mediator complexes that could be augmented by increasing the cellular abundance of limiting subunits.
FIG. 1.
(A) IFN-induced transcription is enhanced by expression of Mediator subunits. Expression vectors for the DRIP proteins indicated (2 μg) were transfected with ISRE-luciferase, and cells were treated for 6 h with 1,000 U of IFN-α/ml. Extracts were normalized for total protein. Values shown are the averages plus standard deviation (SD) from triplicate experiments. (B) Dose-dependent enhancement of ISRE-luciferase activity by DRIP150 expression. Cells were transfected with reporter genes and increasing amounts of DRIP150 expression plasmid as indicated. Dose-dependent enhancement was observed until a point where inhibitory effects were observed, presumably due to squelching by saturation of Mediator complexes at higher DRIP150 levels. Extracts were normalized for total protein. Values shown are averages plus SD from triplicate experiments.
To more carefully examine the ISRE-stimulatory effects of both DRIP130 and DRIP150, dose-response curves for both proteins were evaluated. Consistent with the single dose used in the experiment presented in Fig. 1A, stimulation of IFN-dependent transcription was enhanced by an increase in the expression of either DRIP130 (Fig. 1B) or DRIP150 (Fig. 1C). This enhancement of ISGF3-dependent transcription reached a maximum value at 1 to 2 μg of transfected DRIP130 cDNA and at 2 μg of DRIP150 cDNA, after which an increase in either the DRIP130 or DRIP150 concentration reduced their respective luciferase reporter gene output. This type of saturation effect is typical of proteins that are subunits of multiprotein complexes and may suggest that the transcription enhancement effect of increased DRIP130 and DRIP150 expression is a consequence of allowing STAT2 to have greater access to the multiprotein Mediator complex.
Physical associations between STAT2 and Mediator subunits.
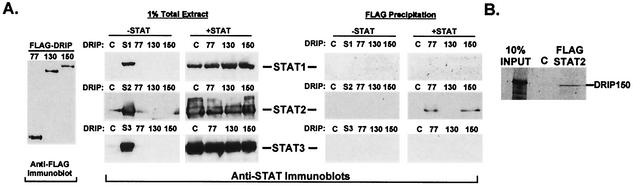
The transcriptional enhancement in the reporter gene assay suggests functional interactions between IFN-activated ISGF3 and the Mediator complex. To more directly test for physical protein-protein interactions between ISGF3 STAT subunits and DRIP proteins, a coimmunoprecipitation assay was performed. Both DRIP130 and DRIP150 stimulated ISRE-dependent transcription and were therefore candidates for ISGF3 interaction. As a control, DRIP77, which did not augment ISRE-luciferase transcription, was also tested. FLAG epitope-tagged DRIP77, DRIP130, and DRIP150 proteins were constructed and expressed in cells with or without cotransfected expression vectors for STAT1, STAT2, or STAT3. Lysates were prepared and immunoprecipitated with immobilized FLAG antibodies. Interactions with STAT proteins were detected by specific immunoblotting (Fig. 2A). Immunoblotting for STAT2 revealed that STAT2 copurified with both DRIP77 and DRIP150, but not DRIP130, in this assay. Importantly, none of the tested DRIP proteins coprecipitated STAT1 or STAT3 in parallel experiments, indicating a specific interaction with STAT2.
FIG. 2.
Interaction of STAT2 with DRIP proteins. (A) STAT2 interacts with DRIP77 and DRIP150. 293T cells were transfected with 5 μg of cDNAs for FLAG-tagged DRIP subunits (DRIP77, DRIP130, and DRIP150) or control FLAG vector (C) in the presence (+STAT) or absence (−STAT) of cotransfected STAT1 (top), STAT2 (middle), or STAT3 (bottom) cDNAs (1.5 μg). At 48 h posttransfection, whole-cell lysates were prepared and subjected to anti-FLAG immunoprecipitation. Immune complexes were separated by SDS-PAGE and then subjected to anti-STAT immunoblotting. The panel at far left shows DRIP expression assayed by anti-FLAG immunoblotting, the set of six panels at center shows STAT levels in 1% of cell extracts, and the set of six panels at right shows FLAG immunoprecipitations probed with anti-STAT sera. (B) In vitro-translated DRIP150 interacts with STAT2. FLAG epitope-tagged STAT2 was expressed in 293T cells, purified and immobilized on FLAG antibody-immobilized Sepharose beads, and incubated with in vitro-translated DRIP150. Sepharose bead-protein complexes were washed extensively and resolved on SDS-PAGE and subjected to autoradiography.
Together, the coimmunoprecipitation and transcription results indicate distinct phenotypes for the tested DRIP proteins in IFN-stimulated transcription: DRIP77 does not enhance transcription but can associate with STAT2, DRIP130 does not bind to STAT1 or STAT2 but enhances IFN-responsive transcription, and DRIP150 both binds to STAT2 and enhances ISGF3 transcription. While these results suggest that DRIP77 and DRIP130 may play significant roles in IFN-mediated transcription, further experiments were focused on the transcription-enhancing interaction identified for DRIP150 and STAT2. To further explore the interaction between STAT2 and DRIP150, the ability of immobilized FLAG-STAT2 to precipitate radiolabeled DRIP150 produced in vitro in rabbit reticulocyte lysates was tested (Fig. 2B). In this assay, DRIP150 was collected on STAT2 beads but not on control beads, which suggests a direct interaction. However, the low binding efficiency in this system might reflect a low binding affinity of the individual subunit that could be enhanced by additional Mediator-ISGF3 contacts or reflect a need for posttranslational modifications.
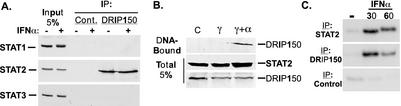
To determine the capacity for physiologically relevant associations, the STAT2-DRIP150 interaction was also evaluated by immunoprecipitation of endogenous proteins from whole-cell extracts. An immunoprecipitation assay was carried out in which IFN-treated and untreated HeLa cell extracts were incubated with polyclonal antiserum specific for DRIP150 and immune complexes were isolated with protein A beads. Immunoblotting for STAT1, STAT2, and STAT3 revealed an interaction pattern that was in agreement with the results shown in Fig. 2A: immunoprecipitated DRIP150 was found to interact exclusively with STAT2 and not with STAT1 or STAT3 (Fig. 3A). No difference in the STAT2 coimmunoprecipitation by DRIP150 was observed following IFN treatment, in agreement with the results of other assays for interaction and in support of the concept that IFN-induced posttranslational modifications are not required for STAT2-DRIP150 association. Instead, their interaction is regulated on the basis of subcellular distribution.
FIG. 3.
ISGF3 interacts with DRIP150. (A) Interaction between endogenous DRIP150 and STAT2. Total extracts prepared from IFNα-treated (6 h) or untreated HeLa cells were immunoprecipitated with DRIP150-specific antibodies, and immune complexes were processed for immunoblotting with STAT1, STAT2, and STAT3 sera. (B) IFN-activated ISGF3 recruits DRIP150. Nuclear extracts prepared from IFNγ-treated (γ; 18 h), IFN-γ- and IFN-α-treated (γ+α; 18 h with IFN-γ plus 30 min with IFN-α), or untreated (C) HeLa cells stably expressing FLAG-DRIP150 were incubated with double-stranded biotinylated oligodeoxynucleotides containing the ISRE from the human ISG15 promoter. DNA-protein complexes were processed for immunoblotting with FLAG antisera for DRIP150. (C) ChIP assay reveals IFN-dependent DRIP150 recruitment to an endogenous promoter. HeLa cells stably expressing FLAG-DRIP150 were either left untreated (−) or treated with 1,000 U of IFN-α/ml as indicated for 30 or 60 min prior to formaldehyde cross-linking, sonication, and immunoprecipitation with indicated antibodies. DNA eluted from immune complexes was used as the template in a 30-cycle PCR with ISG54 primers that centrally span the ISRE within a 200-bp product.
To determine whether DRIP150 is able to bind to the mature IFN-activated ISGF3 transcription complex, double-stranded biotinylated oligodeoxynucleotides encompassing the ISRE element from the human ISG15 promoter were incubated with nuclear extracts prepared from a HeLa cell line that stably expresses FLAG-DRIP150. Prior to lysis, cells were left untreated, treated with IFN-γ for 18 h (a regimen that increases the abundance of IRF9, a limiting factor for ISGF3 activity in HeLa cells [32]), or treated with IFN-γ for 18 h followed by a 30-min treatment with IFN-α to induce formation of the activated heterotrimeric ISGF3 complex. The biotinylated oligodeoxynucleotide-protein complexes were then purified by incubation with streptavidin-conjugated agarose beads, washed, and fractionated by SDS-PAGE. Treatment with IFN-γ plus IFN-α did not significantly alter the amount of STAT2 or FLAG-DRIP150 detected in cell extracts, but immunoblotting of DNA-bound fractions with FLAG antibodies revealed that DRIP150 copurifies with ISGF3 in an IFN-α-dependent fashion (Fig. 3B). This experiment further substantiated the coimmunoprecipitation results by demonstrating interaction between DRIP150 and the intact ISGF3-DNA complex and suggests that ISGF3 might use DRIP150 to recruit Mediator complexes directly to target gene promoters.
The interaction between STAT2 and DRIP150 clearly enables DRIP150 to be recruited to the ISGF3-DNA complex. To directly test whether this interaction facilitates DRIP150 recruitment to endogenous ISGF3 target gene promoters, formaldehyde cross-linking ChIP experiments were conducted to test the coprecipitation of DRIP150 and STAT2, along with endogenous promoter DNA, in an IFN-dependent reaction. HeLa cells stably expressing FLAG-DRIP150 were left untreated or treated with IFN-α for 30 or 60 min, cross-linked with formaldehyde, and then sonicated to yield DNA fragments. The sonicated lysate was subjected to immunoprecipitation with antibodies specific for either STAT2 or DRIP150, and DNA-protein immune complexes were purified with protein A beads. Following reversal of the cross-links, a fraction of the eluted DNA was subjected to PCR analysis using primers specific for the ISRE region of the ISG54 promoter. Copurification of ISG54 DNA was observed with either STAT2 or DRIP150 immunoprecipitation but only in cells that had been stimulated with IFN (Fig. 3C). For both protein immunoprecipitations, the amount of PCR-amplified product decreased between the 30- and 60-min time points, which is consistent with the known transient nature of ISGF3 activation kinetics. These results clearly demonstrate IFN-dependent recruitment of DRIP150 to the ISG54 promoter in a physiologically meaningful context.
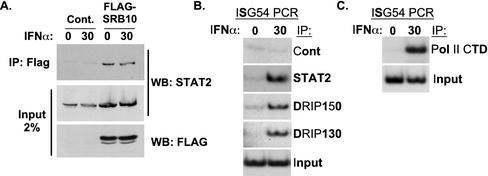
While the results suggest that the Mediator complex might associate with STAT2, it remains formally possible that DRIP150 functions independently in IFN responses as an ISGF3 coactivator. To investigate the possibility of indirect association with other Mediator subunits, the ability of STAT2 to associate with the SRB10 protein was examined. Whole-cell extracts from a stable FLAG-SRB10 cell line (14) were immunoprecipitated with FLAG antibodies, and the immune complexes were analyzed by STAT2 immunoblotting. STAT2 coprecipitation was observed in the SRB10 immune complexes but not in control precipitations (Fig. 4A). The interaction between SRB10 and STAT2 was similar to the DRIP150-STAT2 interaction in that it was independent of IFN signals in these whole-cell extracts. To further support the concept of Mediator complex recruitment by ISGF3, ChIP assays were carried out with antiserum specific for DRIP130 to test IFN-responsive recruitment to a target promoter. As our data indicate that DRIP130 does not directly bind to STAT1 or STAT2, recruitment of this subunit to the promoter would be indicative of an indirect association via the Mediator complex. The ISG54 promoter was detected in DRIP130 immune complexes in an IFN-dependent fashion, as were STAT2 and DRIP150 (Fig. 4B), supporting a model for recruitment of the multiprotein Mediator or a modular subunit of Mediator by ISGF3. The detection of DRIP150 and DRIP130 at the ISG54 promoter correlates with the IFN-dependent appearance of RNA polymerase II at the promoter (Fig. 4C), suggesting a link between Mediator recruitment and transcriptional activation at the ISG54 locus.
FIG. 4.
Association of ISGF3 with other Mediator subunits. (A) SRB10 coprecipitates STAT2. Coimmunoprecipitation analysis was carried out as described in the legend to Fig. 3, except that a FLAG-SRB10 stably transformed cell line was used and immunoprecipitations were performed with FLAG antibody. (B) IFN-dependent, indirect recruitment of DRIP130 to ISG54 promoter. The ChIP assay was carried out as described in the legend to Fig. 3, except that HeLa cells that were unstimulated (0) or treated with IFN for 30 min (30) were used. Cont, control. (C) IFN-dependent RNA polymerase II (Pol II) recruitment to ISG54 promoter. The ChIP assay was carried out with antisera specific for the RNA polymerase II CTD. Input = 5% of DNA prior to precipitation.
STAT2 interaction site maps to DRIP150 amino acids 188 to 566.
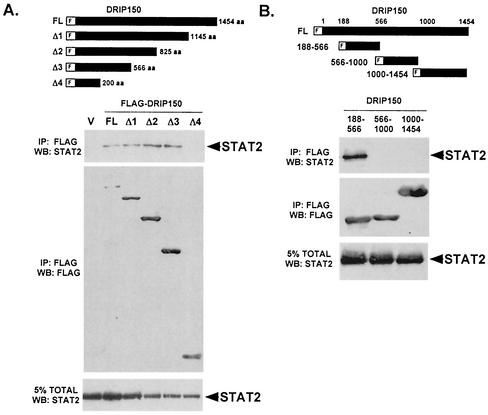
To further characterize the DRIP150-STAT2 interaction and to define the amino acids that mediate this interaction, C- terminally truncated DRIP150 proteins were tested for STAT2 coprecipitation. Amino-terminal fragments of DRIP150 that include amino acids 1 to 566 retained the ability to precipitate STAT2, but an amino-terminal fragment consisting of amino acids 1 to 200 failed to coprecipitate STAT2 (Fig. 5A). This result indicates that amino acids between residues 200 and 566 in the DRIP150 protein are necessary for interaction with STAT2.
FIG. 5.
STAT2 interacts with DRIP150 amino acids 188 to 566. (A) DRIP150 amino acids 200 to 566 are necessary for STAT2 binding. Full-length and C-terminal truncation mutants of FLAG-tagged DRIP150 were expressed in 293T cells along with STAT2, and lysates were immunoprecipitated with immobilized FLAG M2 antibody. Immunoblotting for STAT2 revealed that association is lost with truncation between amino acids 200 and 566. V, FLAG vector with no insert. (B) DRIP150 amino acids 188 to 566 are sufficient for STAT2 binding. The experiment was performed as described for panel A but with isolated DRIP150 segments. Only amino acids 188 to 566 were found to bind STAT2. WB, Western blot.
To determine whether these amino acids constitute a discrete STAT2 binding site, DRIP150 fragments encompassing amino acids 188 to 566, 566 to 1000, and 1000 to 1454 were expressed independently and subjected to STAT2 binding assays (Fig. 5B). Only the fragment encompassing amino acids 188 to 566 coprecipitated STAT2, indicating that this region of DRIP150 contains a modular interaction domain that is necessary and sufficient for binding to STAT2.
STAT2-binding fragment of DRIP150 acts as an ISGF3-specific inhibitor.
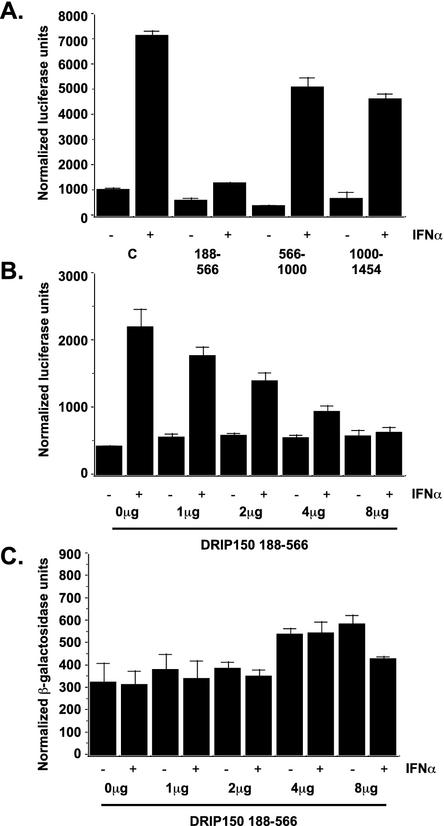
The DRIP150 binding-site mapping results indicate that the STAT2-DRIP150 interaction is mediated by DRIP150 amino acids 188 to 566. To evaluate the functional importance of this interaction in the context of IFN-responsive transcription, experiments were carried out to test the ability of the isolated DRIP150 fragment to disrupt STAT2-dependent transcription in a dominant-negative fashion, a common test for validating protein interactions that has previously been shown to be effective for analysis of STAT-interacting partners (18, 55). Expression of the DRIP150 fragment containing amino acids 188 to 566 in cells blocked IFN-activated transcription, but expression of the DRIP150 fragments containing amino acids 566 to 1000 or 1000 to 1454 had no effect on ISGF3 activity (Fig. 6A). The inhibition of ISGF3-dependent transcription by expression of the DRIP150 fragment containing amino acids 188 to 566 was dose dependent in ISRE-luciferase assays (Fig. 6B), but no inhibition of a cotransfected cytomegalovirus promoter-LacZ reporter gene was detected (Fig. 6C). This finding confirms the specificity of the DRIP150 fragment for inhibition of ISGF3 transcriptional responses and further supports the functional importance of STAT2-DRIP150 interactions in IFN-responsive, ISGF3-dependent transcription.
FIG. 6.
STAT2-binding fragment of DRIP150 inhibits ISGF3 transcription. (A) Expression of the DRIP150 fragment containing amino acids 188 to 566, but not the fragments containing amino acids 566 to 1000 or 1000 to 1454, inhibits IFN induction of ISRE-luciferase reporter. (B and C) The DRIP150 fragment containing amino acids 188 to 566 inhibited ISRE-dependent transcription in a dose-dependent manner (B) but had no effects on cytomegalovirus-LacZ expression (C). Extracts were normalized for total protein. Values shown are averages plus SD from triplicate experiments.
DISCUSSION
Gene regulation by enhancer-binding transcription factors typically requires transcriptional coactivators that function in histone modification, chromatin remodeling, and/or communication with RNA polymerase II (31, 42). The importance of some HAT activities in IFN-responsive, ISGF3-mediated transcription has been established previously (4, 43-45). In the present study, we demonstrate that IFN-activated ISGF3 also utilizes contact with DRIP150 to promote target gene transcription. Apparently, ISGF3-dependent transcription of IFN-stimulated genes relies on the recruitment of distinct types of transcriptional coactivators to the target promoter. In one model, a subset of coactivator proteins with associated histone-modifying activity is initially recruited by ISGF3 to repackage the chromatin template and/or reposition nucleosomes. This reorganization could then facilitate further coactivator recruitment by ISGF3 to induce transcriptional initiation and RNA polymerase II holoenzyme processivity, both of which are activities that have been associated with Mediator complexes (38, 40, 46).
Several distinct ISGF3-directed DRIP protein activities were identified in our experiments. DRIP77 bound to STAT2 but did not enhance transcription. As the inability to enhance IFN-responsive transcription is inconsistent with a direct coactivation role for DRIP77, it is possible that DRIP77 represents a Mediator subunit dispensable for ISGF3 activity, perhaps playing a more central supporting or structural role to stabilize the interactions between ISGF3 and the Mediator complex. Conversely, DRIP130 potentiates IFN-driven transcription but does not interact with STAT2 in binding assays. In this case, rather than contributing to specific interactions with ISGF3, DRIP130 may represent a limiting factor needed for Mediator function or may play a distinct role in delivering activating signals to the RNA polymerase. The function of DRIP150 in IFN signaling that was revealed by our study can be explained more directly. DRIP150 not only interacts with STAT2 and ISGF3 but also potentiates IFN-mediated transcription; therefore, it was analyzed in more detail in this study. Together, these findings suggest that DRIP150 is a STAT2-specific coactivator and might represent an important Mediator interface for ISGF3-specific transcription.
As a complex of more than 25 polypeptides, the Mediator possesses multiple surfaces for protein interactions, some of which have been demonstrated to contact transcriptional activators and others that more directly regulate RNA polymerase. The ability to contact a variety of transcription regulators is a key feature of the Mediator that provides this complex with the capacity to influence a wide array of transcription activation events. In the case of IFN signaling mediated by ISGF3, our results indicate that DRIP150 provides a specific connection between the Mediator and STAT2, which is supported by the finding of STAT2 within SRB10 immune complexes and the indirect IFN-dependent recruitment of DRIP130 to ISGF3 target promoter DNA.
Interestingly, neither STAT1 nor STAT3 could interact with any of the DRIP proteins tested in coimmunoprecipitation assays, suggesting that target genes regulated by these STAT proteins might access the Mediator through untested subunits or might recruit Mediator indirectly through other promoter-binding regulators. Recent studies have suggested that the latter possibility may indeed be the case for some examples of STAT-dependent gene transcription. Both VDR and GR require interactions with Mediator for optimal function. STAT1 interactions with the ligand-bound VDR were reported to be important in the regulation of IFN-γ signaling (61), while in a similar manner, STAT3 and STAT5 have been demonstrated to act synergistically with GR on target promoters (56, 57, 62). Thus, while ISGF3 directly recruits Mediator via STAT2-DRIP150 interaction, other STAT transcription factors may communicate with the Mediator complex indirectly through interactions with other promoter-specific activators.
The results of the immunoprecipitation assays demonstrate that the interaction between ISGF3 and DRIP150 can occur in the absence of IFN stimulation, suggesting that the STAT2-DRIP150 interaction is regulated at the level of nuclear-cytoplasmic distribution. In unstimulated cells, STAT2 is exclusively cytoplasmic, but the Mediator complex has been purified from and functions in the nucleus. Therefore, STAT2 and DRIP150 are compartmentally separated until ISGF3 translocates to the nucleus in response to IFN stimulation. Once STAT2 and DRIP150 are both present in the nucleus, functional interaction can take place, as demonstrated by our observation that DRIP150 can bind to intact ISGF3 in its DNA-bound state in vitro and is recruited to endogenous loci in vivo in response to IFN. This mechanism for regulation of Mediator interactions is distinct from that used by some NRs that are constitutively nuclear and bound to DNA. In these cases, ligand binding reverses interactions with repressor complexes and promotes the recruitment of positive coactivators, including Mediator (15, 31).
The DRIP150 protein is one of the phylogenetically conserved subunits that is common to yeast and metazoan Mediators. The yeast DRIP150 orthologue, RGR1, possesses ∼22% overall identity and ∼41% similarity to mammalian DRIP150. RGR1, first identified in a genetic screen for genes important in the regulation of glucose metabolism, is essential for both positive and negative regulation of most transcriptional events in S. cerevisiae (11, 30, 33). The recruitment of an additional subset of Mediator proteins is also dependent on RGR1, and accordingly, null mutants for this locus have been found to be lethal (30), demonstrating the importance of this protein for basic regulation required for cell viability. More recently, RNA interference of the DRIP150/RGR1 homologue in the nematode Caenorhabditis elegans has been shown to disrupt general transcriptional events required for embryonic development, suggesting that this Mediator subunit might also be required for diverse transcription events in higher organisms (54). The human DRIP150 protein has been purified in several distinct Mediator complexes, highlighting again the central role of this protein. Our investigation has localized the STAT2 interaction site on DRIP150 between residues 188 and 566. This region can interact with STAT2 in isolation and, when expressed alone, can act as an effective dominant-negative inhibitor of IFN-mediated transcription. These results specifically define a region within a single Mediator subunit that is a critical interface for IFN signaling. Computer-aided informatic analysis of this region of DRIP150 did not reveal any homologies to known proteins or protein interaction domains and predicted a mostly alpha-helical structure with leucine repeats within residues 280 to 310. The lack of homology to known proteins suggests a novel STAT2 interaction domain within this region of DRIP150 that is distinct from the region demonstrated to interact with the GR, DRIP150 residues 1360 to 1454 (16). Through these separate interaction surfaces, DRIP150 connects at least two distinct activating signals to the Mediator complex, a reasonable means by which to increase the diversity of Mediator-requiring events while maintaining a high degree of specificity for multiple transcription activators. By facilitating interactions with more than one transcription regulator, Mediator subunits may serve as platforms for integrating various signaling pathways for transcriptional output. In addition, recent findings from structural studies and targeted disruptions suggest a modular nature for the murine Mediator, creating further potential for increased combinatorial diversity (6, 21). Connecting the Mediator, a coactivation complex newly linked to IFN-dependent transcription, to the control of specific target genes, particularly those involved in innate antiviral immunity, represents an important challenge for future investigations.
Acknowledgments
We thank members of the Horvath laboratory for helpful discussions. Initial investigations into STAT-Mediator complexes were inspired by discussions with Bob Roeder and Jim Darnell and members of their laboratories. We are grateful to Wei Gu for SRB10 reagents and to Martin Teichmann for critical comments on the manuscript.
This work was supported by American Cancer Society Research Scholar grant GMC 103079 and NIH grant AI48722 to C.M.H.
REFERENCES
- 1.Aaronson, D. S., and C. M. Horvath. 2002. A road map for those who don't know JAK-STAT. Science 296:1653-1655. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Berk, A. J. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 11:330-335. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon α. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 5.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 6.Boube, M., L. Joulia, D. L. Cribbs, and H. M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 8.Burakov, D., L. A. Crofts, C. P. Chang, and L. P. Freedman. 2002. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J. Biol. Chem. 277:14359-14362. [DOI] [PubMed] [Google Scholar]
- 9.Burakov, D., C. W. Wong, C. Rachez, B. J. Cheskis, and L. P. Freedman. 2000. Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J. Biol. Chem. 275:20928-20934. [DOI] [PubMed] [Google Scholar]
- 10.Darnell, J. E., Jr., I. M. Kerr, and G. M. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 11.Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson, Y. W. Jiang, Y. Li, R. D. Kornberg, and F. J. Asturias. 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97:14307-14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 14.Gu, W., S. Malik, M. Ito, C. X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3:97-108. (Erratum, 3:541.) [DOI] [PubMed] [Google Scholar]
- 15.Hermanson, O., C. K. Glass, and M. G. Rosenfeld. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 16.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath, C. M. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 25:496-502. [DOI] [PubMed] [Google Scholar]
- 18.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath, C. M., Z. Wen, and J. E. Darnell, Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9:984-994. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, K., T. Stuehler, and M. Meisterernst. 2002. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells 7:49-58. [DOI] [PubMed] [Google Scholar]
- 21.Ito, M., H. J. Okano, R. B. Darnell, and R. G. Roeder. 2002. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 21:3464-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, M., and R. G. Roeder. 2001. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12:127-134. [DOI] [PubMed] [Google Scholar]
- 23.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 24.Kang, Y. K., M. Guermah, C. X. Yuan, and R. G. Roeder. 2002. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl. Acad. Sci. USA 99:2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler, D. S., S. A. Veals, X.-Y. Fu, and D. E. Levy. 1990. Interferon-α regulates nuclear translocation and DNA-binding affinity of ISGF-3, a multimeric transcriptional activator. Genes Dev. 4:1753-1765. [DOI] [PubMed] [Google Scholar]
- 26.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 27.Kingston, R. E. 1999. A shared but complex bridge. Nature 399:199-200. [DOI] [PubMed] [Google Scholar]
- 28.Lau, J. F., and C. M. Horvath. 2002. Mechanisms of type I interferon cell signaling and STAT-mediated transcriptional responses. Mt. Sinai J. Med. 69:156-168. [PubMed] [Google Scholar]
- 29.Lau, J. F., J.-P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 97:7278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y. J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 32.Levy, D. E., D. J. Lew, T. Decker, D. S. Kessler, and J. E. Darnell, Jr. 1990. Synergistic interaction between interferon-α and interferon-γ through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 9:1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane, D. J. Stillman, and R. D. Kornberg. 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92:10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorch, Y., J. Beve, C. M. Gustafsson, L. C. Myers, and R. D. Kornberg. 2000. Mediator-nucleosome interaction. Mol. Cell 6:197-201. [DOI] [PubMed] [Google Scholar]
- 35.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5:753-760. [DOI] [PubMed] [Google Scholar]
- 36.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 37.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 38.Mittler, G., E. Kremmer, H. T. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller, M., C. Laxton, J. Briscoe, C. Schindler, T. Improta, J. E. Darnell, Jr., G. R. Stark, and I. M. Kerr. 1993. Complementation of a mutant cell line: central role of the 91-kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathway. EMBO J. 12:4221-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 41.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 42.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 43.Park, C., M. J. Lecomte, and C. Schindler. 1999. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 27:4191-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paulson, M., S. Pisharody, L. Pan, S. Guadagno, A. L. Mui, and D. E. Levy. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343-25349. [DOI] [PubMed] [Google Scholar]
- 45.Paulson, M., C. Press, E. Smith, N. Tanese, and D. E. Levy. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140-147. [DOI] [PubMed] [Google Scholar]
- 46.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 47.Qureshi, S. A., S. Leung, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1996. Function of Stat2 protein in transcriptional activation by IFN-α. Mol. Cell. Biol. 16:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rachez, C., and L. P. Freedman. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13:274-280. [DOI] [PubMed] [Google Scholar]
- 49.Rachez, C., M. Gamble, C. P. Chang, G. B. Atkins, M. A. Lazar, and L. P. Freedman. 2000. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol. Cell. Biol. 20:2718-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 51.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 53.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shim, E. Y., A. K. Walker, and T. K. Blackwell. 2002. Broad requirement for the mediator subunit RGR-1 for transcription in the C. elegans embryo. J. Biol. Chem. 277:30413-30416. [DOI] [PubMed] [Google Scholar]
- 55.Shuai, K., C. M. Horvath, L. H. Tsai-Huang, S. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 56.Stocklin, E., M. Wissler, F. Gouilleux, and B. Groner. 1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383:726-728. [DOI] [PubMed] [Google Scholar]
- 57.Stoecklin, E., M. Wissler, R. Moriggl, and B. Groner. 1997. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 17:6708-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 59.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides, that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 60.Veals, S. A., C. Schindler, D. Leonard, X.-Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding protein. Mol. Cell. Biol. 12:3315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vidal, M., C. V. Ramana, and A. S. Dusso. 2002. Stat1-vitamin D receptor interactions antagonize 1,25-dihydroxyvitamin D transcriptional activity and enhance Stat1-mediated transcription. Mol. Cell. Biol. 22:2777-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Z., S. Jones, J. S. Hagood, N. L. Fuentes, and G. M. Fuller. 1997. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J. Biol. Chem. 272:30607-30610. [DOI] [PubMed] [Google Scholar]