Abstract
The nuclear pore complex (NPC) is a permeable sieve that can dilate to facilitate the bidirectional translocation of a wide size range of receptor-cargo complexes. The binding of receptors to FG nucleoporin docking sites triggers channel gating by an unknown mechanism. Previously, we used deoxyglucose and chilling treatments to implicate Nup170p and Nup188p in the control of NPC sieving in Saccharomyces cerevisiae. Here, we report that aliphatic alcohols increase the permeability of wild-type and nup170Δ NPCs. In conjunction with increases in permeability, aliphatic alcohols, deoxyglucose, and chilling trigger the reversible dissociation of several nucleoporins from nup170Δ NPCs. These results are consistent with the hypothesis that NPC gating occurs when molecular latches composed of FG repeats and structural nucleoporins dissociate.
The nuclear pore complex (NPC) catalyzes the nuclear localization signal (NLS)- or nuclear export signal (NES)-directed transport of macromolecules across the nuclear envelope (28, 33). Transport substrates are targeted to the NPC by soluble receptors, most of which belong to the karyopherin family of importins and exportins. The directionality of transport is dictated by the high nuclear concentration of Ran-GTP, which promotes the formation of outgoing exportin-cargo complexes and the disassembly of incoming importin-cargo complexes.
The NPC is an octagonally symmetric wheel-shaped structure composed of ∼30 nucleoporins (nups), most of which are present in 8, 16, or 32 copies per pore (7, 15, 39). The structural framework of the NPC is comprised of spoke-ring subunits surrounding a hub that has been variously called the central transporter assembly, central channel, or central plug. In addition to the massive channel-forming framework, distinctive cytoplasmic filaments and nuclear basket structures protrude from the NPC. Although the overall architectures of the Saccharomyces cerevisiae and vertebrate NPCs are similar, the structure of the yeast NPC is smaller and seemingly less complex (5, 49). Still, proteomic analyses revealed a high degree of similarity between the protein compositions of yeast and rat NPCs (10, 39), which supports the conclusion that the molecular mechanism of NPC function is conserved among eukaryotes.
The NPC is an open sieve with a resting diameter of ∼10 nm that can dilate during translocation to admit cargos with masses in the millions of daltons. The diffusion channel provides molecules with Stokes' radii of less than ∼10 nm an unfettered route across the nuclear envelope (16). An in vitro analytical study has indicated that mammalian NPCs harbor a single diffusion channel with an accessible diameter of 10.7 nm and a length of 89 nm (22). By monitoring the distribution of microinjected colloidal gold particles, Feldherr and Akin concluded that the diffusion occurs through the middle of the NPC, coincident with the gated translocation channel (16). The results of these studies are consistent with the notion that the diffusion channel is a physical property of the gated translocation channel, which dilates by an unknown mechanism to accommodate receptor-cargo complexes containing large cargos such as 60S ribosomes (2.8 mDa) (however, see reference 47). Other channel-like features located around the periphery of the NPC-nuclear envelope border may facilitate the trafficking of membrane proteins between the inner and outer nuclear membranes (4, 20).
The vectorial translocation of receptor-cargo complexes through the central channel involves their binding to the phenylalanine-glycine (FG)-rich repeats of the FG nups. Exactly how the FG nups are displayed around or within the translocation channel is not known (39). The Brownian affinity gate (39), hydrophobic exclusion (37, 37a), and affinity gradient (8, 17) models address various aspects of how the FG nups might facilitate channel gating and/or vectorial translocation.
Nup170p and Nup188p have previously been implicated in the control of NPC permeability (42). Whereas nup170Δ and nup188Δ NPCs do not exhibit strong defects in either NLS- or NES-directed transport, they are permeable to reporters that are larger than those able to diffuse across wild-type (wt) NPCs (42). The stoichiometries of several FG nups are altered in NPCs isolated from nup170Δ cells (23), which also exhibit defects in chromosome segregation (24). In this study, we report that aliphatic alcohols increase the sieving capacity of wt and nup170Δ NPCs and, in nup170Δ cells, induce the reversible dissociation of several structural and FG nups. Deoxyglucose and chilling produced similar effects. We propose that channel gating is facilitated by molecular latches that open and close to trigger large configurational shifts, which distinguish open and closed channels. Nup170p is required either directly or indirectly to tether latch-forming nups to the NPC when the channel is open.
MATERIALS AND METHODS
Strains, plasmids, and cell culture.
Except where noted, all yeast strains used in this study were based in a W303 (strain WHY12) genetic background (MATaade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100). DF5 cells containing integrated versions of NUP1::green fluorescent protein (GFP), NUP2::GFP, NUP49::GFP, NUP53::GFP, NUP59::GFP, NUP60::GFP, NUP100::GFP, and NUP159::GFP were provided by R. Wozniak (University of Alberta). Integrated versions of NUP1::GFP, NUP2::enhanced yellow fluorescent protein (EYFP), NUP53::EYFP, NUP59::EYFP, NUP82::EYFP, NUP84::EYFP, NUP85:: EYFP, NUP116::EYFP, NUP120::EYFP, NUP133::EYFP, NUP145-C::EYFP, and NUP188::EYFP in FY23 cells were provided by P. Silver (Dana-Farber Cancer Institute). Cells containing both the nup170Δ allele and various integrated NUP::GFP and NUP::EYFP reporters were obtained by mating, sporulation, and tetrad analysis. Genotypes were confirmed by PCR.
GFP-NES (42), classical NLS (cNLS)-GFP (44), and GAL1-SSA1 (43) plasmids were used as previously described. Rich (yeast extract-peptone-dextrose), standard synthetic complete (SC), and drop-out media were used as described previously (41).
In vivo transport assays and microscopy.
Assays to assess the passive nucleocytoplasmic equilibration of NLS-GFP and NES-GFP reporters were performed basically as reported previously (42, 44) with the modifications described in the legends to Fig. 1 and 2. Briefly, about 2 optical density (A600) units of logarithmically growing cells were pelleted and resuspended in 50 μl of glucose-containing synthetic medium (SC-Glu) in the presence or absence of alcohols and incubated at room temperature to allow equilibration. After 10 min, 2 μl of cells was removed, mounted under coverslips on glass slides, and scored at room temperature by using an Olympus BH-2 microscope with an Olympus SPlan 100 oil immersion objective. GAL1-SSA1 expression from the YCpGAL1-SSA1 vector was induced by resuspension of glucose-grown cells (SC-Glu) in SC-Raf/Gal (1% raffinose and 1% galactose) and incubation in a shaking bath at 30°C for 2 h. Equilibration and reimport kinetics were quantified as described previously (44). Recovery from alcohol treatments was performed by washing cells in 1 ml of SC-Glu, pelleting and then resuspending them in 1 ml of fresh SC-Glu, and incubating them for 30 min at 30°C. Images of cells were obtained using a Leica TCS NT confocal microscope equipped with UV, Ar, Kr-Ar, and He-Ne lasers. Confocal images were processed using Adobe Photoshop software.
FIG. 1.
Aliphatic alcohols promote equilibration of cNLS-GFP. (A) Dose-dependent capacity of various alcohols to induce the equilibration of cNLS-GFP across wt nuclear envelopes. ○, methanol; ◊, ethanol; ▪, isopropanol; ▴, HD; •, n-butanol. Cells were exposed to the alcohols mixed with SC-Glu, and the percentage of cells exhibiting predominately nuclear localization of cNLS-GFP (% Nuclear cells) after 10 min was plotted as a function of alcohol concentration (vol/vol). The inset shows the relationship between the relative hydrophobicities (log P) (29) of five alcohols and the molar concentrations at which each induced 50% delocalization of cNLS-GFP in 10 min. Data for cyclohexanol (▵) are included instead of data for HD, for which a partition coefficient is not available. (B) Confocal images of wt cells treated with 0, 1, and 2% HD for 10 min and of cells after a 30-min recovery after 10 min in 5% alcohol. (C) Time course of cNLS-GFP localization in wt cells incubated in either 5% ethanol (▴) or 2% HD (⋄). Also shown are data for cells expressing GAL1::SSA1 in 5% ethanol (▵) or 2% HD (⋄). (D) Adaptive response of wt cells continuously incubated for 6 h in either 2% HD or 5% ethanol. Untreated control cells (white bars), HD-pretreated cells (grey bars), and ethanol-pretreated cells (black bars) were subsequently challenged with either 2% HD or 5% ethanol (EtOH) for 10 min before cNLS-GFP localization was scored.
FIG. 2.
Alcohol-induced increases in the sieving diameters of wt and nup170Δ NPCs. (A) Cells expressing NES-GFP reporters of various sizes were incubated for 10 min in the presence of 5% concentrations of ethanol, 2-propanol, or HD or 2% butanol. Minus signs indicate that the reporter was not excluded from the large majority of nuclei, plus signs indicate that the reporter was excluded from nuclei, and +− indicates partial equilibration. (B) Confocal images of wt and nup170Δ cells expressing the 66-kDa NES-GFP reporter and treated for 10 min with each of the indicated alcohols at the concentrations stated above. Arrowheads indicate location of nuclei. Scoring of cells is indicated in the lower right corner of each panel. The large black vacuolar compartments serve as a control for complete exclusion of the GFP reporter.
RESULTS AND DISCUSSION
Aliphatic alcohols disrupt the permeability barrier of wt NPCs.
In vivo kinetic assays have been developed to assess NPC permeability under different conditions and in various genetic backgrounds (42). These assays employ sized NLS- and NES-GFP reporters whose capacity to diffuse across the nuclear envelope is assessed after their karyopherin-mediated transport is inhibited by chilling or deoxyglucose (42, 44). In chilled cells, wt yeast NPCs are permeable to a 30-kDa NES-GFP reporter but not to larger NES- or NLS-GFP reporters (Fig. 1 and 2) (42). In contrast, nup170Δ NPCs admitted the largest, 126-kDa reporter (42). The NPCs in chilled nup188Δ cells were also more permeable than wt NPCs but less so than nup170Δ NPCs. In this study, we provide evidence that increases in the functional diameters of translocation channels in wt and nup170Δ NPCs are mediated by the reversible breaking of complexes between nups. The results presented here support the hypothesis that central-channel gating is governed by molecular latches which, when closed, maintain the normal permeability barrier of resting channels and, when released, widen the channel to accommodate receptor-cargo complexes.
Here, we extend previous studies on the effects of deoxyglucose and chilling (42) to include the effects of aliphatic alcohols. As shown in Fig. 1A, methanol, ethanol, isopropanol, n-butanol, cyclohexanol, and 1,6-hexanediol (HD) all promoted the leakage of cNLS-GFP down its concentration gradient into the cytoplasm (Fig. 1A and B). Interestingly, the effectiveness of the alcohols was roughly proportional to their relative hydrophobicity (Fig. 1A, inset). These results are most easily explained by a disruptive effect of aliphatic alcohols on key interactions between nups involved in maintaining the permeability barrier. Recently, Ribbeck and Gorlich (37a) suggested that trans-cyclohexane-1,2-diol increased the permeability barrier of mammalian NPCs by wreaking havoc with hydrophobic interactions between FG repeats. This conclusion was supported by the finding that the effects of the alcohol on NPC permeability were thwarted by wheat germ agglutinin, which is known to cross-link mammalian FG nups (37a).
The long-term effects of ethanol and HD on cNLS-GFP localization proved to be interesting. After the rapid ethanol- or HD-induced dumping of cNLS-GFP into the cytoplasm, the reporter reaccumulated into nuclei within ∼8 h (Fig. 1C). The capacity of cells to adapt to the presence of ethanol or HD underscores the generally mild effects of these alcohols on cell physiology. The spontaneous reimport of cNLS-GFP reflects a genuine physiological response, since alcohol-treated cells were tolerant to subsequent exposures (Fig. 1D). In particular, the development of ethanol tolerance in S. cerevisiae is well known and involves, in part, the increased gene expression of molecular chaperones such as Hsp70 (36). This is relevant because it has previously been shown that the ectopic expression of a cytosolic Hsp70, GAL1::SSA1, suppresses the steady-state permeability defects of nup188Δ and nup170Δ cells (42). Thus, it was not unexpected that the overexpression of GAL1::SSA1 prevented the alcohol-induced delocalization of cNLS-GFP in wt cells (Fig. 1C). An “empty” vector lacking the SSA1 open reading frame afforded no protection (data not shown). Since ethanol in cultures of yeasts growing on fermentable carbon sources normally rises to levels which, when added at once, are sufficient to induce the passive equilibration of cNLS-GFP, this tolerance pathway probably plays a physiologically relevant role in protecting the NPC. Stochaj et al. (46) observed a similar adaptation response to heat shock, which also induces Hsp70 expression (36).
Aliphatic alcohols used at the concentrations described here will certainly dissolve into and alter, at least subtly, the physical properties of biological membranes. However, the protective effect of SSA1 overexpression is inconsistent with the idea that cNLS-GFP might have escaped the nucleus through pores created by the alcohols in the lipid phase of the nuclear membranes. To control for chemical effects on the integrity of internal membranes, we tested whether HD affected the uptake and staining of vacuole membranes by the vital dye FM4-64. FM4-64 is delivered to the vacuole via the secretory pathway and requires a vacuole membrane potential for proper staining (48). The uptake and staining of vacuole membranes by FM4-64 were not affected by 2% HD (data not shown). Moreover, if the alcohols acted by punching large holes in cellular membranes, then the GFP reporters we used would equilibrate across the vacuole membrane and also leak out of the cell through the plasma membrane, which they clearly did not (Fig. 1B and 2B).
Treatments other than with aliphatic alcohols also affect NPC permeability. It has previously been reported that incubation of nup170Δ cells at 0°C induced the rapid equilibration of both cNLS-GFP and the 126-kDa NES-GFP reporter (42). Deoxyglucose promoted the equilibration of the 51-kDa NES-GFP reporter in wt cells and the partial equilibration of the largest, 126-kDa reporter in nup170Δ cells (data not shown). Hypertonic shock, which causes the Pkc1p kinase-dependent relocalization of some nuclear proteins to the cytoplasm (35), did not affect the localization of cNLS-GFP in wt cells (data not shown) and thus probably does not act by disrupting NPC permeability.
Aliphatic alcohols increase the maximum sieving capacity of wt and nup170Δ NPCs.
In addition to measuring the active and passive transport of cNLS-GFP (43, 44), we and our coworkers have also used 30- to 126-kDa NES-GFP reporters to estimate the abnormally high sieving capacity of nup188Δ and nup170Δ NPCs (42). NES reporters offer a better assessment of nucleocytoplasmic diffusion than NLS-GFPs because the only way NES reporters can enter the nucleus is by diffusion. In contrast, the nuclear targeting of NLS-GFP is often inefficient, which results in a degree of background cytoplasmic fluorescence whether or not the reporter is able to diffuse out of the nucleus.
Here, we extend our previous study on the sieving capacity of NPC to include the effects of aliphatic alcohols in wt and nup170Δ cells. Each alcohol was used at a concentration sufficient to equilibrate cNLS-GFP across the nuclear envelopes of ∼100% of the cell population in 10 min (Fig. 1A). Figure 2B shows a representative subset of the confocal images used to score the cells and compile the data in Fig. 2A. Untreated wt nuclei were impermeable to 36-kDa and larger NES-GFP reporters (Fig. 2A). Treatment with ethanol, isopropanol, butanol, and HD affected the sieving capacity of wt cells in proportion to their relative hydrophobicity (Fig. 1A, inset). Untreated nup170Δ NPCs were more permeable to larger reporters than were wt cells, admitting the 51-kDa but not the 66-kDa reporter (Fig. 2A) (42). This limit was significantly increased by each of the alcohols, which, with the exception of isopropanol, promoted the passive exchange of the largest, 126-kDa reporter (Fig. 2A). Curiously, ethanol was not found by this assay to enhance the permeability of wt cells, even though it induced cNLS-GFP equilibration (Fig. 1A). Ethanol did, however, have a disproportionately large effect on NPC sieving in nup170Δ cells. This suggests that the effects of ethanol may not be limited to NPC permeability or, possibly, that it has specific effects on the function of the nuclear transport apparatus.
In this regard, it is not clear why 5% ethanol induced the rapid equilibration of a 43-kDa cNLS-GFP reporter (Fig. 1A) but not that of a smaller 36-kDa NES-GFP reporter (Fig. 2A). Rather than revealing something fundamental about NPC channel structure, this particular inconsistency is most likely due to technical factors such as differences in the shapes and Stokes' radii of the cNLS and NES reporters or the differential effects of ethanol on the targeting phases of importin- and exportin-mediated transport. The important conclusion to note here is that simple aliphatic alcohols have strikingly large effects on NPC permeability. Finally, these results underscore the important role that Nup170p plays in NPC permeability.
HD causes the reversible dissociation of nups from nup170Δ NPCs.
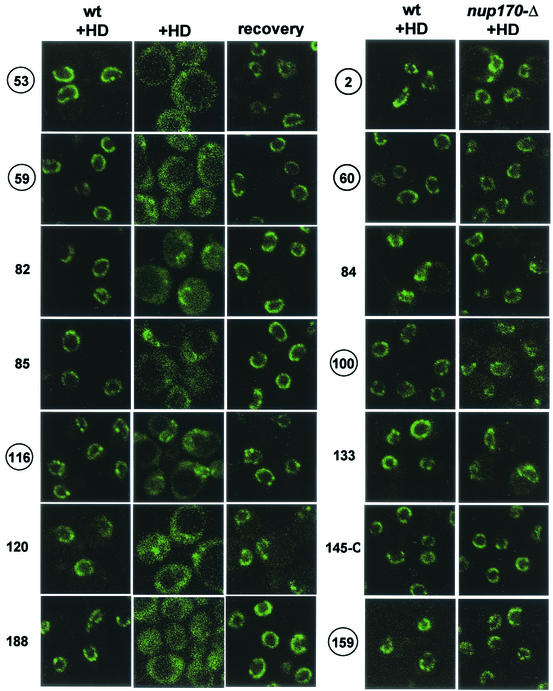
To investigate the possible role of Nup170p in the molecular mechanics of channel permeability, we assessed the effects of 5% HD on the association of 14 representative FG and structural nups with the NPC. Each NUP gene was tagged in the chromosome with either GFP or EYFP in both wt and nup170Δ cells. All 14 nup-GFP reporters localized in the expected pattern in wt and nup170Δ NPCs, and their binding to wt NPCs was not significantly affected by 5% HD (Fig. 3). Remarkably, 5% HD induced the reversible dissociation of seven nups from nup170Δ NPCs (Fig. 3), including both FG nups (Nup53p, Nup59p, and Nup116p) and structural nups (Nup82p, Nup85p, Nup120p, and Nup188p). The binding of seven other nups, including both FG nups (Nup2p, Nup60p, Nup84p, Nup100p, and Nup159p) and structural nups (Nup145-Cp, Nup133p, and Nup84p), was unaffected. Among the FG nups that did not dissociate were those that localize to the cytoplasmic face (Nup159p), central channel (Nup100p), and nuclear basket (Nup60p and Nup2p) (6, 39). The stable association of Nup2p and EYFP is noteworthy, since Kenna et al. (23) have reported that Nup2p was depleted from nuclei isolated from nup170Δ cells. Also, since Nup2p is a mobile nup that normally shuttles between the NPC and the nucleoplasm, its tendency to dissociate may be expected (13). Using confocal imaging of living cells, we observed excellent localization of Nup2p-EYFP to both wt and nup170Δ NPCs (Fig. 3). It is possible that the stability of the association of Nup2p with the NPC is reduced in nup170Δ cells but not in a fashion that makes it susceptible to dissociation in HD.
FIG. 3.
Reversible dissociation of nups in nup170Δ cells is induced by HD. Shown are confocal images revealing the localization of 14 different Nup-EYFP and Nup-GFP reporters in wt and nup170Δ cells after 10 min in the presence of 5% HD or after recovery from the alcohol (see Material and Methods). Nups are indicated by the numbers associated with their gene designations (e.g., 53 indicates Nup53p-EYFP), and the numbers for Nups containing FG repeats are circled.
These results provide a molecular basis for the effects of aliphatic alcohols on the sieving capacities of wt and nup170Δ NPCs. The dissociation of nups from HD-treated NPCs indicates that the increases in NPC permeability are the direct result of space created in the NPC by the disruption of specific interactions between nups. Ribbeck and Gorlich (37a) have made a compelling case for the role of alcohol-sensitive hydrophobic interactions in maintaining the permeability barrier of mammalian NPCs. They concluded that interactions between hydrophobic FG repeats created a meshwork-like barrier to diffusion that is disrupted when receptor-cargo complexes partition into the environment of the meshwork. The dissociation of Nup188p, Nup120p, Nup85p, and Nup82p suggests that binding dynamics involving structural nups may be as critical, or more so, to channel gating as the proposed interactions among FG nups. Taken together, our results and those of others support the hypothesis that HD mimics channel gating by disrupting interactions between nups that normally function to maintain the permeability barrier of the central channel.
There remains the possibility that nup170Δ NPCs are structurally defective in such a way as to make them sensitive to the effects of aliphatic alcohols. Although it is possible that the effects we observed are unrelated to channel gating, we think that this possibility is unlikely since low concentrations of aliphatic alcohols also disrupt the permeability barrier of wt NPCs (Fig. 1 and 2). Still, it is reasonable to ask whether other treatments that affect permeability also induce the dissociation of nups in nup170Δ cells. To address this issue, we investigated the effects of deoxyglucose and chilling on several representative Nup-EYFP reporters. Similar to the effects of HD (Fig. 3), deoxyglucose and chilling induced the dissociation of Nup188p-, Nup53p-, and Nup120p-EYFP reporters from nup170Δ NPCs (Fig. 4). These reporters did not dissociate in either deoxyglucose-treated or chilled wt cells, and the effects in nup170Δ cells were reversible (data not shown). The NPC localization of Nup133p-EYFP, which was unaffected by HD (Fig. 3), was also unaffected by deoxyglucose and chilling (Fig. 4). The effects of chilling could be due to the fact that hydrophobic interactions are generally weaker at lower temperatures, but no such argument can be made for the effects of deoxyglucose, which has the effect of reducing cellular ATP levels (40). Experiments are under way to determine exactly how aliphatic alcohols, deoxyglucose, and chilling increase NPC permeability and induce the dissociation of nups from nup170Δ NPCs.
FIG. 4.
Dissociation of nups in nup170Δ cells induced by deoxyglucose and chilling. Shown are confocal images revealing the localization of four Nup-EYFP reporters in nup170Δ cells in SC-Glu medium before (untreated) and after treatment with 20 mM deoxyglucose for 30 min at 30°C (Deoxyglucose) or chilled on ice for 30 min (0°C). The nups are designated as described in the legend to Fig. 3.
The simplest dissociation events to understand are those of the related proteins Nup53p and Nup59p, since Nup53p is known to associate directly with Nup170p (30). It is likely that binding dynamics involving Nup53p and Nup170p would be mirrored in some fashion by analogous reactions involving Nup59p and Nup157p, even though nup157Δ NPCs do not exhibit increased permeability (42). The dissociation of Nup59p from nup170Δ NPCs (Fig. 3) indicates that there is a functional parallel between the Nup53p/Nup59p and Nup170p/Nup157p pairs of homologous proteins (1). Animal NPCs contain only one version of each pair, designated Nup35 and Nup155, respectively (10).
The case of the dissociation of Nup53p is complicated by the dual role that Kap121p-Nup53p complexes play in NPC assembly and transport (31). The assembly of Nup53p into nascent NPCs appears to be directed by Kap121p, which binds an NLS-like Kap binding domain in Nup53p (26). Once Nup53p is incorporated into NPCs, it likely serves as a docking site for Kap121p- and Kap95p-cargo complexes (14, 31). The case for the involvement of Nup53p and Nup170p in gating is supported by biochemical evidence indicating that Nup53p participates in dynamic binding relationships with Kap121p, Nup170p, and possibly Nic96p (14, 31; R. Wozniak, personal communication). Specifically, the fact that Nup53p exists in mutually exclusive complexes with Nup170p and Kap121p suggests that the binding of Nup53p to Nup170p may trigger the release of Kap121p-cargo from Nup53p to downstream docking sites (26). No matter which nup(s) is responsible for tethering Nup53p to the NPC in nup170Δ cells, the HD sensitivity of the interaction underscores its potential role as a molecular latch governing channel gating.
It has been suggested that some nups function as mobile receptor-cargo carriers that shift between docking stations within the NPC and between the NPC and the nucleus (15, 32, 50). For example, Nup98 shuttles between the NPC and an intranuclear structure called the GLFG body (19). Mammalian Nup50 and its yeast homologue Nup2p also shuttle between the NPC and the nucleoplasm (13, 25a). CAN/Nup214 and Nup153 also appear to be mobile and exchange rapidly with nuclear pools (11, 34). The localization of Nsp1p and Nic96p to multiple sites within the yeast NPC is consistent with this type of mobility (15). Given the gathering body of evidence that dynamic interactions of nups within the NPC may be the norm rather than the exception, channel gating and translocation might be best understand in terms of cascades of partner exchanges initiated by the binding of receptor-cargo complexes to their cognate FG repeat domains. The present findings suggest that several structural nups may participate in a novel class of binding dynamics central to channel gating vis-à-vis permeability.
The dissociation of Nup188p from nup170Δ NPCs is intriguing because, among the null mutants we examined, nup188Δ and nup170Δ NPCs are unique with respect to their abnormally high permeability to NES-GFP reporters (42). Since the sieving capacity of nup170Δ NPCs is greater than that of nup188Δ NPCs and a number of nups including Nup188p-EYFP dissociate from nup170-Δ cells, we think that it is likely that Nup170p functions as a kind of master scaffold for a spectrum of nups, including Nup188p. In turn, Nup188p may itself serve as the direct, or secondary, scaffold for a subset of the nups that dissociate from nup170Δ NPCs. Alternatively, it is possible that the increased sieving of nup188Δ NPCs is due to a hole created only by the absence of the estimated 16 copies of Nup188p, which amounts to a missing mass of 3 mDa or about 5% of the ∼60-mDa yeast NPC (39). Experiments are under way to investigate these possibilities.
The HD-induced dissociation of Nup120p and its binding partner Nup85p from nup170Δ NPCs is consistent with the notion that the Nup84p complex plays a role in the conformational dynamics of the NPC. The Nup84p complex contains seven structural nups (45), all of which are conserved in mammals (10). Nup85p/Seh1p and Nup120p form the forked-shaped head of the Y-shaped complex (27). Nup145-Cp/Sec13p, Nup84p, and Nup133p form an elongated handle-like structure that is connected to the fork subunits through an interaction between Nup120p and Nup145-Cp. Since Nup85p does not assemble into nup120Δ NPCs (18), its dissociation from nup170Δ NPCs in HD is probably an indirect result of the disruption of the Nup85/Nup120p-Nup145-Cp/Sec13p complex. Nup145-Cp, Nup84p, and Nup133p remain bound to HD-treated nup170Δ NPCs (Fig. 3).
The massive interconnected structure of the NPC dictates that various binding hierarchies will determine how and in what order individual nups are incorporated during the assembly of nascent NPCs. For example, a C-terminal truncation of Nup57p that prevented its own assembly into NPCs also reduced the incorporation of its binding partner Nup49p as well as Nsp1p and Nup116p (9). Two of the nups that dissociate from nup170Δ NPCs might do so because their stable binding partners are linked to the NPC via HD-sensitive interactions. As discussed above, the dissociation of Nup85p from nup170Δ NPCs is probably an indirect result of its association with Nup120p. Since Ho et al. (21) have shown that Nup116p does not assemble into nup82Δ NPCs, the HD-induced dissociation of Nup116p might also be a consequence of its interaction with Nup82p.
A novel role for nucleoporin binding dynamics in channel gating.
Current models for NPC channel function make the assumption that the permeability barrier is created by FG nups, whose repeat domains are probably structurally disorganized and therefore capable of occupying lots of space within the channel (12). The Brownian affinity gate (39) and hydrophobic exclusion (37, 37a) models suggest ways by which the binding of receptor-cargo complexes to FG repeats might overcome the permeability barrier and, for all intents and purposes, induce gating. Both of these models view FG repeat domains as flexible filaments attached to a rigid scaffold composed of structural nups (28). If this is correct, and the spoke-ring annulus remains rigid and immobile during transport, then the binding dynamics we observed among structural nups may contribute to gating by mobilizing FG nups within the confines of the central channel. Alternatively, since the only known function of the NPC is channel gating and translocation, it seems reasonable to expect that the entire structure would likely participate in gating and translocation dynamics. In support of this notion, the massive spoke-ring assembly, like the central transporter (2, 3, 25) and nuclear basket structures (47), can be captured by electron microscopy in multiple conformations (3, 4).
The reversible dissociation of structural nups in HD-treated nup170Δ cells suggests that labile binding interactions involving structural nups are at least as important in gating as those postulated to occur between FG nups (37a, 39). The nups that dissociate in HD behave like gates that are normally held in place by stable Nup170p-dependent “hinges” formed during NPC assembly and maintained over the pore's lifetime. Without hinges, as is the case in nup170Δ cells, the ectopic release of latching complexes by HD, deoxyglucose, or chilling causes the gates to fall off the NPC. This analogy does not rule out the equally likely possibility that the disruption of latching complexes which define the configuration of closed channels leads directly to the formation of secondary complexes (i.e., partner exchanges) that characterize open-channel configurations.
In summary, reversible increases in NPC permeability can be correlated with a novel class of nup binding dynamics involving both structural and FG nups. We believe that these phenomena are indicative of bona fide gating reactions that are suited to biochemical analysis.
Acknowledgments
We thank Rick Wozniak for his support and critical insight.
This work was supported by grants to D.S.G. from the National Cancer Society (BE-104C) and National Institutes of Health (GM40362).
REFERENCES
- 1.Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel, and R. W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akey, C., and D. S. Goldfarb. 1989. Nuclear import through the nuclear pore complex is a multi-step process. J. Cell Biol. 109:971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akey, C. W. 1990. Visualization of transport-related configurations of the nuclear pore transporter. Biophys. J. 58:341-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akey, C. W. 1995. Structural plasticity of the nuclear pore complex. J. Mol. Biol. 248:273-293. [DOI] [PubMed] [Google Scholar]
- 5.Akey, C. W., and M. Radermacher. 1993. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen, N. P., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268-29274. [DOI] [PubMed] [Google Scholar]
- 7.Allen, T. D., J. M. Cronshaw, S. Bagley, E. Kiseleva, and M. W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651-1659. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucci, M., and S. R. Wente. 1998. A novel fluorescence-based genetic strategy identifies mutants of Saccharomyces cerevisiae defective for nuclear pore complex assembly. Mol. Biol. Cell 9:2439-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning, D. P., V. Uversky, S. S. Patel, A. L. Fink, and M. Rexach. 2002. The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J. Biol. Chem. 277:33447-33455. [DOI] [PubMed] [Google Scholar]
- 13.Dilworth, D. J., A. Suprapto, J. C. Padovan, B. T. Chait, R. W. Wozniak, M. P. Rout, and J. D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahrenkrog, B., W. Hubner, A. Mandinova, N. Pante, W. Keller, and U. Aebi. 2000. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol. Biol. Cell 11:3885-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahrenkrog, B., D. Stoffler, and U. Aebi. 2001. Nuclear pore complex architecture and functional dynamics. Curr. Top. Microbiol. Immunol. 259:95-117. [DOI] [PubMed] [Google Scholar]
- 16.Feldherr, C. M., and D. Akin. 1997. The location of the transport gate in the nuclear pore complex. J. Cell Sci. 110:3065-3070. [DOI] [PubMed] [Google Scholar]
- 17.Gilchrist, D., B. Mykytka, and M. Rexach. 2002. Accelerating the rate of disassembly of karyopherin cargo complexes. J. Biol. Chem. 277:18161-18172. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, A. L., C. A. Snay, C. V. Heath, and C. N. Cole. 1996. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell 7:917-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffis, E. R., N. Altan, J. Lippincott-Schwartz, and M. A. Powers. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13:1282-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinshaw, J. E., B. O. Carragher, and R. A. Milligan. 1992. Architecture and design of the nuclear pore complex. Cell 69:1133-1141. [DOI] [PubMed] [Google Scholar]
- 21.Ho, A. K., T. X. Shen, K. J. Ryan, E. Kiseleva, M. A. Levy, T. D. Allen, and S. R. Wente. 2000. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol. 20:5736-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keminer, O., and R. Peters. 1999. Permeability of single nuclear pores. Biophys. J. 77:217-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenna, M. A., J. G. Petranka, J. L. Reilly, and L. I. Davis. 1996. Yeast Nle3p/Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol. Cell. Biol. 16:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerscher, O., P. Hieter, M. Winey, and M. A. Basrai. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157:1543-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiseleva, E., M. W. Goldberg, T. D. Allen, and C. W. Akey. 1998. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J. Cell Sci. 111:223-236. [DOI] [PubMed] [Google Scholar]
- 25a.Lindsay, M. E., K. Plafker, A. E. Smith, B. E. Churman, and I. G. Macara. 2002. Npap60/Nup50 is a tri-stable switch that stimulates importin-α-β-mediated nuclear protein import. Cell 110:349-360. [DOI] [PubMed] [Google Scholar]
- 26.Lusk, C. P., T. Makhnevych, M. Marelli, J. D. Aitchison, and R. W. Wozniak. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutzmann, M., R. Kunze, A. Buerer, U. Aebi, and E. Hurt. 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay, D., W. Y. Shiu, and K. C. Ma. 1992. Illustrated handbook of physical-chemical properties and environmental fate for organic chemicals. Lewis Publishers/CRC Press, Boca Raton, Fla.
- 30.Marelli, M., J. D. Aitchison, and R. W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marelli, M., C. P. Lusk, H. Chan, J. D. Aitchison, and R. W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marelli, M., D. J. Dilworth, R. W. Wozniak, and J. D. Aitchison. 2001. The dynamics of karyopherin-mediated nuclear transport. Biochem. Cell Biol. 79:603-612. [PubMed] [Google Scholar]
- 33.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 34.Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18:1982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanduri, J., and A. M. Tartakoff. 2001. Perturbation of the nucleus: a novel Hog1p-independent, Pkc1p-dependent consequence of hypertonic shock in yeast. Mol. Biol. Cell 12:1835-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piper, P. W. 1995. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 134:121-127. [DOI] [PubMed] [Google Scholar]
- 37.Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Ribbeck, K., and D. Gorlich. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenblum, J. S., and G. Blobel. 1999. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA 96:11370-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwoebel, E. D., T. H. Ho, and M. S. Moore. 2002. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 57:963-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 42.Shulga, N., N. Mosammaparast, R. Wozniak, and D. S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shulga, N., P. James, E. Craig, and D. S. Goldfarb. 1999. A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J. Biol. Chem. 274:16501-16507. [DOI] [PubMed] [Google Scholar]
- 44.Shulga, N., P. Roberts, Z. Gu, L. Spitz, M. M. Tabb, M. Nomura, and D. S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siniossoglou, S., M. Lutzmann, H. Santos-Rosa, K. Leonard, S. Mueller, U. Aebi, and E. Hurt. 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149:41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stochaj, U., R. Rassadi, and J. Chiu. 2000. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 114:2130-2132. [DOI] [PubMed] [Google Scholar]
- 47.Stoffler, D., B. Fahrenkrog, and U. Aebi. 1999. The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 11:391-401. [DOI] [PubMed] [Google Scholar]
- 48.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, Q., M. P. Rout, and C. W. Akey. 1998. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1:223-234. [DOI] [PubMed] [Google Scholar]
- 50.Zolotukhin, A. S., and B. K. Felber. 1999. Nucleoporins Nup98 and Nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73:120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]