Abstract
The RelA (p65) subunit of NF-κB is an important regulator of inflammation, proliferation, and apoptosis. We have discovered that the large subunit, p140, of replication factor C (RFC) can function as a regulator of RelA. RFC is a clamp loader, facilitating the addition and removal of proliferating-cell nuclear antigen from DNA during replication and repair but can also interact directly with the retinoblastoma tumor suppressor protein and the transcription factor C/EBPα. We find that RFC (p140) interacts with RelA both in vitro and in vivo and stimulates RelA transactivation. In contrast, coexpression of fragments of RFC (p140) that mediate the interaction with RelA results in transcriptional inhibition. The significance of this regulation was confirmed by using short interfering RNA oligonucleotides targeted to RFC (p140). Down regulation of endogenous RFC (p140) inhibits expression from a chromosomally integrated reporter plasmid induced by endogenous, TNF-α-activated NF-κB. Dominant negative fragments of RFC (p140) also cooperate with overexpressed RelA to induce cell death. Interestingly, RFC (p140) also interacts with the tumor suppressor p53. Taken together, these observations suggest that, in addition to its previously described function in DNA replication and repair, RFC (p140) has an important role as a regulator of transcription and NF-κB activity.
The Rel/NF-κB complex consists of homo- or heterodimers formed from a pool of five proteins, RelA (p65), c-Rel, RelB, p50/p105, and p52/p100 (35, 36). Within their amino termini, these proteins all contain a Rel homology domain (RHD) of approximately 300 amino acids (aa) that mediates both their DNA binding and dimerization. NF-κB1 and NF-κB2 are both synthesized as precursor proteins and remain in the cytoplasm until proteolytic processing yields the DNA-binding isoforms termed p50 and p52, respectively. In contrast, RelA, RelB, and c-Rel do not require proteolytic processing and contain nonhomologous transactivation domains within their carboxy termini. In most cell types, NF-κB complexes are held in an inactive form in the cytoplasm through interactions with one of a family of inhibitory proteins, IκBα, -β, and -ɛ. Activation of NF-κB in response to cellular stimulation involves the rapid phosphorylation and degradation of IκB, allowing NF-κB, to translocate to the nucleus (36). The role of NF-κB in disease stems primarily from circumstances in which it becomes inappropriately activated and constitutively localized in the nucleus. Although it is often associated with inflammatory diseases such as rheumatoid arthritis and asthma, it has become apparent that NF-κB also has a critical role in both tumorigenesis and the response to cancer therapy (4, 6, 36).
Activation of NF-κB is rapidly induced by many stimuli, including UV light, ionizing radiation, cytokines such as tumor necrosis factor (TNF) and interleukin-1, growth factors, bacterial lipopolysaccharide, phorbol esters, and hypoxia (33). NF-κB subunits have many functions and induce the expression of a diverse array of gene products, such as cytokines, growth factors, adhesion molecules, immunoreceptors, and other transcription factors that mediate the cellular response to the initial stimulus (33, 35, 36). These genes will vary, however, dependent on the cell type and the nature of the stimulation (35, 36).
Although more generally associated with regulation of immune function and inflammation, NF-κB subunits have been shown to be important regulators of cellular proliferation and apoptosis. NF-κB induces the expression of cyclin D1, as well as the proto-oncogene that encodes c-Myc (17, 20, 23, 33). Under some circumstances, however, NF-κB activation is associated with cell cycle arrest and differentiation (46, 47). In a large number of cell lines, inhibition of NF-κB stimulates programmed cell death (3). For example, RelA-containing complexes have been found to be antiapoptotic in response to TNF-α stimulation and DNA damage induced by ionizing radiation and chemotherapeutic agents (3, 36). Furthermore, because of extensive apoptosis in the liver, the RelA knockout is embryonically lethal (5). Significantly, under some circumstances, NF-κB has been reported to also promote cell death (3). NF-κB has been described as being proapoptotic following DNA damage by UV light, serum withdrawal, glutamate stimulation, and activation of the tumor suppressor p53 (3, 15, 16, 24, 36, 41). Furthermore, in T cells, NF-κB can be either pro- or antiapoptotic, depending on the stimulus (3), while overexpression of RelA in the pro-B-cell line 220-8, but not in WEHI 231 B cells, results in apoptosis and cell cycle arrest (47). Consistent with these effects, NF-κB can activate the expression of Fas and Fas ligand (33), both of which are potent inducers of apoptosis.
It appears, therefore, that dependent upon the circumstances, NF-κB can have apparently opposing effects upon the cell. The underlying mechanisms that result in these distinct effects are unclear, however. The specificity with which RelA stimulates gene expression can be dependent upon its interactions with transcriptional coactivators and other DNA-binding proteins (35, 36). This suggests that the presence, absence, or modification state of the proteins that regulate the ability of RelA to selectively stimulate gene expression might determine which aspect of RelA function is observed. Examples of such regulatory mechanisms include the p300 and CBP coactivator proteins, which interact with RelA and can be regulated by the cyclin-dependent kinase inhibitor p21WAF1/CIP1 (38, 48). Furthermore, RelA and the tumor suppressor p53 can each inhibit the other's activity in a manner consistent with competition for binding to p300 and CBP (51). Since it is probable that other, similar, regulatory mechanisms exist, we have been actively attempting to identify other proteins capable of modulating NF-κB transcriptional activity. In this study, we investigated the ability of the large subunit of replication factor C (RFC), p140, to function as a regulator of RelA.
RFC was originally described as a pentameric complex that, during the process of DNA replication and repair, functions as a clamp loader, facilitating the addition and removal of proliferating-cell nuclear antigen (PCNA) from sites of DNA synthesis (30). Recent results, however, have suggested more dynamic and diverse cellular functions for RFC (p140) and other RFC subunits. In Saccharomyces cerevisiae, RFC has been shown to function as a regulator of both the G1/S and G2/M checkpoints (25, 43). RFC subunits, including RFC (p140), have been observed in a large complex, termed BASC, that contains the breast cancer susceptibility gene BRCA1 and components of the DNA repair machinery (50). RFC (p140) also contains an LXCXE motif through which it binds the retinoblastoma tumor suppressor protein (Rb) and has a prosurvival function following UV stimulation (34). Rb and BRCA1 are important regulators of transcription and interact directly with DNA-binding proteins such as E2F and p53, respectively (18, 45, 52). Interestingly, RFC (p140) has recently been shown to interact with the b-ZIP domain of the C/EBPα transcription factor and regulate its transcriptional activity (21). Taken together, these observations suggested that RFC (p140) can be a regulator of both DNA replication/repair and transcription. In this report, we describe the regulation of RelA function by RFC (p140).
MATERIALS AND METHODS
Plasmids.
The Rous sarcoma virus (RSV) RelA, glutathione S-transferase (GST) RelA RHD, and pCDNA3 Gal4 plasmids have been previously described (7, 37). GST RelA (1 to 97), GST RelA (1 to 196), and GST RelA (61 to 307) were created by PCR in the laboratory of Gary J. Nabel. The pCDNA3 p53 expression plasmid was created by subcloning the p53 cDNA from plasmid pC53-SN3. The p53 cDNA was inserted into the BamHI site of the pCDNA3 polylinker. The pGL3 Bax luciferase reporter plasmid was supplied by T. Crook and originated in J. Reed's laboratory (Burnham Institute, La Jolla, Calif.). The pVR1012 RelA plasmid was obtained from Gary Nabel (National Institutes of Health). pVR1012 was used with the permission of Vical Inc. The full-length RFC (p140) and RFC (p37) cDNAs were isolated from human foreskin fibroblast cell RNA by reverse transcription-PCR and inserted into the KpnI site of pCDNA3 or pCGN, which inserted a hemagglutinin (HA) tag at the amino terminus. Fragments of RFC (p140) were isolated by PCR from the original clone and also inserted into the KpnI site of pCGN. The A20 chloramphenicol acetyltransferase (CAT) reporter plasmid has been described previously (26). The 3 × κB concanavalin A (ConA) luciferase reporter plasmid and ConA luciferase control were provided by Ron Hay (University of St. Andrews).
Cell culture.
Human embryonic kidney (HEK) 293 cells, human osteosarcoma U-2 OS cells, and HeLa 57A cells were maintained at 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 1% penicillin-streptomycin (Sigma), and 1% l-glutamine (Sigma). HeLa 57A cells containing an integrated copy of the 3 × κB ConA luciferase reporter plasmid have been described previously (40). Except where indicated otherwise, all of the serum used in this study was filtered through a 0.22-μm-pore-size Stericup vacuum filter with a Millipore Express polyethersulfone membrane (Millipore) before use.
Antibodies.
The polyclonal RFC (p140) antibody was generated by using a purified, His-tagged fragment of the protein (aa 1 to 369) expressed in Escherichia coli. The antibody was raised in sheep by the Scottish Antibody Production Unit. RelA (p65) Western blots and immunoprecipitations were performed with Santa Cruz Biotechnology antibodies sc-372 and sc-109, respectively. p300 Western blots were performed with Pharmingen antibody 14991A. The Rad50 antibody was obtained from GeneTex (MS-RAD10-PX1). The Rb, MSH2, and MSH6 antibodies (sc-50, sc-494, and sc-1243, respectively) were obtained from Santa Cruz Biotechnology. The PCNA antibody (P8825) was from Sigma. The HA tag antibody was obtained from Barbara Spruce (Dundee, Scotland).
Transfections and reporter assays.
Calcium phosphate transfections of U-2 OS and HEK 293 cells have been described previously (51). In this study, U-2 OS cells were split the night before transfections took place and harvested after 30 h. HEK 293 cells were split 2 h before transfection and harvested and analyzed after 48 h. HEK 293 cells for cell death studies were grown in DMEM with 10% unfiltered FBS. Transfections for CAT and luciferase assays were performed by using 10- and 6-cm-diameter dishes, respectively. CAT activity was assayed on 10 to 100 μg of protein prepared from whole-cell lysates. For luciferase assays, lysates were prepared by using passive lysis buffer (Promega). Luciferase assays were performed in accordance with the manufacturer's (Promega) instructions. All experiments were performed separately a minimum of three times before calculation of the means and standard errors shown in the figures. Relative luciferase levels were calculated as the level of activity per microgram of protein extract. Internal control reference plasmids (such as those encoding β-galactosidase or Renilla luciferase) were not included. During the investigation of transcription factor function, the promoters driving the expression of such internal controls are often affected by other components of the experiment and this can lead to the incorporation of errors when data are calculated relative to their levels of expression.
siRNA transfections.
HeLa 57A cells were seeded at a density of 5 × 104 per well of a 24-well dish and transfected with short interfering RNA (siRNA) oligonucleotides on the following day. A 3-μl volume of oligofectamine (Invitrogen) was mixed with 12 μl of Optimem (Invitrogen) for 7 to 10 min at room temperature. In another tube, 3 μl of a 20 μM siRNA duplex solution was added to 50 μl of Optimem. The two mixtures were then combined and incubated for a further 20 min at room temperature, and then a further 32 μl of Optimem was added. The medium was then removed from the cells to be transfected, which were then washed once with Optimem before addition of the transfection mixture. The cells were then incubated for 5 h at 37°C. A 500-μl volume of DMEM with 10% FBS (without antibiotics) was then added to the cells, which were left to grow overnight. On the following day, the cells were trypsinized and cells from each well were seeded into a corresponding well of a six-well dish with DMEM containing 10% FBS (without antibiotics). On the following day, the cells were transfected again by using a scaled-up version of the procedure used on day 2 (tube 1, 16.2 μl of oligofectamine and 64.8 μl of Optimem incubated for 7 to 10 min; tube 2, 16.2 μl of 20 μM siRNA duplex solution and 270 μl of Optimem; after mixing and incubation, 173 μl of Optimem was added before addition to the cells). Cells were then left for a further 2 days and then either harvested for whole-cell extracts or luciferase assays. Where indicated, TNF-α (10 ng/ml; Sigma) treatments were done for 6 h.
All siRNA duplex oligonucleotides were synthesized by Dharmacon Research Inc. The sequences used were as follows (sense strand only): RFC (p140) A, GAAGGCGGCCUCUAAAUCA; RFC (p140) B, UGAUGAAGCCAUCGCCAAG; RelA, GCUGAUGUGCACCGACAAG; Scramble siRNA, CAGUCGCGUUUGCGACUGG.
Nuclear protein extracts and EMSAs.
Nuclear protein extracts were prepared essentially as described previously (2), except for the experiment whose results are shown in Fig. 2D, for which nuclei were extracted in 150 mM NaCl. Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (7).
FIG. 2.
RelA and p53 interact with RFC (p140). (A) Immunoprecipitated RelA binds in vitro-translated RFC (p140). RelA was immunoprecipitated from nuclear protein extracts (200 μg) prepared from HEK 293 cells transfected with a RelA expression plasmid. The immunoprecipitated complex was then used in a pull-down assay with reticulocyte lysate-translated RFC (p140). A sample of input material (10%) is shown in this and subsequent panels. IP, immunoprecipitating. (B) RFC (p140) binds the RHD of RelA. Purified GST, GST RelA (RHD), or GST RelA (428 to 551), expressed in E. coli and bound to glutathione agarose, was used in a pull-down assay with reticulocyte lysate-translated RFC (p140). (C) Overexpressed RelA coimmunoprecipitates with RFC (p140). Endogenous RFC (p140) was immunoprecipitated from nuclear protein extracts (200 μg) prepared from HEK 293 cells transfected with either an RSV RelA expression plasmid or a control plasmid. The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted with an anti-RelA antibody. PI, preimmune serum. (D) Endogenous RelAcoimmunoprecipitates with RFC (p140). Endogenous RFC (p140) was immunoprecipitated from U-2 OS cell nuclear protein extracts (300 μg) that had been stimulated with TNF to activate endogenous NF-κB. The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted with an anti-RelA antibody. (E) Endogenous p53 from HEK 293 cells coimmunoprecipitates with RFC (p140). Endogenous RFC (p140) was immunoprecipitated from HEK 293 cell nuclear protein extracts (400 μg). The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted with an anti-p53 antibody. (F) UV-activated p53 coimmunoprecipitates with RFC (p140). Endogenous RFC (p140) was immunoprecipitated from U-2 OS cell nuclear protein extracts (120 μg) that were either prepared 8 h after UV light stimulation (40 J/m2) or left untreated. The immunoprecipitated complex was then resolved by SDS-PAGE and immunoblotted with anti-p53, DO-1, antibody. (G) RFC (p140) is specifically retained in a GST RelA (RHD) column. HEK 293 cell nuclear protein extracts were passed over a GST RelA (RHD) or a GST control column. Each column was then washed before stepwise elution with buffer containing 75, 150, 300, 600, and 1,000 mM NaCl. Eluates were precipitated with TCA, resolved by SDS-PAGE, and analyzed by Western blotting with the antibodies indicated.
GST pull-down assays and immunoprecipitations.
In vitro pull-down assays and immunoprecipitations were performed as described previously (7, 38).
Affinity chromatography.
HEK 293 cell nuclear protein extracts in incubation buffer (IB) (20 mM HEPES [pH 7.9], 2.5 mM MgCl2, 1 mM dithiothreitol, 0.1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml, 1 μg of pepstatin A per ml) containing 75 mM NaCl were passed over 0.4-ml GST RelA (RHD) or GST control columns (prewashed with 20 volumes of IB containing 75 mM NaCl and 2 volumes of IB containing 1 M NaCl). The columns were then washed with 20 volumes of IB containing 75 mM NaCl before stepwise elution in 500 μl of IB containing 75, 150, 300, 600, and 1,000 mM NaCl. Eluates were precipitated with trichloroacetic acid (TCA), resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by Western blotting.
RESULTS
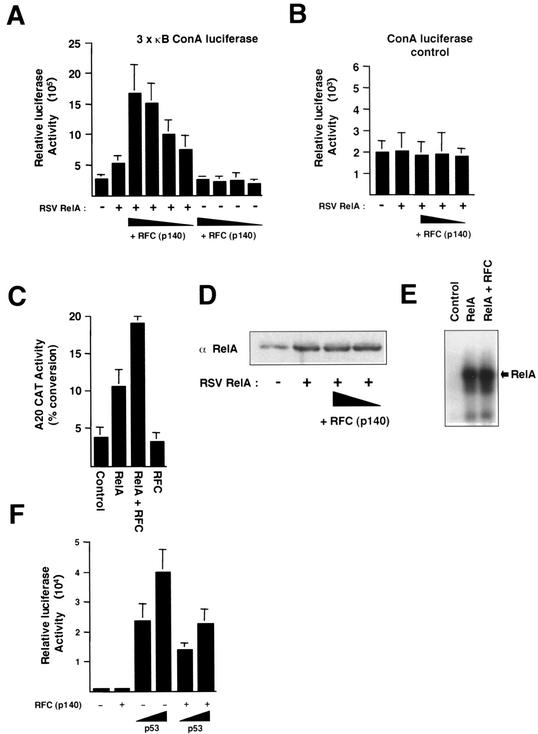
To establish whether RFC (p140) might regulate NF-κB, we initially investigated whether the former might have any effect on RelA transcriptional activity. U-2 OS cells were transfected with RFC (p140) and RelA expression plasmids together with a 3 × κB luciferase reporter plasmid, which contains three copies of the consensus Ig/HIV NF-κB binding site upstream of a minimal ConA promoter. A significant and dose-dependent increase in RelA transactivation was observed (Fig. 1A), comparable to that typically seen with the p300 protein, a known coactivator of NF-κB (38). No effect of RelA or RFC (p140) on a ConA luciferase control plasmid lacking κB elements was seen (Fig. 1B). We next investigated whether a similar effect would be seen with a cellular promoter known to be regulated by NF-κB. The A20 protein is an inhibitor of apoptosis whose promoter contains two NF-κB binding sites (26). Upon cotransfection with the A20 CAT reporter plasmid, full-length RFC (p140) again significantly enhanced RelA transactivation, although to a lesser extent than that seen with the artificial promoter (Fig. 1C). RFC (p140) did not affect RelA expression levels and did not stimulate RelA DNA binding in an EMSA (Fig. 1D and E). These observations were consistent, therefore, with previous reports suggesting that RFC (p140) could function as a coactivator protein (21). We were interested, therefore, in whether RFC (p140) would stimulate the transactivation function of any transcription factor. To investigate this, we examined the effect of RFC (p140) on the p53 tumor suppressor protein. In contrast to the results seen with RelA, however, RFC (p140) repressed p53 transactivation of the Bax promoter (Fig. 1F). RFC (p140) is not a universal coactivator protein, therefore. Moreover, stimulation of RelA and repression of p53 are consistent with both the proproliferative and antiapoptotic functions of RFC (p140) (34).
FIG. 1.
RFC (p140) regulates RelA transcriptional activity. (A) RFC (p140) stimulates RelA transactivation. U-2 OS cells were transfected with the 3 × κB ConA luciferase reporter plasmid (1.5 μg) and the indicated RSV RelA (1 μg) or pCDNA3 RFC (p140) expression plasmid (0.1, 0.5, 1, and 2 μg). Control RSV or pCDNA3 plasmids were included in all transfections such that each condition had the same level of each type of plasmid. Cells were harvested after 30 h. Results shown are the means of three separate experiments. Standard deviations are shown. (B) RelA and RFC (p140) do not affect the activity of a control reporter plasmid. U-2 OS cells were transfected as described for panel A, except that a control ConA luciferase reporter plasmid lacking κB elements was used. (C) RFC (p140) stimulates RelA transactivation of the A20 promoter. U-2 OS cells were transfected as described for panel A with A20 CAT reporter plasmid (5 μg) and the indicated RSV RelA (5 μg) or pCDNA3 RFC (p140) expression plasmid (0.5 μg). Results shown are the means of three separate experiments. Standard deviations are shown. (D) RFC (p140) does not affect transfected RelA protein levels. U-2 OS cells were transfected as described for panel A; however, after 30 h, whole-cell lysates were prepared and analyzed by Western blot analysis for RelA protein levels. (E) RFC (p140) does not affect RelA DNA binding. HEK 293 cells were transfected with a RelA or RFC (p140) expression plasmid (5 μg of each), either alone or in combination, as indicated. Nuclear protein extracts were prepared and analyzed by EMSA with a 32P-labeled oligonucleotide containing the Ig/HIV NF-κB binding site. The position of the RelA-DNA complex is indicated. (F) RFC (p140) represses p53 transactivation. U-2 OS cells were transfected as described for panel A with the Bax luciferase reporter plasmid (1.5 μg) and the indicated pCDNA3 p53 (100 and 500 ng) or pCDNA3 RFC (p140) (0.5 μg) expression plasmid. Results shown are the means of three separate experiments. Standard deviations are shown.
Stimulation of RelA transactivation by RFC (p140) could conceivably occur through a number of different mechanisms. Some of these, such as indirect cell cycle effects or interactions with other coactivators, would not require a direct interaction between the two proteins. We next determined, therefore, whether RelA and RFC (p140) can physically associate with each other. Significantly, both immunoprecipitated RelA complexes and bacterially expressed GST RelA bound to reticulocyte lysate-translated RFC (p140) (Fig. 2A and B). This interaction was mediated by the amino-terminal RHD of RelA, which has been shown to bind many heterologous transcription factors and coactivators (7, 35, 53). This GST RelA (RHD) protein has been previously used to characterize interactions with heterologous transcription factors (7, 37). It also strongly interacts with IκBα (data not shown). This interaction was not affected by the inclusion of ethidium bromide or DNase I (to disrupt protein-DNA interactions), indicating that it does not result from fortuitous colocalization on the same DNA fragment during incubation (data not shown). No interaction was seen with the carboxy terminus of RelA (aa 428 to 551), which contains a strong transactivation domain (44). Confirming the significance of this in vitro interaction, transiently transfected RelA coimmunoprecipitated with endogenous RFC (p140) (Fig. 2C). p300, a known RelA coactivator (38), did not coprecipitate with the RFC (p140)/RelA complex, suggesting that these proteins independently regulate NF-κB activity (Fig. 2C). Importantly, endogenous RelA and RFC (p140) were also seen to associate (Fig. 2D). In this experiment, U-2 OS cells were stimulated with TNF-α, which induced NF-κB translocation to the nucleus, allowing levels of RelA in the nuclear protein extracts to be detectable. Interestingly, endogenous p53 from nuclear extracts prepared from either unstimulated HEK 293 cells or UV light-stimulated U-2 OS cells also coimmunoprecipitated with RFC (p140) (Fig. 2E and F). While these results, together with those in Fig. 1, suggest that RFC (p140) also regulates p53 function, the rest of this report focuses on the significance and consequences of the interaction with RelA.
RFC (p140) can associate with a number of other cellular proteins. To determine if these can also bind RelA, we passed nuclear protein extracts prepared from unstimulated HEK 293 cells over a GST RelA (RHD) affinity column. As expected, endogenous RFC (p140) was retained on the column (Fig. 2G). No interaction was seen with Rb, PCNA, and components of the BASC complex, such as MSH2, MSH6, and Rad50 (Fig. 2G). Although the possibility that these proteins can associate with RelA through RFC (p140) cannot be excluded, this experiment demonstrates both the specificity of this interaction and the potential for RFC (p140) to independently regulate NF-κB.
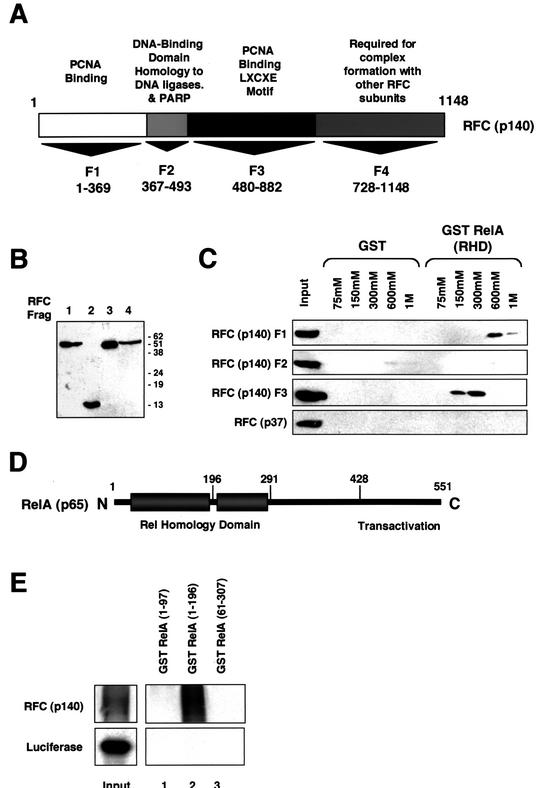
To study the role of RFC (p140) in RelA-induced gene expression further, a series of plasmids encoding RFC (p140) fragments fused to the HA epitope, which might be expected to function as dominant negative inhibitors, were constructed (Fig. 3A). RFC (p140) F1 (aa 1 to 369) encodes the amino terminus of the protein and has been shown to have a PCNA binding domain (29). RFC (p140) F2 (aa 367 to 493) encodes a domain with homology to DNA ligases and has a BRCT domain, also found in BRCA1 and other proteins involved in DNA repair (30). RFC (p140) F3 (aa 480 to 882) also binds PCNA, contains the domain homologous to other RFC subunits, and has an LXCXE motif required for binding to Rb together with a caspase 3 cleavage site (30, 34, 39). RFC (p140) F3 has been previously shown to function as a dominant negative inhibitor of DNA replication in U-2 OS cells (12). RFC (p140) F4 (aa 728 to 1148) contains a domain required for association with other RFC subunits and also has two caspase 3 cleavage sites (30, 39). Western blot analysis with an anti-HA antibody demonstrated that all RFC fragments were expressed equivalently, apart from RFC (p140) F4, where levels were consistently reduced (Fig. 3B). The reason for this effect is unknown, but since this made meaningful comparisons hard, it was decided to focus experiments on RFC (p140) F1 to F3.
FIG. 3.
RelA interacts with two distinct domains of RFC (p140). (A) Schematic diagram showing different domains of RFC (p140). PARP, poly(ADP-ribose) polymerase. (B) Western blot analysis of RFC (p140) fragments (Frag) F1 to F4. Nuclear extracts were prepared from HEK 293 cells transfected with 5 μg of pCGN RFC (p140) fragment expression plasmid. Samples were resolved by SDS-PAGE and immunoblotted with anti HA-antibody 12CA5. The values on the right are molecular sizes in kilodaltons. (C) RelA interacts with RFC (p140) fragments F1 and F3. HEK 293 cell nuclear protein extracts were prepared from cells transfected with the indicated HA-tagged RFC (p140) fragments or RFC (p37). Protein affinity chromatography was then performed with the GST RelA (RHD) or control GST protein essentially as described for Fig. 2G, with 700 μg of the nuclear protein extracts. Eluates were precipitated with TCA, resolved by SDS-PAGE, and analyzed by Western blotting with an anti-HA antibody. (D) Schematic diagram showing different domains of RelA. (E) RFC (p140) binds the amino-terminal subdomain of the RelA RHD. Purified GST RelA (1 to 97), GST RelA (1 to 196), or GST RelA (61 to 307), expressed in E. coli and bound to glutathione agarose, was used in a pull-down assay with reticulocyte lysate-translated RFC (p140) or a luciferase control protein, as indicated.
To determine if these fragments could bind RelA independently, we passed nuclear protein extracts prepared from HEK 293 cells separately transfected with RFC (p140) F1, F2, and F3 over a GST RelA (RHD) affinity column. Both RFC (p140) F1 and F3 bound to GST RelA, although interestingly, RFC (p140) F1 eluted at a higher salt concentration, suggesting a stronger interaction (Fig. 3C). No interaction with RFC (p140) F4 was seen (data not shown). However, as stated above, because of its lower expression levels, it was hard to draw meaningful comparisons with the other RFC (p140) fragments. An equivalent experiment was performed in parallel with RFC (p37), another subunit of the RFC (p140) complex. RFC (p37) did not bind the GST RelA (RHD) column (Fig. 3C). This indicates that RelA can interact with two separate domains of RFC (p140) independently. It also suggests that RelA has the potential to interact with RFC (p140) when not part of the pentameric RFC complex. Further investigation is required before it can be concluded that a separate RFC (p140) transcriptional coactivator complex exists in addition to the classical RFC clamp loader complex, however. We next investigated which domain of the RelA RHD mediates the interaction with RFC (p140). The crystallographic structure of RelA bound to DNA has revealed that the RHD consists of two distinct subdomains joined by a short linker region (8) (Fig. 3D). GST pull-down analysis revealed that the most amino terminal of these subdomains (aa 1 to 196) bound RFC (p140) in vitro (Fig. 3E). No binding was seen with other RelA RHD fragments (aa 1 to 97 and 61 to 307).
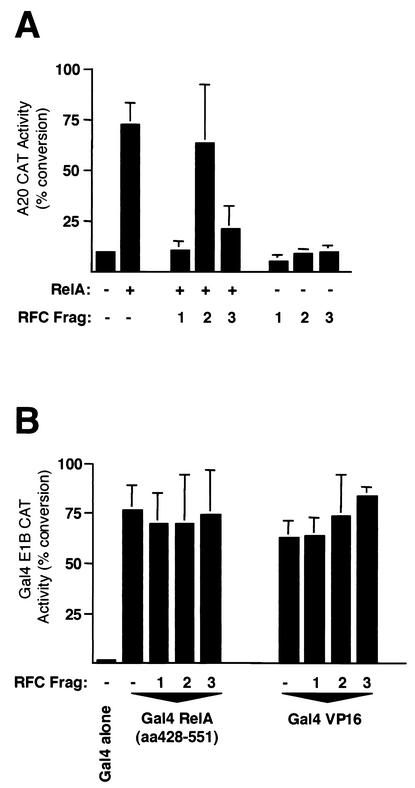
We next determined whether the RFC (p140) fragments that interact with RelA can independently stimulate its transcriptional activity. In contrast to the results seen with full-length RFC (p140) (Fig. 1), and consistent with a dominant negative role, RFC (p140) F1 and F3 were both strong repressors of RelA transactivation (Fig. 4A). In contrast, RFC (p140) F2 had no effect on RelA transactivation (Fig. 4A). Demonstrating the specificity of these effects, no inhibition by these RFC (p140) fragments was seen with Gal4 VP16 or Gal4 RelA (aa 428 to 551) (Fig. 4B). Gal4 RelA (aa 428 to 551) contains the RelA carboxy-terminal transactivation domain but lacks the RHD seen to interact with RFC (p140) in Fig. 2B. Full-length RFC (p140) is therefore required for stimulation of RelA transactivation. These results also suggested that inactivation of endogenous RFC (p140) inhibits RelA transcriptional activity.
FIG. 4.
Fragments (Frag) of RFC (p140) specifically repress RelA-dependent transactivation. (A) U-2 OS cells were transfected with A20 CAT reporter plasmid (5 μg) and the indicated RSV RelA (5 μg) and pCGN RFC (p140) fragment (5 μg) expression plasmids. Control RSV or cytomegalovirus plasmids were included in all transfections such that each condition had the same level of each type of plasmid. Cells were harvested after 30 h and assayed for CAT activity. Results shown are the means of three separate experiments. Standard deviations are shown. (B) Dishes (10-cm diameter) of U-2 OS cells were transfected with Gal4 E1B CAT reporter plasmid (5 μg), pCGN RFC (p140) fragment expression plasmids (5 μg), and 0.15 ng of pCDNA3 Gal4, Gal4 RelA (aa 428 to 551), and Gal4 VP16, as indicated. Control pCDNA3 plasmids were included in all transfections such that each condition had the same level of each type of plasmid. Cells were harvested after 30 h and assayed for CAT activity. Results shown are the means of three separate experiments. Standard deviations are shown.
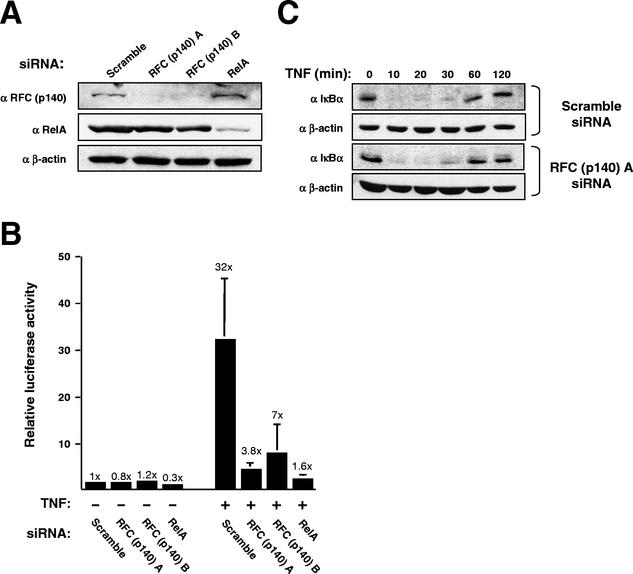
Since the functional experiments described above (Fig. 1 and 4) had relied on overexpressed proteins and transiently transfected reporter plasmids, it was important to demonstrate that this regulatory mechanism functioned for the endogenous proteins. To facilitate this, we took advantage of a recent report demonstrating that short double-stranded RNA oligonucleotides homologous to a target mRNA (siRNAs) specifically down regulate their target proteins in mammalian cells (11). siRNAs were therefore designed to target RFC (p140) and RelA. Both RFC (p140) siRNAs and the single RelA siRNA selectively and strongly down regulated their target proteins in HeLa 57A cells (Fig. 5A). HeLa 57A cells contain an integrated copy of the 3 × κB ConA luciferase reporter plasmid described earlier (Fig. 1A). We therefore investigated the effect of down regulation of RFC (p140) and RelA on the activity of this reporter. In unstimulated cells, the RelA siRNA still had a slight inhibitory effect on expression from this plasmid relative to a Scramble siRNA control (Fig. 5B). No significant effect was seen with either of the two RFC (p140) siRNAs. Significantly, both RFC (p140) siRNAs strongly inhibited TNF activation of the reporter plasmid. Surprisingly, this inhibition was comparable to, although not quite as strong as, inhibiting RelA itself. No effect of these treatments on protein yields was seen (data not shown). In addition, crystal violet staining of samples treated in parallel indicated that there was no significant effect on proliferation or cell death during the time course of these experiments or after the 6-h TNF treatment (data not shown). Inhibition of the NF-κB reporter plasmid does not, therefore, result from cell death. To confirm that the NF-κB activation pathway itself was unaffected by these treatments, we prepared whole-cell lysates from cells treated in parallel and analyzed IκBα levels by immunoblotting (Fig. 5C). No effect on IκBα degradation by RFC (p140) inhibition was observed. Significantly, IκBα resynthesis was also unaffected. This effect is regulated by NF-κB (9, 22, 27). This indicates, therefore, that the inhibition of NF-κB seen is promoter specific and that NF-κB transcriptional activity per se is not inhibited.
FIG. 5.
siRNA-mediated down regulation of RFC (p140) levels inhibits endogenous NF-κB transactivation. (A) siRNAs directed against RFC (p140) and RelA specifically inhibit the expression of their target proteins. A Western blot shows the effect on the RFC (p140), RelA, and β-actin proteins of treating HeLa 57A cells with the indicated siRNAs. (B) Effect of siRNA treatment on stimulation of the integrated 3 × κB ConA luciferase reporter plasmid by TNF-α. HeLa 57A cells were treated with the indicated siRNAs. Cells were either left unstimulated or subjected to TNF-α stimulation (10 ng/ml) for 6 h, as indicated. Luciferase activity is expressed as fold activation relative to the level of activity seen in unstimulated cells with the Scramble siRNA control. The results shown are means of three separate experiments and the standard deviations together with the fold activations. (C) Treatment with anti-RFC (p140) siRNA does not affect IκBα degradation and resynthesis. HeLa 57A cells were treated as described for panel B, and whole-cell lysates were prepared and subjected to Western blot analysis with the indicated antibodies.
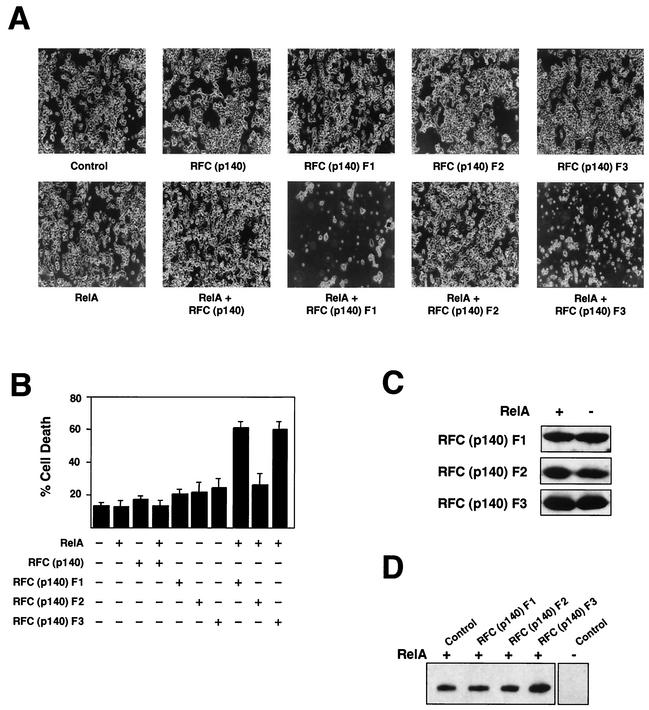
Although no effects on cell viability had been seen in the above-described experiments, RFC (p140) had previously been shown to stimulate cell survival following UV treatment of cells, suggesting that it can function as a regulator of apoptosis (34). Since RelA is also an important regulator of apoptosis (3, 36), we were interested in whether interference with RFC (p140) function might affect this aspect of its behavior. As discussed above, no significant effect of the RFC (p140) siRNAs on cell death following TNF treatment was observed in HeLa cells (data not shown). We investigated, therefore, whether coexpression of RFC (p140) or the RFC (p140) fragments, together with RelA, might have an effect when overexpressed in highly transfectable HEK 293 cells. Initially, no effect was seen, however. Routinely, the FBS used to grow these cells (and that used in the experiments described above) was filtered through a 0.22-μm-pore-size membrane prior to use. We had observed elsewhere, however, that some cell types are more susceptible to stimuli inducing cell death when cultured in unfiltered serum (data not shown). These experiments were repeated, therefore, with HEK 293 cells cultured in unfiltered FBS. There was still no observable effect on cell viability when full-length RFC (p140) and RelA, either alone or in combination, were expressed in HEK 293 cells (Fig. 6A). Similarly, when expressed alone, none of the RFC (p140) fragments significantly affected cell viability (Fig. 6A and B). When RFC (p140) F1 and F3 were coexpressed with RelA, however, a dramatic increase in cell death was observed (Fig. 6A and B). Morphologically, the dead cells had the appearance of having undergone apoptosis, being shrunken and fragmented. These effects were seen over a range of transfected plasmid concentrations, and 10-fold less plasmid produces an effect identical to that seen in Fig. 6A (data not shown). Moreover, no significant effect on the RelA or RFC (p140) F1 to F3 protein level is seen when these proteins are cotransfected (Fig. 6C and D). This effect on cell viability therefore represents a true cooperative effect and does not result from toxic effects resulting from overexpression of either protein alone. Surprisingly, no effect of overexpression of RFC (p140) F1 and F3 upon TNF treatment of these cells was seen (data not shown). Therefore, while inhibition of RFC (p140) can result in RelA-dependent cell death, this effect appears to be highly selective and requires overexpressed RelA. It is possible that for RFC (p140) regulation of endogenous NF-κB-dependent cell death, other stimuli or other cell lines need to be utilized.
FIG. 6.
Disruption of RFC (p140) function results in RelA-dependent induction of cell death. (A) HEK 293 cells were transfected with the indicated pVR1012 RelA (5 μg) and pCGN RFC (p140) (5 μg) expression plasmids. Control pVR1012 or cytomegalovirus plasmids were included in all transfections such that each condition had the same level of each type of plasmid. Representative fields of view are shown. Cell death occurred between 48 and 72 h after transfection. (B) HEK 293 cells were transfected as described above with 5 μg of expression plasmids. At 72 h after transfection, cells were harvested and stained with trypan blue. Living and dead cells were then counted in triplicate with a hemocytometer. The results of two separate experiments are shown. (C and D) HEK 293 cells were transfected as described above with 5 μg of expression plasmids, and whole-cell lysates were prepared after 48 h but prior to the occurrence of cell death. Extracts were resolved by SDS-PAGE and immunoblotted with either anti-HA antibody to detect RFC protein fragment expression levels (C) or anti-RelA antibody (D).
DISCUSSION
In this report, we have demonstrated that RFC (p140) interacts with and regulates the transcriptional activity of the RelA NF-κB subunit. Coexpression of full-length RFC (p140) stimulates the transcriptional activity of transiently transfected RelA (Fig. 1). Conversely, expression of the fragments of RFC (p140) that interact with RelA (Fig. 3) represses its transactivation (Fig. 4). Importantly, down regulation of endogenous RFC (p140) with siRNAs results in inhibition of endogenous, TNF-activated NF-κB activation of an integrated reporter gene, demonstrating that this is a biologically relevant regulatory mechanism (Fig. 5). NF-κB regulates a large number of genes in many cell types with great selectivity. The diversity of these genetic programs and their often apparently contradictory cellular effects have suggested that interactions with other proteins might provide part of the mechanism through which this specificity can be achieved. NF-κB has been previously linked with cell cycle regulation and can directly stimulate proliferation via the activation of proto-oncogenes such as those that encode c-Myc and cyclin D1 (33, 36). Conversely, NF-κB function is also sometimes associated with cellular differentiation and the cyclin-dependent kinase inhibitor p21WAF1/CIP1 can stimulate RelA transactivation indirectly through the p300 and CBP coactivator proteins (38, 48). This study therefore provides an additional and direct link between NF-κB and the proteins that control cell division.
While we cannot completely rule out the possibility that indirect effects of RFC (p140) contribute to the regulation of RelA transactivation, the physical association between these proteins suggests that they are mediated, at least in part, by direct effects on transcriptional activity. How RFC (p140) accomplishes this is not known, however. A number of proteins, including p53, BRCA1, TFIIH, and p300 (10, 13, 19, 28), have multiple roles in transcription, replication, and DNA repair. Interestingly, no p300 was observed coprecipitating with RelA/RFC (p140), suggesting that these complexes may have distinct regulatory functions (Fig. 2). It will therefore be of interest to determine the nature of the RFC (p140) complex that regulates RelA activity. This complex need not be the pentameric RFC complex. Recent papers, this report, and our other preliminary observations suggest that RFC (p140) is a much more complex and multifunctional protein than previously thought. RFC (p140) has now been reported to interact with Rb, C/EBP α, histone deacetylase 1 (HDAC1), a BRCA1 complex, RelA, and p53 (2, 21, 34, 50). It is entirely possible that RFC (p140) exists in complexes independently of the smaller RFC subunits. Consistent with this, a number of recent publications demonstrate that the smaller RFC subunits can form complexes lacking RFC (p140) (14, 31, 32). The presence of such complexes lacking RFC (p140) suggests possible functional redundancy and might explain the surprising lack of effect on cell viability of inhibition of RFC (p140) function in the absence of RelA expression (Fig. 5 and 6). Alternatively, inhibition under these circumstances might not be complete and sufficient RFC (p140) could still be present to allow it to fulfill its role in DNA replication.
Regulation of RelA transactivation by RFC (p140) appears to be promoter specific. siRNA-mediated down regulation of RFC (p140) significantly inhibited activation of the integrated 3 × κB ConA promoter but does not inhibit resynthesis of IκBα (Fig. 5). The IκBα promoter is immediately occupied by NF-κB following its activation, and it has been suggested that this reflects a minimal requirement for chromatin remodeling (42). The integrated 3 × κB ConA luciferase reporter plasmid used here is strongly activated by the HDAC inhibitor trichostatin A, suggesting that it does require chromatin-remodeling activities (R. Evans and R. T. Hay, personal communication). These effects may therefore reflect the differential requirement of NF-κB-regulated promoters for different chromatin-remodeling activities, possibly recruited by interaction with RFC (p140). We have recently reported that RFC (p140) can interact with HDAC1 (2). While such a complex is obviously not consistent with transcriptional activation, since HDAC1 would be expected to repress transcription, it does indicate that RFC (p140) can interact with chromatin remodeling activities. Further investigation is required to elucidate the exact nature of the mechanism through which RFC (p140) coactivates RelA and identify endogenous genes regulated by this activity.
We also found that coexpression of two RFC (p140) fragments, F1 and F3, can result in RelA-dependent cell death (Fig. 6). A role for RFC (p140) as a regulator of programmed cell death has been previously suggested (30, 34, 39). Moreover, RFC (p140) is a substrate for caspases and is proteolytically cleaved during apoptosis (39). Surprisingly, the effects we observed correlate with the ability of these RFC (p140) fragments to inhibit its transcriptional activity (Fig. 4). Furthermore, we also found that to observe this effect on cell viability, it was important not to filter the FBS used to grow the cells. These effects on cell death were observed with multiple batches of serum purchased from different companies (data not shown), indicating that this is not an aberrant result. This observation suggests that coexpression of RelA and RFC (p140) F1 or F3 sensitizes the cells to a factor present in the unfiltered serum. This sensitization cannot be due to just the repression of NF-κB activity by the RFC (p140) fragments, since a strong cooperative effect was seen. It is possible that under these conditions, where the proteins are overexpressed, RelA is still capable of inducing the expression of selective endogenous genes, some of which will be capable of facilitating an apoptotic response. Coexpression of the RFC (p140) fragments, followed by TNF treatment, in HEK 293 cells did not result in an increase in apoptosis (data not shown). Similarly, no obvious effects on cell viability were seen following TNF treatment of HeLa 57A cells treated with RFC (p140) siRNAs (data not shown). Therefore, while these results suggest that RelA-dependent cell death can be regulated by RFC (p140), it is likely to be only under specific conditions. RFC (p140) can regulate the transcriptional activity of TNF-induced NF-κB (Fig. 5). Our results suggest that this is promoter specific, however. It is probable that inhibition of RFC (p140) under these circumstances only modulates the NF-κB response rather than inhibiting it completely, which would be required for significant cell death to be observed. Nonetheless, this demonstrates that regulation of RelA by RFC (p140) can have profound biological consequences.
We also observed that endogenous RFC (p140) coprecipitated with the tumor suppressor p53 (Fig. 2). Moreover, while RFC (p140) stimulated the activity of RelA, it repressed the activity of p53 (Fig. 1). Under many circumstances, the two proteins have contrasting functions, with RelA being proproliferative and antiapoptotic while p53 is antiproliferative and proapoptotic (36, 49). This observation would therefore be consistent with the reported antiapoptotic and proproliferative functions of RFC (p140). Further investigation is required to determine the significance of this interaction.
Taken together, our results reveal a new function for RFC (p140) and a novel mechanism of NF-κB regulation. Given the complexity of the NF-κB response, it is not surprising that multiple factors control its transcriptional activity. It is likely that distinct subsets of genes regulated by NF-κB require different coactivator complexes. Analysis of the beta interferon enhancer, for example, has indicated that multiple coactivator complexes function cooperatively to facilitate gene expression (1). We propose that RFC (p140) is a component of a transcriptional coactivator complex that selectively regulates the expression of genes controlled by NF-κB and other transcription factors. Regulation of RFC (p140), possibly by posttranslational modification, would serve to integrate the expression of these genes with events controlling DNA replication and repair. It is therefore possible that reagents capable of manipulating RFC (p140) function have potential for the treatment of cancer and other diseases.
Acknowledgments
We thank Nicola Wiechens, Donna Bumpass, Neil Chapman, David Gregory, Sonia Rocha, Kevin Roche, Alison Sleigh, Elaine Waller, Stewart McWilliams, Alan Prescott, Niall McTavish, Julian Blow, Barbara Spruce, Tom Owen-Hughes, Stefan Roberts, and Joost Zomerdijk for help during this project.
L.A. is funded by the Wellcome Trust. N.D.P. is the recipient of a Royal Society University Fellowship. This project was also funded by the BBSRC ICR Initiative.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. M. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L. A., and N. D. Perkins. 2002. The large subunit of replication factor C interacts with the histone deacetylase, HDAC1. J. Biol. Chem. 277:29550-29554. [DOI] [PubMed] [Google Scholar]
- 3.Barkett, M., and T. D. Gilmore. 1999. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18:6910-6924. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, P. J., and M. Karin. 1997. Mechanisms of disease: nuclear factor κB, a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336:1066-1071. [DOI] [PubMed] [Google Scholar]
- 5.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 6.Bours, V., M. Bentires-Alj, A. C. Hellin, P. Viatour, P. Robe, S. Delhalle, V. Benoit, and M. P. Merville. 2000. Nuclear factor κB, cancer, and apoptosis. Biochem. Pharmacol. 60:1085-1089. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, N. R., and N. D. Perkins. 2000. Inhibition of the RelA (p65) NF-κB subunit by Egr-1. J. Biol. Chem. 275:4719-4725. [DOI] [PubMed] [Google Scholar]
- 8.Chen, F. E., D. B. Huang, Y. Q. Chen, and G. Ghosh. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391:410-413. [DOI] [PubMed] [Google Scholar]
- 9.Chiao, P. J., S. Miyamoto, and I. M. Verma. 1994. Autoregulation of IκBα activity. Proc. Natl. Acad. Sci. USA 91:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, C. X., and S. G. Brodie. 2000. Roles of BRCA1 and its interacting proteins. Bioessays 22:728-737. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 12.Fotedar, R., R. Mossi, P. Fitzgerald, T. Rousselle, G. Maga, H. Brickner, H. Messier, S. Kasibhatla, U. Hubscher, and A. Fotedar. 1996. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant-negative inhibitor of DNA replication in mammalian cells. EMBO. J. 15:4423-4433. [PMC free article] [PubMed] [Google Scholar]
- 13.Frit, P., E. Bergmann, and J. M. Egly. 1999. Transcription factor IIH: a key player in the cellular response to DNA damage. Biochimie 81:27-38. [DOI] [PubMed] [Google Scholar]
- 14.Griffith, J. D., L. A. Lindsey-Boltz, and A. Sancar. 2002. Structures of the human Rad17-replication factor C and checkpoint Rad9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J. Biol. Chem. 277:15233-15236. [DOI] [PubMed] [Google Scholar]
- 15.Grilli, M., M. Pizzi, M. Memo, and P. Spano. 1996. Neuroprotection by aspirin and sodium salicylate through blockade of NF-κB activation. Science 274:1383-1385. [DOI] [PubMed] [Google Scholar]
- 16.Grimm, S., M. K. A. Bauer, P. A. Baeuerle, and K. Schulze-Osthoff. 1996. Bcl-2 down-regulates the activity of transcription factor NF-κB induced upon apoptosis. J. Cell Biol. 134:13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttridge, D. C., C. Albanese, J. Y. Reuther, R. G. Pestell, and A. S. Baldwin. 1999. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 19.Hasan, S., P. O. Hassa, R. Imhof, and M. O. Hottiger. 2001. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature 410:387-391. [DOI] [PubMed] [Google Scholar]
- 20.Hinz, M., D. Krappmann, A. Eichten, A. Heder, C. Scheidereit, and M. Strauss. 1999. NF-κB function in growth control: regulation of cyclin D1 expression and G0G1-to-S-phase transition. Mol. Cell. Biol. 19:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong, S. H., S. J. Park, H. J. Kong, J. D. Shuman, and J. H. Cheong. 2001. Functional interaction of bZIP proteins and the large subunit of replication factor C in liver and adipose cells. J. Biol. Chem. 276:28098-28105. [DOI] [PubMed] [Google Scholar]
- 22.Ito, C. Y., A. G. Kazantsev, and A. S. Baldwin. 1994. 3 NF-κB sites in the IκBα promoter are required for induction of gene expression by TNF-α. Nucleic Acids Res. 22:3787-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaltschmidt, B., C. Kaltschmidt, S. P. Hehner, W. Droge, and M. L. Schmitz. 1999. Repression of NF-κB impairs HeLa cell proliferation by functional interference with cell cycle checkpoint regulators. Oncogene 18:3213-3225. [DOI] [PubMed] [Google Scholar]
- 24.Kasibhatla, S., T. Brunner, L. Genestier, F. Echeverri, A. Mahboubi, and D. R. Green. 1998. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-κB and AP-1. Mol. Cell 1:543-551. [DOI] [PubMed] [Google Scholar]
- 25.Kim, H. S., and S. J. Brill. 2001. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:3725-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laherty, C. D., N. D. Perkins, and V. M. Dixit. 1993. Human T cell leukemia virus type I Tax and phorbol 12-myristate 13-acetate induce expression of the A20 zinc finger protein by distinct mechanisms involving nuclear factor κB. J. Biol. Chem. 268:5032-5039. [PubMed] [Google Scholar]
- 27.Lebail, O., R. Schmidtullrich, and A. Israel. 1993. Promoter analysis of the gene encoding the IκBα/Mad3 inhibitor of NF-κB-positive regulation by members of the Rel/NF-κB family. EMBO J. 12:5043-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May, P., and E. May. 1999. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18:7621-7636. [DOI] [PubMed] [Google Scholar]
- 29.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mossi, R., and U. Hubscher. 1998. Clamping down on clamps and clamp loaders: the eukaryotic replication factor C. Eur. J. Biochem. 254:209-216. [PubMed] [Google Scholar]
- 31.Naiki, T., T. Kondo, D. Nakada, K. Matsumoto, and K. Sugimoto. 2001. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21:5838-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohta, S., Y. Shiomi, K. Sugimoto, C. Obuse, and T. Tsurimoto. 2002. A proteomics approach to identify PCNA binding proteins in human cell lysates: identification of the human CHL12/RFCs2-5 complex as a novel PCNA binding protein. J. Biol. Chem. 277:40362-40367. [DOI] [PubMed] [Google Scholar]
- 33.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 34.Pennaneach, V., I. Salles-Passador, A. Munshi, H. Brickner, K. Regazzoni, F. Dick, N. Dyson, T. T. Chen, J. Y. J. Wang, R. Fotedar, and A. Fotedar. 2001. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell 7:715-727. [DOI] [PubMed] [Google Scholar]
- 35.Perkins, N. D. 1997. Achieving transcriptional specificity with NF-κB. Int. J. Biochem. Cell Biol. 29:1433-1448. [DOI] [PubMed] [Google Scholar]
- 36.Perkins, N. D. 2000. The Rel/NF-κB family: friend and foe. Trends Biochem. Sci. 25:434-440. [DOI] [PubMed] [Google Scholar]
- 37.Perkins, N. D., A. B. Agranoff, E. Pascal, and G. J. Nabel. 1994. An interaction between the DNA-binding domains of RelA (p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 14:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Y. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 39.Rheaume, E., L. Y. Cohen, F. Uhlmann, C. Lazure, A. Alam, J. Hurwitz, P. P. Sekaly, and F. Denis. 1997. The large subunit of replication factor C is a substrate for caspase-3 in vitro and is cleaved by a caspase-3-like protease during Fas-mediated apoptosis. EMBO. J. 16:6346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez, M. S., J. Thompson, R. T. Hay, and C. Dargemont. 1999. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J. Biol. Chem. 274:9108-9115. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-κB in p53-mediated programmed cell death. Nature 404:892-897. [DOI] [PubMed] [Google Scholar]
- 42.Saccani, S., S. Pantano, and G. Natoli. 2001. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 193:1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, S. L. G., A. L. Pautz, and P. M. J. Burgers. 2001. ATP utilization by yeast replication factor C. IV. RFC ATP-binding mutants show defects in DNA replication, DNA repair, and checkpoint regulation. J. Biol. Chem. 276:34792-34800. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz, M. L., M. A. dos Santos Silva, H. Altmann, M. Czisch, T. A. Holak, and P. A. Baeuerle. 1994. Structural and functional analysis of the NF-κB p65 C terminus. J. Biol. Chem. 269:25613-25620. [PubMed] [Google Scholar]
- 45.Scully, R., S. F. Anderson, D. M. Chao, W. J. Wei, L. Y. Ye, R. A. Young, D. M. Livingston, and J. D. Parvin. 1997. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 94:5605-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seitz, C. S., Q. Lin, H. Deng, and P. A. Khavari. 1998. Alterations in NF-κB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-κB. Proc. Natl. Acad. Sci. USA 95:2307-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheehy, A. M., and M. S. Schlissel. 1999. Overexpression of RelA causes G1 arrest and apoptosis in a pro-B cell line. J. Biol. Chem. 274:8708-8716. [DOI] [PubMed] [Google Scholar]
- 48.Snowden, A. W., L. A. Anderson, G. A. Webster, and N. D. Perkins. 2000. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol. Cell. Biol. 20:2676-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 51.Webster, G. A., and N. D. Perkins. 1999. Transcriptional cross talk between NF-κB and p53. Mol. Cell. Biol. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, H. B., K. Somasundaram, Y. Peng, H. Tian, H. X. Zhang, D. K. Bi, B. L. Weber, and W. S. El-Deiry. 1998. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 16:1713-1721. [DOI] [PubMed] [Google Scholar]
- 53.Zhong, H. H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]