Abstract
The yeast MADS-box transcription factor Mcm1p plays an important regulatory role in several diverse cellular processes. In common with a subset of other MADS-box transcription factors, Mcm1p elicits substantial DNA bending. However, the role of protein-induced bending by MADS-box proteins in eukaryotic gene regulation is not understood. Here, we demonstrate an important role for Mcm1p-mediated DNA bending in determining local promoter architecture and permitting the formation of ternary transcription factor complexes. We constructed mutant mcm1 alleles that are defective in protein-induced bending. Defects in nuclear division, cell growth or viability, transcription, and gene expression were observed in these mutants. We identified one likely cause of the cell growth defects as the aberrant formation of the cell cycle-regulatory Fkh2p-Mcm1p complex. Microarray analysis confirmed the importance of Mcm1p-mediated DNA bending in maintaining correct gene expression profiles and revealed defects in Mcm1p-mediated repression of Ty elements and in the expression of the cell cycle-regulated YFR and CHS1 genes. Thus, we discovered an important role for DNA bending by MADS-box proteins in the formation and function of eukaryotic transcription factor complexes.
Members of the MADS-box family of transcription factors play pivotal roles in regulating key biological processes in a diverse range of eukaryotic organisms, including yeasts, plants, lower vertebrates, and mammals (reviewed in reference 30). In Saccharomyces cerevisiae, there are four MADS-box proteins, ArgRIp, Rlm1p, Smp1p, and Mcm1p. Mcm1p is involved in controlling several different processes, including arginine metabolism, mating type determination, cell type-specific gene expression (reviewed in references 10 and 30), and the cell cycle (15, 16, 19, 24, 43). In addition, Mcm1p plays an important role in controlling transcription via TY transposable elements (11, 41).
In order to achieve these diverse functions, Mcm1p interacts with a series of different coregulatory proteins in a promoter-specific manner (reviewed in reference 30). For example, mating type control is exerted by interacting with the activator protein α1 or the repressor protein α2, which act in α-cells to turn on α-cell- and turn off a-cell-specific genes, respectively (3, 14). More recently, the forkhead transcription factor Fkh2p has been shown to be a key partner for Mcm1p and forms the Mcm1p-SFF complex, which drives the expression of genes involved in the G2 and M phases of the cell cycle (15, 16, 24, 43).
A combination of biochemical and structural studies have uncovered the molecular mechanisms by which MADS-box transcription factors function. The MADS-box comprises the N-terminal part of the minimal DNA-binding domain of these proteins, and an additional C-terminal extension to the MADS-box is required for efficient dimerization (reviewed in reference 30). A series of biochemical studies (reviewed in reference 30) and the structures of SRF, MEF2A, and Mcm1p bound to DNA (13, 22, 26, 35) indicate that DNA binding is mediated by amino acids in the MADS-box, with an α-helix providing the key structural feature. In Mcm1p, additional DNA contacts are made by an N-terminal extension to this α-helix and residues in a loop between the two β-strands (35).
DNA bending by transcription factors is thought to be an important facet of their function and plays a role in both the DNA recognition process and determining the correct architecture of nucleoprotein complexes at promoters and enhancers (reviewed in reference 23). Mcm1p induces considerable DNA bending in its binding sites (33, 35) In contrast, other MADS-box proteins such as MEF2A, Rlm1p, and Smp1p induce minimal DNA bending (13, 26, 37, 39). DNA bending likely plays a key role in the regulation of gene expression by MADS-box proteins, although to date, little evidence has been gathered to support this hypothesis. In the case of Mcm1p, the bending propensity of a binding site correlates well with the ability of Mcm1p to correctly regulate transcription (1), but mutational analysis of Mcm1p failed to conclusively demonstrate a link between DNA bending and aberrant gene regulation (2).
The mechanism of DNA bending by MADS-box proteins appears to be evolutionarily conserved and is mediated by residues in the MADS box (39). Residues which are located at the N-terminal end of the DNA-binding α-helix and in the β-loop region play important roles in mediating the differential bending observed for different family members (37, 39). Further studies have identified a role for additional conserved MADS-box residues in contributing to the DNA bending mediated by SRF and Mcm1p (2, 39). The sequence of the binding site also plays a major role in dictating the degree of bending observed by a subset of MADS-box proteins, including Mcm1p and SQUA (1, 2, 38, 39).
In this study, we investigated the mechanisms of DNA bending employed by the yeast MADS-box protein Mcm1p and the functional consequences of reductions in DNA bending. We demonstrate that residues in the β-loop region and at the N-terminal end of the recognition helix play a key role in determining the magnitude of DNA bending. Mcm1p mutants that are defective in DNA bending were analyzed in vitro and in vivo. We demonstrate the importance of Mcm1p-mediated DNA bending in permitting the formation of ternary transcription factor complexes in vitro and for regulating transcription, normal cell growth and viability, and nuclear division and determining correct gene expression profiles in vivo.
MATERIALS AND METHODS
Plasmid constructions and mutagenesis.
The pBluescript-KS+-derived plasmids pAS37 (encoding METcoreSRF; amino acids 142 to 222) (27), and pAS704 (encoding coreMcm1; amino acids 1 to 98) (39) have been described previously. pAS1247 (encoding Fkh2p amino acids 254 to 458 fused to Flag and His tags) was constructed by inserting an NcoI- and XhoI-cleaved PCR fragment into the same sites in pAS798.
For coreMcm1 mutants, plasmids encoding single point mutants, pAS715 (R19A), pAS717 (K24E), pAS719 (K29E), pAS744 (K40A), pAS746 (T66E), and pAS748 (L68E), were constructed with fragments obtained from a two-step PCR protocol (29) with two flanking primers, FOR and REVL, the template pAS704, and the mutagenic primers ADS452, ADS453, ADS414, ADS500, ADS501, and ADS502, respectively.
Plasmids encoding double mutants, pAS716 (R19A/K29E), pAS718 (K24E/K29E), pAS743 (K29E/K38R), pAS747 (K29E/T66E), and pAS749 (K29E/L68E), were constructed with the template pAS719 and the mutagenic primers ADS452, ADS453, ADS499, ADS501, and ADS502, respectively. PCR products were cleaved with NcoI and XbaI and ligated into pAS37 cleaved with the same enzymes.
A series of Mcm1p mutants were generated with a mutagenic primer that allowed random amino acids to be inserted at T66 (ADS869) or L68 (ADS687). PCR products were cleaved with NcoI and XbaI and inserted into the same sites in pAS704. The following Mcm1p mutants were identified by sequencing: pAS1235 (L68D), pAS1237 (L68H), pAS1238 (L68M), pAS1239 (L68S), pAS1240 (L68Y), pAS1250 (T66A), pAS1601 (T66G), pAS1602 (T66L), pAS1603 (T66S), pAS1604 (T66V), and pAS1605 (T66W).
The following yeast expression vectors were created to express coreMcm1 derivatives: pAS797 (Mcm1 amino acids 1 to 98; also called pSL2190 [7]) and Mcm1(1-98), which is under the control of the MCM1 upstream promoter region in vector pRS314, a CEN/ARS-TRP1 yeast shuttle vector (31). The mutant Mcm1p clones pAS1210 (L68E), pAS1231 (K29E), and pAS1245 (T66E) were created by replacing the EcoRI/BglII fragment from pAS797 with identically cleaved PCR fragments from pAS748, pAS719, and pAS746, respectively.
The following bacterial expression vectors were created: pAS1223 (encoding MATα1 amino acids 1 to 176 fused to Flag and His tags) and pAS1232 (encoding MATα2 amino acids 1 to 211 fused to Flag and His tags) were created by inserting NcoI/XhoI fragments from pAS1216 and pAS1217, respectively, into the same sites in pETnef-PFH (42). The intermediary plasmids pAS1216 and pAS1217 were created by inserting NcoI- and XhoI-cleaved PCR fragments into pAS1209. pAS1752 (encoding Fkh2p amino acids 254 to 458 fused to glutathione S-transferase [GST]) was constructed by inserting an EcoRI- and XhoI-cleaved PCR fragment into the same sites in pGEXKG (12).
The circular permutation vectors pAS720 (SRET7), pAS722 (QPSTE3), pAS724 (QPPAL) (39), pAS76 (serum response element [SRE]) (28), and pAS152 (N10) (37) have been described previously.
The phasing vectors pAS525 to -529, which contain four phased A:T tracts and the N10 binding site, with distances between the center of the last A:T tract and the center of the N10 binding site of 30, 32, 35, 37, and 40 bp, respectively, have been described previously (39).
Details of PCR primers and mutagenic oligonucleotides can be supplied upon request. The sequences of all plasmids encoding mutant proteins and PCR-derived sequences were confirmed by automated or manual dideoxy sequencing.
Protein production and pulldown assays.
Wild-type and mutant Mcm1p proteins and Fkh2p derivatives were produced by coupled or sequential in vitro transcription and translation and subsequently analyzed and quantified as described previously (37). The polyhistidine tagged-proteins α1 and α2 were expressed in Escherichia coli BL21(DE3)/pLysS and purified by nickel affinity chromatography according to standard procedures (40). GST pulldown assays were carried out as described previously (29) with GST-Fkh2 and in vitro-translated Mcm1p derivatives.
Gel retardation, circular permutation, and phasing analysis.
In this study, we probed the mechanism of protein-induced DNA bending by Mcm1p and the functional consequences of loss of bending with the core DNA-binding domain of Mcm1p (amino acids 1 to 98). Yeast cells harboring this truncated Mcm1p protein are viable, and this core domain exhibits the majority of the molecular characteristics observed with the full-length protein (6, 9, 25). This core domain represents a globular part of the protein (35) (Fig. 1) and is thus less likely to generate anomalously migrating protein-DNA complexes in nondenaturing gels due to protein shape. Similar constructs have been used previously to analyze protein-induced DNA bending by other MADS-box proteins (37, 39).
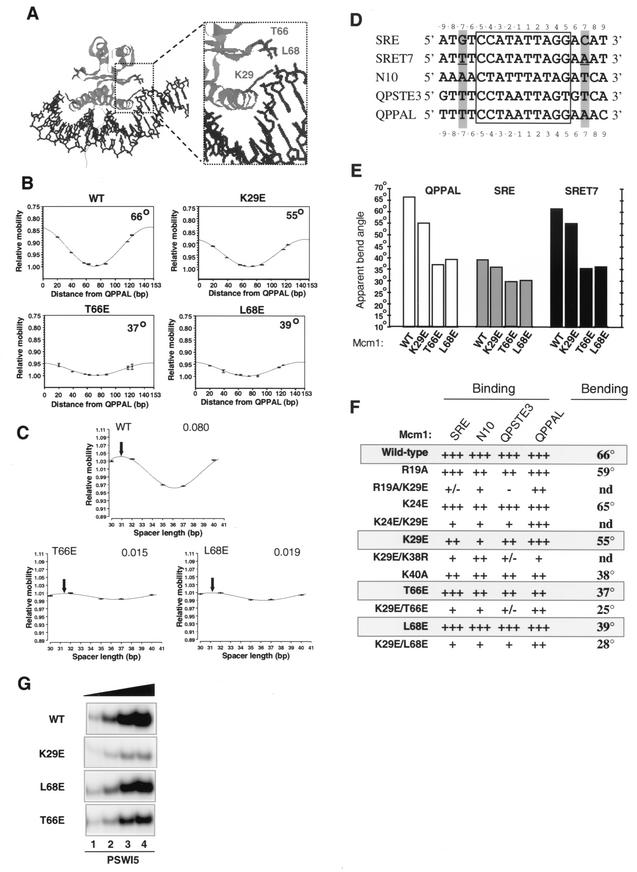
FIG.1.
T66E and L68E mutations abrogate Mcm1p-induced DNA bending. (A) Structural features of the Mcm1p-DNA complex (35). Ribbon representation of Mcm1p DNA-binding domain (cyan) bound to DNA (blue). The location of K29 at the end of the DNA-binding α-helix and T66 and L68 the end of the β-loop region are highlighted (residues are shown in red). (B) Circular permutation analysis of the indicated wild-type (WT) and mutant coreMcm1 proteins complexed with the QPPAL binding site. The data from each circular permutation experiment are fitted to a cosine function and shown graphically. The apparent bend angles calculated from these data are shown within each graph. R2 values for the curve fits were all ≥0.91. (C) Phasing analysis of wild-type and mutant coreMcm1p proteins bound to probes containing the N10 site. Data are shown fitted to a cosine phasing function, as described for Fig. 2. The spacer length where the protein-induced and intrinsic bends theoretically cooperate to generate the minimum mobility complex is indicated by a black arrow for each particular data set. The amplitudes of the phasing functions are presented for each data set. (D) Sequences of the core regions of the binding sites used in this study. The CArG box is boxed, residues at the −7 and +7 positions are highlighted, and the mutations introduced into SRET7 are underlined. (E) Graphic summary of apparent DNA bend angles observed in complexes between wild-type and mutant coreMcm1p proteins with the c-fos SRE, SRET7, and QPPAL sites calculated by circular permutation analysis. (F) Summary of DNA bend angles on the QPPAL site and binding affinities relative to wild-type Mcm1p on each of the c-fos SRE, N10, QPSTE3, and QPPAL sites. Relative binding affinities: +++, >70% wild-type value; ++, 10 to 70%; +, 1 to 10%; +/−, <1%; and −, not detectable. (G) Gel retardation analysis of increasing relative concentrations of the Mcm1p(1-98) mutants K29E, T66E, and L68E (1, lane 1; 3, lane 2; 10, lane 3; 20, lane 4) on the SWI5 promoter site.
Gel retardation assays were carried out essentially as described previously (37) on the c-fos SRE (29), the N10 site (28), the SRET7, QPSTE3, and QPPAL sites (39), the STE6 site (20), and the SWI5 site (34). The STE6 site was made by annealing ADS684 (5′-CTAGTCGACATGTAATTACCTAATAGGGAAATTTACACGCTCGA-3′) and ADS685 (5′-CTAGTCGAGCGTGTAAATTTCCCTATTAGGTAATTACATGTCGA-3′) (20). Mcm1p and α1, α2, and Fkh2 derivatives used in Fig. 1, 3, and 4 were made by in vitro transcription-translation or purified from E. coli (see above).
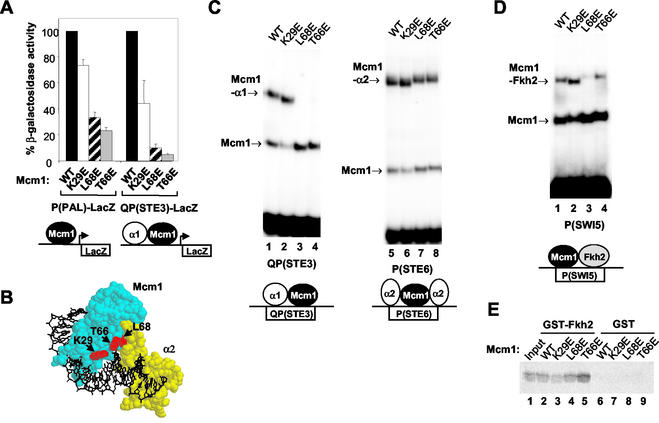
FIG. 3.
Ternary transcription factor complex formation by Mcm1p DNA-bending mutants. (A) Reporter gene assays. The activities of the indicated lacZ reporter genes were analyzed in strains harboring wild-type (WT) and mutant mcm1 alleles. The schematic shows the complexes which formed on the P(PAL) and QP(STE3) sites. (B) Spacefilling representation of the ternary Mcm1p-α2-DNA complex. Mcm1p (cyan) and α2 (yellow) are shown bound to DNA (black). The locations of Mcm1p residues K29, T66, and L68 are shown in red. Figures were prepared with RasMol version 2.6 based on the coordinates of the Mcm1p-α2-DNA complex (35). (C and D) Gel retardation analysis of the indicated Mcm1p proteins and bacterially expressed α1, bacterially expressed α2 (C), and in vitro-translated Fkh2p (D) on the QP(STE3) (lanes 1 to 4), P(STE6) (lanes 5 to 8), and P(SWI5) (lanes 1 to 4) sites, respectively. The amount of each Mcm1p protein was normalized to give equal amounts of DNA-binding activity on each particular site. Diagrams below each panel depict the complexes formed on each site. (E) Pulldown analysis of the indicated Mcm1p mutant proteins with GST and GST-Fkh2p(254-458); 10% input of wild-type Mcm1p is shown.
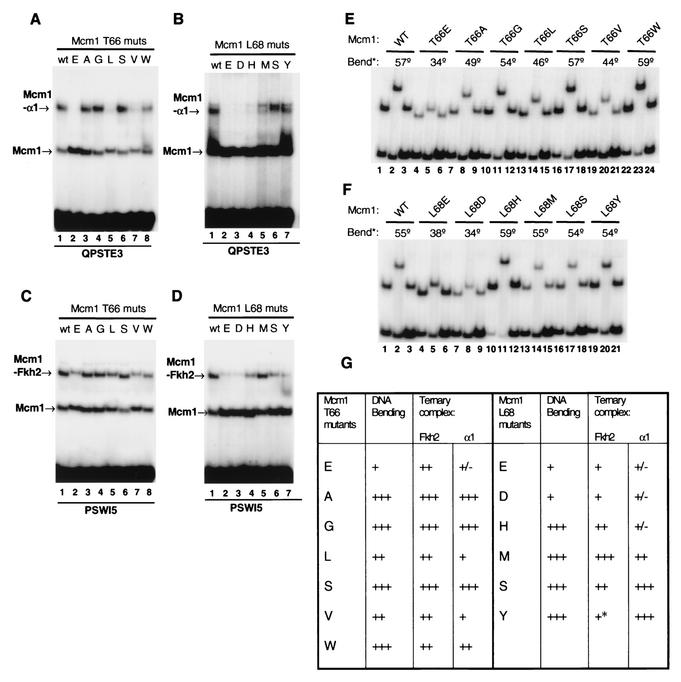
FIG. 4.
Role of positions T66 and L68 in mediating DNA bending and ternary complex formation. (A to D) Ternary complex formation with α1 (A and B) and Fkh2p (C and D) was monitored for a panel of T66 (A and C) and L68 (B and D) Mcm1p mutants on the PSWI5 and QPSTE3 binding sites. The amount of each Mcm1p protein was normalized to give equal amounts of DNA-binding activity on each particular site. (E and F) Three-point circular permutation assays of a series of mutated coreMcm1p proteins containing different substitutions at positions T66 (E) or L68 (F) in comparison to the wild-type (WT) protein. Binding sites that positioned the center of the QPPAL binding site either towards the middle (RV; lanes 2, 5, 8, 11, 14, 17, 20, and 23) or towards either end (Ml, lanes 1, 4, 7, 10, 13, 16, 19 and 22, and BH, lanes 3, 6, 9, 12, 15, 18, 21 and 24) of the DNA fragment were used. Relative bend angles (bend*) were calculated with an empirical equation rather than by curve fitting (36). (G) Summary of DNA bending (determined by three-point circular permutation analysis) and ternary complex-forming ability of Mcm1p mutants with Fkh2p. Relative DNA bending: +++, 49 to 59°; ++, 40 to 48°; and +, <39°. Relative binding affinities of Fkh2p in ternary complexes (compared to wild-type Mcm1): +++, >80%; ++, 35 to 60%; +, 10 to 25%; and ±, <5%. The asterisk indicates that dissociation of L68Y complexes can be seen during electrophoresis.
Yeast extracts for gel retardation analysis (Fig. 2E) were made as described previously (6). Relative DNA-binding affinities were calculated by phosphorimager analysis of DNA-protein complexes (FUJI BAS1500; TINA 2.08e software). Experiments were carried out to achieve ≤50% total DNA binding in protein-DNA complexes. Under these conditions, relative binding affinities within an experiment can be calculated by direct quantification of DNA-protein complexes. The scoring of these relative binding affinities is indicated in the figure legends.
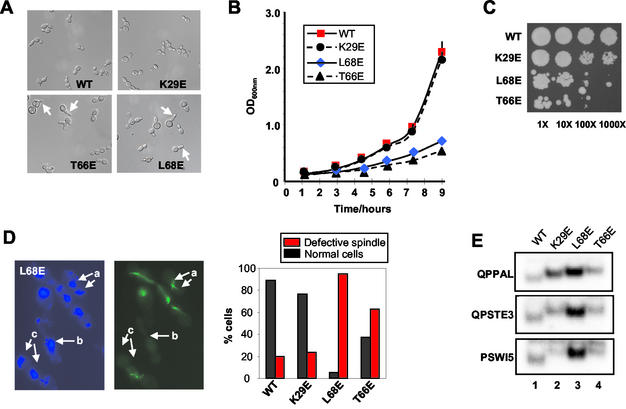
FIG. 2.
Growth and nuclear spindle defects of mcm1 DNA-bending mutants. Yeast strains harboring the indicated wild-type (WT) and mutant mcm1 expression constructs were analyzed. (A) Strain phenotypes. Arrows indicate elongated cells found in the mutant stains. (B) Growth curves. Each point represents the average of three independent experiments. (C) Viability tests. Serial dilutions of exponentially growing cultures (1- to 1,000-fold dilutions) of the indicated mutants on YPAD plates. (D) Spindle defects. The total proportion of cells exhibiting spindle defects in each of the indicated strains is shown graphically, and a representative field is shown for the L68E mutant (4′,6′-diamidino-2-phenylindole stain, left, and fluorescein isothiocyanate-antitubulin, right). Typical spindle defects are arrowed (a, elongated spindle; b, no spindle; c, broken spindle). (E) Gel retardation analysis of equal amounts of total cell extract on the indicated binding sites. Only the protein-DNA complexes are shown.
For circular permutation analysis, DNA fragments were produced by appropriate restriction enzyme digestion of PCR products derived from the vectors pAS76 (containing the c-fos SRE), pAS152 (N10), pAS720 (SRET7), pAS722 (QPSTE3), and pAS724 (QPPAL) and subsequently purified as described previously (28, 39). For details of the production of a full series of restriction fragments containing the circularly permuted sites used in Fig. 1, see West and Sharrocks (39). Circular permutation analysis was carried out on 5% polyacrylamide gels cast in 1× Tris-borate-EDTA (TBE).
Curve fitting and apparent DNA bend angles were calculated as described previously (37). Bend angles are not absolute but are given with respect to that of wild-type Mcm1p under the conditions used and are referred to as apparent bend angles. Bend angle values are quoted as the average of three independent experiments. Error bars shown on the graphs represent standard deviations from three independent experiments and in most cases are barely visible due to the high reproducibility of the results. Standard deviations (n − 1) of bend angles are in the range from 0.5 to 1.6°. Estimated relative bend angles were calculated from three-point bending assays with the simplified equation μM/μE = cosα/2, where μM and μE are the relative mobilities of complexes on fragments where the center of the binding site is located at the middle or at the end, respectively, and α is the bend angle (36). These bend angles are typically lower in this assay than from complete circular permutation assays, as curve fitting permits theoretical maxima and minima to be extrapolated beyond the values obtained from the measurable parameters (where the center of the binding site cannot be located at the end of the fragments).
For phasing analysis, DNA fragments were produced by PCR amplification of the inserts from pAS525 to -529 with the primer pair ADS262/ADS346. The resulting probes contain the N10 binding site with its center located between 30 and 40 bp from the center of the last A:T tract. The fragment ends are 71 bp and 75 bp from the center of the N10 site and the last A:T tract, respectively, and the magnitude of the intrinsic bend (72°) is identical to that observed for Mcm1p (35). These parameters were optimized for this study as described previously (39). Binding sites were purified and phasing experiments were carried out as described for the circular permutation experiments. Data are shown fitted to a cosine function as described previously for circular permutation analysis (28).
All figures were generated electronically from either phosphorimager files or scanned images of autoradiographic images with Picture Publisher (Micrografx) or Adobe PhotoDeluxe (Adobe) and Powerpoint (Microsoft) software. Final images are representative of the original autoradiographic images.
Yeast strain generation, growth and viability assays, spindle defects, and reporter gene assays.
The yeast reporter strains were derived from YY1901 [MATα P(PAL)-lacZ::fus1/pSL1574], YY2049 [MATα QP(STE3)-lacZ::fus1/pSL1574], and YY2052 [MATa P(PAL)-lacZ::fus1/pSL1574], where pSL1574 is a plasmid which contains MCM1 on a CEN/ARS-URA3 vector (7). Plasmids pAS797 (also called pSL2190 [7]), pAS1210, pAS1231, and pAS1245, expressing wild type Mcm1(1-98) or the L68E, K29E, or T66E mutant, respectively, were introduced into each of the reporter strains, and pSL1574 was lost by the 5-fluoroorotic acid plasmid shuffle technique (4). Yeast extracts for reporter gene assays were prepared according to the method described previously (6), and β-galactosidase activity was measured with Galacton plus (Tropix).
To analyze growth rates, yeast cultures were grown to 0.3 to 0.9 optical density at 600 nm (OD600) units and diluted to 0.1 to 0.3 OD600 units in YPAD. Cultures were incubated at 30°C; three samples of each culture were taken at the times indicated, and their OD600 was measured. Cell viability was determined by carrying out spot tests. Yeast cultures were grown to 0.3 to 0.9 OD600 and diluted to 0.1 OD600 in YPAD. Then 10 μl each of the undiluted culture and 10-fold, 100-fold, and 1,000-fold dilutions were spotted on YPAD agar and incubated at 30°C for 2 days.
Spindle defects were analyzed as described previously (39).
Microarray analysis.
YY2052-derived yeast strains containing wild-type or mutant Mcm1(1-98) alleles were grown in batch or chemostat cultures on carbon-limiting minimal medium. Cells were harvested and disrupted with a Mikro-dismembrator. RNA was extracted from the disrupted cells with the Trizol reagent (Gibco-BRL), chloroform extracted twice, and precipitated in isopropanol. cDNA probes labeled with indocarbocyanine and indodicarbocyanine were generated by reverse transcribing 60 μg of total RNA with Superscript II (Gibco-BRL) reverse transcriptase in the presence of oligo(dT)20 primer and indocarbocyanine- or indodicarbocyanine-dUTP (5-amino-propargyl-2′-deoxyuridine 5′-triphosphate; Amersham Pharmacia Biotech), and purified with the Qiaquick PCR purification kit (Qiagen).
Microarray hybridizations were performed as described on the Consortium for Functional Genomics of Microbial Eukaryotes (COGEME) website (http://www.cogeme.man.ac.uk). Qualitative assessment of incorporation of fluorescent label was achieved by agarose gel electrophoresis of the targets, followed by fluorescent scanning of the gel. During the scanning of the hybridized arrays, the photo-multiplier tube voltages for the GenePix 4000A scanner were independently adjusted to achieve visually equivalent levels of fluorescent signal between the two channels. Data for the L68E mutant represent the averages of five experiments, whereas the T66E values are the averages of duplicate experiments.
RESULTS
Identification of residues in Mcmlp involved in DNA bending.
In order to study the functional consequences of Mcm1p-induced DNA bending, we first attempted to create mutant Mcm1p proteins that were defective in protein-induced DNA bending but were still able to bind DNA efficiently. Mcmlp and SRF both induce substantial DNA bending. However, unlike SRF, Mcm1p bends DNA in a site-dependent manner. This suggests that the residues responsible for dictating protein-induced DNA bending may differ between these proteins. Based on our results with SRF (37, 39), the roles of residues at positions 14, 51, and 53 in the MADS-box of Mcm1p (K29, T66, and L68; Fig. 1A) were investigated in detail. Each of these residues was mutated either individually or in combination to negatively charged glutamate residues to disrupt protein-DNA contacts. In addition, further residues in Mcmlp were mutated due to either their potential or proposed interactions with the DNA backbone (K38 and K40 [35]) or differences with residues at identical positions in SRF (R19 and K24). In the case of R19, K38, and K40, neutral or conservative substitutions were employed because the introduction of negatively charged residues at these positions would be predicted to severely affect DNA-binding affinity.
The ability of the wild-type and mutant Mcmlp proteins to bend DNA was analyzed by circular permutation analysis on the QPPAL site (Fig. 1B, summarized in Fig. 1F). DNA bending by the mutant proteins T66E and L68E was severely compromised, whereas a small reduction was observed for K29E (Fig. 1B). Of the other mutants, DNA bending by K24E was virtually identical to that by the wild-type protein, whereas a small reduction in DNA bending was observed for R19A. However, considerable reductions were observed for K40A. Double mutations of K29E in addition to either T66E or L68E led to further decreases in DNA bending by Mcmlp (Fig. 1F).
Phasing analysis was used to confirm that the T66E and L68E mutant proteins resulted in a reduction in Mcmlp-induced DNA bending. In comparison to wild-type Mcmlp, the amplitudes of the phasing function were significantly reduced with mutant proteins (Fig. 1C), further demonstrating the effect of these mutations in neutralizing DNA bending induced by Mcmlp.
A DNA-binding site dependence for Mcm1p-mediated DNA bending has been determined (1, 39). In order to investigate this phenomenon further, protein-induced bending was analyzed on a panel of CA/T-rich G (CArG) box-containing sites (Fig. 1D and E). Mcmlp induced significant DNA bending of the QPPAL and QPSTE3 sites, but considerably reduced DNA bending was observed on the SRE and N10 sites (Fig. 1B and data not shown). Differential DNA bending of synthetic binding sites by Mcmlp was previously attributed to the presence of a thymidine residue at the −7 position and an adenine residue at the symmetrically related +7 position which enhanced DNA bending (1). The QPPAL site contains both of these bases, the QPSTE3 site contains one of these bases, but the SRE and N10 sites lack appropriately positioned A:T base pairs.
In order to examine whether bending of the SRE sites by Mcmlp could be enhanced, a mutant site was produced with symmetrically related A:T base pairs at positions −7 and +7 (Fig. 1D). As predicted from this model, DNA bending by Mcmlp was significantly enhanced by introducing these base pairs (SRET7; Fig. 1E). Thus, Mcmlp induces DNA bending in a sequence-dependent manner, and the base pairs at positions −7 and +7 in its binding sites play a major role in dictating the magnitude of the resulting bends.
In order to probe the molecular mechanism underlying the site-dependent DNA bending by Mcm1p, we tested the ability of a series of mutant Mcm1p proteins to bend the QPPAL, SRE, and SRET7 sites by circular permutation analysis (Fig. 1E). DNA bending on the QPPAL site varied greatly between mutants, whereas little difference was seen with the same mutants on the SRE (Fig. 1E). A notable difference between the QPPAL site and the SRE is the lack of symmetrically oriented A:T base pairs at the −7 and +7 positions. We therefore tested binding to SRET7 and found that the wild-type protein and the K29E mutant induced significantly more bending on this site than the T66E and L68E mutants (Fig. 4B). This closely mirrors the situation on the QPPAL site. Thus, T66 and L68 are important for sensing the presence of an A:T base pair at the −7 and +7 positions.
In order to detect whether the changes in DNA bending were accompanied by changes in DNA-binding affinity, the ability of the panel of Mcm1p mutants to bind to a series of different binding sites was tested (Fig. 1G and summarized in Fig. 1F). Mutation of K29 caused only a moderate reduction in DNA binding at most sites but virtually abolished binding to the N10 site. Mutation of T66 and L68 had little effect on the efficiency or specificity of Mcmlp binding. However, the K29E mutation in combination with either T66E or L68E caused a general reduction in DNA-binding affinity, whereas in combination with K24E, a clear specificity change was elicited. The K24E/K29E double mutant protein retained high-affinity binding to the QPPAL site, but binding to the other sites tested was severely reduced (Fig. 1F). Mutation of residues R19 and K24 had little effect on the binding affinity of Mcmlp for the four binding sites. However, mutations at positions K38 (in combination with K29E) and, to a lesser extent, K40 caused a reduction in DNA-binding affinity to all the sites tested (Fig. 1F).
We also tested the binding of Mcm1p K29, T66, and L68 mutants on the PSWI5 site, as this became the focus of ensuing experiments (see below). Of these mutants, K29E exhibited the weakest binding affinity (Fig. 1G, second panel). The two DNA bending-defective mutants T66E and L68E exhibited slightly reduced affinity in comparison to the wild-type protein but much higher binding affinity than the K29E mutant (Fig. 1G).
In summary, the Mcm1p mutants T66E and L68E show much reduced protein-induced DNA bending while retaining nearly wild-type DNA-binding efficiencies. In contrast, the K29E mutant exhibited much reduced DNA-binding affinities to a range of sites but nearly normal DNA bending. The K29E mutant version of Mcmlp could therefore be used as a control in subsequent experiments to rule out effects due to reduced DNA-binding affinity. Thus, together, these mutant proteins represent useful tools with which to study the functional consequences of abrogating Mcm1p-mediated DNA bending. This differential DNA bending occurs at sites containing palindromically orientated A:T base pairs at positions −7 and +7. Other residues such as K40 were also identified as important mediators of DNA bending but exhibited more substantial reductions in binding efficiency.
Mcm1p DNA-bending mutants cause transcriptional and growth defects in vivo.
To study the phenotypic consequences of inhibiting DNA bending by Mcm1p, yeast strains that harbored K29E, T66E, and L68E mutant mcm1 alleles in place of the wild-type MCM1 gene were constructed. First, we examined the effect of Mcm1p mutant proteins on cellular morphology. Morphological abnormalities such as budding defects, including elongated and misshapen buds, were observed in all the mutant strains but were most pronounced in the T66E and L68E mutants (Fig. 2A). Such defects suggest that these mcm1 mutations affect the cell cycle. We therefore analyzed the growth rate and viability of the different strains. Both the T66E and L68E mutant strains exhibited greatly reduced growth rates and reduced viability (Fig. 2B and C), while the K29E mutant grew at a rate similar to the wild-type rate and appeared to have only a slight increase in the number of inviable cells.
Defects in the Fkh2p component of the Mcm1p-Fkh2p complex have been shown to affect spindle morphology and nuclear division (24). Hence, we also probed for possible defects in nuclear division and the mitotic spindle that might arise from Mcm1p mutations. Both the T66E and L68E mutants demonstrated defects in nuclear migration and division (Fig. 2D, a and c, and data not shown). In addition, multiple spindle defects were detected in the T66E and L68E strains (Fig. 2D). Common defects included elongated spindles, broken spindles, and the absence of a spindle (illustrated for the L68E strain in Fig. 2D). The defects in the L68E strain were the most severe. Indeed, no normal spindle morphology was detected in the L68E mutant cells examined in the S and G2 phases of the cell cycle compared to the wild type (>50%; data not shown). Moreover, although the defects in the T66E mutant were less severe, microscopic analysis indicated that the percentages of cells with normal spindle morphology in the S and G2 phases of the cell cycle were greatly reduced compared to the wild-type cells (data not shown).
The amount of active Mcm1p contained in each strain was analyzed by gel retardation analysis to determine whether alterations in Mcm1p levels might account for the altered phenotypes that we observed. We tested their relative binding efficiencies in vitro and saw little difference (Fig. 1). However, to confirm that equal amounts of binding activity were present in each strain, we tested total cell extracts on a series of different Mcm1p binding sites (Fig. 2E). In comparison to the wild-type protein, each strain contained equal or enhanced Mcm1p DNA-binding activity. The enhanced binding activity observed in several cases, which was especially apparent for L68E, suggests that its expression is upregulated to compensate for the defects introduced by the mutation. A decrease in Mcm1p DNA-binding activity is therefore not what underlies the phenotypic defects that we observed.
Taken together, these data demonstrate a strong correlation between the severity of DNA-bending defects and the severity of the phenotypes introduced into strains harboring mutant mcm1 alleles. Strains harboring the mild Mcm1p bending mutation K29E showed mild defects, while the severe DNA-bending Mcm1p mutations T66E and L68E exhibited serious phenotypic defects.
Mcmlp DNA-bending mutants are defective in complex formation with coregulatory proteins.
Mcm1p participates in the activation and repression of several different promoters in conjunction with coregulatory proteins. To probe for potential transcriptional defects, we tested the ability of the mutant Mcm1p proteins to regulate one such promoter with the QP(STE3)-lacZ reporter, which Mcm1p activates in conjunction with α1. In order to assess the effect of Mcm1p in the absence of other DNA-bound coregulatory proteins, we also used the artificial P(PAL)-lacZ reporter strain, which lacks flanking binding sites (Fig. 3A) (7).
The absolute activity of the P(PAL)-lacZ reporter was approximately threefold higher than that of the QP(STE3)-driven reporter in cells harboring wild-type Mcm1(1-98) proteins. In comparison to the wild-type protein, Mcm1p (K29E) exhibited moderately reduced activation properties, whereas both the L68E and T66E mutant proteins showed severely abrogated transactivating ability on these reporters (Fig. 3A). Problems in DNA bending were clearly correlated with reduced Mcm1p activity, although the loss of transcriptional activity was more apparent on the QP(STE3)-lacZ reporter gene, suggesting that DNA bending might be more important in the context for ternary complexes involving Mcm1p.
One possible consequence of defects in DNA bending would be an inability to form ternary DNA-bound complexes with coregulatory proteins due to alterations in local promoter architecture and resulting disruptions in the spatial positioning of partner proteins. Alternatively, interactions might be disrupted by alterations to the interaction interfaces. For example, the structure of the Mcm1p-α2 complex (35) indicates that both T66 and L68 are located close to the Mcm1p-α2 interaction interface (Fig. 3B). We therefore tested the ability of the DNA-bending mutants K29E, T66E, and L68E to form complexes with several coregulatory proteins.
Initially, we looked for defects in the mating pathways, which are in part regulated by Mcm1p-containing complexes. Defects were observed in the mating pathway, as MATa cell-types containing the T66E and L68E mutant alleles were arrested less efficiently than wild-type cells by α-factor (data not shown). The transcription factors α1 and α2 regulate mating type determination in combination with Mcm1p. First, we examined complex formation between Mcm1p and α1, as Mcm1p bending mutants exhibited greater transcriptional defects on composite α1-Mcm1p elements than on a simple Mcm1p-regulated promoter (Fig. 3A). In contrast to the wild-type and K29E proteins, the L68E and T66E mutant proteins were unable to form complexes with α1 (Fig. 3C, lanes 1 to 4). However, all four proteins were able to form ternary complexes with α2 (Fig. 3C, lanes 5 to 8), demonstrating that not all complexes were affected by these mutations.
The growth defects that we observed were unlikely to arise from alterations in Mcm1p interactions with α1 and α2 in the mating type pathway. We therefore focused on Fkh2p, a different partner protein, which plays an important role in cell cycle control (15, 16, 24, 43). Indeed, the spindle defect phenotypes observed with the Mcm1p mutants T66E and L68E were reminiscent of those observed in an Fkh2Δ mutant, suggesting that these mutations may affect the formation of Fkh2p-Mcm1p complexes. Complex formation between Fkh2p and the L68E mutant Mcm1p was severely reduced (Fig. 3D, lane 3). The efficiency of Mcm1p-Fkh2p complex formation was also reduced in the T66E mutant, although the defect was not as severe as with the L68E mutant (Fig. 3D, lane 4).
To establish whether the disruption of ternary complex formation was due to changes in DNA bending or to alterations in protein-protein interactions, we carried out pulldown analysis with a GST-Fkh2p fusion and in vitro-translated Mcm1p derivatives. The DNA bending-defective mutants T66E and L68E bound to Fkh2p with an efficiency equivalent to or greater than that of the wild-type protein (Fig. 3E, lanes 4 and 5), indicating that protein-protein interactions were unaffected by these mutations in the absence of DNA. Importantly, another mutant Mcm1p protein that affected Mcm1p-Fkh2p complex formation, V69E, showed clear reductions in interactions with Fkh2p with this assay (A. D. Sharrocks, unpublished data), thus validating the use of this assay to detect changes in protein-protein interactions.
Collectively, these data indicate that the DNA bending-defective T66E and L68E mutants exhibit reduced ternary DNA-bound complex formation with α1 and Fkh2p. The defects in ternary complex formation with Fkh2p are not due to reductions in protein-protein interactions and hence likely result directly from changes in local DNA architecture. Such alterations in Mcm1p-Fkh2p complex formation are also consistent with the growth and nuclear division defects observed in the T66E and L68E mcm1 mutants.
Role of T66 and L68 in β-loop of Mcmlp in DNA bending.
The insertion of negatively charged residues in the β-loop of Mcm1p caused a severe reduction in bending induced by this protein. In addition, these insertions could also affect ternary complex formation with coregulatory proteins (Fig. 3). To investigate whether it was the nature of the mutated residue or the ability of the protein to bend DNA that was affecting complex formation, random mutagenesis was used to further examine the role of β-loop residues T66 and T68 in regulating DNA bending and complex formation.
The resulting Mcm1p mutants were tested for their ability to form ternary complexes with α1 (Fig. 4A and B) and Fkh2p (Fig. 4C and D) and compared with their ability to bend DNA (Fig. 4E and F; summarized in Fig. 4G). Ternary complex formation with both α1 and Fkh2p was reduced upon the introduction of E, L, V, and, to a lesser degree, W into position 66 (Fig. 4A and C). Large reductions in complex formation were observed upon the introduction of E or D at position 68 with both α1 and Fkh2p (Fig. 4B and D). However, more moderate reductions in Mcm1p-Fkh2p complexes were observed upon the introduction of H, S, or Y into position 68 in Mcm1p (Fig. 4D, lanes 4, 6, and 7). Differences were observed in complex formation with α1, with the introduction of S at this position having no effect, H having a very severe effect, and M a moderate effect on ternary complex formation (Fig. 4B, lanes 4 to 6).
To investigate how these effects on ternary complex formation correlate with DNA bending defects, a three-point circular permutation assay was used to compare the relative bending induced by mutant coreMcm1p proteins. Here, the difference in relative mobility of complexes formed with a centrally and terminally located binding site can be used to calculate an approximate relative bend angle (36). In addition to the T66E mutation, the introduction of either A, L, or V led to reductions in bending by Mcm1p, indicating that the introduction of a hydrophobic side chain at this position is also detrimental to bending (Fig. 4E). However, none of these mutations caused as severe an effect on DNA bending as T66E. The introduction of the conservative S or G and W had little effect on DNA bending (Fig. 4E; summarized in Fig. 4G). Position 68 was less sensitive to mutations (Fig. 4F), and virtually wild-type bending was observed upon the introduction of either an H, M, S, or Y residue. However, the introduction of a negatively charged D residue at this position severely reduced DNA bending. The proximity of T66 to the DNA likely explains why less extreme mutations at this position can also have effects on DNA bending, whereas L68 is located further from the DNA (Fig. 1) (35).
The severity of the reductions observed in ternary complex formation of the Mcm1p T66 mutants with either α1 or Fkh2p and correlated well with reductions in DNA bending caused by these mutations (Fig. 4G). At the L68 position, the correlation was not as tight, but the weakest binding of the ternary complex corresponded to the mutants with the biggest defects in DNA bending. However, some mutations that do not affect DNA bending still affected complex formation, albeit to a lesser extent than those which disrupted DNA bending. Thus, effects in the local structure and/or protein-protein interactions with coregulatory partners are also likely to contribute to the loss of interactions seen with this class of mutants. However, overall, these data are consistent with the hypothesis that the loss of protein-induced DNA bending is responsible for the defects in the formation of ternary transcription factor complexes that we observed.
Changes in gene expression profiles in strains harboring T66E or L68E Mcm1p mutant.
The above data indicate that loss of DNA bending contributes to the altered regulation of artificial reporter genes by Mcm1p (Fig. 3). Defects in ternary transcription factor complex formation with Mcm1p imply that gene expression profiles will be altered in strains harboring defective mcm1 alleles. Hence, microarray analysis was used to assess the global changes in gene expression caused by DNA-bending-defective mcm1 mutants.
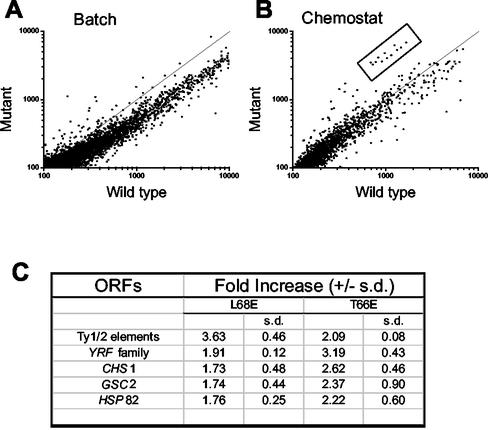
Initial experiments with batch cultures suggested the misregulation (>2-fold) of many (ca. 960) different genes in the L68E mutant strain (Fig. 5A). However, many of these changes may be secondary consequences of the reduction in growth rate. This was especially likely because the genes showing the greatest degree of downregulation were those encoding ribosomal proteins. In order to focus on the primary consequences of the mcm1 mutations, mutant and wild-type cells were grown in chemostat cultures in which their growth rates were equalized. Under these conditions, many fewer genes (ca. 103) were misregulated (Fig. 5A and B). Further repeat experiments resolved these into 28 misregulated genes. Interestingly, Ty elements figured prominently among the upregulated genes (see website http://www.cogeme.man.ac.uk). Mcm1p has previously been implicated in repressing retrotransposon gene expression in the absence of its known accessory proteins (11, 41).
FIG. 5.
Microarray analysis of altered gene expression profiles in DNA-bending mutants. (A and B) Relative gene expression profiles of the L68E mutant versus wild-type strain grown in batch (A) and chemostat (B) cultures. The boxed area indicates a group of upregulated genes revealed by analysis of chemostat cultures. (C) Table of genes that are commonly upregulated in the T66E and L68E mutant strains.
We also analyzed the changes in global gene expression in the T66E mutant. It is likely that by comparing the genes deregulated in strains containing different DNA-bending mutant versions of Mcm1p, genes whose expression is critically dependent upon Mcm1p-induced DNA bending will be identified. A comparison of the gene expression profiles of the T66E and L68E mutants revealed an overlap in deregulated genes and gene families (Fig. 5C). Of the 50 genes upregulated by >1.77-fold in the L68E mutant strain, 17 were also among the 50 most upregulated genes in the T66E mutant (the complete datasets are available at the COGEME website [http://www.cogeme.man.ac.uk]). Among these were numerous Ty elements, indicating that DNA bending by Mcm1p likely plays a key role in maintaining these elements in a repressed state.
In addition, it was found that the YRF genes (a telomere-associated gene family encoding ATP-dependent DNA helicases [17]) were upregulated in both mutants, the effect being particularly marked in the T66E mutant. Finally, a significant effect was seen on the expression of three single-copy genes, two of which, CHS1 and GSC2, are involved in cell wall biogenesis (8, 18). Both the YRF genes and CHS1 are under cell cycle control (34), and both are bound by the cell cycle regulator Swi5p in vivo (32). As SWI5 is a direct target of the Fkh2p-Mcm1p complex, this provides a direct link to the molecular defects that we observed.
DISCUSSION
The role of transcription factor-induced DNA bending in regulating eukaryotic gene expression is still poorly understood. However, roles in the DNA recognition process and in determining the correct architecture of nucleoprotein complexes at promoters and enhancers have been shown (reviewed in reference 23). In common with several MADS-box transcription factors, Mcm1p induces DNA bending into its recognition site (33, 35). Here, we probed the mechanism of DNA bending and showed that it exhibits both similarities to and differences from other MADS-box proteins.
By using mutant Mcm1p proteins in vitro and strains harboring mutant mcm1 alleles that encode DNA bending-defective proteins, we demonstrated an important role for DNA bending in transcription factor complex assembly and downstream gene activation. These defects in gene expression led to malfunctional nuclear division and reduced cell growth. Importantly, we used the K29E protein, which retained substantial DNA bending but exhibited a significant decrease in DNA-binding affinity (Fig. 1) and showed nearly normal Mcm1p function both in vitro and in vivo. This rules out the possibility that small decreases in DNA-binding affinity in the DNA-bending mutants T66E and L68E gave rise to the phenotypic defects that we observed. Thus, Mcm1p-mediated DNA bending is an important facet of its function.
Role of Mcm1p-mediated DNA bending in transcription factor complex formation and gene expression.
One function of DNA bending by MADS-box proteins appears in some cases to be to provide additional specificity determinants (see below). However, a major role for DNA bending is thought to be to correctly juxtapose transcription factors located on neighboring binding sites. Mcm1p forms complexes with numerous other transcription factors in a promoter-dependent manner, including MATα1, MATα2, Ste12p, and Fkh2p (reviewed in references 5, 10, and 30). Structural studies indicate that DNA bending might be essential for the function of Mcm1p in complexes with MATα2 on the STE6 promoter (35).
The propensity of a site to bend correlates with the ability of Mcm1p to activate transcription and potentially for MATα2 to repress transcription via DNA-bound Mcm1p in vivo, although in the latter case, a potential effect on DNA binding by MATα2 was not ruled out (1). However, strict correlations between DNA bending and transcriptional repression through MATα2 could not be deduced from studies on mutant mcm1 alleles (2). Indeed, from our studies, none of the mutant Mcm1p proteins that we have investigated significantly affected the formation of ternary Mcm1p-α2 complexes irrespective of the degree of bending elicited (Fig. 3C and data not shown). This suggests that formation of this complex is relatively flexible in its DNA-bending requirements. However, Mcm1p-induced DNA bending is an important determinant in the formation of other nucleoprotein complexes involving α1 and Fkh2p (Fig. 3 and 4). This is particularly apparent for mutations in Mcm1p at the T66 position, for which loss of DNA bending correlated with a loss in complex formation.
Interestingly, at the T66 position, the T66A and T66L mutants, which bent DNA by 49° and 46°, respectively, exhibited different amounts of ternary complex formation with α1 and Fkh2p (Fig. 4). This suggests that there might be a threshold for DNA bending below which complex formation is inhibited. This is consistent with our hypothesis that DNA bending affects transcription factor complex topology, as a point will be reached where the proteins can no longer interact and form a stable complex without imparting additional strain on the DNA duplex.
The observation that Mcm1p-Fkh2p complex formation is abrogated in DNA-bending mutants is consistent with the defects in the growth rate of yeast cells harboring the T66E and L68E mutant alleles (Fig. 2) (2). fkh2 null mutant strains also exhibit growth defects due to effects on the expression of genes whose products are involved in the G2 and M phases of the cell cycle (15, 16, 24, 43). Moreover, fkh2 null strains exhibit spindle defects that are reminiscent of those linked with the mcm1 T66E and L68E alleles (24), although the severity of the defects are more akin to those of the fkh2-fkh1 double mutant. The Mcm1p and Fkh2p binding sites exhibit a strict spacing requirement to permit efficient ternary complex formation (J. Boros and A. D. Sharrocks, unpublished data); thus, alterations in bending and juxtapositioning of sites would be predicted to affect complex formation.
Interestingly, all of the composite Mcm1p-Fkh2p binding elements identified by global chromatin precipitation analysis contain A:T base pairs at the +7 position relative to the Mcm1p site that precedes the Fkh2p binding sites (e.g., in the SWI5 promoter) (32). This base pair is a critical determinant of Mcm1p-mediated DNA bending (Fig. 1) (1, 39) and further underscores the likely importance of DNA bending at these sites.
Microarray analysis of altered gene expression profiles in the T66E and L68E strains identified five classes of genes that are upregulated in both strains and hence most likely due to a common defect in these strains, i.e., Mcm1p-mediated DNA bending defects. None of the misregulated genes were identified as direct Mcm1p targets in a genomic chromatin immunoprecipitation analysis (32). Further analysis of the regions immediately upstream from the open reading frames did not reveal any obvious Mcm1p binding sites (with the exception of the TY elements; see below). However, of these identified genes, the single-copy CHS1 and multiple YRF genes are under cell cycle control and are bound in vivo by the cell cycle regulator Swi5p (32). SWI5 is itself regulated by Mcm1p-Fkh2p complexes (15, 16, 24, 43). In fkh2 null strains, the basal level of SWI5 expression rises slightly but its cell cycle regulation is lost, indicating a role for Fkh2p in repressing basal expression as well as cell cycle-dependent activation. This effect is more pronounced in fkh1/fkh2 strains (16, 24). This rise in basal levels might also be achieved by disruption of Mcm1p-Fkh2p complexes in the mcm1 mutant strains that we have analyzed and lead to enhanced SWI5 expression and hence effects on CHS1 and YRF genes. This in turn implies that Mcm1p may in fact be a critical player in the nucleation of higher-order promoter-bound complexes containing both Fkh1p and Fkh2p. Loss of Mcm1p-Fkh2p interactions might then be propagated throughout the promoter. In the unsynchronized chemostat cultures, changes in SWI5 expression were not elevated above the thresholds that we set, but it might be the loss of periodicity rather than overall steady-state levels that leads to changes in downstream gene expression.
The continuous chemostat culture used, while essential for uncovering changes in gene expression from general “noise,” probably negates finding direct targets involved in cell cycle regulation. Attempts to synchronize cells with α-factor failed due to defects in the mating pathway. Thus, we were unable to analyze gene expression defects in the cell cycle-regulated CLB2 cluster and M/G1 genes or in the mating pathway. However, our novel application of microarray analysis with a molecularly defined genetic system and a controlled growth environment is likely to be of general significance in dissecting primary from secondary effects on the control of gene transcription in different complex systems.
In addition to the overlap in the alterations of gene expression, other changes appeared to be unique to each mcm1 mutant. This suggests that the mutations might also affect specific interactions of Mcm1p either with coregulators directly or at specific promoters where DNA bending might have alternative effects depending on the specific mutations. Indeed, the degree of DNA bending elicited by the T66E and L68E mutants was not identical (Fig. 1). Furthermore, the specificity of the Mcm1p mutations is also reflected in differences in the observed nuclear and spindle defects (Fig. 2 and data not shown). For example, the defects in the L68E mutant were more severe than in the T66E strain. For example, in the L68E strain, very few cells were normal and large numbers of cells showed no spindle (14%) or elongated spindles with fragmented nuclei (16%) compared to 2% and 6%, respectively, in the T66E mutant. Thus, common phenotypes are present that most likely result from the common DNA-bending defects, while unique features were observed that are mutation specific.
Further inspection of the commonly deregulated genes identified the Ty elements. These genes are bound by Mcm1p (11) and are repressed through the action of Spt13 (41). The role of Mcm1p-mediated DNA bending is unclear in this context but could be involved in the formation of higher-order enhanceosome-like complexes that permit other repressors such as a1-α2 complexes to be recruited to the Ty elements (11). Links between Mcm1p and GSC2 and HSP82 upregulation are likely to be indirect, through deregulation of other transcription factors and signaling molecules, as these genes are not known to be direct targets of either Mcm1p or regulators encoded by Mcm1p target genes. Mcm1p also plays a role in regulating arginine metabolism in complex with ARGRI to -III (21). However, no significant changes in the expression of genes associated with arginine metabolism and catabolism, such as ARG1 and ARG3 and CAR2, were observed in the mutant strains, probably due to maintenance of constant nutrient levels in the chemostat cultures.
Mechanisms of DNA bending.
The structure of the Mcm1p-α2-DNA complex has provided important clues to the mechanism by which Mcm1p mediates protein-induced DNA bending (35). For example V34 and K38 have been suggested to contribute DNA bending around Thy +7, and S37 and T66 have been proposed to stabilize this bend. Several mutagenic studies on other MADS-box proteins have provided a detailed understanding of how MADS-box proteins (37, 38, 39), including Mcm1p (2), elicit differential DNA bending. Here, we have uncovered further details of the Mcm1p-mediated DNA-bending mechanism.
The substitution of negatively charged amino acids for residues in the β-loop, T66 and L68, also severely affected DNA bending by Mcmlp (Fig. 1). However, the residue located at position 14 towards the N-terminal end of the recognition helix in the MADS-box (K29) played a lesser role in Mcm1p than was observed in the human MADS-box protein SRF. In SRF, residues located in both the β-loop and position 14 play important roles in regulating DNA bending (38). These observations are consistent with an Mcm1p-DNA structure in which T66 and L68 are either close to or contact DNA but K29 is not in contact with the DNA (Fig. 1 and 3) (35). A further residue in Mcm1p, K40, is directly implicated in inducing DNA bending. This residue makes DNA backbone contacts, and a loss-of-contact mutation (K40A) caused significant reductions in DNA bending (Fig. 1 and 2). A previous study with loss-of-contact mutants suggests that V34 and S37 also play a role in mediating DNA bending, although combinatorial mutations are required to cause a severe decrease in DNA bending (2).
Mcm1p exhibits DNA sequence-specific DNA-bending activity in which the presence of a palindromically oriented A:T base pair at the −7 and +7 positions in the binding site results in higher degrees of bending (Fig. 1) (1, 39). V34 in Mcm1p has previously been suggested to play a role in sensing sequence-specific bending mediated by the −7 and +7 positions (2). However, in this study, the authors demonstrated that while mutation of the A:T base pairs at the −7 and +7 positions caused a reduction in DNA bending by the wild-type protein, the V34A protein exhibited no such reduction. One interpretation of this result, therefore, is that it is the presence of the side chain in V34 that prevents bending of sites lacking the −7 and +7 position A:T base pairs, and in the absence of this side chain, Mcm1p is able to bend DNA. Here, we have taken the opposite approach and shown that mutating residues K40, T66, and L68 resulted in a reduction in DNA bending and that these residues were required to permit enhanced bending of sites containing the −7 and +7 position A:T base pairs (Fig. 1 and data not shown). Thus, residues in or close to the β-loop are implicated as important sensors of the base pairs at the −7 and +7 positions in the binding sites for Mcm1p.
Collectively, these results show that a complex network of residues exists in Mcm1p which act in concert to either mediate or permit DNA bending and direct DNA bending in a sequence-specific manner.
In summary, we have provided important mechanistic insights into how DNA bending by Mcm1p is achieved. With this information, we generated tools to demonstrate the importance of DNA bending in gene regulation by Mcm1p. We have shown that DNA bending is an important determinant of local promoter architecture and that loss of DNA bending results in alterations in gene expression that lead to growth defects.
Acknowledgments
We thank Margaret Bell, Linda Shore, Phil Butler, and Bharat Rash for excellent technical assistance. We are grateful to members of our laboratories for helpful discussions. We are grateful to Jason Kahn and George Sprague for reagents.
This work was supported by the BBSRC (by grants from the Genes and Development Committee to A.D.S. and the COGEME project of the IGF initiative to S.G.O.), the Cancer Research United Kingdom, and the Hitachi Corporation, as well as by an MRC Studentship to A.G.W. A.D.S. is a Research Fellow of the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Acton, T. B., H. Zhong, and A. K. Vershon. 1997. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS-box protein. Mol. Cell. Biol. 17:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acton, T. B., J. Mead, A. M., Steiner, and A. K. Vershon. 2000. Scanning mutagenesis of Mcm1: residues required for DNA binding, DNA bending, and transcriptional activation by a MADS-box protein. Mol. Cell. Biol. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, A., and G. F. Sprague, Jr. 1987. MATα1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell type-specific genes. Cell 50:681-691. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 5.Breeden, L. L. 2000. Cyclin transcription: timing is everything. Curr. Biol. 10:R586-R588. [DOI] [PubMed] [Google Scholar]
- 6.Bruhn, L., J. J. Hwang-Shum, and G. F. Sprague, Jr. 1992. The N-terminal 96 residues of MCM1, a regulator of cell type-specific genes in Saccharomyces cerevisiae, are sufficient for DNA binding, transcription activation, and interaction with α1. Mol. Cell. Biol. 12:3563-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruhn, L., and G. F. S. Sprague, Jr. 1994. MCM1 point mutants deficient in expression of α-specific genes: residues important for interaction with α1. Mol. Cell. Biol. 14:2534-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabib,E., A. Sburlati, B. Bowers, and S. J. Silverman. 1989. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 108:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christ,C., and B. K. Tye. 1991. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 5:751-763. [DOI] [PubMed] [Google Scholar]
- 10.Dolan, J. W., and S. Fields. 1991. Cell-type-specific transcription in yeast. Biochim. Biophys. Acta 1088:155-169. [DOI] [PubMed] [Google Scholar]
- 11.Errede, B. 1993. MCM1 binds to a transcriptional control element in Ty1. Mol. Cell. Biol. 13:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan, K., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 13.Huang, K., J. M. Louis, L. Donaldson, F.-L. Lim, A. D. Sharrocks, and G. M. Clore. 2000. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 19:2615-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keleher, C. A., S. Passmore, and A. D. Johnson. 1989. Yeast repressor α2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell. Biol. 9:5228-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koranda, M., A. Schleiffer, L. Endler, and G. Ammerer. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406:94-98. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, R. D. M. Reynolds, A. Shevchenko, A. Shevchenko, S. D. Goldstone, and Dalton,S. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 17.Louis, E. J., and J. E. Haber. 1992. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 131:559-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazur, P., N. Morin, W. Baginsky, M. el-Sherbeini, J. A. Clemas, J. B. Nielsen, and F. Foor. 1995. Differential expression and function of two homologous subunits of yeast 1,3-beta-d-glucan synthase. Mol. Cell. Biol. 15:5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer, and L. L. Breeden. 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11:1277-1288. [DOI] [PubMed] [Google Scholar]
- 20.Mead, J., H. Zhong, T. B. Acton, and A. K. Vershon. 1996. The yeast α2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol. Cell. Biol. 16:2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messenguy, F., and E. Dubois. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellegrini, L., S. Tan, and T. J. Richmond. 1995. Structure of serum response factor core bound to DNA. Nature 376:490-498. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Martin, J., and V. de Lorenzo. 1997. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 51:593-628. [DOI] [PubMed] [Google Scholar]
- 24.Pic, A., F.-L. Lim, S. J. Ross, A. L. Johnson, R. A. Sultan, A. G. West, L. H. Johnston, A. D. Sharrocks, and B. A. Morgan. 2000. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19:3750-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primig, M., H. Winkler, and G. Ammerer. 1991. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 10:4209-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santelli, E., and T. J. Richmond. 2000. Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution. J. Mol. Biol. 297:437-449. [DOI] [PubMed] [Google Scholar]
- 27.Sharrocks, A. D., F. Von Hesler, and P. E. Shaw. 1993. The identification of elements determining the different DNA binding specificities of the MADS-box proteins p67SRF and RSRFC4. Nucleic Acids Res. 21:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharrocks, A. D., and P. Shore. 1995. DNA bending in the ternary nucleoprotein complex at the c-fos promoter. Nucleic Acids Res. 23:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shore, P., and A. D. Sharrocks. 1995. The MADS-box family of transcription factors. Eur. J. Biochem. 229:1-13. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 33.Smith, D. L., A. B. Desai, and A. D. Johnson. 1995. DNA-bending by the a1 and α2 homeodomain proteins from yeast. Nucleic Acids Res. 23:1239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan, S., and T. J. Richmond. 1998. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 391:660-666. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West, A. G., P. Shore, and A. D. Sharrocks. 1997. DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell. Biol. 17:2876-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West, A. G., B. E. Causier, B. Davies, and A. D. Sharrocks. 1998. DNA binding and dimerisation determinants of the Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 26:5277-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, A. G., and A. D. Sharrocks. 1999. MADS-box transcription factors adopt different mechanisms to bend DNA. J. Mol. Biol. 286:1311-1323. [DOI] [PubMed] [Google Scholar]
- 40.Yang, S-H., P. R. Yates, A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. The Elk-1 ETS-domain transcription factor contains a MAP kinase targeting motif. Mol. Cell. Biol. 18:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, G., and J. S. Fassler. 1993. SPT13 (GAL11) of Saccharomyces cerevisiae negatively regulates activity of the MCM1 transcription factor in Ty1 elements. Mol. Cell. Biol. 13:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, L. J., and O. Narayan. 1993. A gene expression vector useful for protein purification and studies of protein-protein interaction. Gene 137:345-346. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]