Abstract
The Tor pathway mediates cell growth in response to nutrient availability, in part by inducing ribosomal protein (RP) gene expression via an unknown mechanism. Expression of RP genes coincides with recruitment of the Esa1 histone acetylase to RP gene promoters. We show that inhibition of Tor with rapamycin releases Esa1 from RP gene promoters and leads to histone H4 deacetylation without affecting promoter occupancy by Rap1 and Abf1. Genetic and biochemical evidence identifies Rpd3 as the major histone deacetylase responsible for reversing histone H4 acetylation at RP gene promoters in response to Tor inhibition by rapamycin or nutrient limitation. Our results illustrate that the Tor pathway links nutrient sensing with histone acetylation to control RP gene expression and cell growth.
The rapamycin-sensitive Tor signaling pathway couples nutrient availability with cell growth in Saccharomyces cerevisiae, Drosophila melanogaster, and mammalian cells (36, 38, 41). In addition to a well-established role in the control of translation initiation, the Tor proteins play a dynamic role in controlling transcription in response to nutrient signals. Inhibition of the Tor kinases by rapamycin or nitrogen limitation results in the marked induction of genes subject to nitrogen catabolite repression (NCR), genes associated with the retrograde response, and genes induced by various environmental stresses such as sodium toxicity and carbon starvation (2, 4, 6, 9, 13, 19). In contrast, inhibition of Tor results in the rapid repression of genes involved in ribosome biogenesis, including tRNAs and rRNAs transcribed by Pol I and Pol III as well as ribosomal proteins expressed by Pol II (6, 25, 35, 49). Significant progress has been made in understanding how the genes that are induced by Tor inhibition are controlled. In these cases, under optimal nutrient conditions the Tor pathway prevents the nuclear import of the corresponding transcription factors, including Gln3, Gat1, Msn2, Msn4, Rtg1, and Rtg3 (2, 4, 19). However, the molecular mechanism(s) by which Tor regulates expression of the RP genes remains poorly understood.
The RP genes are subject to stringent regulation in order to couple protein synthesis and growth to the availability of nutrients and the physiological status of the cell (48). In addition to the Tor pathway, two other important signaling pathways regulate RP gene expression. The nutrient-sensing protein kinase A (PKA) pathway is required to activate RP gene expression while the PKC pathway mediates repression of RP genes in response to perturbations of the cell integrity pathway (18, 31). Additional signaling programs are also thought to regulate RP gene expression in response to nutrients (30). The majority of RP gene promoters contain binding sites for two transcription factors of partially overlapping function: Abf1 and Rap1 (21, 23; reviewed in reference 33). The most prominent of these factors, Rap1, activates RP gene expression and also functions in silencing telomeres and the silent mating type loci HML and HMR (15, 47).
Gene activation in eukaryotic cells requires mechanisms that overcome the repressive effects of chromatin at specific promoters. A growing body of evidence suggests that this is accomplished by the recruitment of chromatin-remodeling complexes by site-specific transactivators (46). Recent work has demonstrated a strong correlation between recruitment of the Esa1 histone acetylase and transcription from RP gene promoters (37). Furthermore, recruitment of Esa1 to RP gene promoters requires a binding site for Rap1 and/or Abf1 (37). Esa1 is the catalytic subunit of the NuA4 histone acetylase complex that acetylates histones H4 and H2A (1). The NuA4 complex is recruited to DNA by acidic activators such as VP16 and Gcn4 (5).
In this work we examined whether Tor signaling is required for the occupancy of known regulatory factors at the RP gene promoters by using chromatin immunoprecipitation assays. We found that Tor signaling is required for the maintenance of Esa1 at RP gene promoters. Repression of RP genes in response to nutrient depletion or rapamycin treatment requires components of the Rpd3-Sin3 histone deacetylase complex. Our results establish a link between Tor-mediated nutritional signaling and histone acetylation and illustrate a novel mechanistic paradigm by which the Tor pathway controls gene expression.
MATERIALS AND METHODS
Saccharomyces cerevisiae strains, plasmids, and growth conditions.
Strain MCY47 was obtained by introducing a three-hemagglutinin (HA) epitope-tagged Esa1 in a two-step gene replacement (with plasmid YIplac211 HA-Esa1, a generous gift from Kevin Struhl) into strain MLY41 Σ1278b MATaura3-52 (37). Strains JRY16a, JRY17a, and JRY18a were derived from MLY41a by replacing the entire open reading frame of RPD3, SIN3, and SAP30, respectively, with kanMX. Gene disruptions were all verified by PCR.
Chromatin immunoprecipitation and quantitative PCR.
Exponentially growing cultures of strain MCY47 containing HA3 epitope-tagged Esa1 were treated with 100 nM rapamycin for 0, 15, 30, and 60 min. Cultures were adjusted to 1% formaldehyde and incubated for 20 min at room temperature with gentle shaking. Glycine was added to a final concentration of 150 mM, and incubation continued for 5 min. Cells were harvested in prechilled bottles and washed twice with ice-cold phosphate-buffered saline. Chromatin was prepared essentially as described previously with the following modifications, and the entire procedure was performed at 4°C (14, 20). Cells were resuspended in 0.8 ml of lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 25 mM β-glycerophosphate, 25 mM NaF, and protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin ml−1, 1 mM benzamidine, 1% aprotinin]), an equal volume of 0.5-mm-diameter glass beads was added, and cells were disrupted by vigorous shaking. Glass beads were discarded as indicated (14), and cell lysates were centrifuged at 200,000 × g for 20 min to recover the cross-linked chromatin. The chromatin was resuspended in 0.8 ml of lysis buffer, sonicated to produce fragments with an average size of 350 bp (6 times for 10 s at setting 3 in a Bradson sonicator fitted with a microtip), and centrifuged at 10,000 × g for 15 min. The protein concentrations from the different samples were determined, and 1.8 mg was incubated with antibodies for 2 h. The antibodies used were anti-HA monoclonal antibody F-7, anti-Rap1 (yC-19), anti-Abf1 (yC-20), anti Rpd3 (yN-19 and yC-19) from Santa Cruz Biotechnology and anti-K5, -K8, -K12, and -K16-acetylated histone H4, chromatin immunoprecipitation grade, from Upstate Biotechnology. Immunocomplexes were recovered by adding 30 μl (bead volume) of protein A-Sepharose or protein G-Sepharose. Following incubation for 1 h on a rotator, beads were washed twice with lysis buffer, twice with lysis buffer containing 0.5 M NaCl, twice with 10 mM Tris-HCl (pH 8.0)-0.25 M LiCl-1 mM EDTA-0.5% NP-40-0.5% sodium deoxycholate, and once with Tris-EDTA (TE). All washes were done with a volume of 1.5 ml and for a period of 5 min on a rotator. Immunocomplexes were eluted by incubating the beads in 100 μl of TE-1% sodium dodecyl sulfate at 65°C for 10 min. To reverse the cross-links, the samples were incubated at 65°C overnight and protein was removed by digestion with 100 μg of proteinase K for 2 h at 37°C. Following extraction with phenol-chloroform-isoamyl alcohol, 5 μg of glycogen and 1/10 volume of 5 M LiCl-50 mM Tris-HCl (pH 8.0) were added and DNA was ethanol precipitated overnight at −20°C.
Quantitative PCR in the linear range for each set of primers and DNA was performed as indicated previously (20) with the following modifications. The amount of DNA used for the PCRs for the immunoprecipitates was 1/50 to 1/1,000 of the total sample, and that for the inputs was 1/5,000 to 1/10,000 of the total sample. PCRs were carried out with 15 μl containing 1 μM primers, 125 μM deoxynucleoside triphosphates, 0.1 μCi of [32P]dCTP ml−1 (specific activity, 3,000 Ci mmol−1), 0.37 U of Ex-Taq polymerase, and 5 μl of DNA template. PCR amplification was for 28 to 30 cycles consisting of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C, and finally 5 min at 72°C. For amplification of longer PCR products (see Fig. 4C), PCR amplification was for 35 cycles consisting of 30 s at 94°C, 30 s at 55°C, 2 min at 72°C, and finally 5 min at 72°C. PCR products (typically, 275 to 300 bp) were quantified with a Storm 860 PhosphorImager (Molecular Dynamics), and promoter occupancy was calculated by dividing the amount of PCR product obtained with immunoprecipitated DNA by the amount obtained with total DNA (input).
FIG. 4.
The Rpd3-Sin3 complex is required for adaptation to nutrient limitation. (A) Isogenic wild-type (MLY41a) and rpd3 (JRY16a), sin3 (JRY17a), and sap30 (JRY18a) mutant strains were grown to early exponential phase in YEPD. Cultures were collected by centrifugation, washed, and resuspended in SD-N medium. Samples were collected at 0, 15, 60, and 120 min as indicated. RNA was prepared and analyzed by Northern blotting with radioactive probes that hybridize to the genes indicated at left. Hybridization to the ACT1 message served as a loading control. (B) Isogenic wild-type (MLY41a), and rpd3 (JRY16a), sin3 (JRY17a), and sap30 (JRY18a) mutant strains were grown on YEPD solid medium overnight at 30°C to achieve confluent growth. Cells were replica plated to SD-N medium, incubated for 12 days, replica plated back to YEPD, grown for 24 h, and photographed.
Starvation assays.
Nitrogen starvation medium (SD-N) contained 0.17% Bacto yeast nitrogen base without amino acids or ammonium sulfate, 2% glucose, and 0.2% uracil. Yeast strains were grown on yeast extract-peptone-dextrose (YEPD) plates at 30°C for 24 h and replica plated to SD-N media. After 12 days at 30°C, cells were replica plated back to YEPD and grown at 30°C for 24 h and photographed.
Northern and Western blotting.
RNA isolation and Northern blot analysis were performed as described previously (6). Protein extracts were quantitated, and 100 μg of protein was analyzed by Western blotting with the antibodies indicated above for chromatin immunoprecipitation and standard techniques.
RESULTS
Maintenance of Esa1 histone acetylase at RP promoters is rapamycin sensitive.
The Tor pathway responds to nutrients by controlling expression of RP genes (6, 35). Recent work has revealed that the coordinate expression of RP genes is associated with recruitment of the histone acetylase Esa1 to the RP gene promoters (37). Furthermore, conditions that repress ribosomal protein gene expression such as heat shock or amino acid deprivation lead to a loss of Esa1 from RP gene promoters (37). Here we tested if Tor regulation of ribosomal protein gene expression involves targeted recruitment of Esa1 to these promoters.
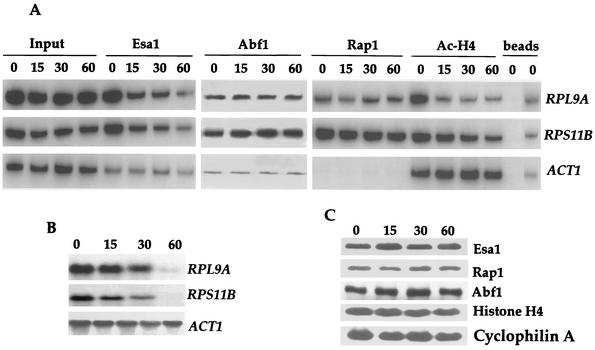
We examined promoter occupancy by Esa1 in untreated or rapamycin-treated cells by chromatin immunoprecipitation. Rapamycin treatment resulted in a marked release of Esa1 from the promoters of two RP genes, RPL9A and RPS11B, whose expression is known to be inhibited by rapamycin treatment (Fig. 1A and B). In addition, the kinetics of Esa1 release are similar to the kinetics of RP mRNA disappearance. Following 60 min of treatment, Esa1 promoter occupancy was reduced to the background level of detection (see control lanes [beads]) and RP gene mRNAs were nearly undetectable (compare Fig. 1A and B). In agreement with earlier results (37), recruitment of Esa1 to RP gene promoters was specific and only a background level of Esa1 promoter occupancy was observed at the ACT1 promoter, which is not an Esa1 target (Fig. 1A) (37). In accordance with these results, the amount of acetylated histone H4, the major Esa1 substrate (8, 44) diminished at the RP gene promoters while it remained unchanged at the ACT1 promoter (Fig. 1A). These results demonstrate that Tor controls the occupancy of Esa1 at RP gene promoters.
FIG. 1.
Rapamycin-sensitive maintenance of Esa1 at RP gene promoters. (A) Exponentially growing cultures of strain MCY47 containing HA3-tagged Esa1 were treated with 100 nM rapamycin for 0, 15, 30, and 60 min and cross-linked with formaldehyde. Chromatin was prepared and immunoprecipitated with antibodies specific for HA3-Esa1, Rap1, Abf1, and K5, K8, K12, and K16-acetylated histone H4. PCR was performed in the total chromatin (Input) or the immunoprecipitated DNA with specific primers for the RPL9A, RPS11B, and ACT1 gene promoters indicated on the right. Controls were performed with protein G for goat antibodies against Rap1 and Abf1 (left lane) and protein A for mouse antibodies against Esa1 and AcH4 (right lane) beads. PCR products were quantified with a PhosphorImager, and promoter occupancy was calculated by dividing the amount of PCR product obtained with immunoprecipitated DNA by the amount obtained with total DNA (input). These values are not shown, because the intensity of the PCR products directly reflects the quantified results. (B) Strain MCY47 was grown and treated with rapamycin as indicated for panel A. RNA was prepared and analyzed by Northern blotting with radioactive probes that hybridize to the genes indicated at the right. (C) Rapamycin treatment does not affect Esa1, Rap1, Abf1, or histone H4 protein stability. Samples from the cell extracts prepared as described for panel A were adjusted to 0.5 M 2-mercaptoethanol, incubated at 95°C for 30 min to reverse protein-protein cross-links, and analyzed by Western blotting with antibodies against the proteins indicated at the right. The results shown in panels A, B, and C are representative of three independent experiments.
Tor regulates Esa1 maintenance at RP gene promoters without affecting Rap1 or Abf1 binding.
The majority of the RP gene promoters contain binding sites for the transcription factors Rap1 and Abf1 (21). Moreover, evidence suggesting that Esa1 is recruited to RP gene promoters by Rap1 and Abf1 has been presented (37). We examined the effect of rapamycin on RP gene promoter occupancy by Rap1 and Abf1. The levels of Rap1 and Abf1 at the RPL9A and RPS11B promoters remained unchanged during the course of rapamycin treatment (Fig. 1A). By contrast, Rap1 and Abf1 were not detected at the ACT1 promoter, which lacks binding sites for these factors (Fig. 1A). These results illustrate that the Tor pathway regulates RP gene promoter occupancy by Esa1 without affecting the binding of Rap1 and Abf1 to these promoters. Similarly, previous results have shown that amino acid deprivation induces release of Esa1 from RP gene promoters yet has no effect on Rap1 or Abf1 binding (37).
An involvement of the Tor pathway in regulating protein stability has been previously reported. Rapamycin treatment leads to a rapid destabilization of the tryptophan amino acid permease Tat2 and the translation initiation factor eIF4G (3, 42). However, the steady-state levels of the Esa1, Rap1, and Abf1 proteins were not affected during the time course of rapamycin treatment in our studies (Fig. 1C). Thus, loss of Esa1 and acetylated histone H4 from RP gene promoters is not attributable to decreases in protein levels.
The Rpd3-Sin3 complex mediates RP gene repression in the absence of Tor signaling.
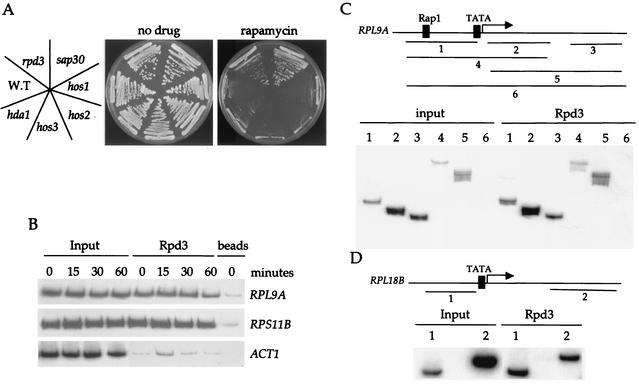
Deacetylation of chromatin by histone deacetylase complexes has been implicated in the repression of genes (reviewed in references 11, 12, and 28). The link between Tor signaling and Esa1-dependent acetylation prompted us to investigate the role of histone deacetylase complexes in RP gene repression. We examined the rapamycin sensitivity of an isogenic set of yeast strains lacking the Hda1, Hos1, Hos2, Hos3, Sir2, or Rpd3 histone deacetylases. Mutations in the Rpd3-Sin3 histone deacetylase complex, which includes Sap30, conferred a clear increase in rapamycin resistance (Fig. 2A), while mutation of the other deacetylases tested had no effect (Fig. 2A and data not shown). In support of these results, we note that deletion of the gene encoding Sap30 or the Sin3-associated protein Sbt3 has recently been shown to confer rapamycin resistance in a large-scale screening of the yeast deletion consortium strain series (7).
FIG. 2.
Rpd3 localizes at RP genes independently of Tor signaling or RP gene activators. (A) Isogenic wild-type (S288C), and hos1, hos2, hos3, hda1, rpd3, and sap30 mutant strains were grown on YEPD (no drug) or YEPD containing 100 ng of rapamycin ml−1. After 3 days of incubation at 30°C the plates were photographed. (B) Exponentially growing cultures of strain MCY47 were treated with 100 nM rapamycin for 0, 15, 30, or 60 min and cross-linked with formaldehyde. Chromatin was prepared and immunoprecipitated with antibodies specific for Rpd3. PCR was performed in the total chromatin (Input) or the immunoprecipitated DNA with specific primers for the RPL9A, RPS11B, and ACT1 gene promoters indicated on the right. Controls were performed with protein G beads. The results shown are representative of two independent experiments. (C and D) The Rpd3 immunoprecipitated DNA from rapamycin-untreated cells in the experiment shown in Fig. 2B was amplified with primers specific for RPL9A (panel C) or RPL18B (panel D). The PCR products encode the following sequences: RPL9A, −398 to −98 (1), 35 to 300 (2), 917 to 1154 (3), −398 to 300 (4), 35 to 1154 (5), −398 to 1154 (6) in RPL18B, −410 to −151 (1) and 1216 to 1500 (2). For a control, we performed PCR with the DNA obtained from the mock immunoprecipitation (with no antibody but beads from the same experiment) and the different sets of primers. The results obtained were very similar to those of the beads control presented in Fig. 2B.
The Rpd3-Sin3 histone deacetylase complex has not been previously linked to RP gene promoter regulation. Thus, we examined the occupancy of Rpd3 at RP gene promoters in the presence of rapamycin. Chromatin immunoprecipitation reveals that Rpd3 specifically occupies the promoters of RPL9A and RPS11B, and this occupancy is unaffected by rapamycin treatment (Fig. 2B and data not shown). We found that Rpd3 occupancy is not limited to the RP gene promoters and that Rpd3 is also present in the coding regions of the RPL9A and RPL18B genes (Fig. 2C and D). More-extensive PCR analysis excluded the possibility that association of Rpd3 is an artifact of binding to the promoter present in long pieces of immunoprecipitated DNA that also contain the coding region. This analysis revealed that Rpd3 associates with both the 5′ promoter region of RPL9A and the most distal 3′ coding region (fragment 3) even though no PCR products spanning both regions were detected (Fig. 2C). Moreover, the RPL18B promoter lacks Rap1 and Abf1 binding sites (37), indicating that Rpd3 is not targeted to RP genes by these transcription factors. These data are in agreement with a recent report that demonstrates that the Sin3 complex constitutively associates with chromatin in a nontargeted fashion (22).
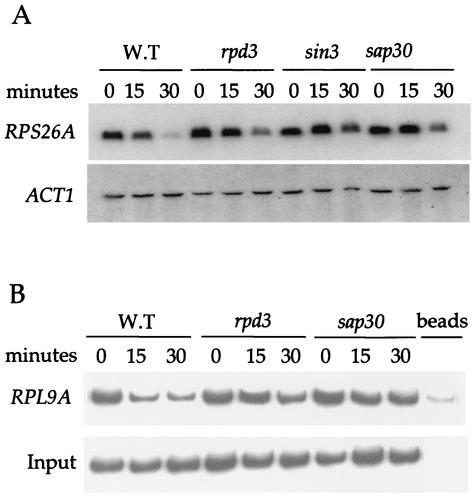
The functional significance of Rpd3 occupancy at RP gene promoters was tested by Northern blotting and chromatin immunoprecipitation. Mutations in the RPD3, SIN3, or SAP30 genes resulted in a defect in rapamycin-induced repression of RP genes (Fig. 3A). While this effect is modest (three- to fivefold as measured by phosphorimaging), it is highly reproducible in a set of mutants constructed in two different strain backgrounds (Σ1278b and S288C). Importantly, deletion of the RPD3 or SAP30 genes also largely prevents the loss of acetylated histone H4 from RP gene promoters that accompanies rapamycin treatment in wild-type cells (Fig. 3B). Taken together, our data indicate that the Rpd3-Sin3 histone deacetylase complex contributes to RP gene repression in the absence of Tor signaling.
FIG. 3.
The Sin3-Rpd3 complex is required for proper rapamycin-induced repression of RP genes. (A) Isogenic wild-type (MLY41a), and rpd3 (JRY16a), sin3 (JRY17a), and sap30 (JRY18a) mutant strains were grown to early exponential phase and treated with or without 200 ng of rapamycin ml−1 for 15, 30, and 60 min as indicated. RNA was prepared and analyzed by Northern blotting with radioactive probes that hybridize to the genes indicated at the left. (B) Following the rapamycin treatment as indicated above in Fig. 3A, cultures were cross-linked with formaldehyde. Chromatin was prepared and immunoprecipitated with antibodies specific for hyperacetylated histone H4. PCR was performed in the total chromatin (Input) or the immunoprecipitated DNA with specific primers for the RPL9A promoter. The results shown are representative of two independent experiments.
The Rpd3-Sin3 complex is required for proper adaptation to nutrient limitation.
The transcriptional program induced by rapamycin is strikingly similar to that seen in cells subjected to a nutritional downshift (6, 13; reviewed in reference 38). In addition, the Rpd3-Sin3 complex has been implicated in the repression of the CAR1 and CAR2 genes that are subject to nitrogen catabolite repression (27). We reasoned that mutations in genes encoding the Rpd3-Sin3 complex might therefore also cause an aberrant response to nutrient limitation, similar to that observed with rapamycin treatment. Subjecting rapidly growing cells to a nutritional downshift (YEPD to SD-N media) faithfully reproduced the transcriptional program imposed by rapamycin. Wild-type cells induced the expression of the NCR gene GAP1, and the rpd3, sin3, and sap30 mutant strains exhibited a similar NCR gene expression profile (Fig. 4A). In contrast, whereas wild-type cells rapidly repressed RP gene expression, cells bearing rpd3, sin3, or sap30 mutations exhibited a modest reduction in the ability to repress RP gene expression (Fig. 4A). We next tested the idea that these mutant strains may be more susceptible to nitrogen starvation-induced death. Indeed, we find that rpd3, sin3, and sap30 mutations dramatically impair the ability of cells to survive nitrogen starvation (Fig. 4B). In summary, the Rpd3-Sin3 histone deacetylase complex is required for proper adaptation to nutrient limitation. We suggest that proper regulation of RP gene transcription mediated by the Rpd3-Sin3 complex is important for adaptation to nutritional stress.
DISCUSSION
The expression of RP genes represents a fundamental question in biology: how do cells coordinately regulate a large set of genes in response to multiple signals? RP gene expression must be sufficient to satisfy the demands of cell growth during favorable conditions, while at the same time be subject to rapid repression in response to a myriad of conditions that are unfavorable for growth. The Tor kinases transduce signals that promote or repress growth via the RP genes. Tor signaling is intimately linked to other signaling programs also implicated in the regulation of RP genes, including the PKC-cell integrity pathway, the nutrient-sensing PKA pathway, and the nitrogen-sensing pathway. In higher organisms, Tor is known to respond to a variety of mitogenic signals, including amino acid availability, and it has recently been suggested that the Tor kinases may act as direct sensors of ATP (10).
Under optimal growth conditions, the Tor kinases mediate inactivation of genes subject to nitrogen catabolite repression, the retrograde response-induced genes, and stress-induced genes by regulating the phosphorylation status and thereby preventing the nuclear import of the corresponding transcription factors (2, 4, 6, 19). Here we show that, in response to nutrients, Tor signaling activates the expression of RP genes by promoting maintenance of the Esa1 histone acetyltransferase complex to RP gene promoters. Accordingly, our results demonstrate that the association of Esa1 with the RP gene promoters is rapamycin sensitive and that the loss of Esa1 from these promoters mimics the kinetics of RP gene repression upon addition of rapamycin. A formal possibility is that a decrease in RP gene expression contributes to the observed loss of Esa1 from chromatin. However, because there is no precedent for this model we do not favor this interpretation.
We find that Rap1 and Abf1 remain bound to the RP gene promoters even after inactivation of the Tor kinases with rapamycin. Analogous observations were reported when RP gene expression was repressed following amino acid starvation or heat shock (37). Moreover, recruitment of Esa1 to RP gene promoters was mapped to the Rap1 binding sites, leading to the proposal that Esa1 is recruited by Rap1 (37). Our results indicate that maintenance of Esa1 at RP gene promoters is regulated by Tor signaling. It remains to be established if the targets of this regulation are Rap1 and Abf1 and/or components of the Esa1 histone acetylase complex.
Supporting the idea that histone acetylation is linked to Tor signaling, we find that deletion of the genes encoding subunits of the Rpd3-Sin3 complex results in a defect in rapamycin-induced repression of RP genes. Furthermore, these strains also display a defect in the repression of RP genes in response to nutrient limitation. Importantly, the Rpd3-Sin3 complex preferentially deacetylates histone H4, which is the major substrate of Esa1 (1, 39, 40). Although the Rpd3-Sin3 complex is required for rDNA silencing, it has not been previously implicated in the repression of RP genes (45). Our results demonstrate that Rpd3 is present at both active and inactive RP gene promoters. Additionally, Rpd3 is present at the promoter regions as well as the coding regions of RP genes and the localization of Rpd3 to these genes does not require binding sites for Rap1 or Abf1. These results are consistent with recent reports demonstrating that untargeted and global deacetylase activity at promoters and coding regions is important for rapidly reversing the effects of localized transcription responses (17, 22).
Our results are consistent with the following model. In response to ample nutrients, Tor signaling favors the occupancy of the Esa1-histone acetylase complex at RP gene promoters resulting in histone H4 acetylation and alterations in chromatin conformation that activate transcription. In response to nutrient limitation or rapamycin treatment, Tor signaling is inhibited, leading to the release of the Esa1-histone acetylase complex from RP gene promoters. The chromatin is returned to the repressed state by the Rpd3-Sin3 histone deacetylase complex that is already resident at RP gene promoters. Our ongoing studies are focused towards understanding the signaling events between the Tor kinases and the transcription machinery at the RP gene promoters.
We suggest that histone acetylation is an important mechanism for adaptation to growth in nutrient-limiting conditions. Recent years have seen a flurry of reports establishing a connection between the Sir2 histone deacetylase complex and aging (12, 24). Sir2 is thought to regulate life span by blocking inappropriate gene expression. Most interesting is that the NAD-dependent activity of Sir2 may provide a link between chromatin acetylation and the overall metabolism of a cell. We therefore find it intriguing that, in mammalian cells, rapamycin treatment results in a gene expression profile that resembles one seen with amino acid limitation (32).
Rapamycin is having a dramatic impact on clinical medicine in several arenas. Approved for use in cases of transplant rejection in 1999, rapamycin is now being developed to prevent restenosis following cardiac stent surgery and most recently as a novel chemotherapy agent (16, 26, 43). The promise of rapamycin as a cancer drug is being explored in phase II and III clinical trials and recent reports have demonstrated its remarkable antitumor activity in cells where the phosphatidylinositol 3-kinase-AKT pathway is upregulated (16, 29, 34). A more complete understanding of mTor signaling should enhance the clinical utility of rapamycin. Our studies elucidating novel roles for Tor in controlling transcription in response to nutrients may provide insights into mTor signaling in human cells.
ADDENDUM
While this paper was in preparation, it was reported that Rpd3 is highly enriched at actively transcribed genes, including the RP genes (S. Kurdistani, D. Robyr, S. Tavazoie, and M. Grunstein, Nat. Genet. 31:248-254, 2002).
Acknowledgments
We thank Kevin Struhl for the generous gift of plasmid YIplac211 HA-Esa1. We also thank Joseph Heitman, Miguel Arevalo-Rodriguez, Robin Wharton, John McCusker, and Bryan Cullen for suggestions and critical reading of the manuscript.
This work was supported by K22 award CA94925-01 from the NCI (to M.E.C.).
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Côté. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 3.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 6.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. D. Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, T. F., J. Carvalho, L. Riles, and X. F. Zheng. 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl. Acad. Sci. USA 97:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo, J. L., K. Daicho, T. Ushimaru, and M. N. Hall. 2001. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 276:34441-34444. [DOI] [PubMed] [Google Scholar]
- 10.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 11.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 3:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 13.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304:399-414. [DOI] [PubMed] [Google Scholar]
- 15.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383:92-96. [DOI] [PubMed] [Google Scholar]
- 16.Hidalgo, M., and E. K. Rowinsky. 2000. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 19:6680-6686. [DOI] [PubMed] [Google Scholar]
- 17.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone actylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 21.Lascaris, R. F., W. H. Mager, and R. J. Planta. 1999. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics 15:267-277. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., Q. Lin, W. Wang, P. Wade, and J. Wong. 2002. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 16:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 24.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan, P. B. 1994. Modulation of transcription of rRNA genes by rapamycin. Int. J. Immunopharmacol. 16:711-721. [DOI] [PubMed] [Google Scholar]
- 26.Marx, S. O., and A. R. Marks. 2001. Bench to bedside: the development of rapamycin and its application to stent restenosis. Circulation 104:852-855. [DOI] [PubMed] [Google Scholar]
- 27.Messenguy, F., F. Vierendeels, B. Scherens, and E. Dubois. 2000. In Saccharomyces cerevisiae, expression of arginine catabolic genes CAR1 and CAR2 in response to exogenous nitrogen availability is mediated by the Ume6 (CargRI)-Sin3 (CargRII)-Rpd3 (CargRIII) complex. J. Bacteriol. 182:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moazed, D. 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13:232-238. [DOI] [PubMed] [Google Scholar]
- 29.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuman-Silberberg, F. S., S. Bhattacharya, and J. R. Broach. 1995. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell. Biol. 15:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nierras, C. R., and J. R. Warner. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274:13235-13241. [DOI] [PubMed] [Google Scholar]
- 32.Peng, T., T. R. Golub, and D. M. Sabatini. 2002. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 22:5575-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planta, R. J. 1997. Regulation of ribosome synthesis in yeast. Yeast 13:1505-1518. [DOI] [PubMed] [Google Scholar]
- 34.Podsypanina, K., R. T. Lee, C. Politis, I. Hennessy, A. Crane, J. Puc, M. Neshat, H. Wang, L. Yang, J. Gibbons, P. Frost, V. Dreisbach, J. Blenis, Z. Gaciong, P. Fisher, C. Sawyers, L. Hedrick-Ellenson, and R. Parsons. 2001. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proc. Natl. Acad. Sci. USA 98:10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raught, B., A. C. Gingras, and N. Sonenberg. 2001. The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. USA 98:7037-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 38.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The Tor kinases link nutrient sensing to cell growth. J. Biol. Chem. 276:9583-9586. [DOI] [PubMed] [Google Scholar]
- 39.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 41.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shapiro, A. M., J. R. Lakey, E. A. Ryan, G. S. Korbutt, E. Toth, G. L. Warnock, N. M. Kneteman, and R. V. Rajotte. 2000. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 343:230-238. [DOI] [PubMed] [Google Scholar]
- 44.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, J. S., E. Caputo, and J. D. Boeke. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19:3184-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Struhl, K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98:1-4. [DOI] [PubMed] [Google Scholar]
- 47.Sussel, L., and D. Shore. 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. USA 88:7749-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 49.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]