Abstract
Interstrand cross-links (ICLs) make up a unique class of DNA lesions in which both strands of the double helix are covalently joined, precluding strand opening during replication and transcription. The repair of DNA ICLs has become a focus of study since ICLs are recognized as the main cytotoxic lesion inflicted by an array of alkylating compounds used in cancer treatment. As is the case for double-strand breaks, a damage-free homologous copy is essential for the removal of ICLs in an error-free manner. However, recombination-independent mechanisms may exist to remove ICLs in an error-prone fashion. We have developed an in vivo reactivation assay that can be used to examine the removal of site-specific mitomycin C-mediated ICLs in mammalian cells. We found that the removal of the ICL from the reporter substrate could take place in the absence of undamaged homologous sequences in repair-proficient cells, suggesting a cross-link repair mechanism that is independent of homologous recombination. Systematic analysis of nucleotide excision repair mutants demonstrated the involvement of transcription-coupled nucleotide excision repair and a partial requirement for the lesion bypass DNA polymerase η encoded by the human POLH gene. From these observations, we propose the existence of a recombination-independent and mutagenic repair pathway for the removal of ICLs in mammalian cells.
A DNA interstrand cross-link (ICL) is formed when both strands of the double helix are covalently joined by a single molecule. Since ICLs effectively prevent strand separation, essential metabolic functions of DNA such as transcription, replication, and recombination are severely blocked by these lesions. The formation of DNA ICLs appears to be an essential prerequisite for the potent cytotoxicity and antitumor activity of a large array of chemotherapeutic compounds used in cancer treatment (41).
In Escherichia coli and lower eukaryotes, the repair of ICLs is carried out primarily by a combination of the nucleotide excision repair (NER) and homologous recombination pathways. In a model proposed by Cole et al. (9, 10) based on genetic evidence, the NER mechanism introduces incisions flanking the site of the cross-link on the same strand. The resulting gap is then repaired by using a lesion-free homologous chromosome as a donor via the recA-dependent homologous recombination pathway. Subsequent biochemical analyses fully confirmed that the removal of ICLs in E. coli is mediated by both NER and homologous recombination (39, 44, 45). Similarly, with Saccharomyces cerevisiae, a group of RAD3 mutants (deficient in NER) and a group of RAD52 mutants (deficient in homologous recombination) are hypersensitive to the killing of bifunctional alkylating agents, suggesting that both pathways are essential for the repair of ICLs (21, 28, 30, 38). These observations also indicated the presence of a combination of NER and homologous-recombination mechanisms in ICL repair. More recently, direct evidence of psoralen ICL-induced homologous recombination in budding yeast has been demonstrated (16). While the combined NER-homologous-recombination mechanism appears to be the predominant error-free pathway for ICL repair in E. coli and yeast, homology-independent ICL repair has also been observed in both organisms. In E. coli, a moderate level of ICL repair takes place in recombination-deficient backgrounds and is likely mediated by a lesion bypass process in which the gapped intermediate created by the Uvr(A)BC excinuclease is resynthesized by DNA polymerase II (polB) (4, 5) independently of undamaged homologous sequences. In S. cerevisiae, the pso1 mutant exhibits profound sensitivity to psoralen cross-links. Identification of the gene responsible for such sensitivity revealed that the pso1/rev3 locus encodes the catalytic subunit of polymerase ζ, a lesion bypass DNA polymerase (7, 31, 34). A possible role for polymerase ζ may be the resynthesis of the gap after the initial uncoupling of the cross-link. Consistent with this notion, the pso1 mutant was found to be defective in ICL processing in stationary-phase yeast cells (28). More recently, mutagenic repair of DNA ICLs was also detected in repair-proficient yeast cells (16).
Several mammalian mutants defective in homologous recombination are highly sensitive to bifunctional alkylating agents, which indicates an essential role for recombination in the repair of ICLs in higher eukaryotes (22, 32, 36). In contrast, most mammalian NER mutant cell lines display only moderate sensitivity to the cross-linking agents, suggesting that the NER mechanism may have a limited participation in the removal of DNA ICLs (3, 19). However, since ERCC1 and ERCC4/XPF mutants exhibit profound hypersensitivity to cross-linking agents, it has been suggested that the endonuclease activity of ERCC1-XPF may provide unhooking activity at ICL-stalled replication forks (25). These findings also imply that a pathway other than NER may recognize and process ICLs into recombinogenic substrates. The observation that nitrogen mustard treatment generates double-strand breaks (DSBs) in mammalian cells provides a connection between ICL repair and homologous recombination (12). Interestingly, a recent study of ICL repair as a function of the cell cycle showed that the introduction of psoralen ICLs during late S or G2 phase of the cell cycle did not activate the G2-M checkpoint, suggesting that mammalian cells are able to tolerate the presence of unrepaired ICLs until they are encountered by the DNA replication machinery (2). This suggests that ICLs can be converted into replication-induced DSBs that are subject to homologous recombination. As is the case with E. coli and yeast, an error-prone repair pathway exists in mammalian cells and appears to be dependent on NER and a lesion bypass mechanism (47). These findings may explain the observation that bifunctional alkylating agents are more mutagenic than their monofunctional derivatives (50).
As a model lesion, photoreactive psoralen derivatives have been used in most studies of ICL repair. The formation of psoralen-induced ICLs leads to drastic distortion of the double helix (13), which may be detected by a variety of damage recognition complexes that sense the integrity of the helical structure. Conversely, ICLs induced by mitomycin C (MMC) result in minimum disruption of the helix structure and also have a distinct sequence preference (35, 48). In a previous study, psoralen-induced ICLs were used to demonstrate the recombination-independent repair (RIR) of cross-links in a reporter-based reactivation assay (47). Here, we present data showing that repair of ICLs formed with MMC is similar to that of the psoralen ICLs, which involves a lesion bypass process. We also present evidence that transcription-coupled repair (TCR) plays an important role in the removal of ICLs located in the actively transcribed region. These results further substantiate a model of combined NER and lesion bypass mechanisms for error-prone ICL removal.
MATERIALS AND METHODS
Preparation of MMC ICLs and lesion-defined luciferase reporter substrates.
Two complementary oligonucleotides with the following sequences were synthesized and annealed: 5′-TAGATATCATCGATATAGT and 5′-TAGACTATATCGATGATAT. Bold letters indicate the guanine residues involved in the MMC cross-link, and a BspDI recognition sequence in the oligonucleotides is underlined. Annealing of these oligonucleotides created identical 3-nucleotide 5′ cohesive ends at both ends of the duplex. The annealed duplex oligonucleotides were incubated with MMC in a sodium dithionite-catalyzed reaction as previously described (48). The ICL DNA was purified in a single step by high-temperature Sephadex chromatography and characterized by a combined P1 nuclease-T4 kinase assay and by denaturing polyacrylamide gel electrophoresis (PAGE) to ensure that the purity of the cross-linking percentage was at least 97% (49). To insert the cross-linked oligonucleotide, pCMV-LUC was digested by NheI and filled in with a single dCMP residue. The latter step prevents self-ligation of the plasmid by creating ends that can only be ligated to the cross-linked oligonucleotide. After ligation of the cross-linked oligonucleotide to the vector, covalently closed cross-linked plasmid was purified by CsCl-ethidium bromide gradient centrifugation. For the construction of the undamaged luciferase control plasmid, an unmodified oligonucleotide was cloned into the NheI site of the pCMV-LUC plasmid as described for the cross-linked oligonucleotide. The purity of each cross-linked plasmid was determined by the release of a cross-link-containing fragment by restriction enzyme digestion and examination of the fragment with denaturing PAGE (see Fig. 1).
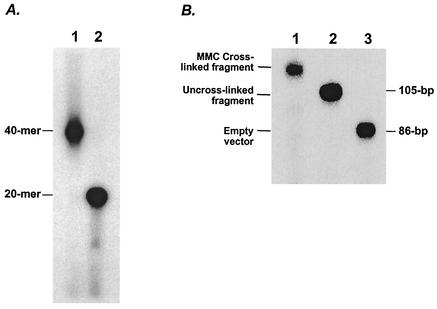
FIG. 1.
Preparation of an MMC-cross-linked plasmid substrate. (A) Preannealed oligonucleotides with a single central CpG sequence were incubated with activated MMC, and cross-linked oligonucleotides were purified by high-temperature size exclusion column chromatography. A purified cross-linked oligonucleotide (lane 1) and an unmodified oligonucleotide (lane 2) were resolved by denaturing 20% PAGE. (B). Purity of the site-specific MMC-cross-linked plasmid substrate. Results for pCMV-LUC plasmid with an MMC-cross-linked oligonucleotide inserted at the NheI site (lane 1), pCMV-LUC with an unmodified oligonucleotide inserted at the NheI site (lane 2), and pCMV-LUC vector only (lane 3) are shown. All three sample DNAs were digested with ScaI and HindIII to release a 105-bp fragment (86 bp in the case of vector only) and resolved by denaturing 15% PAGE. End labeling was used to visualize the DNA by autoradiography.
Expression of the XPV/POLH gene.
Full-length POLH cDNA was released as a 2.6-kb BamHI-SalI fragment from the pECUh6-XPV plasmid (51) (kindly provided by Zhigang Wang, University of Kentucky) and cloned in frame into the pcDNA4.0HisMaxC vector (Invitrogen). Polymerase η (PolH) expressed from the resulting construct carried the Xpress tag (DLYDDDDK) at the N terminus. The expression of PolH was verified by immunoblotting with a monoclonal antibody against the Xpress tag (Invitrogen).
Cell lines and tissue culture conditions.
Mammalian cell lines used in this study were purchased from either the American Type Culture Collection (Manassas, Va.) or the Human Genetics Mutant Cell Repository (Camden, N.J.) unless otherwise stated. Xeroderma pigmentosum (XP) fibroblast cell lines XP2OS (XPA), XP4PA (XPC), XP6BE (XPD), and XP30RO (XPV) and Cockayne syndrome (CS) fibroblast cell line CS3BE (CSA) were maintained in minimal essential medium (MEM) supplemented with 15% fetal calf serum (FCS). The human repair-proficient cell lines HT-1080 (fibrosarcoma) and RKO (colon cancer epithelial) and the Chinese hamster lung fibroblast cell line V79 were cultured in MEM supplemented with 10% FCS. The CHO AA8 cell line and its derived mutants UV24 (XPB), UV61 (ERCC6/CSB), UV24 (XPB), UV41 (XPF), and UV135 (XPG) were grown in Dulbecco MEM supplemented with 10% FCS. The E1KO7-5 (ERCC1KO) and E1KO-47 (ERCC1OK) CHO lines were maintained under similar conditions. The irs1 (XRCC2) and irs1SF (XRCC3) mutants were maintained in MEM supplemented with 10% FCS.
Repair reactivation assay.
In order to introduce the cross-linked reporter substrates into cultured cells, transient transfections were performed with the FuGENE-6 reagent (Roche Molecular Biochemicals) according to the recommendations of the manufacturer. When needed, carrier DNA was used to equalize the total amount of plasmid DNA. For the luciferase reactivation assay, 0.1 to 2.5 ng of cross-linked or unmodified control substrate was used for transfections of 1.5 × 105 cells seeded in 35-mm-diameter plates. Cells were harvested for the preparation of protein extracts 30 h after transfection. In the experiments where TCR mutants were examined, cells were harvested 18 h after transfection. The luciferase activity was determined by using the Luciferase Assay System (Promega) and measured on a Moonlight 3010 luminometer (Pharmingen). The linear ranges of the luciferase assay, both in terms of the amount of transfected DNA and the amount of protein extract used in the luciferase assay, were established individually for each cell line. Each transfection was performed at least three times, and the standard deviation is provided for each data point.
Mutation analysis.
MMC cross-linked plasmid (150 ng) was transfected into 5 × 105 cells/60-mm-diameter plate and harvested 30 h after transfection. Plasmid DNA was recovered by a modified alkaline lysis procedure (14) and subsequently electroporated into the E. coli AB2480 (uvrA recA) mutant (17), which is defective in both NER and homologous recombination. The resulting colonies were lysed and directly amplified by PCR to generate a 255-bp fragment flanking the cross-linked region. The PCR products were digested with BspDI to identify colonies containing plasmid that was resistant to BspDI cleavage. Plasmids that were found resistant to BspDI cleavage were further analyzed by DNA sequencing.
RESULTS
Generation of site-specific MMC cross-linked DNA substrates.
The nucleotide target for MMC ICL formation has been determined to be a 5′ CpG 3′ sequence (42, 48). We have designed an oligonucleotide that has only one CpG motif in the duplex region. The duplex oligonucleotide was subjected to an MMC cross-linking reaction, and the cross-linked products were purified by high-temperature gel filtration. Analysis of the purified oligonucleotide by denaturing PAGE indicated that there were no detectable levels of un-cross-linked oligonucleotide (Fig. 1A).
Repair of MMC ICL in NER-proficient cells.
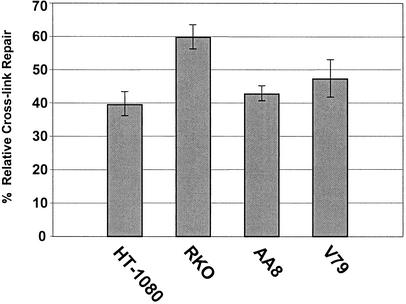
We prepared plasmid substrate containing a site-specific MMC cross-link by using in vitro ligation to insert the cross-linked oligonucleotide into the plasmid vector as described previously (26). The MMC cross-link was positioned between the CAP (transcription initiation) site of a cytomegalovirus promoter and the translation initiation site of the downstream firefly luciferase reporter gene. As shown (Fig. 1B), the purified cross-linked plasmid substrate contained no detectable amount of un-cross-linked plasmid or empty vector. As a result of the presence of the MMC ICLs, the transcription of the luciferase reporter gene becomes dependent upon the removal of the cross-links. To quantify the level of ICL repair, we constructed an identical reporter plasmid without cross-links. The efficiency of the ICL repair was determined by normalizing luciferase activities from cells transfected with cross-linked plasmid against those of cells transfected with an unmodified plasmid. Initially, two human repair-proficient cell lines (HT-1080 and RKO) were examined, and the reactivation of luciferase activity in these cell lines was found to range between 40 and 60% of that observed with unmodified plasmid (Fig. 2). These results suggest that a substantial portion of the MMC ICLs in the reporter plasmid were removed to allow the expression of the luciferase gene. Also, the repair of the cross-links apparently took place in the absence of undamaged homologous sequences, since there is no genomic sequence exhibiting significant homology to the pCMV-LUC plasmid according to the results of a BLAST search of the complete human sequence from the Celera database. Moreover, a sequence alignment analysis of the pCMV-LUC vector showed no tandem repeats flanking the cross-linked region to support a single-strand annealing type of intramolecular recombination that could lead to the reactivation of luciferase expression. To examine this RIR of ICLs in other repair-proficient cells, we tested two normal hamster cell lines, AA8 and V79. Both cell lines generated levels of reactivation similar to those of the human repair-proficient cells (Fig. 2).
FIG. 2.
Reactivation of the luciferase reporter gene by repair of a single site-specific MMC ICL in the NER-proficient cell lines HT-1080, RKO, AA8, and V79. The relative efficiencies of ICL repair were calculated as the percentage of luciferase activity of the cross-linked reporter normalized to that of the unmodified reporter.
NER is essential for the RIR of MMC ICLs.
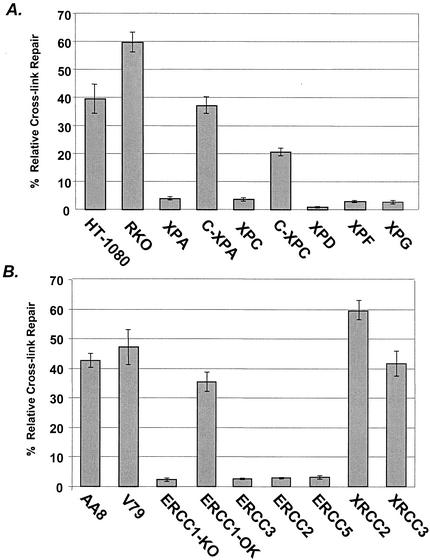
The NER pathway is the principal pathway for the repair of bulky DNA adducts. Two NER mutants, ERCC1 and XPF mutants, are hypersensitive to bifunctional alkylating agents, while other NER mutants exhibit moderate sensitivity (3, 19). To investigate whether the RIR of ICLs described above requires NER components, we used the MMC-cross-linked luciferase reporter to assay a panel of representative human NER mutants. As shown (Fig. 3A), the XPA mutant, XP2OS, was extremely deficient in reactivating the luciferase reporter. Upon reintroduction of a full-length XPA cDNA expression vector via cotransfection (C-XPA), the repair reactivation was restored to near the normal level. This result indicates that the function of XPA as a DNA damage-binding protein (24) and the incision complex assembly scaffold (46) are required in the RIR of MMC ICLs. The XPG and XPF mutants, which are defective in the 3′ and 5′ excinuclease activities of NER, respectively, were also defective in reactivating the luciferase gene (Fig. 3A), indicating that both incision nucleases are required in the RIR of ICLs. Additionally, ICL repair was also greatly reduced in the XPC and XPD mutants, indicating that the DNA damage recognition activity of XPC (40) and the helicase activity of XPD are important for the RIR of ICLs in human cells.
FIG. 3.
NER mutants are defective in repair of the cross-linked luciferase reporter. (A) Repair efficiency of XP mutants compared to that of repair-proficient HT-1080 cells. C-XPA, complemented XPA; C-XPC, complemented XPC. (B) Repair efficiency of hamster NER-deficient mutant cells compared to that of parental AA8 and V79 cells.
Confirmatory results were obtained upon a similar examination of a series of Chinese hamster mutants that harbor defective NER genes. ERCC1KO is an ERCC1-null cell line generated via a targeted knockout in repair-proficient CHO-ATS49 cells, and ERCC1OK is a stable transformant of ERCC1KO obtained upon integration of a wild-type ERCC1 cDNA expression vector, resulting in full complementation of both UV- and MMC-sensitive phenotypes (1, 47). The reactivation assay results showed (Fig. 3B) that ERCC1KO cells were severely defective in the RIR of MMC ICLs but that complemented ERCC1 cells exhibited a level of ICL repair comparable to that of the repair-proficient cells. An analysis of additional hamster NER mutants confirmed that the 5′ incision activity of the ERCC1-XPF complex, the helicase activity of the TFIIH complex (ERCC2 and ERCC3), and the 3′ incision activity (ERCC5) are involved in the RIR of MMC ICLs in mammalian cells. Taken together, these results indicate that an intact NER pathway is necessary for the removal of MMC ICLs in the absence of undamaged homologous sequences.
Lesion bypass PolH plays a role in the repair of MMC ICLs.
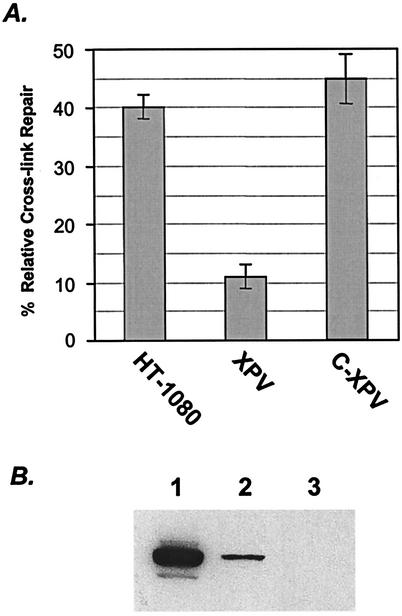
In yeast, loss of the translesion polymerase ζ leads to severe sensitivity to cross-linking agents (31, 34). In mammals, XPV mutant cells exhibited a high mutation frequency and altered mutation spectrum when triplex-forming oligonucleotide-directed psoralen adducts were introduced (37). In light of these observations, we tested an XP-variant mutant cell line, XP30RO, which is defective in the human lesion bypass PolH (23, 27). The result (Fig. 4A) showed a drastic reduction in the reactivation of the luciferase reporter, suggesting an important role for the POLH gene product in the RIR of MMC ICLs.
FIG. 4.
Participation of PolH in the RIR of MMC ICLs. (A). The ectopic expression of POLH in XP30RO complemented its ICL repair defect. C-XPV, complemented XPV. (B) Immunoblot of tagged PolH with a monoclonal antibody against the Xpress tag. Results demonstrate the expression of exogenous PolH in 293T cells (lane 1) and in XP30RO cells (lane 2) compared to that in XP30RO cells transfected with control vector only.
To verify whether the defective ICL repair in XPV cells was indeed caused by the loss of the PolH function, we constructed a cDNA expression vector, which produces full-length PolH in transient-transfection studies (Fig. 4B). When this construct was cotransfected with the cross-linked luciferase substrate, the repair ability of the XP30RO mutant was fully restored (Fig. 4A). This result provided additional support for the idea that PolH has a role in the RIR of ICLs, presumably during a lesion bypass step.
Lack of XRCC2 and XRCC3 functions does not affect RIR of ICLs.
The pivotal role of recombinational ICL repair mechanisms is reflected by the extreme hypersensitivity of the irs1 and irs1SF mutants to cross-linking agents (6). The mechanistic basis for the cross-link hypersensitivity has been identified as a defect in gene conversion (22, 36). We tested both the irs1 and the irs1SF mutant to see whether the loss of recombinational ICL repair function influences reactivation in our assay system. Our results (Fig. 3B) showed that the repair of the MMC ICLs was normal in both the irs1 and the irs1SF mutant compared to that in their isogenic parental cell lines V79 and AA8, respectively. These observations provide additional evidence that the repair of MMC ICLs occurs in the absence of homologous recombination.
Involvement of transcription-coupled NER in the RIR of MMC ICLs.
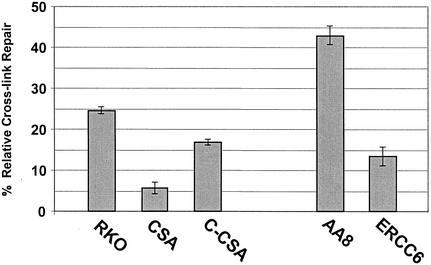
The site-specific MMC ICL was placed in an actively transcribed region in the luciferase reporter plasmid. Therefore, TCR may play a role in the recombination-independent removal of cross-links. We tested two cell lines, CS3BE and UV61, that are deficient in the TCR genes CSA and CSB, respectively. Based on the kinetics of TCR, the cells were harvested 18 h after transfection for the luciferase assay instead of 30 h after transfection as in the experiment described above. By shortening the incubation time, the involvement of TCR, if any, would not be masked by the global repair pathway, as may occur when longer incubation times are allowed. As shown (Fig. 5), both mutants have substantially reduced levels of ICL repair compared to that of repair-proficient cells, albeit the reduction is not to the same extent as that in XPA and ERCC1 mutants. Complementing the CS3BE mutant with wild-type CSA cDNA through transient transfection resulted in the substantial restoration of ICL repair. Similar results were obtained when the above-mentioned cell lines were tested with psoralen-cross-linked luciferase reporter plasmid (data not shown). These results, taken together, suggest that the TCR pathway plays a significant role in the repair of the MMC ICLs in the cross-linked luciferase reporter substrate.
FIG. 5.
TCR mutants are defective in the repair of the MMC-cross-linked luciferase reporter. Results show the repair of MMC cross-links in CSA, complemented CSA (C-CSA), and ERCC6/CSB mutants as determined by the luciferase reporter assay.
Mutagenic effect of the RIR pathway.
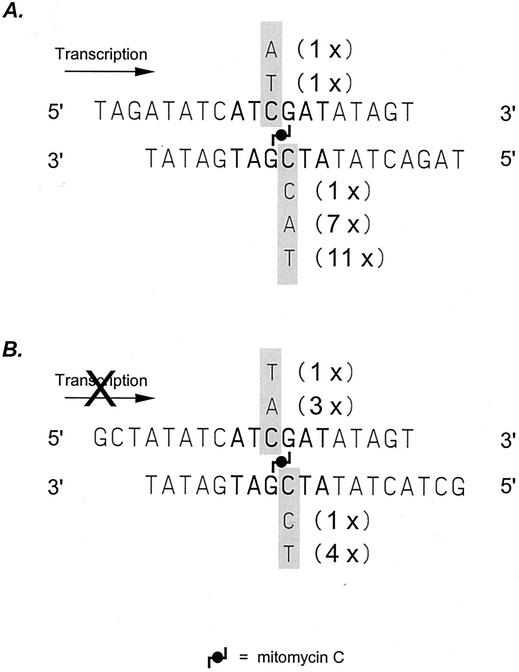
In the absence of a lesion-free homologous donor, the recombination-independent removal of MMC ICLs may lead to mutations at the site of the cross-links. To determine whether mutations are generated as a result of the RIR of ICLs, MMC cross-linked luciferase reporter substrates were transfected into the HT1080 or AA8 cell line and incubated for 30 h to allow the cross-links to be repaired in vivo. Subsequently, plasmid DNA was recovered and transformed into the E. coli AB2480 (recA uvrA) strain. Because the AB2480 strain is deficient in both NER and homologous recombination (17), only lesion-free plasmids were able to give rise to colonies. In our luciferase reporter substrate, the guanine residues attached to the MMC cross-link were located within a BspDI restriction sequence. Any mutations at and adjacent to the cross-linked guanine residues would result in the inactivation of the BspDI site. With primers flanking the cross-linked site, we used PCR to amplify this region and examined the resulting fragment by digestion with BspDI. Ninety-six colonies were analyzed, and 21 were found to be refractory to BspDI cleavage, which indicated sequence alterations within the recognition site. Plasmid DNAs were then isolated from these identified clones, and DNA sequencing was performed to reveal the nature of each mutation. As shown in Fig. 6A, all point mutations identified were found to have occurred at the cytosine position opposing a cross-linked guanine. Among the base substitutions, 12 were C-to-A transversions, 8 were C-to-T transitions, and 1 was a C-to-G transversion. One of the regions with a C-to-T transition also had a C-to-A transversion 9 bp upstream of the cross-linked guanine residue. Interestingly, a profound strand bias was observed in that 19 out of 21 base substitutions were found opposite the MMC-linked guanine residue in the nontranscribed strand, indicating that the initial incisions occurred in the transcribed strand. This result is consistent with our observation that ICL repair was significantly reduced in TCR mutants. In a parallel experiment, a promoterless luciferase reporter gene with MMC ICL placed at the same location was used to allow the repair of the ICLs in vivo and recovered plasmid DNA was again analyzed for mutations. In all, nine mutations, distributed evenly between the transcribed and nontranscribed strands, were identified from 103 colonies (Fig. 6B). This result indicated that in the absence of transcription through the cross-linked region, strand-specific mutation formation was abolished, providing additional support for the idea that TCR plays an important role in the RIR of MMC ICLs located in actively transcribed regions.
FIG. 6.
Mutation spectrum and strand-specific induction of mutations as a result of MMC RIR. Boldface indicates the BspDI recognition site. (A) In the presence of active transcription through the cross-linked site, mutations occurred predominantly at the C position in the transcribed strand, suggesting that the initial NER incisions took place on the transcribed strand. (B) In the absence of transcription, no strand bias in mutation induction was observed, as indicated by nearly equal numbers of mutations on the two strands.
A parallel mutation analysis was performed to ensure that the observed mutagenesis at the cross-linked site was a direct consequence of passage through mammalian cells as opposed to generation in the repair- and recombination-deficient AB2480 strain. The cross-linked luciferase substrate was electroporated directly into AB2480 cells. The transformation efficiency of the MMC cross-linked plasmid was approximately 1.5% of the unmodified luciferase reporter plasmid. PCR amplification and analysis of 108 individual colonies revealed that 84 did not contain the cross-linked oligonucleotide and that 24 clones that had the oligonucleotide insert were cleavable by BspDI. The latter colonies presumably arose from a low level of contamination of uncross-linked oligonucleotides. These results indicate that the observed mutations were generated as a result of repair in the mammalian cells.
We also studied mutation induction in the cross-linked pCMV-LUC plasmid recovered from XPV cells. Analysis of 75 recovered colonies produced 11 independent clones with mutations around the cross-linked site. The resulting mutation frequency (15%) may indicate a reduction in the mutation rate compared to the 22% mutation rate (21 out of 96) for repair-proficient HT-1080 and AA8 cells. The positions and nature of base substitutions, primarily A for C and T for C, are similar to those of XPV-positive repair-proficient cells. These results suggest that PolH contributes to the ICL-induced mutation but that other lesion bypass polymerases may have redundant roles in the RIR mechanism to remove MMC ICLs.
DISCUSSION
We studied the removal of position-defined MMC ICLs in mammalian cells through a repair-mediated reporter reactivation assay. Our data suggest that mammalian cells are able to remove MMC ICLs by an NER-dependent mechanism, which does not require lesion-free homologous sequences or elements involved in the recombinational repair of ICLs. The identification of mutations as a result of this RIR of ICLs indicates that it is an error-prone pathway that may be, at least in part, responsible for the observed bifunctional alkylating-agent-induced mutagenesis.
Model for the RIR of ICLs.
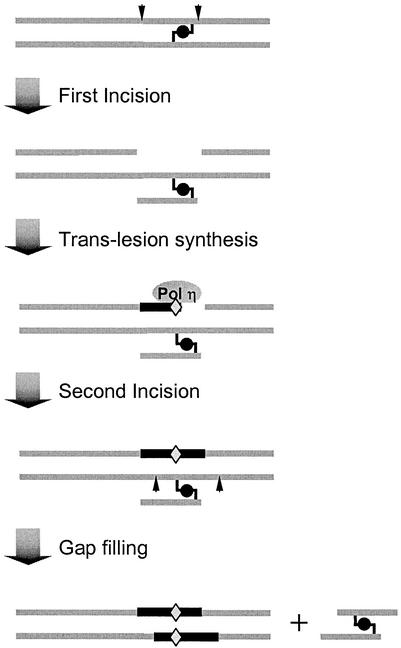
The incision step of the NER pathway consists of a number of essential activities: the excinuclease activities of ERCC1-XPF and XPG, the helicase activity of TFIIH (XPB and XPD), and the damage-binding activities of XPA and XPC-human RAD23B. We found that all five factors were required for the RIR of the MMC ICLs, a result that is consistent with the results of previous studies of photoactivated psoralen-mediated ICLs (47). A number of in vitro studies have suggested the involvement of NER incision activities in ICL processing. We have shown that the ERCC1-XPF complex is essential in ICL-induced repair synthesis (26). Dual incisions 5′ to the cross-link were observed and postulated to result in a futile cycle of DNA synthesis (33). Also, the ERCC1-XPF heterodimer was shown to exhibit an endonuclease activity capable of introducing incisions on the unpaired 3′ tail of the cross-link placed in a duplex 4 to 6 bases from a junction with unpaired DNA (25). These incision reactions, however, do not lead to the uncoupling of the ICLs, which is a crucial step in ICL removal. Conversely, if the NER dual incisions flanked the cross-linked base on the same strand, the resulting gap structure would effectively provide an uncoupling mechanism. The observed participation of Polη in the experiments in our study can be best explained by a lesion bypass gap-filling process, which is substantiated by the identification of mutations at the site opposing the cross-linked base. These data, collectively, support a model proposed in Fig. 7. Moreover, ICLs formed with psoralen and MMC differ drastically in their degrees of helix distortion and their ICL configurations and have distinct base preferences (13, 35, 48). Hence, the RIR model may apply to ICLs in general.
FIG. 7.
Model of lesion bypass mutagenic repair of MMC DNA interstrand cross-links. The first incision step places two nicks on the same strand, flanking the cross-linked base. The gap structure is created by the unwinding of the incised oligonucleotide and subsequently filled in by Pol η and other lesion bypass polymerases. Misincorporation likely occurs when DNA synthesis passes the still cross-linked base. Subsequently, the remaining lesion is repaired by a second round of regular NER reaction. Arrows indicate incisions. Diamonds indicate the positions of potential misincorporations, and dark lines depict repair synthesis patches.
Role of TCR in strand-specific formation of mutations.
The TCR pathway repairs lesions located in actively transcribed regions in the genome more rapidly than those in nontranscribed regions. In addition, the transcribed strand is preferentially repaired over the nontranscribed strand (8, 29). In mammalian cells, two gene products, CSA and CSB, in addition to the basic NER factors, are essential for TCR function (18, 43). In our study, both the CSA and the CSB mutant were significantly defective in the repair of MMC ICLs located in the actively transcribed region of the reporter substrate. These observations are consistent with the report that TCR is involved in the repair of psoralen cross-links in hamster cells (20). In our study, a profound strand bias toward the transcribed strand was found, whereas the loss of transcription completely abolished this effect. Apparently, the TCR pathway is able to direct the NER incisions toward the transcribed strand even though two opposing lesions are in virtually the same location. Our observations provide additional evidence that the transcription elongation complex may be a strong determinant of strand-specific repair in the case of ICLs.
TCR is evidently a major contributor in the RIR of MMC ICLs. Our results with the XP4PA mutant, however, showed that the repair of the MMC ICLs was also affected in an XPC-deficient cell line. Additional studies may be required to understand why the loss of XPC also affects ICL repair in the XPC mutants, which presumably have an intact TCR function.
Recombination-independent ICL repair is likely a minor pathway in mammalian cells.
Two types of DNA lesions, DSBs and ICLs, affect both strands of the double helix and can be repaired through either error-free mechanisms that involve homologous recombination with undamaged homologous sequences as donors or error-prone mechanisms. In the case of radiation-induced DSBs, the nonhomologous-end-joining pathway appears to be the predominant mechanism in mammals, as demonstrated by the severe radiation sensitivity of Ku mutants, while mutants such as XRCC2 and XRCC3 that are defective in the gene conversion mechanism exhibit minor sensitivities to ionizing radiation (11). These same XRCC mutants are, nevertheless, extremely sensitive to ICL-inducing agents, indicating that homologous recombination is likely part of the major pathway for ICL repair. In mammals, the loss of NER function results in moderate sensitivity to cross-linking agents (3, 19). Thus, the NER lesion bypass mechanism does not seem to be the predominant pathway for ICL removal from chromosomal DNA in mammalian cells, despite the relatively high efficiency of ICL repair in our plasmid reporter constructs.
A number of factors may limit the participation of the RIR mechanism. At the site of an ICL, strand separation is severely restricted by the cross-link, which consequently inhibits the formation of the bubble structure required for the dual NER incisions. As a result, the efficiency and accuracy of introducing flanking incisions may be limited. In budding yeast, the RIR of psoralen ICLs has recently been characterized (16) and a rev3/polymerase ζ-dependent repair of nitrogen mustard-mediated ICLs has been shown to function mostly in stationary-phase yeast cells (28). Similarly, in mammalian cells, specific stages of the cell cycle in which ICLs are encountered may further limit the role of the RIR pathway (15). Although the mechanism for the RIR of ICLs may not contribute extensively to the overall removal of ICLs genome-wide, its error-prone nature may be an important source of induced mutagenesis by cross-linking agents.
Acknowledgments
We thank Errol Friedberg, Zhigang Wang, Qingyi Wei, Lawrence Grossman, James Cleaver, Larry Thompson, and Nigel Jones for providing constructs and cell lines. We also acknowledge the staff of the DNA Sequencing Core Facility of the M. D. Anderson Cancer Center (supported by core grant CA16672) for their assistance in sequence analysis.
This work was supported by National Cancer Institute grants CA75160 (R.J.L.), CA76172 (L.L.), and CA91029 (L.L.).
H. Zheng and X. Wang contributed equally to this work.
REFERENCES
- 1.Adair, G. M., R. L. Rolig, D. Moore-Faver, M. Zabelshansky, J. H. Wilson, and R. S. Nairn. 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19:5552-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkari, Y. M. N., R. L. Bateman, C. A. Reifsteck, S. B. Olson, and M. Grompe. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 20:8283-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, B. S., T. Sadeghi, M. J. Sicilano, R. J. Legerski, and D. Murray. 1996. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosamide analogs. Cancer Chemother. Pharmacol. 38:406-416. [DOI] [PubMed] [Google Scholar]
- 4.Berardini, M., P. L. Foster, and E. L. Loechler. 1999. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 181:2878-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berardini, M., W. Mackay, and E. L. Loechler. 1997. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry 36:3506-3513. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott, K., and P. Jeggo. 1991. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res. 255:111-121. [DOI] [PubMed] [Google Scholar]
- 7.Cassier-Chauvat, C., and E. Moustacchi. 1988. Allelism between pso1-1 and rev3-1 mutants and between pso2-1 and snm1 mutants in Saccharomyces cerevisiae. Curr. Genet. 13:37-40. [DOI] [PubMed] [Google Scholar]
- 8.Christians, F. C., and P. C. Hanawalt. 1992. Inhibition of transcription and strand-specific DNA repair by alpha-amanitin in Chinese hamster ovary cells. Mutat. Res. 274:93-101. [DOI] [PubMed] [Google Scholar]
- 9.Cole, R., S. D. Levitan, and R. R. Sinden. 1976. Removal of psoralen interstrand crosslinks from DNA of Escherichia coli: mechanism and genetic control. J. Mol. Biol. 103:39-59. [DOI] [PubMed] [Google Scholar]
- 10.Cole, R. S. 1973. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl. Acad. Sci. USA 70:1064-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, A. R. 1993. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat. Res. 293:99-118. [DOI] [PubMed] [Google Scholar]
- 12.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichman, B. F., B. H. Mooers, M. Alberti, J. E. Hearst, and P. S. Ho. 2001. The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions. J. Mol. Biol. 308:15-26. [DOI] [PubMed] [Google Scholar]
- 14.Faruqi, A. F., M. M. Seidman, D. J. Segal, D. Carroll, and P. M. Glazer. 1996. Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell. Biol. 16:6820-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara, Y. 1982. Defective repair of mitomycin C crosslinks in Fanconi's anemia and loss in confluent normal human and xeroderma pigmentosum cells. Biochim. Biophys. Acta 699:217-225. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, R. B., M. Alberti, J. E. Hearst, M. A. Chua, and W. A. Saffran. 2001. Recombinational and mutagenic repair of psoralen interstrand cross-links in Saccharomyces cerevisiae. J. Biol. Chem. 276:31551-31560. [DOI] [PubMed] [Google Scholar]
- 17.Gurzadyan, G. G., H. Gorner, and D. Schulte-Frohlinde. 1995. Ultraviolet (193, 216 and 254 nm) photoinactivation of Escherichia coli strains with different repair deficiencies. Radiat. Res. 141:244-251. [PubMed] [Google Scholar]
- 18.Henning, K. A., L. Li, N. Iyer, L. D. McDaniel, M. S. Reagan, R. Legerski, R. A. Schultz, M. Stefanini, A. R. Lehmann, and L. V. Mayne. 1995. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555-564. [DOI] [PubMed] [Google Scholar]
- 19.Hoy, C. A., L. H. Thompson, C. L. Mooney, and E. P. Salazar. 1985. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 45:1737-1743. [PubMed] [Google Scholar]
- 20.Islas, A. L., F. J. Baker, and P. C. Hanawalt. 1994. Transcription-coupled repair of psoralen cross-links but not monoadducts in Chinese hamster ovary cells. Biochemistry 33:10794-10799. [DOI] [PubMed] [Google Scholar]
- 21.Jachymczyk, W. J., R. C. Von Borstel, M. R. A. Mowat, and P. J. Hastings. 1981. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems of DNA repair: the RAD3 system and the RAD51 system. Mol. Gen. Genet. 182:196-205. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, R. D., N. Liu, and M. Jasin. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401:397-399. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 24.Jones, C. J., and R. D. Wood. 1993. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry 32:12096-12104. [DOI] [PubMed] [Google Scholar]
- 25.Kuraoka, I., W. R. Kobertz, R. R. Ariza, M. Biggerstaff, J. M. Essigmann, and R. D. Wood. 2000. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 275:26632-26636. [DOI] [PubMed] [Google Scholar]
- 26.Li, L., C. A. Peterson, X. Lu, P. Wei, and R. J. Legerski. 1999. Interstrand cross-links induce DNA synthesis in damaged and undamaged plasmids in mammalian cell extracts. Mol. Cell. Biol. 19:5619-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 28.McHugh, P. J., W. R. Sones, and J. A. Hartley. 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 30.Miller, R. D., L. Prakash, and S. Prakash. 1982. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol. Cell. Biol. 2:939-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison, A., R. B. Christensen, J. Alley, A. K. Beck, E. G. Bernstine, J. F. Lemontt, and C. W. Lawrence. 1989. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 171:5659-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moynahan, M. E., T. Y. Cui, and M. Jasin. 2001. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61:4842-4850. [PubMed] [Google Scholar]
- 33.Mu, D., T. Bessho, L. V. Nechev, D. J. Chen, T. M. Harris, J. E. Hearst, and A. Sancar. 2000. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol. 20:2446-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson, J. R., D. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 35.Norman, D., D. Live, M. Sastry, R. Lipman, B. E. Hingerty, M. Tomasz, S. Broyde, and D. J. Patel. 1990. NMR and computational characterization of mitomycin cross-linked to adjacent deoxyguanosines in the minor groove of the d(T-A-C-G-T-A)·d(T-A-C-G-T-A) duplex. Biochemistry 29:2861-2875. [DOI] [PubMed] [Google Scholar]
- 36.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raha, M., G. Wang, M. M. Seidman, and P. M. Glazer. 1996. Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc. Natl. Acad. Sci. USA 93:2941-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, J. A., P. Szankasi, D. K. Nguyen, C. Ludlow, H. M. Dunstan, C. J. Roberts, E. L. Jensen, L. H. Hartwell, and S. H. Friend. 2000. Differential toxicities of anticancer agents among DNA repair and checkpoint mutants of Saccharomyces cerevisiae. Cancer Res. 60:328-333. [PubMed] [Google Scholar]
- 39.Sladek, F. M., M. M. Munn, W. D. Rupp, and P. Howard-Flanders. 1989. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 264:6755-6765. [PubMed] [Google Scholar]
- 40.Sugasawa, K., J. M. Ng, C. Masutani, S. Iwai, P. J. van der Spek, A. P. Eker, F. Hanaoka, D. Bootsma, and J. H. Hoeijmakers. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2:223-232. [DOI] [PubMed] [Google Scholar]
- 41.Theicher, B. A. 1997. Antitumor alkylating agents, p. 405-414. In V. T. DeVita, Jr., S. Hellman, and S. A. Rosenberg (ed.), Cancer: principles and practice of oncology, 5th ed. Lippincott-Raven, Philadelphia, Pa.
- 42.Tomasz, M. 1994. DNA adducts of the mitomycins, p. 349-357. In K. Hemminki, A. Dipple, D. E. G. Shuker, D. Segerback, F. F. Kadlubar, and H. Bartsch (ed.), DNA adducts: identification and biological significance, 1st ed. IARC, Lyon, France.D. A. K.D. A.
- 43.Troelstra, C., A. van Gool, J. de Wit, W. Vermeulen, D. Bootsma, and J. H. Hoeijmakers. 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71:939-953. [DOI] [PubMed] [Google Scholar]
- 44.Van Houten, B., H. Gamper, J. E. Hearst, and A. Sancar. 1986. Construction of DNA substrates modified with psoralen at a unique site and study of the action mechanism of ABC excinuclease on these uniformly modified substrates. J. Biol. Chem. 261:14135-14141. [PubMed] [Google Scholar]
- 45.Van Houten, B., H. Gamper, S. R. Holbrook, J. E. Hearst, and A. Sancar. 1986. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. USA 83:8077-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 47.Wang, X., C. A. Peterson, H. Zheng, R. S. Nairn, R. J. Legerski, and L. Li. 2001. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell. Biol. 21:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren, A. J., and J. W. Hamilton. 1996. Synthesis and structural characterization of the N2G-mitomycin C-N2G interstrand cross-link in a model synthetic 23 base pair oligonucleotide DNA duplex. Chem. Res. Toxicol. 9:1063-1071. [DOI] [PubMed] [Google Scholar]
- 49.Warren, A. J., A. E. Maccubbin, and J. W. Hamilton. 1998. Detection of mitomycin C-DNA adducts in vivo by 32P-postlabeling: time course for formation and removal of adducts and biochemical modulation. Cancer Res. 58:453-461. [PubMed] [Google Scholar]
- 50.Wijen, J. P., M. J. Nivard, and E. W. Vogel. 2000. The in vivo genetic activity profile of the monofunctional nitrogen mustard 2-chloroethylamine differs drastically from its bifunctional counterpart mechlorethamine. Carcinogenesis 21:1859-1867. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Y., F. Yuan, X. Wu, O. Rechkoblit, J. S. Taylor, N. E. Geacintov, and Z. Wang. 2000. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 28:4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]