Abstract
Acyclovir (ACV), like many antiviral drugs, is a nucleoside analog. In vitro, ACV triphosphate inhibits herpesvirus DNA polymerase by means of binding, incorporation into primer/template, and dead-end complex formation in the presence of the next deoxynucleoside triphosphate. However, it is not known whether this mechanism operates in vivo. To address this and other questions, we analyzed eight mutant polymerases encoded by drug-resistant viruses, each altered in a region conserved among α-like DNA polymerases. We measured Km and kcat values for dGTP and ACV triphosphate incorporation and Ki values of ACV triphosphate for dGTP incorporation for each mutant. Certain mutants showed increased Km values for ACV triphosphate incorporation, suggesting a defect in inhibitor binding. Other mutants showed reduced kcat values for ACV triphosphate incorporation, suggesting a defect in incorporation of inhibitor into DNA, while the rest of the mutants exhibited both altered km and kcat values. In most cases, the fold increase in Ki of ACV triphosphate for dGTP incorporation relative to wild-type polymerase was similar to fold resistance conferred by the mutation in vivo; however, one mutation conferred a much greater increase in resistance than in Ki. The effects of mutations on enzyme kinetics could be explained by using a model of an α-like DNA polymerase active site bound to primer/template and inhibitor. The results have implications for mechanisms of action and resistance of antiviral nucleoside analogs in vivo, in particular for the importance of incorporation into DNA and for the functional roles of conserved regions of polymerases.
Drugs can be valuable both for treating human disease and for probing biological and biochemical processes. The antiviral drug acyclovir (ACV) is the leading therapy for herpes simplex virus (HSV), an important human pathogen. ACV is a nucleoside analog that is a prototype of many other anti-herpesvirus and anti-HIV drugs. ACV consists of guanine linked to an acyclic sugar moiety that, in effect, is deoxyribose lacking its 2′ and 3′ moieties. The mechanism of ACV action (1) entails preferential phosphorylation in HSV-infected cells by a viral-encoded thymidine kinase. The resulting monophosphate (ACV monophosphate) is converted to ACV triphosphate (ACV-TP) by cellular enzymes. As is true with most other antiviral nucleoside analogs, the drug triphosphate is a more potent inhibitor of viral DNA polymerase than of cellular DNA polymerases (2); inhibition of DNA replication results in antiviral activity. Accordingly, HSV can become resistant to ACV by means of mutations affecting viral thymidine kinase or the catalytic subunit of viral DNA polymerase (Pol), and both kinds of mutants have been associated with failures in ACV therapy [reviewed in (3)].
In vitro, ACV-TP has been found to inhibit HSV DNA polymerase by means of a three-step mechanism (4). In the first step, ACV-TP binds to Pol, where it can act as a competitive inhibitor of the natural substrate, dGTP. Then, ACV-TP itself acts as a substrate and ACV monophosphate is incorporated into the growing DNA chain, where it is an obligate chain terminator. In a third step, the dNTP complementary to the next template position acts as a potent inhibitor of the polymerase, freezing it onto the ACV-terminated primer/template in a dead-end complex. Although this is an elegant mechanism, it has not been shown that it operates in virus-infected cells. Indeed, only indirect evidence exists to suggest that ACV is incorporated into HSV DNA in infected cells (5), where, in principle, ACV action could depend solely on competitive inhibition, drug incorporation causing chain termination, dead-end complex formation, or some combination of these. If one could find a drug-resistance pol mutation that altered one of these parameters specifically, that would argue for the importance of that parameter in vivo.
Aside from being a target for antiviral drugs, HSV Pol shares several discrete regions of sequence similarity with the catalytic subunits of α-like DNA polymerases, which include eukaryotic DNA polymerases α, δ, ɛ, and ζ. Seven of these regions (I–VII) are shared by most members of the family (6); others (e.g., δ-region C) are shared with only certain members of the family exemplified by DNA polymerase δ (7). HSV Pol has served as a model for α-like polymerases, especially because it is amenable to genetic, pharmacological, biochemical, and biophysical studies. The involvement, direct or indirect, of several conserved regions [I, II, III, V, VII, and δ-region C (formerly designated region A)] in substrate recognition was initially inferred from studies of HSV pol mutants that exhibit altered sensitivity to ACV and other antiviral drugs [reviewed in (8)]. Subsequent studies with other DNA polymerases have provided evidence for the specific roles of regions I, II, and III in polymerase function; for example, mutations that alter region I of DNA polymerase α specifically impair the rate of polymerase catalysis rather than substrate affinity (9). Recently, the crystal structure of a member of the α-like polymerase family, bacteriophage RB69 DNA polymerase, was solved, and certain conserved residues were located relative to a model of its active site (10). However, the specific roles of numerous conserved residues in polymerase function and the specific role of any conserved residue in ACV action and resistance have not been thoroughly explored.
To address these issues of drug mechanism and roles of conserved polymerase regions, we sought to determine how ACV inhibition is affected by eight HSV pol mutations representing each of six conserved regions by using a modified form of the assay used by Reardon and Spector (4) to define the three-step mechanism of polymerase inhibition in vitro by ACV-TP. We correlated these results with the effects of the mutations on susceptibility of HSV to ACV in cell culture and the locations of mutant residues in a model of the structure of an α-like DNA polymerase active site bound to primer/template and inhibitor. The results have implications for mechanisms of nucleoside analog action—particularly for the importance of incorporation of drug into DNA—and for how conserved regions function in polymerase action and drug resistance.
METHODS
Cells and Viruses.
Spodoptera frugiperda Sf9 and Sf21 cells were obtained from Invitrogen and maintained in Grace’s insect medium (BioWhittaker) supplemented with 10% fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin. Recombinant baculoviruses derived from Autographa californica nuclear polyhedrosis virus (AcNPV) were propagated by using standard methods (11).
Construction of Recombinant Baculoviruses.
For each recombinant baculovirus, a transfer plasmid encoding the mutant DNA polymerase was constructed. For most mutants, the BglII-NotI fragment of pBlue-pol (12) or pHDP1 (C.B.C.H., unpublished results), each of which contains the wild-type (wt) pol gene under the control of the baculovirus polyhedrin promoter, was replaced with the corresponding fragment of the mutant pol gene. In no case was PCR or site-directed mutagenesis used. To construct recombinant baculoviruses, each transfer plasmid was cotransfected into Sf9 cells with AcNPV DNA (Invitrogen or PharMingen) by using either cationic liposomes (Invitrogen), calcium phosphate, or the BaculoGold kit (PharMingen). Plaques expressing β-galactosidase were detected by using 5-bromo-4-chloro-3-indolyl-D-galactoside (X-gal) and then purified multiple times. The relevant portions of the pol genes of transfer plasmids and recombinant baculoviruses were sequenced on both strands to ensure that the desired mutation was introduced. Relevant information regarding the mutations is provided in Table 1.
Table 1.
Properties of mutant HSV Pols
| Kinetic constants for incorporation of
|
Fold ACVr | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ACV-TP
|
dGTP
|
||||||||
| Class | Name | Change | Region | Km, μM | kcat, min−1 | Km, μM | kcat, min−1 | Ki ACV-TP, μM | |
| KOS (wt) | — | — | 0.81 ± 0.13 | 37 ± 3.1 | 0.14 ± 0.02 | 14 ± 4.9 | 0.99 ± 0.12 | 1 | |
| Km | tsD9 | E597K | δ-C | 3.8 ± 0.6 | 13 ± 1.6 | 0.15 ± 0.03 | 9 ± 0.5 | 4.1 ± 0.45 | 3 |
| PFAr1 | R700G | II | 5.1 ± 1.4 | 23 ± 8.7 | 0.47 ± 0.15 | 11 ± 1.6 | 5.2 ± 1.5 | 2 | |
| PAAr5 | R842S | III | 3.1 ± 0.41 | 23 ± 2 | 0.24 ± 0.1 | 13 ± 4.3 | 2.5 ± 0.34 | 20 | |
| kcat | PFAr2 | R605V | δ-C | 1.1 ± 0.23 | 1.8 ± 0.6 | 0.53 ± 0.12 | 40 ± 19 | 3.5 ± 0.33 | 4 |
| F891C | F891C | I | 1.3 ± 0.35 | 0.61 ± 0.07 | 0.33 ± 0.03 | 9.6 ± 3.8 | 6.1 ± 2.5 | 10 | |
| Km, kcat | PFAr5 | S724N | II | 2.5 ± 0.51 | 11 ± 5.1 | 0.19 ± 0.14 | 9.9 ± 4.1 | 8.9 ± 1.1 | 8 |
| 615.8 | Y941H | VII | 4.7 ± 1.3 | 10 ± 0.59 | 0.33 ± 0.06 | 15 ± 1.4 | 3.7 ± 0.3 | 7 | |
| AraAr9 | N961K | V | 2.7 ± 1.1 | 9.5 ± 4.7 | 0.22 ± 0.05 | 4.9 ± 1.3 | 3.3 ± 0.63 | 4 | |
The leftmost column indicates the class of each mutant Pol defined in this study. The next column gives the name of the virus from which each mutant Pol derives. The amino acid change of each mutant is indicated in the next column in single letter code and the conserved region in which the altered residue lies is designated in the next column. The kinetic constants (± standard errors) derived in this study are presented in the next five columns. The rightmost column indicates the fold-resistance of mutant virus due to the pol mutation. These values were derived by dividing the dose of drug that reduces plaque formation of the mutant by 50% by the dose that reduces plaque formation of virus lacking the pol mutation by 50%. Fold-resistance values are derived from data from plaque reduction assays summarized in Refs. 15, 19, 20, and 35.
Purification of HSV Pol.
Each wt and mutant HSV Pol was overexpressed from 8 × 109 Sf21 cells infected with the corresponding recombinant baculovirus and purified as described (12) with the following modifications: The hypotonic buffer used during cell lysis was supplemented with one complete protease inhibitor mixture tablet (Boehringer Mannheim) and fractions containing Pol from the ssDNA-agarose column were pooled, diluted, and then applied to a Mono Q HR5/5 column (Amersham Pharmacia). The column was equilibrated with buffer A (50 mM Hepes–NaOH, pH 7.6/1 mM DTT/0.5 mM EDTA/10% glycerol) and eluted by a 15-ml linear gradient of 50–500 mM NaCl in buffer A. Each of the wt and mutant Pols eluted as a single peak and was apparently homogeneous as determined by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. Concentrations of HSV Pol were determined as described (13).
Synthesis of [3H] ACV-TP.
To synthesize radiolabeled ACV-TP, 800 μCi of 8-[3H] ACV monophosphate (18 Ci/mmol, Moravek Biochemicals, Brea, CA) was incubated with 50 mM Tris acetate, pH 7.5/10 mM MgCl2/1 mM potassium ATP (Sigma)/10 mM potassium phosphoenolpyruvate (Boehringer Mannheim)/6 units/ml of guanylate kinase (Sigma)/560 units/ml of pyruvate kinase (Boehringer Mannheim) at 37°C for 16 hr. The radiolabeled ACV-TP was purified and stored as described previously (4).
Enzyme Assays.
Enzyme assays were performed based on the methods of Reardon and Spector (4) in 25 μl volumes at 22°C and contained 50 mM Tris⋅HCl, pH 8.5/6 mM MgCl2/1 mM DTT/100 μg/ml BSA/50 mM (NH4)2SO4/4% glycerol/0.1-1 pmol of wt or mutant HSV-Pol, and 100 pmols of a hairpin primer/template, which was a 40-base oligonucleotide 5′-TTTTTTATGCGAGGTGGCTGGGTTTTTTCCCAGCCACCTC-3′ (purchased from Genosys, The Woodlands, TX). Varying amounts of [3H]-labeled ACV-TP or 8-[3H] dGTP (26 Ci/mmol, Dupont/NEN) were added to measure the Km and kcat with these substrates, respectively. Varying amounts of unlabeled ACV-TP (generously provided by Wayne Miller, Glaxo Wellcome) and radiolabeled dGTP were added to measure the Ki of ACV-TP for dGTP incorporation. Reactions were started by adding enzyme and stopped by adding 30 μl 0.1 M EDTA at times when incorporation was linear with time and multiple turnovers had occurred. Each sample was then passed through a Sephadex G-50 minicolumn (Amersham Pharmacia) to remove unincorporated labeled nucleotides, and the radioactivity was measured by using a scintillation counter.
Measurement of Kinetic Constants.
Initial velocities were calculated by plotting incorporation of substrate vs. time. Km and Vmax values were determined from Lineweaver–Burk plots. kcat values were determined by dividing Vmax by the enzyme concentration. Ki values were derived from Dixon plots. These values were measured based on two to four independent assays.
Model Building.
The structure of T7 DNA polymerase with primer/template and ddGTP bound (14) was superimposed onto the structure of bacteriophage RB69 DNA polymerase (10) by using the superposition option in the program quanta, version 4.0 (Molecular Simulations, Waltham, MA). The superimposition was greatly facilitated by the very close alignment (10) of regions II and I of RB69 polymerase with motifs A and C from the DNA polymerase I family, which includes T7 DNA polymerase as well as a β-strand in the “palms” of the two polymerases. The T7 polymerase was removed, as were extraneous segments of the RB69 polymerase, leaving a model of the active site of RB69 polymerase bound to primer/template and ddGTP. The ddGTP was then converted to ACV-TP by removing the 2′ and 3′ moieties. Residues in the RB69 structure corresponding to residues of interest in HSV Pol were identified by using the sequence alignment of Wang et al. (10).
RESULTS AND DISCUSSION
Mutant HSV Pols.
To address mechanisms of ACV action and resistance and the roles of amino acids within conserved regions of HSV Pol, we studied eight different mutant HSV Pols encoded by drug-resistant mutants of HSV. Each of the six different conserved regions to which ACV-resistance mutations map was represented (Table 1). The mutations have been shown previously to confer varying degrees of resistance to ACV in cell culture ranging from 2-fold to 20-fold (Table 1, rightmost column). The mutations also vary in conferring other phenotypes, such as cross-resistance or hypersensitivity to other drugs, pathogenesis, and mutation frequency (15–21).
To obtain sufficient quantities of enzyme, we purified wt and mutant HSV Pols from insect cells that had been infected with recombinant baculoviruses in which expression of each Pol was under the control of the strong polyhedrin promoter. For the wt Pol, ≥90% of the enzyme is active, as assayed by extension of a synthetic primer/template (J. Randell and D.M.C., unpublished results). The specific activities of the wt and mutant Pols assayed by using activated calf thymus DNA as primer/template and the four dNTPs as substrates were all very similar. Certain data that are not shown in this paper because of space constraints can be accessed on the Internet at coen.med.harvard.edu. Also, all of the enzymes exhibited similar maximal rates of incorporation of dGTP on a synthetic primer/template (see below).
A Modified Assay System for Studying ACV-TP Incorporation and Inhibition.
To analyze the different mutant Pols, we adapted the assays used by Reardon and Spector to elucidate a three-step mechanism of inhibition of HSV Pol in vitro by ACV-TP (4, 22). [Although it is theoretically possible that resistance could arise by acquisition of the ability to remove ACV monophosphate from primer/template, none of the mutant enzymes contains alterations in residues conserved among exonuclease active sites and none exhibited exonuclease activity on an ACV-terminated primer/template (L.H. and D.M.C, unpublished results).] The assays use a synthetic primer/template designed to accept either dGTP or ACV-TP as the first incorporated nucleotide opposite a C on the template strand. The original study used a 20-mer template annealed to a 9-mer primer; however, in our hands, this primer/template did not work well, perhaps because of inefficient or unstable annealing (data not shown). We therefore devised a 40-mer hairpin primer/template (see Methods), which, like the original template, contained an 11 base single-stranded template region. The primer region is annealed to the template to form a 12 base pair double-stranded region, which is connected by a six-base loop of thymine residues.
The assays devised by Reardon and Spector are performed under conditions of primer/template excess and multiple turnovers, and thus the rate of synthesis per polymerase molecule (kcat) depends on the ability of the polymerase to bind an initial primer/template, then bind and incorporate dGTP or ACV-TP, then dissociate after incorporation, and then bind a second primer/template and continue the cycle (4, 22). The original study (4) used HSV DNA polymerase holoenzyme isolated from HSV-infected cells, which is a heterodimer of Pol and its processivity factor, UL42 (23, 24) that dissociates from primer/template more slowly than Pol alone (25). To reduce the contribution of dissociation rate to kcat and to simplify our purification of Pols from recombinant baculovirus-infected insect cells, we modified the assay by omitting UL42. In the absence of other dNTPs, as expected from its more rapid dissociation rate (25), Pol by itself incorporated ACV-TP into primer/template at about a 3-fold higher rate than Pol plus UL42. Also, as expected, the presence of other dNTPs drastically reduced incorporation of ACV-TP by Pol alone just as they did for Pol plus UL42 (L.H. and D.M.C., unpublished results), showing that Pol alone formed dead-end complexes on ACV-terminated primer/template as does holoenzyme. Thus, except for the advantage of the higher incorporation rate, Pol alone behaved just as did holoenzyme on ACV-terminated primer/template (4). Similar results were obtained with certain mutant Pols with or without UL42 (data not shown). We therefore performed the remaining analyses with wt or mutant Pols in the absence of UL42.
A Class of Mutants Specifically Affected for Km of ACV-TP.
Using the modified assay system, we measured the Km and kcat values for incorporation of dGTP and ACV-TP by wt and mutant Pols in the absence of any other dNTP substrates (Table 1). Wt Pol exhibited a Km of 0.81 μM for ACV-TP and Km of 0.14 μM for dGTP (Table 1). The former value is only ≈3-fold less than the value obtained by Reardon and Spector by using a 20:9 mer primer/template (4). The latter value is >10-fold less than that observed by Reardon and Spector (4); this may be because the primer/template used in this study is different than that used by Reardon and Spector as the Km for dGTP of HSV DNA polymerase varies considerably with primer/template (4).
By using the criteria that Km or kcat values that differ by >3-fold from those of wt are meaningful, three classes of mutant Pols were discerned (Table 1). The first class of mutants (tsD9, PFAr1, and PAAr5), with alterations in three different conserved regions (δ-region C and regions II and III) exhibited >3-fold higher Km values for ACV-TP than did wt Pol, but <3-fold decreases in kcat for ACV-TP incorporation. None of these mutants exhibited a meaningful decrease in kcat for dGTP incorporation. For mutant tsD9, there was little or no increase in the Km for dGTP. For mutants PAAr5 and PFAr1, there were slight increases in Km for dGTP relative to wt Pol, similar to those previously described for these polymerases isolated from HSV-infected cells on activated DNA (26, 27), but these were less than the corresponding increases in Km for ACV-TP. Thus, this class of mutants (Km) was specifically affected for the Km of ACV-TP.
In the assays used here and by Reardon and colleagues, DNA synthesis is distributive because of “forced termination” after the incorporation of only one residue per primer/template (22). In the case of dGTP, this is because of the lack of other dNTPs; in the case of ACV-TP, this is because of the lack of other dNTPs and to chain termination. Under these circumstances, the Km of ACV-TP is expected to be a function of both the affinity (KD) of ACV-TP for the enzyme–primer/template complex and the ratio of the rate of dissociation of enzyme from primer/template (koff) to the rate of catalysis (kp) (including both conformational and chemical steps), which together determine the kcat observed (22). Thus, in theory, a higher Km could be caused by a higher KD for ACV-TP, a higher koff, or a lower kp. However, as the kcat values of the mutants for ACV-TP differed only slightly from that of wt enzyme, the simplest interpretation of the data is that the Km mutants bind ACV-TP with lower affinity than does wt virus. This interpretation is consistent with the locations of the PFAr1 and PAAr5 mutations in a model of an α-like DNA polymerase active site (see below).
A Class of Mutants Affected Specifically for kcat of ACV-TP.
The second class of mutants (Table 1) included mutants from δ-region C and region I (PFAr2 and F891C). These exhibited large decreases (>20-fold) in kcat for ACV-TP with little or no increase in Km for ACV-TP. There was little or no decrease in kcat for dGTP incorporation. The two mutants exhibited modest increases in Km values for dGTP; in the case of PFAr2, this was consistent with previous results with this polymerase isolated from HSV-infected cells on activated DNA (27). Thus, this class of mutants (kcat) was specifically impaired for kcat in reactions with ACV-TP.
As discussed above, the observed kcat is determined by the rate of catalysis, kp, and, the rate of dissociation, koff. Thus, in theory, the decreases in kcat of the mutants could be caused by decreases in kp, koff, or both. If the reductions in kcat of these mutant Pols were caused by reductions in koff, that would be expected to cause increased inhibition of Pol by ACV-TP, much as reductions in koff caused by inclusion of the dNTP complementary to the next template position traps wt enzyme in a dead-end complex, resulting in very low apparent Ki values for ACV-TP (4). However, these mutant Pols exhibited decreased inhibition of dGTP incorporation by ACV-TP both in the absence (Table 1) and presence (L.H. and D.M.C., unpublished results) of other dNTPs, as expected from the ACV resistance of the corresponding mutant viruses. Moreover, reduced koff values would be expected to result in reduced Km values, which were not observed. It is much more likely, then, that the reductions in kcat observed are caused by reductions in kp. With wt DNA polymerases, normally, under conditions of forced termination, koff is expected to be rate limiting (22). Thus, the fold reductions in kcat observed with these mutants relative to wt may well underestimate the fold reduction in kp; i.e., kp is probably rate limiting for the kcat mutant Pols rather than koff. Regardless, the analysis suggests that the kcat mutants are specifically defective for catalytic incorporation of drug into DNA. This interpretation is consistent with the location of the F891C mutation in a model of an α-like DNA polymerase active site presented below.
A Class of Mutants Affected for Both Km and kcat.
The three remaining mutants (PFAr5, 615.8, and AraAr9) from regions II, VII, and V, exhibited >3-fold increases in Km for ACV-TP and >3-fold decreases in kcat for ACV-TP incorporation (although none were as drastically impaired for kcat as the kcat mutants; Table 1). They exhibited increases in Km for dGTP, but these were substantially less than the corresponding increases in Km for ACV-TP. In the case of the region II mutant, PFAr5, a modest increase in Km for dGTP had been observed previously with this polymerase isolated from HSV-infected cells on activated DNA (27). Of the three mutants, only one, AraAr9, exhibited a meaningful decrease in kcat for dGTP; this decrease was ≈3-fold. Possible reasons for the behaviors of these mutants are discussed below.
Inhibition of dGTP Incorporation by ACV-TP in the Absence of Other dNTPs.
We then measured the ability of ACV-TP to inhibit dGTP incorporation in the absence of other dNTPs; i.e., without dead-end complex formation (Table 1, second column from the right). For five of the mutant Pols— the three Km mutants and two of the Km, kcat mutants—the Kis for ACV-TP inhibition of dGTP incorporation were very similar to the Kms for ACV-TP, as would be expected normally for an inhibitor that is an alternate substrate. For the three other mutant Pols, however, the Ki values for ACV-TP inhibition of dGTP incorporation were ≥3-fold higher than the Km values for ACV-TP. These latter mutant Pols include those that exhibited the lowest kcat values for ACV-TP, so that kp is probably more rate limiting for incorporation of ACV-TP than for dGTP. Thus, for these mutants, the Km for ACV-TP incorporation likely reflects occupancy of half of the enzyme molecules, but the Ki of ACV-TP for dGTP incorporation, where off-rate is expected to be rate limiting, would require occupancy of more than half of the enzyme molecules and would thus be higher.
Implications for Mechanisms of Nucleoside Analog Action.
In principle, nucleoside analogs such as ACV could inhibit viral DNA replication by either or both of two general mechanisms: (i) They could simply inhibit viral polymerases competitively with their analogous dNTP (dGTP for ACV-TP), or (ii) they could inhibit after incorporation by viral polymerase into DNA. If the former mechanism were sufficient, all drug-resistant pol mutants would be expected to affect affinity for drug triphosphate. In contrast, the latter mechanism predicts the existence of drug-resistance mutations that specifically impair catalytic incorporation of drug into DNA without impairing affinity for the drug. Thus, our results identifying the ACV-resistant kcat mutants argue that competitive inhibition is not sufficient to explain the mechanism of ACV inhibition of viral DNA replication. Rather, incorporation into DNA appears to be critical for ACV action in vivo. Similar results have been obtained recently by means of a presteady-state kinetic analysis of the effects of an HIV pol mutation that confers resistance to 3′-thiacytidine and, to a lesser extent, dideoxynucleosides. The resulting mutant reverse transcriptase exhibited a decreased rate of incorporation without an effect on affinity (28). We suggest that these results point to a critical role for incorporation of those anti-HIV drugs in vivo.
Incorporation of a nucleoside analog into DNA could inhibit subsequent viral DNA replication, (i) by inhibiting viral polymerase at a step beyond competitive inhibition or (ii) by rendering the viral DNA unable to be replicated (e.g., by chain-termination). For ACV, the first mechanism has been proposed; i.e., that ACV-terminated primer/template traps HSV polymerase in the presence of the next dNTP, causing dead-end complex formation (4). This mechanism suggests the possibility of a drug-resistance mutation that would relieve trapping of Pol by ACV-terminated primer/template in the presence of the dNTP. Because all of the mutants that we examined were impaired for incorporation of ACV-TP incorporation, either by higher Km, lower kcat, or both, they also required higher concentrations of the next dNTP to cause dead-end complex formation (L.H. and D.M.C., unpublished results). Thus, no simple mutation of this type was identified. Therefore, we asked whether the degree of resistance exhibited by a mutant in vivo was similar to the degree of resistance of the mutant Pol in vitro to inhibition by ACV-TP of dGTP incorporation in the absence of other dNTPs; i.e., in the absence of dead-end complex formation. For seven of the mutants, the fold resistance in vivo was no more than 2-fold higher than the increase in Ki (Table 1, last two columns). Thus, for these mutants, there was no need to invoke an effect on dead-end complex formation to explain the degree of resistance. However, mutant PAAr5 exhibits a 20-fold increase in ACV-resistance, but exhibited only a 2.5-fold increase in Ki in the absence of other dNTPs. Further studies are required to address this discrepancy; however, it raises the possibility that the PAAr5 mutation affects inhibition of viral replication at a step subsequent to ACV incorporation.
Locations of Mutations in a Model of an α-like DNA Polymerase Active Site.
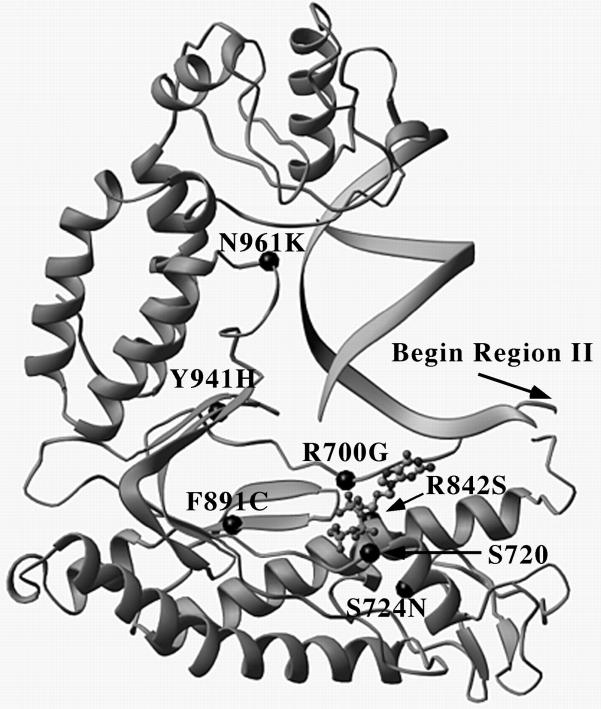
To try to understand the structural basis for the effects of the pol mutations on binding and incorporation of dGTP and ACV-TP, we constructed a model of an α-like DNA polymerase active site. Toward this end, we used the coordinates derived from the crystal structure of the bacteriophage RB69 DNA polymerase (10), which is a member of the α-like DNA polymerase family (29). Wang et al. (10) built a model of duplex DNA and dCTP into the RB69 active site; however, the duplex used was too short to reach residues corresponding to some of those mutated here, which made it difficult to use this model to understand the effects of certain mutations. To overcome this difficulty, we made use of the recent crystal structure of T7 DNA polymerase, which contains not only a longer primer/template than does the RB69 model, but also contains ddGTP, which is more closely related to ACV-TP than is dCTP (14). We took advantage of the very close superimposition of conserved structural elements in the “palms” of the RB69 and T7 polymerases to build the longer primer/template and ACV-TP into the RB69 structure (Fig. 1). The resulting model placed ACV-TP into the active site and provided more information about protein–DNA contacts than the earlier model. The primer/template orientations are slightly different in the two models, but this does not affect our interpretations.
Figure 1.
Cutaway diagram of an α-like DNA polymerase active site containing primer/template and ACV-TP. The α-carbon backbone of the protein chain derived from the structure of RB69 DNA polymerase (10) is presented as a gray ribbon diagram. The location of the N terminus of conserved region II is indicated with an arrow. The positions of amino acids altered by HSV drug resistance mutations (e.g., R700G) as well as a conserved serine (S720) relative to the RB69 sequence are indicated with black dots and were aligned according to Wang et al. (10). Primer/template is presented as two ribbons in the upper right quadrant of the figure and ACV-TP is presented as gray dots connected by gray lines.
Six of the amino acid changes whose effects we had analyzed enzymologically could be located in this model active site by comparisons of the HSV and RB69 DNA polymerase sequences. [The other two, E597K and R605V, one Km and one kcat, are within residues that are conserved between DNA polymerase δ and HSV DNA polymerase (δ-region C), but are not found in RB69 DNA polymerase.] Of the Km mutants that could be visualized, the locations of both the R700G (PFAr1) and the R842S (PAAr5) substitutions in regions II and III, respectively, were very close to the binding site for the acyclic sugar of ACV-TP. These locations readily lead to the suggestion that these mutations impair binding of ACV-TP more dramatically than that of dGTP, which is more constrained and contains 2′ and 3′ moieties that can stabilize its binding in the presence of the mutations. The behavior of these mutants is consistent with that of certain region II and III mutants of related polymerases, e.g., human DNA polymerase α (30, 31), which are affected for dNTP affinity. However, previous studies have not examined residues corresponding to R700 or R842 of HSV Pol.
Of the kcat mutants, the F891C substitution lies at the base of the loop that includes region I. The most conserved residues in region I, which include two aspartates, are directly involved in catalysis by means of Mg2+ in related polymerases, e.g., human DNA polymerase α (9). These residues lie adjacent to both the 3′ end of the primer and the phosphates of ACV-TP in our model. We suggest that the mutation alters the conformation of this loop, so that, although the enzyme can readily catalyze incorporation of dGTP, it is less able to catalyze incorporation of ACV-TP, whose phosphate positions are less constrained. The M184V mutation of HIV reverse transcripase (RT) that specifically decreases 3-thiacytidine triphosphate incorporation (28) may operate in a similar manner. Indeed, it has been suggested that the wt methionine residue may be needed to position precisely aspartate residues, which are homologous to those in region I of α-like DNA polymerases, for catalysis (32).
Of the Km, kcat mutations (none of which exerted large effects on kcat), the S724N (PFAr5) mutation in region II lies too far away in the model to make a direct contact with ACV-TP. However, this residue ordinarily interacts with the other serine (S720) in the SLYPSII motif in region II, which in turn interacts with the phosphates of ACV-TP. Thus, the mutation might affect S720, which could reduce both affinity for ACV-TP and its incorporation, thereby accounting for the effects on both Km and kcat.
The other two Km, kcat mutations are in conserved regions that lie in the thumb subdomain in the RB69 polymerase structure and whose roles have not been examined previously by enzymological studies of α-like polymerase mutants. In the model, the tyrosine corresponding to that affected by Y941H (615.8) in region VII is buried in a hydrophobic pocket that includes the phenylalanine altered in F891C. This could explain its effects on kcat. The tyrosine is also adjacent to basic residues that interact with the primer strand near its terminus. Similarly, the residue affected by N961K (AraAr9) in region V comes in close proximity to the template strand in our model; indeed, the model predicts a role for this conserved region in template binding. Interestingly, the mutation might be predicted to increase binding to primer/template by placing a lysine near the phosphate backbone and might thereby reduce koff. Consistent with this possibility, this mutant exhibits reduced kcat for dGTP as well as ACV-TP incorporation (Table 1). How these changes in primer/template binding could lead to drug resistance is not clear. One possibility, similar to that proposed for HIV RT mutations (33, 34), is that these mutations result in a repositioning or change in the conformation of primer/template that alters the ability of the enzyme to select or reject ACV-TP. This explanation, however, is not highly explicit. Moreover, a recent structure of HIV RT bound to primer/template and a nucleoside analog indicates that residues formerly thought to position primer/template actually lie near the dNTP binding site in the ternary complex (36). Our studies shed light on the functions of conserved regions in α-like DNA polymerases. Nevertheless, further enzymological and structural studies are required to fully understand these functions and the participation of conserved regions in antiviral drug action and resistance. The existence of multiple mechanisms for ACVr in vivo may help in the design of novel antiviral compounds to combat resistant strains of HSV.
Acknowledgments
We thank W. Miller for generous provision of unlabeled ACV-TP, J. Reardon for advice on synthesis of labeled ACV-TP and encouragement; T. Ellenberger for help with HPLC purification of labeled ACV-TP; K. Anderson, K. Grove, and C. Walsh for helpful comments, especially on enzymological issues; S. Doublié for help with model building; and S. Harrison for communicating results before publication. This work was supported by research grants AI 19838 and AI26077 to D.M.C. and DE10051 to C.B.C.H. from the National Institutes of Health. The Coen laboratory has received grants and gifts at various times from Glaxo Wellcome. L.H. was also supported by National Institutes of Health postdoctoral training grant AI07245 and A.M.G. was a predoctoral fellow of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HSV
herpes simplex virus
- ACV
acyclovir
- ACV-TP
acyclovir triphosphate
- Pol
catalytic subunit of HSV DNA polymerase
- wt
wild type
- kp
rate of catalysis
- koff
dissociation rate
- RT
reverse transcriptase
Footnotes
Data deposition: The model of an α-like polymerase active site bound to primer/template and inhibitor has been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1b1f).
References
- 1.Elion G B. In: History, Mechanism of Action, Spectrum and Selectivity of Nucleoside Analogs. Mills J, Corey L, editors. New York: Elsevier; 1986. pp. 118–137. [Google Scholar]
- 2.Martin J L, Brown C E, Matthews-Davis N, Reardon J E. Antimicrob Agents Chemother. 1994;38:2743–2749. doi: 10.1128/aac.38.12.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coen D M. Antiviral Res. 1991;15:287–300. doi: 10.1016/0166-3542(91)90010-o. [DOI] [PubMed] [Google Scholar]
- 4.Reardon J E, Spector T. J Biol Chem. 1989;264:7405–7411. [PubMed] [Google Scholar]
- 5.McGuirt P V, Shaw J E, Elion G B, Furman P A. Antimicrob Agents Chemother. 1984;25:507–509. doi: 10.1128/aac.25.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong S W, Wahl A F, Yuan P M, Arai N, Pearson B E, Arai K, Korn D, Hunkapiller M W, Wang T S. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Chung D W, Tan C K, Downey K M, Davie E W, So A G. Biochemistry. 1991;30:11742–11750. doi: 10.1021/bi00115a002. [DOI] [PubMed] [Google Scholar]
- 8.Coen D M. In: DNA Replication in Eukaryotic Cells. DePamphilis M, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 495–523. [Google Scholar]
- 9.Copeland W C, Wang T S-F. J Biol Chem. 1993;268:11028–11040. [PubMed] [Google Scholar]
- 10.Wang J, Sattar A K M A, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 11.Summers M D, Smith G E. A Manual of Methods for Baculovirus Vectors and Insect Cell Culture Procedures. College Station, TX: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 12.Hwang Y T, Liu B-Y, Coen D M, Hwang C B C. J Virol. 1997;71:7791–7798. doi: 10.1128/jvi.71.10.7791-7798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisshart K, Kuo A A, Hwang C B, Kumura K, Coen D M. J Biol Chem. 1994;269:22788–22796. [PubMed] [Google Scholar]
- 14.Doublié S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 15.Coen D M, Fleming H E, Jr, Leslie L K, Retondo M J. J Virol. 1985;53:477–488. doi: 10.1128/jvi.53.2.477-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field H J, Coen D M. J Virol. 1986;60:286–288. doi: 10.1128/jvi.60.1.286-289.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall J D, Coen D M, Fisher B L, Weisslitz M, Randall S, Almy R E, Gelep P T, Schaffer P A. Virology. 1984;132:26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson J, Kramer M, Rozenberg F, Hu A, Coen D M. Virology. 1995;206:263–268. doi: 10.1016/s0042-6822(95)80041-7. [DOI] [PubMed] [Google Scholar]
- 19.Chiou H C, Kumura K, Hu A, Kerns K M, Coen D M. Antiviral Chem Chemother. 1995;6:281–288. [Google Scholar]
- 20.Pelosi E, Mulamba G B, Coen D M. Antiviral Res. 1998;37:17–28. doi: 10.1016/s0166-3542(97)00054-5. [DOI] [PubMed] [Google Scholar]
- 21.Pelosi E, Rozenberg F, Coen D M, Tyler K L. Virology. 1998;252:364–372. doi: 10.1006/viro.1998.9447. [DOI] [PubMed] [Google Scholar]
- 22.Wilson J E, Porter D J T, Reardon J E. Methods Enzymol. 1996;275:398–424. doi: 10.1016/s0076-6879(96)75024-3. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb J, Marcy A, Coen D M, Challberg M D. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez T R, Lehman I R. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 25.Weisshart K, Chow C S, Coen D M. J Virol. 1999;73:55–66. doi: 10.1128/jvi.73.1.55-66.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.St. Clair M H, Miller W H, Miller R L, Lambe C U, Furman P A. Antimicrob Agents Chemother. 1984;25:191–194. doi: 10.1128/aac.25.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank K B, Derse D D, Bastow K F, Cheng Y C. J Biol Chem. 1984;259:13282–13286. [PubMed] [Google Scholar]
- 28.Krebs R, Immendörfer U, Thrall S H, Wöhrl B M, Goody R S. Biochemistry. 1997;36:10292–10300. doi: 10.1021/bi970512z. [DOI] [PubMed] [Google Scholar]
- 29.Wang C C, Yeh L S, Karam J D. J Biol Chem. 1995;270:26558–26564. doi: 10.1074/jbc.270.44.26558. [DOI] [PubMed] [Google Scholar]
- 30.Dong Q, Copeland W C, Wang T S-F. J Biol Chem. 1993;268:24163–24174. [PubMed] [Google Scholar]
- 31.Dong Q, Wang T S-F. J Biol Chem. 1995;270:21563–21570. doi: 10.1074/jbc.270.37.21563. [DOI] [PubMed] [Google Scholar]
- 32.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 34.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coen D M, Aschman D P, Gelep P T, Retondo M J, Weller S K, Schaffer P A. J Virol. 1984;49:236–247. doi: 10.1128/jvi.49.1.236-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H, Chopra R, Verdine G L, Harrison S C. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]