Abstract
Many endoplasmic reticulum (ER) proteins maintain their residence by dynamic retrieval from downstream compartments of the secretory pathway. In previous work we compared the retrieval process mediated by the two signals, KKMP and KDEL, by appending them to the same neutral reporter protein, CD8, and found that the two signals determine a different steady-state localization of the reporter. CD8-K (the KDEL-bearing form) was restricted mainly to the ER, whereas CD8-E19 (the KKMP-bearing form) was distributed also to the intermediate compartment and Golgi complex. To investigate whether this different steady-state distribution reflects a difference in exit rates from the ER and/or in retrieval, we have now followed the first steps of export of the two constructs from the ER and their trafficking between ER and Golgi complex. Contrary to expectation, we find that CD8-K is efficiently recruited into transport vesicles, whereas CD8-E19 is not. Thus, the more restricted ER localization of CD8-K must be explained by a more efficient retrieval to the ER. Moreover, because most of ER resident CD8-K is not O-glycosylated but almost all CD8-E19 is, the results suggest that CD8-K is retrieved from the intermediate compartment, before reaching the Golgi, where O-glycosylation begins. These results illustrate how different retrieval signals determine different trafficking patterns and pose novel questions on the underlying molecular mechanisms.
INTRODUCTION
KDEL and KKXX are short carboxy-terminal signals that play a crucial role for the localization in the endoplasmic reticulum (ER) of eukaryotic cells of many soluble proteins contained in the cisternal lumen and of type I transmembrane proteins, respectively (Munro and Pelham, 1987; Pelham, 1988; Nilsson et al. 1989; Jackson et al., 1990). These signals determine retrieval from post-ER located compartments in the exocytic pathway by virtue of retrograde transport mechanisms that are only partly understood (Lippincott-Schwartz, et al., 2000). Biochemical and genetic evidence indicates that a key role in these retrieval processes is played by the p26 KDEL receptor and by the proteins forming the COPI coatomer structure (Lewis et al., 1990; Lewis and Pelham, 1990; Semenza et al., 1990; Tang et al., 1993; Cosson and Letourneur, 1994; Letourneur et al., 1994; Cosson et al., 1996; Aoe et al., 1998; Majoul et al., 2001). Retrieval mediated by both signals certainly occurs from an early Golgi complex location, as demonstrated by Golgi-specific modifications of the N- or O-linked carbohydrate side chains of reporter proteins (Pelham, 1988; Pelham et al., 1988; Dean and Pelham, 1990; Jackson et al., 1993; Townsley et al., 1993; Gaynor et al., 1994); however, for the KDEL signal retrograde transport to the ER is possible from stations as far as the trans-Golgi network (Pelham, 1991; Peter et al., 1992; Miesenböck and Rothman, 1995; Jackson et al., 1993; Griffiths et al., 1994). In addition, indirect evidence suggests that retrieval may also take place before the protein has reached the Golgi complex, i.e., from the intermediate compartment (IC) (Martinez-Menàrguez et al., 1999; Shima et al., 1999; Stephens et al., 2000; Oprins et al., 2001). How the cell regulates the relative rates of retrograde transport from all these compartments is presently unknown.
To investigate recycling between the ER and the Golgi, and to compare the behavior of two different retrieval signals, we previously set up a model system based on a single reporter protein, the α chain of human CD8 glycoprotein tagged either with the KDEL or the KKXX signal, stably expressed in heterologous cells (Martire et al. 1996). The CD8-K construct is a soluble protein, corresponding to the lumenal domain of CD8 tagged with KDEL at its C terminus, whereas the CD8-E19 construct consists of the transmembrane form of CD8 in which the cytosolic tail is substituted with the one of adenovirus E19 protein, the latter contributing a KKMP sequence at the C terminus (Nilsson et al. 1989; Jackson et al., 1993). Using this system, we studied the trafficking events of the two constructs and found interesting differences between them, in their rate of initial O-glycosylation (an event reporting passage through the cis-Golgi compartment), in their extent of delivery to the cell surface, and in their steady-state distribution (Martire et al., 1996; Lotti et al., 1999). More precisely, CD8-E19 underwent relatively rapid initial O-glycosylation (t1/2 of 3 h) and at steady state was almost entirely represented by a single, initially O-glycosylated form, widely distributed among ER, IC, and Golgi compartments. In contrast, CD8-K was slowly secreted, but within the cell its distribution was almost completely restricted to the ER and it underwent initial O-glycosylation with slow kinetics (t1/2 of 16 h). Noteworthy, at steady state only 30% of CD8-K was initially O-glycosylated, but the 70% nonglycosylated and 30% initially glycosylated molecules formed a single intracellular pool (Lotti et al., 1999). These results could be explained by postulating that CD8-E19 has easier access to the cis-cisternae of the Golgi stacks from which it is efficiently retrieved, so that its transport further down the secretory pathway is precluded. The CD8-K construct on the other hand would have slower access to the Golgi stack but would be less efficiently retrieved from this compartment, resulting in its slow secretion.
A question that remained open from these studies was the cause of the different rates of transport of the two constructs to the Golgi. This could be explained by a faster exit of the CD8-E19 construct from the ER, by a more efficient retrieval of CD8-K from a pre-Golgi compartment, or by a combination of these two phenomena. Herein, we have addressed this question with a combined in vitro and in vivo approach. We show that CD8-K is recruited much more efficiently into transport vesicles budding from the ER and travels more quickly between the ER and the IC than CD8-E19, suggesting that its retrieval rate from pre-Golgi stations is correspondingly higher. Possible explanations for the different behavior of the two constructs are considered and the implications for the mechanisms of protein recycling between the ER and Golgi are discussed.
MATERIALS AND METHODS
Materials
All culture reagents were supplied by Sigma-Aldrich (Milano, Italy). Solid chemicals and liquid reagents were obtained from Merck (Darmstadt, Germany), Farmitalia Carlo Erba (Milano, Italy), Serva Feinbiochemica (Heidelberg, Germany), and BDH (Poole, United Kingdom). Enhanced chemiluminescence reagents were from Amersham Biosciences (Milano, Italy)
Antibodies
The following antibodies were used: mouse anti-CD8 protein monoclonal antibody OKT8, from Ortho (Raritan, NJ); mouse anti-CD8 protein monoclonal antibody N1 (Martire et al., 1996); polyclonal anti-CD8 (Jackson et al., 1993); polyclonal anti-ribophorin I (Yu et al., 1990); polyclonal anti-ERGIC-p58 (Saraste and Svensson, 1991); and polyclonal antifibronectin (Chemicon International, Temecula, CA).
Cell Culture, Transfection, Radioactive Labeling, and Immunoprecipitation
FRT cell lines stably expressing various CD8 reporter constructs were cultured as described previously (Martire et al., 1996). Parental FRT and HuH7 cells were grown and transiently transfected as described in Iodice et al. (2001). Radioactive labeling and immunoprecipitation were performed as reported previously (Iodice et al., 2001).
Recombinant DNAs
CD8-E19-D4, a construct encoding for a version of CD8-E19 with a deletion in the last four amino acids (KKMP signal), was obtained by introducing an XbaI between nucleotides 1881–1886 of the E3/19K sequence without altering the encoded amino acid sequence. The resulting cDNA, in plasmid pT8, was then used for cassette mutagenesis, by introducing complementary oligonucleotides coding for the deleted E3/19K sequence between the XbaI and the 3′ end BamHI sites.
Immunofluorescence Microscopy
Cells grown on glass coverslips were manipulated for indirect immunofluorescence as described previously (Mottola et al., 2000). Cells were observed under an Axiophot microscope or with an LSM 510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany).
In Vitro Budding Assay
Isolation of microsomal membranes from FRT cells, preparation of rat liver cytosol and in vitro vesicle-formation assay were performed as described previously (Nohturfft et al., 2000). Briefly, monolayers of FRT cells (80% confluent) were scraped in ice-cold buffer B (phosphate-buffered saline [PBS], 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 2 μg/ml aprotinin, 25 μg/ml N-acetyl-leu-leu-norleucinal). The cell pellet was resuspended in 0.4 ml of buffer F [10 mM HEPES-KOH pH 7.2, 250 mM sorbitol, 10 mM KOAc, 1.5 mM Mg(OAc)2, plus protease inhibitors], passed through a 22-gauge needle 20 times, and centrifuged at 1000 × g for 5 min at 4°C. The resulting supernatant was centrifuged at 1.6 × 104 g for 3 min at 4°C in an Eppendorf centrifuge to obtain microsomal membranes. The pellet was resuspended in 80 μl of buffer E [50 mM HEPES-KOH pH 7.2, 250 mM sorbitol, 70 mM KOAc, 2.5 mM Mg(OAC)2, 5 mM potassium EGTA, plus protease inhibitors]. The complete incubation mixtures contained 50 μg of microsomes, 600 μg of rat liver cytosol, 1.5 mM ATP, 0.5 mM GTP, 10 mM creatine phosphate, 4 U/ml creatine kinase in a final volume of 80 μl of buffer E, and were incubated either at 37°C or held on ice for 20 min. Reactions were terminated by transferring tubes to ice, followed by centrifugation for 3 min at 1.6 × 104 g at 4°C to obtain a medium speed pellet (M) and a supernatant fraction. The supernatant was centrifuged for 10 min at 10 × 104 g at 4°C in the Beckman Coulter TL-100 centrifuge to obtain a high-speed pellet (V).
Cell Fractionation and Sucrose Gradient Analysis
Cell fractionation and analysis from cultured cells was performed as detailed previously (Lotti et al., 1999). For in vitro budding analysis, incubated complete reaction mixtures or V fractions were loaded on the top (sedimentation analysis) or on the bottom (flotation analysis) of a discontinuous sucrose gradient (60, 45, 40, 35, 30, 25, 20, 15% [wt/vol] sucrose in 10 mM HEPES/KOH pH 7.3). The gradients were centrifuged at 43,000 rpm in a SW 50.1 Beckman Coulter rotor for 1 h (sedimentation) or 16 h (flotation). Fractions were then collected with the aid of a peristaltic pump.
SDS-PAGE and Western Immunoblot Analysis
Proteins were resolved on linear 12.5% polyacrylamide gels as described previously (Bonatti et al., 1989) and electrophoretically transferred to nitrocellulose filters, which were then incubated with primary antibodies diluted in blocking buffer (5% nonfat dry milk, 0.1% Tween 20 in PBS), followed by peroxidase-conjugated secondary antibodies. After washing, bound antibodies were detected by enhanced chemiluminescence. To quantify the relative amounts of the immunolabeled bands, different exposures of the blots were analyzed with the NIH Image program. More specifically, exposures were chosen so as to have equal, subsaturating, intensities of all analyzed polypeptides in the low-speed pellet (M) fractions. The intensities of the corresponding bands in the V fractions were then determined. After subtraction of the background from the V fraction, taken as the band intensity in the V fraction after incubation at 0°C, the relative amounts in V and M fractions for each protein were calculated after correction for the different proportions of the fractions loaded on the gels.
Electron Microscopy and Immunoelectron Microscopy
For conventional thin section electron microscopy, cell monolayers were fixed with 2% glutaraldehyde in PBS buffer at room temperature. Detached cells were then postfixed in 1% osmium tetroxide in veronal acetate buffer pH 7.4 for 1 h at 25°C, stained with 0.1% tannic acid in the same buffer for 30 min at 25°C and with uranyl acetate (5 mg/ml) for 1 h at 25°C, dehydrated in acetone, and embedded in Epon 812. Thin sections were examined unstained or poststained with uranyl acetate and lead hydroxide.
For immunoelectron microscopy, cells and fractions were fixed with 2% paraformaldehyde, 0.2% glutaraldehyde in 0.1 M phosphate buffer pH 7.4 for 2 h at room temperature, washed, and embedded in 10% gelatin in 0.1 M phosphate buffer. After solidification on ice, gelatin blocks were infused with 2.3 M sucrose overnight at 4°C, frozen in liquid nitrogen, and cryosectioned. Ultrathin cryosections were collected with sucrose and methylcellulose and incubated with polyclonal anti-CD8 antibody followed by 10-nm-diameter protein A-colloidal gold conjugates (British BioCell International, Cardiff, United Kingdom). Control experiments were performed by omission of the primary antibodies from the immunolabeling procedures. All ultrathin cryosections were finally stained with a solution of 2% methylcellulose and 0.4% uranyl acetate.
Quantitative Evaluation of Immunolabeling
The length of ER membranes was calculated using the Sigma Scan Measurement System (Yandel Scientific, Corte Madera, CA).
RESULTS
CD8-K, but Not CD8-E19, Shows High Rate of Export from ER In Vitro
To follow the recruitment of CD8-K and CD8-E19 into ER transport vesicles, we analyzed the export from the ER of the two proteins in vitro, applying to the FRT-derived clones an ER budding assay on isolated microsomal fractions (Rexach and Schekman, 1991; Rowe et al., 1996; Nohturfft et al., 2000; Muniz et al., 2001; see MATERIALS AND METHODS). Briefly, a total microsomal fraction is prepared by differential centrifugation and incubated in the presence of an ATP-regenerating system and rat liver cytosol; microsomes are again recovered by centrifugation, whereas the vesicles generated during the incubation are collected by ultracentrifugation; aliquots of the different fractions are then analyzed by SDS-PAGE and Western blot. As general positive controls for the procedure, i.e., proteins expected to efficiently exit the ER, we followed fibronectin and ERGIC-p58; conversely, ribophorin I was chosen as an example of a protein kept in the ER by static retention mechanisms (Fu et al., 1997, 2000) and that therefore should not be recruited into transport vesicles (or be recruited with low efficiency; Nohturfft et al., 2000). In addition, the nonglycosylated form of wild-type CD8 (CD8 unglycosylated or CD8u), and of its anchor-less counterpart devoid of cytosolic tail and transmembrane region (CD8-S unglycosylated or CD8-Su), were used as specific controls for CD8-E19 and CD8-K, respectively. Parallel incubations were also performed to confirm the requirement in the budding process of cytosol and ATP-regenerating system (our unpublished data).
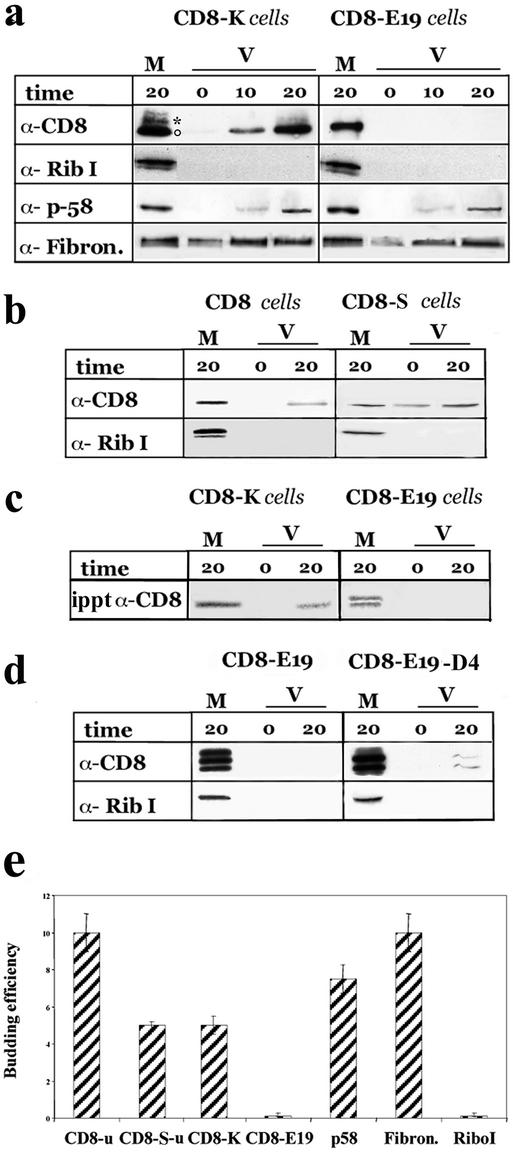
As illustrated in Figure 1, a and e, a clear-cut difference between the two reporter constructs was observed: CD8-K protein was detected in the vesicular fraction (both the nonglycosylated and initially glycosylated forms), whereas CD8-E19 was not. The significance of the negative result obtained for CD8-E19 protein was strengthened by the findings that 1) fibronectin and ERGIC-p58, but not ribophorin I were detected to a similar extent in the vesicular fraction of incubations programmed with microsomes from either CD8-K– or CD8-E19–expressing cells (Figure 1a); and 2) both CD8u and CD8-Su were recovered in the vesicular fraction programmed with microsomes prepared from the corresponding clones (Figure 1, b and e).
Figure 1.
CD8-K, but not CD8-E19, is efficiently exported from the ER in vitro. (a) Total microsomal fractions from cells stably expressing CD8-K or CDE19 were used to program budding assays in vitro as detailed in MATERIALS AND METHODS. The incubations were performed for the indicated times (in minutes; 0 indicates samples incubated on ice for 20 min) and then the microsomal (M) and the vesicular (V) fractions were separated by differential centrifugation and analyzed by SDS-PAGE and Western immunoblot to detect theindicated proteins. One-tenth of the M and 100% of the V fractions were loaded on the gels. The experiment shown is representative of seven and three independent assays for CD8-K and CD8-E19, respectively. ○ and * indicate the position on the gels of the nonglycosylated and initially glycosylated forms of CD8-K, respectively. (b) Total microsomal fraction from cells stably expressing CD8 and CD8-S were used for budding assay as described above. One-tenth of M and one entire V fraction, and one-half of M and three V fractions were loaded on the gels to detect CD8u and CD8-Su, respectively. Only the portions of the blots containing the nonglycosylated forms of the two proteins were analyzed. (c) Cells stably expressing CD8-K and CD8-E19 were radiolabeled for 20 min with [35S]cysteine and methionine and then used for the budding assay. The M and V fractions were lysed with 1% SDS, subjected to immunoprecipitation, and the total immunoprecipitated products were loaded on the gel. The dried gel was exposed for 3 d to a PhosphorImager (Bio-Rad, Hercules, CA) screen. (d) FRT cells were transiently transfected with pT8E19 or pT8D4, as described in MATERIALS AND METHODS and then used for the budding assay. One hundred percent of the M and V fractions were loaded on the gels. (e) Quantitation of budding efficiency of each protein recovered in the V fraction with respect to the total (M + V fractions). Only the results obtained with assays from stably expressing cells analyzed by immunoblotting are reported. Different exposures of the immunoblots were analyzed with the NIH Image program (see MATERIALS AND METHODS). The percentage of protein measured in the V fraction of incubations held on ice was subtracted from the corresponding incubations performed at 37°C. SD is indicated (n = 3).
To rule out the possibility that the budding assay was not detecting the exit from the ER of CD8-E19 only because at steady state an insufficient amount of this protein is localized in the ER compared with CD8-K, we pulse-labeled CD8-K and CD8-E19 cells for 20 min with [35S]methionine and cysteine to load the ER with newly synthesized protein forms, and microsomes were prepared to program budding assays that were analyzed by immunoprecipiatation (Iodice et al., 2001). As documented in Figure 1c, SDS-PAGE-autoradiography analysis of the immunoprecipitates revealed again only CD8-K in the vesicular fraction, whereas CD8-E19 was not detected (newly synthesized CD8-E19 shows as a doublet of nonglycosylated forms; Jackson et al., 1993; Martire et al., 1996).
The low budding efficiency of CD8-E19 compared with the wild-type CD8 suggested that the cytosolic tail of E19, in addition to causing retrieval, also reduces recruitment into transport vesicles. To investigate whether the C-terminal KKMP sequence was responsible for this reduced exit, we analyzed the behavior of a CD8-E19 mutant lacking these four amino acids (CD8-E19-D4). This mutant was previously reported to be transported from the ER to the plasma membrane with a t1/2 close to that of untagged CD8 (Nilsson et al., 1989). We first reproduced this observation in FRT cells (our unpublished data) and then performed parallel budding assays with microsomes prepared from FRT cells transiently transfected with either CD8-E19 or CD8-E19-D4. As shown in Figure 1d, CD8-E19-D4 was indeed detected in the vesicular fraction, whereas again CD8-E19 was not (note that multiple intermediate forms of initially O-glycosylated CD8-E19 and CD8-E19-D4 are present in overexpressing, transiently transfected cells (Jackson et al., 1993; Lotti et al., 1999).
To quantitatively compare the results obtained, the budding efficiency of each protein was calculated as the percentage of total recovered in the vesicular fraction (total being the amount of the protein present in the microsome + vesicular fraction) (Figure 1e): this was 10 and 7.5% for fibronectin and ERGIC-p58, respectively (from microsomes prepared with both CD8-K– and CD8-E19–expressing cells); 10, 5, and 5% for CD8u, CD8-Su, and CD8-K, respectively; and 0.1 and 0.2% for ribophorin I and CD8-E19. CD8-E19-D4 expressed by transient transfection, and pulse-labeled CD8K, showed a 7 and 6% budding efficiency (our unpublished data; Figure 1e). Taken together, the results of the in vitro budding assay suggest that CD8-K and CD8-E19 differ greatly in their rate of ER export, with the former showing the same efficiency as its untagged counterpart (CD8-Su), close to proteins described to be rapidly exported from the ER; and the latter being much more retained in the ER due the presence of the KKMP motif at the carboxyl terminus.
Analysis by Sucrose Gradient Centrifugation of Vesicular Fraction Generated In Vitro
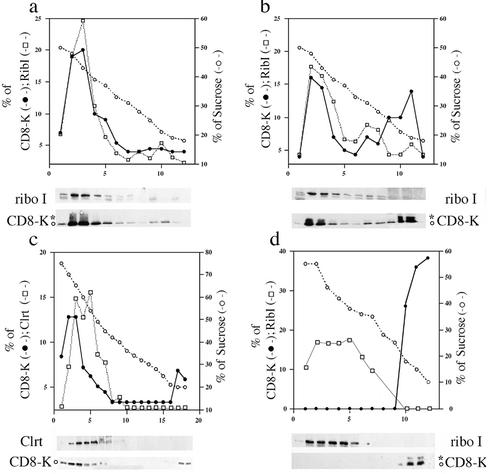
To further characterize the vesicles released during in vitro incubation, mixtures were examined by centrifugation on sucrose gradients. We have previously used a discontinuous sucrose gradient to analyze small amounts of postnuclear supernatant fractions obtained from FRT and other cell lines (Erra et al., 1999; Lotti et al., 1999; Iodice et al., 2001; Martire et al., 2001). The procedure was optimized to separate with a brief ultracentrifugation ER-, Golgi-, and IC-derived elements (banding at sucrose concentrations of ∼45, 35, and 25%, respectively). As shown in Figure 2a, roughly all CD8-K and ribophorin I proteins present in total reaction mixtures held on ice sedimented into the ER-enriched region, toward the bottom of the gradient. On incubation at 37°C (Figure 2b), a portion of both nonglycosylated and initially glycosylated CD8-K protein remained at the top of the gradient (in a region containing 15% sucrose), and a much smaller fraction equilibrated at the center (in a region containing 30–35% sucrose); in contrast, no ribophorin I remained at the top and little moved toward the center of the same gradient. These results indicated that the incubation at 37°C may have effect on the sedimentation of ER membranes, but clearly suggested that the vesicular fraction generated in vitro is of lower buoyant density than the starting microsomes (Muniz et al., 2001). Total reaction mixtures were also analyzed by flotation in sucrose gradients run to equilibrium (see MATERIALS AND METHODS). In these more stringent conditions, a significant portion of CD8-K (∼12% of total) was still detected in membrane fractions floating to the top region of the gradient (Figure 2c), whereas the lumenal ER marker calreticulin, and the transmembrane marker ribophorin I (our unpublished data), were confined to the lower part of the same gradient. Finally, we asked whether all CD8-K recovered in the vesicular fraction in vitro was in vesicles of low buoyant density on sucrose gradients. To answer this question, a vesicular fraction obtained by differential centrifugation of a reaction mixture programmed with microsomes derived from CD8-K–expressing cells was first mixed at 0°C with total nonincubated microsomes from parental FRT cells and then analyzed by sedimentation on a sucrose gradient. As shown in Figure 2d, all CD8-K protein present in the vesicular fraction was recovered in the top, less dense region of the gradient.
Figure 2.
Sucrose gradient analysis supports the evidence that CD8-K protein is exported from the ER to a vesicular fraction during the in vitro assay. See MATERIALS AND METHODS for details. Incubation mixtures of budding reactions programmed with microsomes from cells stably expressing CD8-K were examined by sedimentation on discontinuous sucrose gradients, except for c that reports the result of a flotation gradient. (a) Reaction mixture held on ice. (b–d) Reaction mixtures incubated for 20 min at 37°C. (d) At the end of the incubation, the microsomes were removed by centrifugation and the supernatant first mixed on ice with a corresponding amount of microsomes from parental FRT cells and then loaded on the top of the gradient. For all the gradients, equal aliquots from each fraction were analyzed by SDS-PAGE and Western immunoblot to detect the indicated proteins. The percentage of total protein contained in each fraction is reported on the left scale; on the right scale is the percentage (wt/vol) of sucrose. The relevant part of the immunoblot used for densitometry scanning is shown below each panel. ○ and * indicate the position on the gels of the nonglycosylated and initially glycosylated forms of CD8-K, respectively.
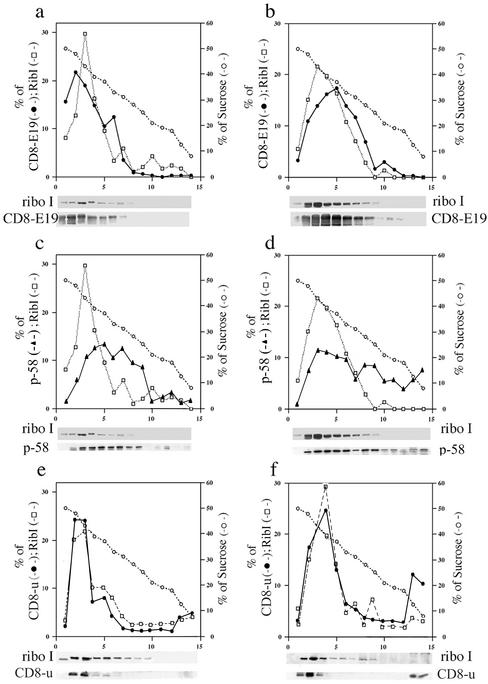
The different efficiency of recruitment of CD8-K and CD8-E19 was confirmed by sucrose gradient analysis. As shown in Figure 3, and in good agreement with the results reported in Figure 1, no CD8-E19 protein was recovered in the top region of the gradient as a result of the incubation at 37°C (Figure 3, a and b), whereas a portion of the internal control p58, and of CD8u, was shifted to the top (Figure 3, c and d and e and f, respectively), as observed for CD8-K (Figure 2). Thus, these results confirm the conclusion based on differential centrifugation, and demonstrated that the vesicular fraction generated in vitro has a characteristic low buoyant density,
Figure 3.
Sucrose gradient analysis confirms that CD8-E19 is not exported from the ER during the in vitro assay. Incubation mixtures of budding reactions programmed with microsomes from CD8-E19 (a–d), or from wild-type CD8 (e and f)-expressing cells, were examined by sedimentation on discontinuous sucrose gradients. (a, c, and e) Reaction mixtures held on ice. (b, d, and f) Reaction mixtures incubated for 20 min at 37°C. Couples a and c and b and d each refer to a single gradient. In e and f, only the portion of the blots containing the unglycosylated form of CD8 was analyzed. Gradient analysis and illustration as in Figure 2.
Trafficking of CD8-K and CD8-E19 Proteins between ER and IC In Vivo
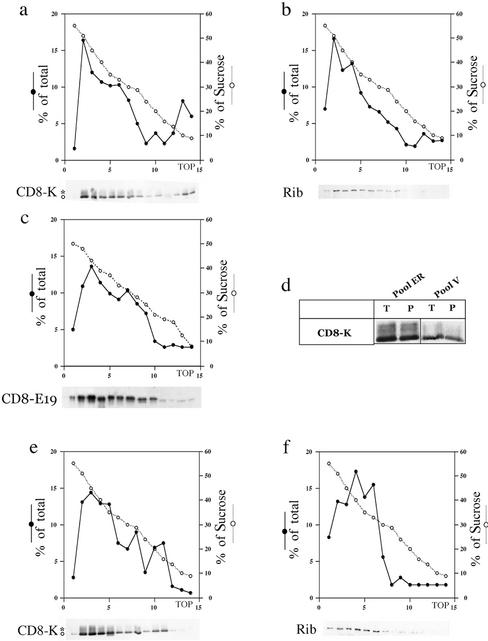
To confirm the evidence coming from the in vitro experiments we returned to investigate the trafficking of CD8-K and CD8-E19 proteins in cultured cells. Exponentially growing cultures were quickly chilled to 0°C to be processed for cell fractionation analysis, and the postnuclear supernatant fractions obtained were examined with the same discontinuous sucrose gradient described above. CD8-K and ribophorin I proteins clearly peaked in the more dense region of the gradient, enriched for ER-derived elements (Figure 4, a and b); and CD8-E19 protein was present, as expected, in the fractions ranging from 50 to 25% sucrose, reflecting its distribution among ER, IC, and Golgi complex (Figure 4c) (Lotti et al., 1999). Strikingly, a portion of CD8-K protein (roughly 10% of total), as well as of CD8 protein (our unpublished data), sedimented in the less dense region of the gradient (Figure 4a). In contrast, both ribophorin I and CD8-E19 were absent from this region (Figure 4, b and c). The CD8-K molecules detected at the top of the gradient were inside vesicular structures and not released in the buffer by the homogenization procedure, because like the material in the denser fractions, they were quantitatively recovered by sedimentation (Figure 4d). Therefore, the cell fractionation analysis of postnuclear supernatant fractions of exponentially growing cells showed the existence at steady state of a vesicular fraction that has an apparent low density in sucrose gradients and contains a portion of the CD8-K protein. This finding suggested that in live cells CD8-K protein, but not CD8-E19, is actively exported from the ER.
Figure 4.
Cell fractionation analysis of postnuclear supernatant fractions obtained from cultured cells reveals that CD8-K has an higher rate of trafficking in vivo than CD8-E19. Postnuclear supernatant fractions were prepared from exponentially growing, subconfluent cultures of CD8-K– and CD8-E19–expressing cells and analyzed by sedimentation on discontinuous sucrose gradients. Couples a and b and e and f each refer to a single gradient and Western blot. (d) Fractions 2 and 3 (pool ER) and 13 and 14 (pool V) of a gradient like the one in a were pooled and divided in two equal aliquots; one aliquot (T), was incubated in 20% trichloroacetic acid and the precipitated proteins collected by centrifugation; the other (P) was diluted fourfold and centrifuged for 30 min at 45,000 rpm in a Beckman Coulter TL-100 centrifuge to recover the membrane fraction. Finally, the four pellets were analyzed by SDS-PAGE and Western immunoblot. (e and f) CD8-K–expressing cultures were incubated for 3 h at 15°C before being processed and analyzed as described above. Gradient analysis and illustration as in Figure 2.
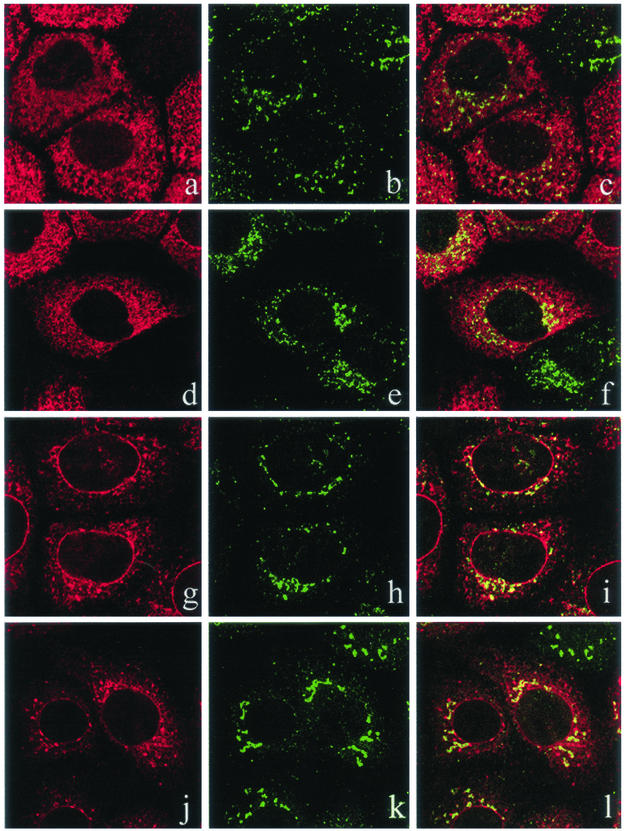
To test this hypothesis, we attempted to interfere with the trafficking of CD8-K protein by incubating the cells at 15°C, a temperature at which the anterograde transport from the ER to the Golgi complex is blocked in the IC (Saraste and Kuismanen, 1984; Bonatti et al., 1989; Klumperman et al., 1998). As shown in Figure 4e, at 15°C the light (∼15% sucrose) fraction of CD8-K was shifted to a slightly denser region (around 20% sucrose; compare a and e), which did not contain ribophorin I (Figure 4f). We have previously shown that this region of the gradient is enriched in IC-derived elements of FRT, HuH-7, Vero, and CV1 cell lines (Erra et al., 1996; Iodice et al., 2001; Martire et al., 2001). In parallel, a confocal immunofluorescence analysis was performed on cells incubated at 37 or 15°C, using ERGIC-p58 as marker of the IC. As already reported (Lotti et al., 1999), CD8-K showed less colocalization with ERGIC-p58 than did CD8-E19 in cells cultured at 37°C (Figure 5, compare a–c with g–i), reflecting its more tight localization in the ER. However, in cells incubated at 15°C, CD8-K showed an increase in the amount of colabeling with ERGIC-p58, whereas CD8-E19 did not (Figure 5, compare d–f with j–l). This finding, together with the evidence obtained by cell fractionation, supports the hypothesis that at steady-state CD8-K actively exits and returns to the ER from the IC, whereas CD8-E19 circulates slowly among ER, IC, and Golgi complex.
Figure 5.
A fraction of CD8-K, but not CD8-E19, moves to the IC when the cells are incubated at 15°C. Single optical sections of a confocal indirect immunofluorescence analysis. (a–c and g–i) CD8-K– and CD8-E19–expressing cells, respectively, incubated at 37°C. (d–f and j–l) CD8-K– and CD8-E19–expressing cells, respectively, incubated for 3 h at 15°C. Left, CD8-K or CD8-E19; middle, ERGIC-p58; and right, merge.
CD8-K Is More Present than CD8-E19 at ER Exit Sites of Transiently Transfected HuH-7 Cells
If CD8-K is exported from the ER at higher rate than CD8-E19, it should be proportionally more associated with the ER exit sites, the specialized subregions of the ER where the budding of transport carriers actually occurs. In exocrine pancreatic cells the exit sites are well defined by morphological criteria (Martinez-Menàrguez et al., 1999), but in tissue-cultured cells they seem less organized (Bannykh et al., 1996) and recognizable mostly as single membrane protrusions, sometimes facing a peripheral IC unit, arising from partly smooth regions of the ER (Lotti et al., 1996). We attempted to visualize the exit sites in FRT cells by conventional transmission electron microscopy, but the investigation was hampered by the nonsuitable organization of the ER in these cells: short cisternae, with a limited number of bound ribosomes, and poor spatial organization (our unpublished data). Conversely, in the human hepatoma cell line HuH-7 (Mottola et al., 2002), the ER is abundant, with long rough cisternae frequently parallel and limited smooth areas with coated or noncoated protrusions facing IC units (Figure 6, a and b). Therefore, we performed an immunogold labeling analysis on ultrathin cryosections of transiently transfected HuH-7 cells. As shown in Figure 6, c, d, g, and h, both CD8-K and CD8E19 immunolabeling were abundant and specific on ER cisternae and nuclear membranes. As previously reported for FRT cells (Lotti et al., 1999), CD8K was clustered within the ER, whereas CD8E19 was more homogeneously distributed, and CD8E19 was much more present on the IC and Golgi complex than CD8K (our unpublished data). Most relevant for our purpose, a close inspection of the specimens revealed that labeling for both proteins was present on membrane buds protruding from the ER (Figure 6, d, e, f, and j). However, the quantitative assessment of the relative amount of this labeling indicates that CD8-K was ∼ 2.2 times more concentrated on protrusions than CD8E-19 (Table 1) (but it is likely that the real difference is larger: a protein present in higher amount in the limited space of a protrusion may be not proportionally detected by the immunogold labeling because of steric hindrance). In addition, 70% of all protrusions were labeled by CD8-K, whereas only 38% were labeled by CD8-E19. Therefore, also an immunoelectron microscopical analysis after transient transfection of a cell line different from the FRT clearly indicated that CD8-K protein is more actively exported from the ER than CD8-E19.
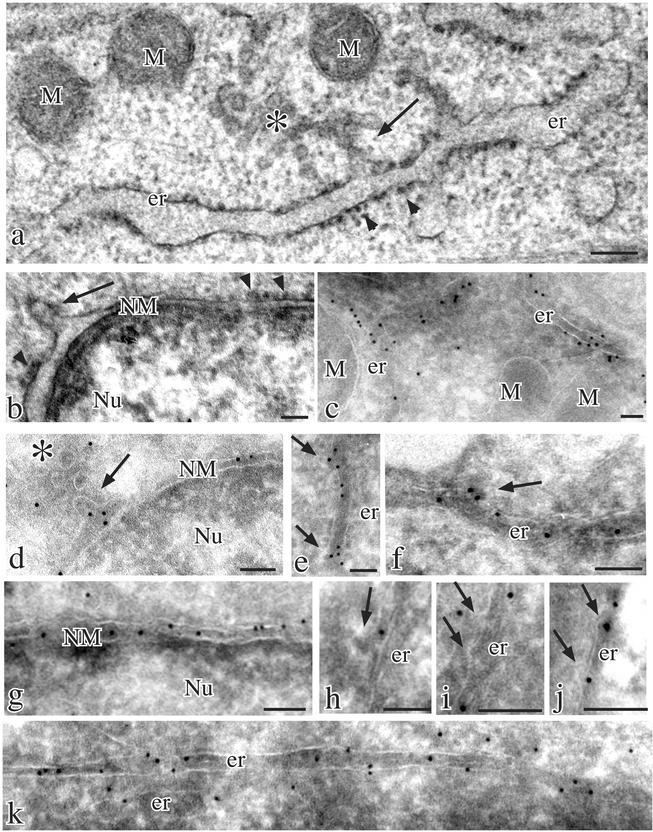
Figure 6.
Different presence of CD8-K and CD8-E19 at ER exit sites in transiently transfected HuH-7 cells. Conventional thin section electron microscopy (a and b) and cryoimmunoelectron microscopy (c–k) of parental (a and b) and transiently transfected HuH-7 cells expressing CD8-K (c–f) and CD8-E19 (g–k) proteins. The ultrastructural analysis revealed the presence of long ER cisternae and outer nuclear membranes mostly studded with ribosomes (arrowheads in a and b) and occasionally showing smooth areas with emerging protrusions (arrows in a and b) facing IC units (asterisk in a). Immunogold labeling with anti-CD8 polyclonal antibody and protein A-colloidal gold conjugates were dense and unevenly distributed over ER cisternae (c), whereas the gold particles frequently were localized on the smooth ribosome-free areas of the membranes and clustered over protrusions extending from them (e and f, arrows). Similar clustering of gold particles was observed in protrusions extending from the outer nuclear membranes (d, arrow). Immunolabeling was also present on IC units (d, asterisk). In contrast, immunogold labeling of CD8-E19–expressing cells was homogeneously distributed over either outer nuclear membranes (g) or ER cisternae (k) and the protrusions extending from the ER (arrows in h–j) were often unlabeled but sometimes labeled (j). M, mitochondria; er, endoplasmic reticulum; NM, nuclear membrane; Nu, nucleus. Bars, 0.1 μm.
Table 1.
Quantitation of CD8-K and CD8-E19 at ER exit sites
| Cell expressing | μm ER | No. of protrusions | Labeled protrusions (% total) | Gold particles in ER (% total) | Gold particles in protrusions (% ER-total) |
|---|---|---|---|---|---|
| CD8-K | 219 | 77 | 54 (70) | 3240 (89) | 78 (2.4) |
| CD8-E19 | 189 | 73 | 28 (38) | 3028 (70) | 33 (1.1) |
Parallel cultures of HuH-7 cells were transfected with CD8-K and CD8-E19 expression plasmids and processed for cryosectioning and immunogold labeling as detailed in the MATERIALS AND METHODS section. Transfection conditions were optimized to obtain roughly the same expression level of the two reporters. Fifteen low magnification micrographs of different expressing cells were taken at random for each transfection. Different enlargements of the micrographs were used to measure the following: the total number of gold particles, the number of gold particles in the ER, the total length of ER membranes, the total number of protrusions (see Figure 5), the number of gold-labeled protrusions, the total number of gold particles in protrusion. As judged by tracing a line to connect the ER membrane at the basis of the protrusion, only gold particles within the space of a protrusion were counted.
DISCUSSION
In this work, we have set up an in vitro budding assay to study the first steps of trafficking of a neutral reporter tagged with two different ER retrieval signals. Surprisingly, our results showed more efficient exit from the ER for the construct (CD8-K) that at steady state is almost completely confined to the ER, implying a more rapid recycling from compartments located downstream in the exocytic pathway. This conclusion was confirmed by cell fractionation and immunofluorescence microscopy of the cells stably expressing the two constructs, and by immunoelectron microscopy of a different cell line after transient transfection. Our results illustrate how the analysis of recycling based only on posttranslational modifications may give an incomplete picture of the underlying phenomena.
Different Trafficking of CD8-K and CD8-E19
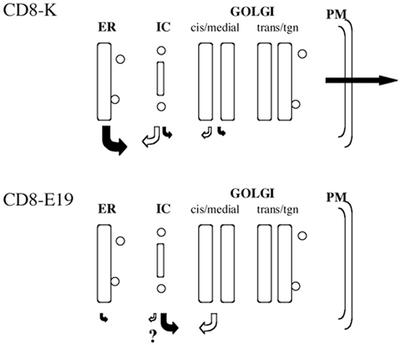
Taking together the results of this and previous studies (Martire et al., 1996; Lotti et al., 1999), the differences in the trafficking between CD8-K and CD8-E19 can be summarized as follows (Figure 7). CD8-E19 exits at low rate from the ER, but is retrieved inefficiently from pre-Golgi stations; therefore, it can reach the early Golgi, but proceeds no further, as indicated by the finding that no mature glycosylated form is present. CD8-E19 is distributed at steady state between ER, IC, and cis-Golgi complex and is represented almost completely by a single, initially O-glycosylated form; thus, a tight retrieval process from the early Golgi must guarantee its distribution.
Figure 7.
Model of the trafficking events of CD8-K and CD8-E19. The size of the arrows indicates the relative rate of the transport event considered. Question mark indicates the possibility that CD8-E19 is retrieved at low rate also from the IC, an event yet to be proven.
CD8-K has a higher rate of exit from the ER, which must be compensated by presumably higher retrieval. We believe that the main site from which CD8-K is retrieved is the IC, because initial O-glycosylation of the protein is very slow. Moreover, the retrieval from the Golgi is partially leaky: a small portion of the initially glycosylated protein proceeds to the trans-Golgi network where it is terminally glycosylated, and eventually secreted with an overall t1/2 of ∼20 h. Nonetheless, our data do not exclude that some of the CD8-K that reaches the cis-Golgi is rapidly retrieved to the ER without undergoing initial O-glycosylation.
Different Rate of Export of CD8-K and CD8-E19
The data from the in vitro budding assay indicate that CD8-K is recruited into vesicles with an efficiency comparable with its KDEL-less counterpart CD8-S, whereas CD8-E19 recruitment is severely reduced compared with the wild-type CD8 transmembrane protein. This reduction is so great as to result in undetectable amounts of CD8-E19 in the budded vesicle fraction; in contrast, of all the constructs tested, wild-type CD8 was the one with highest recovery in the vesicular fraction.
The reduced recruitment of CD8-E19 into budding vesicles can be ascribed to two causes. First, wild-type CD8 has a positive signal in its cytosolic tail, a C-terminal valine, which increases its rate of exit from the ER by approximately four- to fivefold (Iodice et al., 2001). This signal is lost in the CD8-E19 chimera. Second, the KKMP sequence contributed by the E19 tail, seems to have the ability to reduce recruitment into vesicles, as demonstrated herein by the recovery in the vesicular fraction of a mutant CD8-E19 deleted for the KKMP sequence (CD8-E19-D4). Thus, both the loss of a positive export signal and the acquisition of a retention signal probably contribute to the slow exit of CD8-E19 from the ER. The heretofore unreported ability of the KKMP sequence to slow exit from the ER recalls that of some other KKXX signals (Andersson et al., 1999; Zerangue et al., 2001). It should be noted, however, that the KKMP sequence of our CD8-E19 construct, although slowing recruitment into transport vesicles as revealed by our in vitro assay, does allow exit from the ER as demonstrated in vivo by the initial O-glycosylation of the recombinant protein.
In conclusion, the results reported herein indicate that the KDEL sequence has no effect on the recruitment of the lumenal domain of CD8 into transport vesicles, whereas the KKMP sequence retards exit of the transmembrane protein CD8-E19. The mechanism of this retardation remains to be established.
Possible Explanations for Different Site and Rate of Retrieval of CD8-K and CD8-E19
About 90% of total CD8-K is in the ER at steady state, and approximately one-third of it has undergone initial O-glycosylation, i.e., contains GalNAc attached to serine and threonine residues (Spiro, 2002), a process that occurs in an early Golgi region (Pascale et al., 1992). Newly synthesized CD8-K is glycosylated with a t1/2 of ∼16 h; thus, given its rapid exit from the ER demonstrated in the present study, the main station for its retrieval is located most likely upstream to the Golgi. We refer to this station as IC, but we cannot state at present whether the retrieval occurs preferentially from the peripheral or centrally located IC (Klumperman et al., 1998), or from both sites. It should be noted that although morphological investigations have strongly suggested retrieval from the IC (Martinez-Menàrguez et al., 1999; Oprins et al., 2001), a rigorous demonstration of this phenomenon has been difficult, because of the lack of posttranslational modifications characterizing this compartment. The combination of the in vitro and in vivo approaches of the present study gives strong support to the idea that the IC indeed plays an important role in the retrieval process.
In our opinion, the most interesting question that is opened by this work is the basis for the more efficient retrieval to the ER of CD8-K than CD8-E19. Current evidence suggests that both KDEL and KKXX driven retrievals require the interaction with COPI coatomer structures. The KDEL signal would induce oligomerization of the KDELr in the Golgi, recruitment of ArfGAP, and formation of COPI-coated budding complexes (Aoe et al., 1998; Majoul et al., 2001). The KKXX signal would directly bind COPI protein members (Cosson and Letourneur, 1994; Letourneur et al., 1994; Cosson et al., 1996). Thus, the most obvious difference between the two signals is that the KDEL is in the cisternal lumen, and thus interacts indirectly, via the transmembrane KDELr, with the cytosolic COPI coatomer structures, whereas the KKXX signal is in the cytosol and interacts directly with them. Therefore, a simple explanation for the more efficient retrieval of CD8-K to the ER is that within the IC the KDEL-KDELr-COPI interaction occurs with higher affinity than the one between the KKXX motif and COPI. This could allow retrieval of much CD8-K back to the ER before reaching the cis-Golgi, whereas CD8-E19 would proceed to the cis-Golgi to become initially O-glycosylated. In contrast, within the Golgi stack the interaction with COPI mediated by the KDELr would be weaker, causing the secretion of a small fraction of CD8-K. This reduced efficiency could be explained by a lower concentration of KDELr in the Golgi stack, or by its reduced affinity for the KDEL sequence and/or coat components, due to altered ionic conditions within the Golgi stack. Immunocytochemical evidence (Griffiths et al., 1994; Martinez-Menàrguez et al., 1999) and our own results on FRT-derived clones (Lotti et al., 1999) make the first hypothesis unlikely. Conversely, specific ionic conditions in the IC and early Golgi could favor the binding, whereas later in the Golgi a change in the environment could lower the affinity and explain the secretion of a small fraction of CD8-K. Finally, it cannot be excluded that p26 KDELr is not involved in retrieving CD8-K or that it plays a major role only in the retrieval from the Golgi complex. Indeed, the existence of other KDELr proteins has been claimed (Lewis and Pelham, 1992; Dunham et al., 1999), and their role in the retrieval process has not been clarified yet.
Another open question concerns the molecular basis for the more efficient retrieval of CD8-E19 from the early Golgi stack than from the IC, because COPI coats are abundant components of both compartments (Oprins et al., 1993; Griffiths et al., 1995). The simplest explanation is that COPI coatomer proteins are not the only players in retrieval of proteins bearing a KKXX signal. Most likely, other proteins, either more abundant or more functional in an early Golgi region than in the IC, play an important role in modulating the specificity of COPI structures and determine the generation of functionally different COPI coated vesicular carriers.
ACKNOWLEDGMENTS
We thank M. Jackson, G. Kreibich, and J. Saraste for the generous gift of antibodies; L. Nitsch for the confocal analysis; and E. Brescia and B. Mugnoz for excellent technical assistance. This work was supported in part by grants from the European Community (Transfer and Mobility of Researchers program) (to S.B.), Ministero Università Ricerca Scientifica e Tecnologica (PRIN 2000 and 2001) (to S.B., M.R.T., and N.B.), Associazione Italiana Ricerca sul Cancro (to M.R.T.), and Regione Molise (P.O.P. 94/99) (to G.M.).
Abbreviations used:
- ER
endoplasmic reticulum
- IC
intermediate compartment
- KDELr
KDEL receptor
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0468. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0468.
REFERENCES
- Andersson H, Kappeler F, Hauri HP. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J Biol Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- Aoe T, Lee AJ, van Donselaar E, Peters PJ, Hsu VW. Modulation of intracellular transport by transported proteins: insight from regulation of COPI-mediated transport. Proc Natl Acad Sci USA. 1998;95:1624–1629. doi: 10.1073/pnas.95.4.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatti S, Migliaccio G, Simons K. Palmitylation of viral membrane glycoproteins takes place after exit from the endoplasmic reticulum. J Biol Chem. 1989;264:12590–12595. [PubMed] [Google Scholar]
- Cosson P, Demolliere C, Hennecke S, Duden R, Letourneur F. δ- and ξ-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. Embo J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Dean N, Pelham HR. Recycling of proteins from the Golgi compartment to the ER in yeast. J Cell Biol. 1990;111:369–377. doi: 10.1083/jcb.111.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Erra MC, Iodice L, Lotti LV, Bonatti S. Cell fractionation analysis of human CD8 glycoprotein transport between endoplasmic reticulum, intermediate compartment and Golgi complex in tissue cultured cells. Cell Biol Int. 1999;23:571–577. doi: 10.1006/cbir.1999.0420. [DOI] [PubMed] [Google Scholar]
- Fu J, Kreibich G. Retention of subunits of the oligosaccharyl-transferase complex in the endoplasmic reticulum. J Biol Chem. 2000;275:3984–3990. doi: 10.1074/jbc.275.6.3984. [DOI] [PubMed] [Google Scholar]
- Fu J, Ren M, Kreibich G. Interactions among subunits of the oligosaccharyltransferase complex. J Biol Chem. 1997;272:29687–29692. doi: 10.1074/jbc.272.47.29687. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Heesen ST, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in the yeast. J Cell Biol. 1994;127:636–647. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Ericsson M, Krijnse-Locker J, Nilsson T, Goud B, Soling HD, Tang BL, Hong SH, Hong W. Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Pepperkok R, Locker JK, Kreis TE. Immunocytochemical localization of beta-COP to the ER-Golgi boundary and the TGN. J Cell Sci. 1995;108:2839–2856. doi: 10.1242/jcs.108.8.2839. [DOI] [PubMed] [Google Scholar]
- Iodice L, Sarnataro S, Bonatti S. The carboxyl-terminal valine is required for the transport of glycoprotein CD8α from the endoplasmic reticulum to the intermediate compartment. J Biol Chem. 2001;276:28920–28926. doi: 10.1074/jbc.M103558200. [DOI] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nisson T, Peterson PA. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Schweizer A, Clausen H, Tang BL, Hong W, Oorschot V, Hauri HP. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysines-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. A human homologue of the yeast HDEL receptor. Nature. 1990;348:162–163. doi: 10.1038/348162a0. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-s. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Sweet DJ, Pelham HR. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti LV, Mottola G, Torrisi MR, Bonatti S. A different intracellular distribution of a single reporter protein is determined at steady state by KKXX or KDEL retrieval signals. J Biol Chem. 1999;274:10413–10420. doi: 10.1074/jbc.274.15.10413. [DOI] [PubMed] [Google Scholar]
- Lotti LV, Torrisi MR, Erra MC, Bonatti S. Morphological Analysis of the transfer of VSV ts-045 G glycoprotein from the endoplasmic reticulum to the intermediate compartment in Vero cells. Exp Cell Res. 1996;227:323–331. doi: 10.1006/excr.1996.0281. [DOI] [PubMed] [Google Scholar]
- Majoul I, Straub M, Hell SW, Duden R, Soling HD. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic, measurements in living cells using FRET. Dev Cell. 2001;1:139–153. doi: 10.1016/s1534-5807(01)00004-1. [DOI] [PubMed] [Google Scholar]
- Martinez-Menàrguez JA, Geuze HJ, Slot JW, Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- Martire G, Mottola G, Pascale MC, Malagolini N, Turrini I, Serafini-Cessi F, Jackson MR, Bonatti S. Different fate of a single reporter protein containing KDEL or KKXX targeting signals stably expressed in mammalian cells. J Biol Chem. 1996;271:3541–3547. doi: 10.1074/jbc.271.7.3541. [DOI] [PubMed] [Google Scholar]
- Martire G, Viola A, Iodice L, Lotti LV, Gradini R, Bonatti S. Hepatitis C virus structural proteins reside in the endoplasmic reticulum as well as in the intermediate compartment/cis-Golgi complex region of stably transfected cells. Virology. 2001;280:176–182. doi: 10.1006/viro.2000.0733. [DOI] [PubMed] [Google Scholar]
- Miesenböck G, Rothman JE. The capacity to retrieve escaped ER proteins extends to the trans-most cisterna of the Golgi stack. JCell Biol. 1995;129:309–319. doi: 10.1083/jcb.129.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottola G, Cardinali G, Ceccacci A, Trozzi C, Bartholomew L, Torrisi MR, Pedrazzini E, Bonatti S, Migliaccio G. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology. 2002;293:31–43. doi: 10.1006/viro.2001.1229. [DOI] [PubMed] [Google Scholar]
- Mottola G, Jourdan N, Castaldo G, Malagolini N, Lahm A, Serafini-Cessi F, Migliaccio G, Bonatti S. A new determinant of endoplasmic reticulum localization is contained in the juxtamembrane region of the ectodomain of hepatitis C virus glycoprotein E1. J Biol Chem. 2000;275:24070–24079. doi: 10.1074/jbc.M910400199. [DOI] [PubMed] [Google Scholar]
- Muniz M, Morsomme P, Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HRB. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Jackson MR, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Oprins A, Duden R, Kreis TE, Geuze HJ, Slot JW. Beta-COP localizes mainly to the cis-Golgi side in exocrine pancreas. J Cell Biol. 1993;121:49–59. doi: 10.1083/jcb.121.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprins A, Rabouille C, Posthuma G, Klumperman J, Geuze HJ, Slot JW. The ER to Golgi interface is the major concentration site of secretory proteins in the exocrine pancreatic cell. Traffic. 2001;2:831–838. doi: 10.1034/j.1600-0854.2001.21112.x. [DOI] [PubMed] [Google Scholar]
- Pascale MC, Malagolini N, Serafini-Cessi F, Leone A, Bonatti S. Biosynthesis and oligosaccharide structure of human CD8 glycoprotein expressed in a rat epithelial cell line. J Biol Chem. 1992;267:9940–9947. [PubMed] [Google Scholar]
- Pelham HRB. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988;7:913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. Recycling of proteins between the endoplasmic reticulum and Golgi complex (review) Curr Opin Cell Biol. 1991;3:585–591. doi: 10.1016/0955-0674(91)90027-v. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter F, Nguyen VP, Soling HD. Different sorting of Lys-Asp-Glu-Leu proteins in rat liver. J Biol Chem. 1992;267:10631–10637. [PubMed] [Google Scholar]
- Rexach MF, Schekman RW. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T, Aridor M, McCaffery JM, Plutner H, Nuoffer C, Balch WE. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J Cell Biol. 1996;135:895–911. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, Kuismanen E. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell. 1984;38:535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- Saraste J, Svensson K. Distribution of the intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- Semenza JC, Hardwick KG, Dean N, Pelham HR. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- Shima DT, Scales SJ, Kreis TE, Pepperkok R. Segregation of COPI-rich and anterograde-cargo-rich domains in endoplasmic reticulum-to-Golgi transport complexes. Curr Biol. 1999;9:821–824. doi: 10.1016/s0960-9822(99)80365-0. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Lin-Marq N, Pagano A, Pepperkok R, Paccaud JP. COPI-coated ER-to-Golgi transport complexes segregate from the COPII in close proximity to ER exit sites. J Cell Sci. 2000;113:2177–2185. doi: 10.1242/jcs.113.12.2177. [DOI] [PubMed] [Google Scholar]
- Spiro RG. Protein glycosylation, nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- Tang BL, Wong SH, Qi XL, Low SH, Hong W. Molecular cloning, characterization, subcellular localization and dynamics of p23, the mammalian KDEL receptor. J Cell Biol. 1993;120:325–328. doi: 10.1083/jcb.120.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Wilson DW, Pelham HRB. Mutational analysis of the human KDEL receptor, distinct structural requirements for Golgi retention, ligand binding and retrograde transport. EMBO J. 1993;12:2821–2829. doi: 10.1002/j.1460-2075.1993.tb05943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sabatini DD, Kreibich G. Antiribophorin antibodies inhibit the targeting to the ER membrane of ribosome containing nascent secretory polypeptides J. Cell Biol. 1990;111:1335–1342. doi: 10.1083/jcb.111.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Malan MJ, Fried SR, Dazin PF, Jan YN, Jan LY, Schwappach B. Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc Natl Acad Sci USA. 2001;98:2431–2436. doi: 10.1073/pnas.051630198. [DOI] [PMC free article] [PubMed] [Google Scholar]