Abstract
Oxysterol binding protein (OSBP) homologs comprise a family of 12 proteins in humans (Jaworski et al., 2001; Lehto et al., 2001). Two variants of OSBP-related protein (ORP) 1 have been identified: a short one that consists of the carboxy-terminal ligand binding domain only (ORP1S, 437 aa) and a longer N-terminally extended form (ORP1L, 950 aa) encompassing three ankyrin repeats and a pleckstrin homology domain (PHD). We now report that the two mRNAs show marked differences in tissue expression. ORP1S predominates in skeletal muscle and heart, whereas ORP1L is the most abundant form in brain and lung. On differentiation of primary human monocytes into macrophages, both ORP1S and ORP1L mRNAs were induced, the up-regulation of ORP1L being >100-fold. The intracellular localization of the two ORP1 variants was found to be different. Whereas ORP1S is largely cytosolic, the ORP1L variant localizes to late endosomes. A significant amount of ORP1S but only little ORP1L was found in the nucleus. The ORP1L ankyrin repeat region (aa 1–237) was found to localize to late endosomes such as the full-length protein. This localization was even more pronounced for a fragment that additionally includes the PHD (aa 1–408). The amino-terminal region of ORP1L consisting of the ankyrin repeat and PHDs is therefore likely to be responsible for the targeting of ORP1L to late endosomes. Interestingly, overexpression of ORP1L was found to enhance the LXRα-mediated transactivation of a reporter gene, whereas ORP1S failed to influence this process. The results suggest that the two forms of ORP1 are functionally distinct and that ORP1L is involved in control of cellular lipid metabolism.
INTRODUCTION
Cellular sterol homeostasis is achieved through fine-tuned control of endogenous cholesterol biosynthesis, receptor-mediated uptake of low-density lipoprotein cholesterol, storage of cholesterol as fatty acyl esters, and removal of cholesterol to extracellular acceptors (Brown and Goldstein, 1999; Yokoyama, 2000). Oxysterols are naturally occurring oxygenated derivatives of cholesterol that are believed to form part of the regulatory apparatus for cholesterol homeostasis (Schoonjans et al., 2000). They are also involved in a variety of other processes such as cell differentiation (Hanley et al., 2000; Hayden et al., 2002), apoptosis (Panini and Sinensky, 2001), and calcium uptake (Kolsch et al., 1999). The presence of oxysterols in atherosclerotic plaques has implicated these compounds in the pathophysiological processes of atherosclerosis (Brown and Jessup, 1999; Vaya et al., 2001).
Oxysterol binding protein (OSBP) was the first known protein with affinity for oxysterols (Taylor et al., 1984; Taylor and Kandutsch, 1985; Dawson et al., 1989). This protein is distributed between the cytosol and membranes of the Golgi apparatus, depending on the cellular sterol content and transport status (Ridgway et al., 1992; Mohammadi et al., 2001) and influences both cholesterol and sphingomyelin biosynthesis (Lagace et al., 1997, 1999). The membrane association of OSBP is mediated by a pleckstrin homology domain (PHD) close to the amino terminus of the protein (Ridgway et al., 1992; Levine and Munro, 1998) and is a prerequisite for its functional effects (Lagace et al., 1997). Proteins with a ligand binding domain related to that of OSBP have been identified in organisms from yeast to human (Beh et al., 2001; Jaworski et al., 2001; Lehto et al., 2001). Phylogenetic analysis of the OSBP sequences in different organisms suggests that a divergent family of OSBP genes was present already at the early stages of eukaryotic evolution. The seven yeast OSBP homologs, named Osh1–7, are not individually essential but together share a vital function that evidently involves the cellular sterol homeostasis (Beh et al., 2001). Data on one of the yeast genes, KES1/OSH4, indicates a role in transport vesicle biogenesis at the Golgi complex (Fang et al., 1996; Li et al., 2002). A Drosophila OSBP protein has been implicated in cell cycle regulation (Alphey et al., 1998), whereas a Caenorhabditis elegans OSBP homolog called BIP was identified as a tentative modulator of the transforming growth factor-β signaling routes (Sugawara et al., 2001).
The mammalian OSBP homologs are called OSBP-related proteins (ORPs) (Laitinen et al., 1999; Lehto et al., 2001; Xu et al., 2001) or OSBP-like proteins (OSBPL) (Jaworski et al., 2001). The mammalian OSBP and its closest relative ORP4/OSBP2 (Moreira et al., 2001; Wang et al., 2002) have been shown to bind oxysterols, whereas ORP1 and ORP2 were suggested to bind phospholipids (Xu et al., 2001). The ligands of the other family members are at present unknown. Our recent data indicate a role for ORP2 in the control of cellular cholesterol metabolism and in the secretory function of the Golgi complex (Laitinen et al., 2002). Thus, the data presently available on the function of the OSBP gene family in different organisms yields a rather fragmented picture. It is plausible, however, that these gene products are regulatory components involved in various biological processes and use a similar ligand binding domain to accommodate diverse lipid ligands.
We now report on the mRNA expression of two variants of human ORP1, ORP1S and ORP1L, and characterize some of the structural features of ORP1 that determine its membrane association and intracellular localization. Furthermore, the effect of ORP1 overexpression on liver X receptor-mediated transactivation of a reporter gene is determined.
MATERIALS AND METHODS
Cell Culture and Transfection
Chinese hamster ovary (CHO)-K1 (ATCC CCL-61) were grown in Iscove's modified Dulbecco's medium (IMDM; Sigma-Aldrich, St. Louis, MO), 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were transiently transfected for 24 h with cDNA constructs in complete medium using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Stably expressing cells were generated by transfection with the ORP1L/pcDNA3.1 construct using LipofectAMINE 2000 followed by geneticin G418-sulfate selection (400 μg/ml IMDM) (Invitrogen) according to Invitrogen's protocol.
Isolation of Monocytes and Their Differentiation into Macrophages
Mononuclear cells were isolated from blood of healthy normolipidemic donors (thrombopheresis residues). Monocytes isolated by Ficoll gradient centrifugation were suspended in RPMI 1640 medium containing 40 mg/ml gentamicin, 0.05% glutamine (Sigma-Aldrich), and 10% of pooled human serum. Differentiation of monocytes into macrophages occurred spontaneously by adhesion of cells to the culture dishes. Mature monocyte-derived macrophages were used for experiments after 8 d of culture, whereas experiments in monocytes were done using cells isolated after 45 min of adherence to the plastic dish in RPMI 1640 medium containing gentamicin and glutamine.
cDNA Constructs
A full-length ORP1L cDNA was created by combining overlapping ORP1L 5′-end and 3′-end cDNA fragments. One microgram of total RNA from human fetal brain was reverse transcribed with the Superscript II enzyme (Invitrogen) according to the manufacturer's instructions by using primers ORP1N-R (5′-TCCAGTGCTTCTGACAAGATTTTG-3′) and ORP1L-R (5′-ACTCGGATCCGTAGATTAGCCAAACACCCTGAC-3′) for the 5′-end and 3′-end fragments, respectively. The 5′-end cDNA fragment was amplified using ORP1N-R cDNA as template. The 3′ end was amplified using ORP1L-R cDNA as template. All polymerase chain reactions (PCR) were carried out with 2.5 U of Pfu Turbo DNA-polymerase (Stratagene, La Jolla, CA) in a reaction volume of 20 μl containing 2–5 μl of template cDNA, 10× cloned Pfu DNA polymerase reaction buffer, 200 μM dNTPs, 10 pmol of each primer, and 10% dimethyl sulfoxide. The thermal cycling protocol applied for all reactions consisted of denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 3 min, and a final extension at 72°C for 10 min. The PCR products were cloned into pcDNA3.1 (Invitrogen) and transformed into Escherichia coli JM109. The cloned sequences were verified with a cycle-sequencing kit (BIGDYE; Applied Biosystems, Foster City, CA) and an automated ABI377 sequencer. The N-terminal fragment was subsequently ligated using a unique XbaI site in the overlap region into the C-terminal construct, generating the full-length ORP1L construct. The full-length ORP1L/pcDNA3.1 plasmid was used as template for the amplification of all truncated constructs except the ankyrin repeat (ANK) + PHD construct, which was generated by removing a 3′-end fragment by XbaI digestion of ORP1L. Oligonucleotide primers for the generation of cDNA constructs are listed in Table 1. EGFP-Rab5 and -Rab7 expression constructs were kindly provided by Dr. Marino Zerial (Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany) and Dr. Angela Wandinger-Ness (Department of Pathology, University of New Mexico Health Sciences Center, Albuquerque, NM).
Table 1.
Oligonucleotide primer sequences for amplification of ORP1 cDNAs
| Fragment | Forward primer | Reverse primer |
|---|---|---|
| 5′ end of ORP1L | 5′-ACTCGGATCCATGAACACAGAAGCGGAGCAACA-3′ | 5′-TCACGAATTCGGCCAGCGTCTCCAGTGCTTC-3′ |
| 3′ end of ORP1L | 5′-CCAGCATCCTTAGCGAGGACGA-3′ | 5′-ACTCGGATCCGTAGATTAGCCAAACACCCTGAC-3′ |
| ORP1S | 5′-ACTCGGATCCATGTCCGAAGAAAAAGACTGTGGT-3′ | 5′-ACTCGGATCCGTAGATTAGCCAAACACCCTGAC-3′ |
| ANK | 5′-ACTCGGATCCATGAACACAGAAGCGGAGCAACA-3′ | 5′-CTCGGATCCTATCGTTTCAATGCTTTGTAGATGAC-3′ |
| PHD | 5′-TCACGGATCCAAACCTCTTGACCTTGCCCAG-3′ | 5′-ACTCGGATCCTACAGCTGGTCCTGGGAACAGTA-3′ |
| LBD + PHD | 5′-TCACGGATCCAAACCTCTTGACCTTGCCCAG-3′ | 5′-ACTCGGATCCGTAGATTAGCCAAACACCCTGAC-3′ |
Antibodies
A glutathione S-transferase–ORP1 fusion protein corresponding to amino acids 428–950 in the ORP1L protein was expressed in E. coli BL21(DE3), purified by affinity chromatography on glutathione-Sepharose 4B (Pharmacia AB, Uppsala, Sweden), and used for immunization of HsdRIVM ELCO rabbits according to a standard protocol. The antibody (R250) was affinity purified in two steps. First, the ORP1 antiserum (1:5 dilution in phosphate-buffered saline [PBS]) was absorbed using glutathione-Sepharose 4B carrying glutathione S-transferase–ORP2 (Laitinen et al., 2002). Then, the unbound fraction was incubated with a Sepharose 4B column (Pharmacia AB) to which ORP1 (aa 428–950) had been coupled. The antibodies were eluted with 0.2 M glycine pH 2.8, neutralized and dialyzed against PBS. This antibody reacts with both ORP1S and ORP1L. A multiple antigenic peptide (Invitrogen) spanning amino acids 118–137 of ORP1L (not present in ORP1S) was used for raising antibodies (R279) specific for ORP1L, which were affinity purified using a Sepharose 4B column with covalently coupled immunizing peptide. Expression of ORP1 inserts in vector pcDNA4/HisMax was detected with mouse monoclonal antibody against the Xpress epitope (Invitrogen).
Immunofluorescence Microscopy
CHO-K1 cells were fixed with 4% paraformaldehyde, 250 mM HEPES pH 7.4 for 30 min and permeabilized for 30 min with 0.05% Triton X-100 in PBS. Nonspecific binding of antibodies was blocked with 10% FBS/PBS for 30 min, after which cells were incubated with ORP1 antiserum or anti-Xpress antibody in 5% FBS/PBS for 30 min at 37°C. The bound primary antibodies were visualized with tetramethylrhodamine B isothiocyanate (TRITC)-conjugated anti rabbit or TRITC-conjugated anti-mouse secondary antibody (Immunotech, Marseille, France). Cells were mounted in Mowiol (Calbiochem, San Diego, CA) containing 50 mg/ml 1,4-diazocyclo-[2,2,2]octane (Sigma-Aldrich). The specimens were analyzed with a TCS SP laser scanning confocal microscope (Leica, Wetzlar, Germany). In some experiments, cells were treated with 0.05% saponin, 80 mM PIPES, 5 mM EGTA, and 1 mM MgCl2 pH 6.8, for 1 min on ice before fixation.
Electron Microscopy
Cells cultured in complete IMDM medium containing G418-sulfate were fixed for 1 h at room temperature with 2.5% glutaraldehyde in 0.1 M phosphate buffer pH 7.2, postfixed with osmium tetroxide, dehydrated, and embedded in EMBed 812 (Electron Microscopy Sciences, Fort Washington, PA). Sections were cut horizontally and viewed with an EX200 electron microscope (JOEL, Tokyo, Japan).
Analysis of ORP1 Membrane Association
CHO-K1 cells were grown on 10-cm culture dishes and transfected with cDNA constructs for 24 h, after which cells were washed with PBS and scraped in 0.8 ml of 250 mM sucrose; 140 mM KCl; 10 mM HEPES pH 7.4; and 25 μg/ml each of chymostatin, leupeptin, antipain, and pepstatin A. The cells were disrupted by repeated passage through a 21-gauge needle and nuclei were pelleted by centrifugation for 10 min at 500 × g. Sucrose was added to the postnuclear supernatant to a final concentration of 2 M in a total volume of 2.0 ml, which was overlaid with 1.6 ml of 1.7 M sucrose and 0.4 ml of 0.8 M sucrose, 140 mM KCl, and 10 mM HEPES pH 7.4. The gradients were centrifuged in a SW 60 Ti rotor at 37 000 rpm for 20.5 h at 10°C. Fractions of 0.8 ml were collected, and proteins were precipitated with trichloroacetic acid and analyzed by Western blotting.
Western Blotting
Proteins were resolved in 12.5% SDS-polyacrylamide gels and transferred to Hybond-C extra nitrocellulose (Amersham Biosciences, Piscataway, NJ). Nonspecific binding of antibodies was blocked with, and all antibody incubations were carried out in, 5% fat-free powdered milk in 10 mM Tris-HCl pH 7.4, 150 mM NaCl, and 0.05% Tween 20. The bound antibodies were visualized with horseradish peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad) and the enhanced chemiluminescence system ECL (Amersham Biosciences).
Isolation and Quantitation of Total RNA
Total RNA was isolated from freshly isolated monocytes (day 0) and differentiating macrophages (days 4 and 8) with TRIzol reagent (Invitrogen). For tissue-specific expression analysis, seven human total RNA samples were purchased from Stratagene: fetal brain (male, 17 wk), adult liver (male, 73 yr), adult kidney (male, 56 yr), adult skeletal muscle (male, 17 yr), adult heart (male, 52 yr), adult colon (male, 69 yr), and adult lung (male, pooled donors 40, 51, 56, and 68 yr). Contaminating genomic DNA was removed from RNA samples by DNase treatment. The RNA quantity was determined with the RiboGreen RNA quantitation kit (Molecular Probes, Eugene, OR) and a 1420 multilabel counter (PerkinElmer Wallac, Turku, Finland).
Quantitation of ORP mRNA Expression by Real-Time PCR
Before first-strand cDNA synthesis, contaminating genomic DNA was eliminated as described previously with the following modifications (Huang et al., 1996). DNase treatment was performed in a total volume of 10 μl containing 1 μg of total RNA, 50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 1 U of RQ1 RNase-free DNase I, 5 μM random hexamers (Applied Biosystems), and 12 U of RNasin ribonuclease inhibitor. The reaction mixture was incubated at 37°C for 30 min, whereafter the DNase I was heat inactivated at 75°C for 5 min. The first-strand cDNA synthesis was initiated by adding a 10-μl reaction mixture containing 50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 20 mM dithiothreitol, 0.8 mM dNTP mix, 12 U of RNasin ribonuclease inhibitor, and 200 U of SuperScript II RNase H− reverse transcriptase (Invitrogen). After incubation for 1 h at 42°C, the reaction was stopped by keeping the samples at 70°C for 15 min.
Real-time PCR was carried out with the Amplifluor universal amplification and detection system (Intergen, Purchase, NY). The PCR reaction was carried out in a volume of 20 μl containing 25–50 ng of cDNA, 20 mM Tris-HCl pH 8.4, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTP mix, 1 U of Platinum Taq DNA polymerase (Invitrogen), 10 pmol of Amplifluor uniprimer, 10 pmol of untailed specific primer, and 1–2 pmol of tailed specific primer. Each sample was run in triplicate. In addition to primers for 11 ORPs, OSBP, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes, specific primers were designed to detect the two alternatively spliced ORP1 variants (Table 2 and Figure 1). The PCR amplification and fluorescence signal detection were performed with the ABI Prism 7700 sequence detection system (Applied Biosystems). PCR was initiated with denaturation at 94°C for 4 min, followed by 50 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The fluorescence signal was recorded during the annealing step.
Table 2.
Primers used for quantitation of ORP mRNA expression levels
| Forward primer (5′→3′) | Reverse primer (5′→3′) | Size (bp) | E value | ||
|---|---|---|---|---|---|
| ORP1L-F | TGATTGCCTTAATCTCTTCACC | ORP1-R | aACTCAGGGACCTTTCGGACTC | 223 | 1.78 |
| ORP1S-F | GGTCCTCGGATCTGGCCCA | ORP1-R | aACTCAGGGACCTTTCGGACTC | 209 | 1.88 |
| ORP1LS-F | GTGAGGAATTTTAAATTGGAACAA | ORP1-R | aACTCAGGGACCTTTCGGACTC | 189 | 1.57 |
| ORP2-F | aACCACCTGAGAAAGGCCAAGC | ORP2-R | CTCCAGCTCGTTGAGGCTCAC | 197 | 1.65 |
| ORP3-F | aGCATCTAGCTACTACCGAGCT | ORP3-R | CACATGGGTTGTGCCAATTGGA | 237 | 1.58 |
| ORP4-F | aAAGCTCTGGATCGACCAGTCA | ORP4-R | CTGGGACTGCTATGCATGACC | 218 | 1.72 |
| ORP5-F | aGTGCCGCTGGAGGAGCAGAC | ORP5-R | AGGGGCTGTGGTCCTCGTATC | 235 | 1.69 |
| ORP6-F | aGGATACTGCTCCACCTATTTCA | ORP6-R | ACAGGCAGGATTTCCATCGAC | 227 | 1.53 |
| ORP7-F | aCGGCCTACTCCTCCACATACC | ORP7-R | ACTGTTCCCACAGGCACAATC | 238 | 1.49 |
| ORP8-F | aGAAGAACAGGGAGATTTTGAATCA | ORP8-R | TCCTGTGAGTGGATCAAGTTC | 192 | 1.54 |
| ORP9-F | aCATCTTCCACACTAAACCCTTC | ORP9-R | CTCGTTCTGATCTTCCAACTTC | 217 | 1.58 |
| ORP10-F | aCTCAGCGACAGTGATATTCCAC | ORP10-R | AGGTCTGATCTTCTTGGGATAC | 208 | 1.56 |
| ORP11-F | aCACATTTTCTCTACCCTGTGCA | ORP11-R | CCCTTGCACTCTGCATACCAC | 211 | 1.65 |
| OSBP-F | aGATCCATCAGGAAAAGTCCAC | OSBP-R | CAGTGCCACTTTCCCAAGCA | 235 | 1.61 |
| GAPDH-F | CGGAGTCAACGGATTTGGTCGTAT | GAPDH-R | aAGCCTTCTCCATGGTGGTGAAGAC | 307 | 1.56 |
Primer carries UniPrimer target sequence 5′-ACTGAACCTGACCGTACA-3′ at its 5′ end. The sizes of the PCR products are given without this 18-bp extension. Real-time PCR efficiencies for each target gene are shown in the E value column.
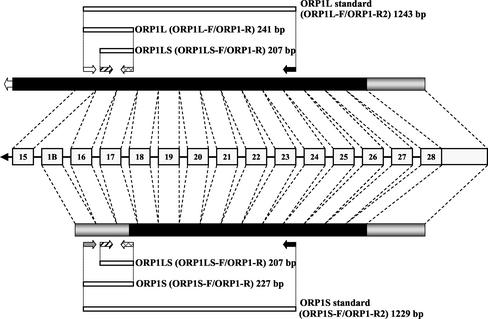
Figure 1.
Detection of the two ORP1 mRNA variants encoding ORP1L and ORP1S. The numbered boxes in the middle denote exons of the ORP1 gene. The bars consisting of black and gray boxes represent the two mRNAs; black indicates translated and gray nontranslated regions. The positions of primers for specific detection of the respective mRNAs are shown with arrows. The primer pairs are indicated within parentheses. The sequences of the primers are listed in Table 2.
To determine ORP mRNA expression levels during monocyte-macrophage differentiation, real-time PCR efficiency (E) values were determined for each target gene. Serial dilutions of cDNA corresponding to 100, 50, 25, 12.5, and 6.25 ng of total RNA were amplified as described above. Threshold cycle (CT) values were standardized by using the same calibrator cDNA during all runs. The slope was determined by plotting CT values against the logarithm of cDNA mass. E values for each target gene were calculated with the equation [E = 10(−1/slope)] (Table 2). The relative expression ratio (R) of the target gene was calculated as follows: R = (Etarget)ΔCTtarget (monocyte−macrophage) (Pfaffl, 2001). The expression levels were normalized for the total amount of RNA.
Tissue-specific Expression of ORP1L and ORP1S
Two DNA calibrators were generated for the quantitation of ORP1L and ORP1S expression in different tissues. ORP1L- and ORP1S-specific DNA templates were PCR amplified with the following primer pairs: ORP1L-F (5′-TGATTGCCTTAATCTCTTCACC-3′)/ORP1-R2 (5′-GCTTCCAGGGATAATGAATACACT-3′) and ORP1S-F (5′-GGTCCTCGGATCTGGCCCA-3′)/ORP1-R2 (Figure 1). After PCR amplification, the ORP1L standard (1243 base pairs, predicted molecular mass of 768 kDa) and the ORP1S standard (1229 base pairs, predicted molecular mass of 759 kDa) were run in a 1% agarose gel, excised, and purified with QIAquick gel extraction columns (QIAGEN, Valencia, CA). Concentrations of DNA fragments were determined with the Picogreen dsDNA quantitation kit (Molecular Probes) and a 1420 multilabel counter (PerkinElmer Wallac). Concentrations of DNA standards and tissue-specific cDNA samples were optimized before quantification of ORP1 variants. Serially diluted DNA standards were used to generate a calibration curve. DNA standards (20, 4, 0.8, 0.16, and 0.032 fg of double-stranded DNA) and tissue samples (50 ng of total RNA transcribed into cDNA as described above) were PCR amplified with three sets of primers: ORP1L-F/ORP1-R (for detection of ORP1L mRNA), ORP1S-F/ORP1-R (for detection of ORP1S mRNA), and ORP1LS-F/ORP1-R (for detection of both ORP1L and ORP1S mRNA) (Figure 1).
Transactivation Assay
The transactivation assays were carried out essentially as described previously (Willy et al., 1995). Twenty-four hours before transfection, COS-1, human embryonic kidney (HEK) 293, or HepG2 cells were seeded on 24-well plates in DMEM supplemented with 10% fetal calf serum at 5 × 104 cells/well. Transfection mixtures contained 50 ng of the luciferase reporter plasmid and 25 ng of LXRα and/or retinoid X receptor (RXR) expression plasmids, or 10 ng of a chimeric Gal4 construct containing the ligand-binding domain of LXRα, and (50–100 ng) of ORP1/pcDNA4HisMax expression plasmid. The expression plasmid for β-galactosidase, pSVβ-gal (50 ng), was added for normalization. Cells were transfected for 2 h by lipofection by using RPR-120535B (Aventis, Strasbourg, France) in serum-free medium. The medium was subsequently replaced by DMEM containing 0.2% fetal calf serum and cells were treated for 36 h with 10 μM 22(R)-hydroxycholesterol ([22(R)-HC]; SigmaAldrich) or 0.1 μM synthetic LXR agonist T0901317 (Tularik, San Francisco, CA). Cell extracts were prepared and assayed for luciferase activity. Data were normalized using the β-galactosidase activity.
RESULTS
Analysis of ORP1S and ORP1L mRNA Tissue Expression
Two mRNA variants of the ORP1 gene have recently been reported (Lehto et al., 2001; Xu et al., 2001). One of these, denoted hereafter as ORP1S (ORP1 “short”), encodes a predicted 50-kDa protein with 437 aa residues that consists of a ligand binding domain only (Xu et al., 2001). The other, ORP1L (ORP1 “long”), encodes a 108-kDa protein with 950 aa. This protein structurally resembles OSBP, which has a PHD (Lemmon and Ferguson, 2000) near its amino terminus. In addition, ORP1L has three ankyrin repeats (Sedgwick and Smerdon, 1999) situated near the N terminus (Lehto et al., 2001). The ORP1L transcript consists of sequences from 28 exons (Lehto et al., 2001). The ORP1S mRNA is apparently generated through the use of an alternative first exon, named exon 1B, which in the genome is located between exons 15 and 16 (Figure 1). The two transcripts have been suggested to arise from the use of two different promoters (Jaworski et al., 2001).
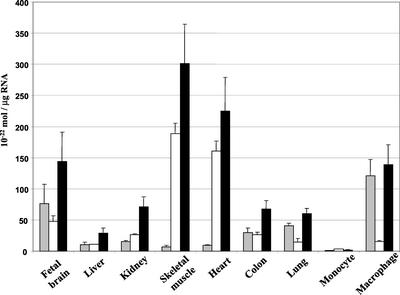
To investigate the expression of the ORP1S and ORP1L transcripts in human tissues we used reverse transcription-PCR with fluorescent on-line detection and primer pairs specific for the two distinct mRNAs (Figure 1). cDNA fragments corresponding to each transcript were used to make standard curves for converting the results to absolute mRNA quantities. A primer pair that detects both mRNAs was used as a control. Total RNA from human tissues reported to express ORP1 at high levels based on Northern blot analysis (Laitinen et al., 1999) were chosen for reverse transcription-PCR analysis: brain, heart, skeletal muscle, colon, lung, kidney, and liver. Primary human monocytes and macrophages differentiated in vitro were also included in the analysis. The tissue expression patterns were markedly different for the two mRNAs (Figure 2). ORP1S is more abundant than ORP1L in skeletal muscle and heart, whereas ORP1L is the predominant transcript in macrophages, brain, and lung. Approximately equal levels of the two mRNAs were found in colon, kidney, and liver. Both messages were detected at extremely low levels in the primary monocytes. The product quantities from reactions (LS) that nonselectively detect both mRNAs were close to the sum of the specific L and S reactions in all cases.
Figure 2.
Real-time PCR analysis of tissue expression of the ORP1 variants. The mRNA source tissues are indicated below the bars. The level of ORP1L and ORP1S expression is indicated with gray and white bars, respectively. Black bars show the level of ORP1 expression determined with primers that detect both variants. The data has been normalized using the total amount of RNA in each sample. Data are given as mean ± SD (n = 2).
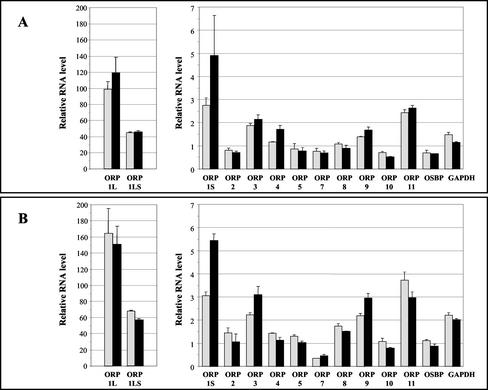
To study whether the increase observed in the ORP1 mRNA levels during monocyte-macrophage differentiation was unique for this ORP gene or whether it also occurs with the other family members, the relative quantities of all OSBP-related gene messages in monocytes and macrophages were determined. Monocytes isolated from buffy coat by incubation on plastic dishes in the absence of serum were differentiated by adding 10% human serum (on day 0), followed by inbubation for 4 or 8 d in the serum-containing medium. The results showing the mRNA quantity in differentiating macrophages on days 4 and 8 relative to that of monocytes (day 0) are presented in Figure 3. Real-time PCR analysis revealed that ORP1L expression was induced 100- to 160-fold during monocyte-macrophage differentiation. The expression of ORP1S was also up-regulated, but to a lesser extent (3- to 5-fold). With primers amplifying both ORP1 variants a 40- to 60-fold up-regulation of ORP1 messages was observed. Of the other ORP genes studied ORP3, ORP9, and ORP11 mRNAs were mildly (2- to 4-fold) up-regulated. The level of ORP6 expression was too low for reliable quantitation. GAPDH mRNA expression was not used for normalization of the data because it was up-regulated 1.5- to 2-fold during monocyte-macrophage differentiation. There was little or no difference between the 4- and 8-d time points. In conclusion, the ORP1L, ORP1S, ORP3, ORP9, and ORP11 messages were consistently up-regulated in cells of the two subjects.
Figure 3.
Real-time PCR analysis of ORP mRNA expression during monocyte-macrophage differentiation. Monocytes isolated from two healthy individuals (A and B) were differentiated into macrophages by incubation for 4 or 8 d in serum-containing medium. Total RNA was isolated from monocytes and the differentiated macrophages. Real-time PCR analysis of the messages for ORP1L, ORP1S, nine other ORP genes, OSBP, and GAPDH was performed with the Amplifluor-system using an ABI7700 analyzer. Expression levels on day 4 (gray bars) and on day 8 (black bars) are given relative to that of monocytes. The data has been normalized using the total amount of RNA in each sample. Data are given as mean ± SD (n = 2).
Detection of ORP1S and ORP1L Proteins
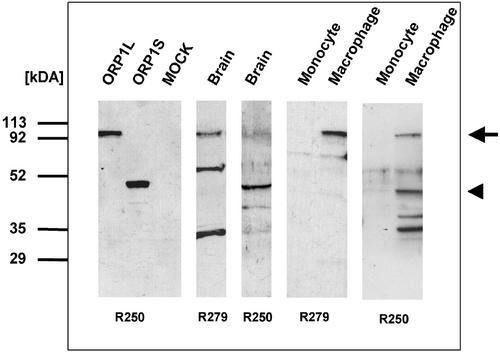
Antibodies were raised against two separate ORP1 antigens for the purpose of detecting the ORP1 proteins predicted from the cDNA sequences. The rabbit R250 antiserum generated against amino acid residues 428–950 of ORP1L was used for detection of both variants of ORP1 because these residues span the ligand binding domain common to both proteins. An ORP1L-specific antibody, R279, was raised against a peptide spanning amino acids 118–137. The antibodies were used for Western blot analysis of human brain, a tissue with high expression of both ORP1 mRNAs. Lysates of CHO cells transiently transfected with ORP1S or ORP1L cDNAs in the pcDNA3.1 expression plasmid were analyzed in parallel for comparison of protein sizes. The proteins expressed from the pcDNA3.1 constructs had the apparent molecular masses of 48 (ORP1S) and 100 (ORP1L) kDa, corresponding well to the masses predicted from the cDNA sequences, 50.2 and 108.5 kDa, respectively (Figure 4). The ORP1L specific antibody R279 indeed detected a band with the mobility of transiently expressed ORP1L (arrow). Additional strong signals were visible in the 55- and 34-kDa regions, which may represent yet undescribed splice variants, proteolytic fragments, or possibly cross-reactivity with unrelated antigens. The R250 antiserum detected a strong 50-kDa band (arrowhead) with a mobility identical to transiently expressed ORP1S and showed relatively weak reactivity against the 100-kDa ORP1L. The results show that both ORP1S and ORP1L proteins are present in brain, but no estimate of their relative quantities can be made based on this Western analysis.
Figure 4.
ORP1L and ORP1S proteins in human brain and during monocyte-macrophage differentiation. Proteins were resolved in 12.5% SDS-PAGE and Western blotted with ORP1-specific antibodies. The cell and tissue specimens are indicated above the lanes. ORP1L and ORP1S refer to nontagged transiently overexpressed proteins in CHO-K1 cells, the MOCK lane was transfected with empty vector pcDNA3.1. R250 refers to the primary antibody against amino acids 428–950 in ORP1L, common to both ORP1 variants. R279 refers to the primary antibody against an N-terminal peptide consisting of amino acids 118–137, present only in ORP1L. The mobilities of ORP1L and ORP1S are indicated on the right with an arrow and an arrowhead, respectively. Molecular weight markers are indicated on the left.
To verify that the up-regulation of the ORP1S and L documented at the mRNA level is also detectable at the protein level, human monocytes and macrophages were included in the analysis. Neither antiserum showed reactivity against the monocyte preparation, whereas both antisera reacted with a protein with the apparent size of ORP1L in the macrophage preparation (Figure 4). In addition, up-regulation of ORP1S was visible with the R250 antibody. The R250 antiserum also reacted with 36- and 40-kDa protein bands, which may represent proteolytic fragments of ORP1.
Intracellular Localization of the ORP1 Variants in CHO Cells
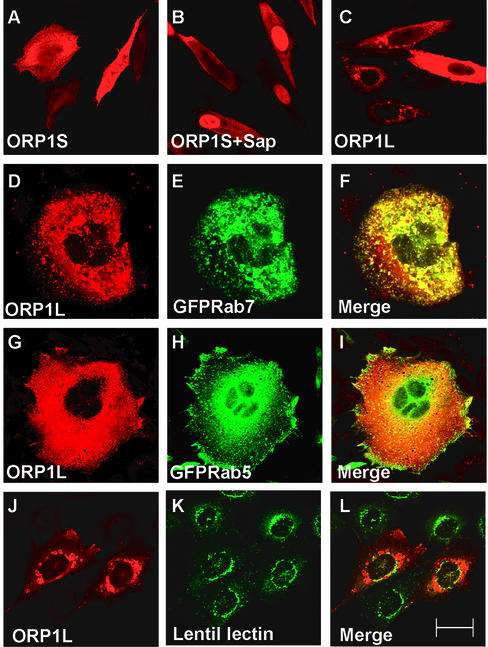
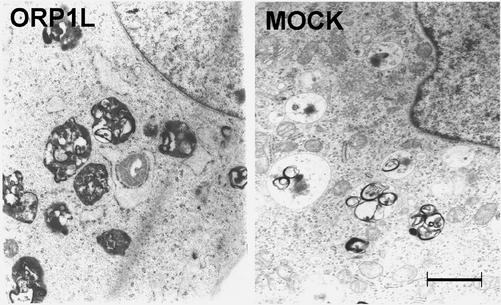
To determine the intracellular localization of ORP1S and L, CHO-K1 cells were transfected with the cDNAs in the pcDNA3.1 expression vector and the distribution of the proteins was assessed by indirect immunofluorescence microscopy. ORP1S was found evenly distributed throughout the cells, and no specific labeling of intracellular structures could be observed (Figure 5A), reflecting a predominantly cytosolic localization. In some experiments, transfected cells were briefly permeabilized with saponin before fixation to wash out cytosolic ORP1S. This treatment revealed prominent staining of the nucleus, with weak membrane or cytosolic staining remaining in the cells (Figure 5B). In some cells ORP1L also displayed an even staining throughout the cytoplasmic compartment, whereas in a majority of cells the protein was associated with vacuolar-appearing cytoplasmic organelles (Figure 5C). After saponin treatment nuclear staining was visible in some cells but not nearly as prominent as in ORP1S-transfected cells (our unpublished data). The vacuole-like ORP1L staining was evident in both transiently and stably transfected cells. This staining colocalized extensively with green fluorescent protein (GFP)-Rab7, a late endosomal marker that was cotransfected into the cells (Figure 5, D–F). The staining showed hardly any overlap with the early endosomal marker GFP-Rab5 (Figure 5, G–I) or fluorescein isothiocyanate-lentil lectin, which was used as a marker of the Golgi apparatus (Figure 5, J–L). Electron microscopy observation of stably transfected CHO-K1 cells expressing ORP1L revealed an abundance of apparently enlarged multivesicular bodies/late endosomes in the perinuclear region that were abnormally full of internal membranes with electron dense aberrant morphology (Figure 6). In addition, many of the cells displayed abnormalities of nuclear morphology (our unpublished data).
Figure 5.
Localization of overexpressed nontagged ORP1L and ORP1S in CHO-K1 cells. The cells were transfected with ORP1L or ORP1S cDNA constructs in pcDNA3.1, fixed with paraformaldehyde, and immunostained with the R250 antiserum. ORP1S is largely cytosolic (A) but can also be seen associated with the nucleus when cells are treated with saponin before fixation (B). ORP1L exhibits a vacuolar-like perinuclear staining pattern in most cells (C). The vacuolar-like ORP1L staining colocalizes extensively with the GFP-tagged late endosomal marker Rab7 (D–F), whereas no significant colocalization is observed with the early endosomal marker GFP-Rab5 (G–I) or the Golgi marker fluorescein isothiocyanate-lentil lectin (J–L). Bar, 10 μm.
Figure 6.
Electron microscopy of endosome morphology in cells stably transfected with ORP1L cDNA (ORP1L) or the empty vector pcDNA3.1 (MOCK). The cells were fixed with glutaraldehyde, processed for electron microscopy using a standard procedure, and horizontal sections were viewed with an EX200 electron microscope (JOEL). The ORP1L-expressing cells display abundant multivesicular body/late endosome-like organelles that are abnormally full of electron dense internal membranes. Bar, 1 μm.
Structural Features That Determine Localization of ORP1L
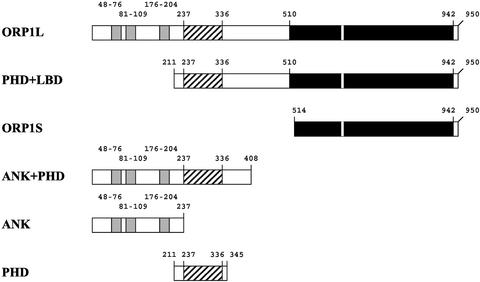
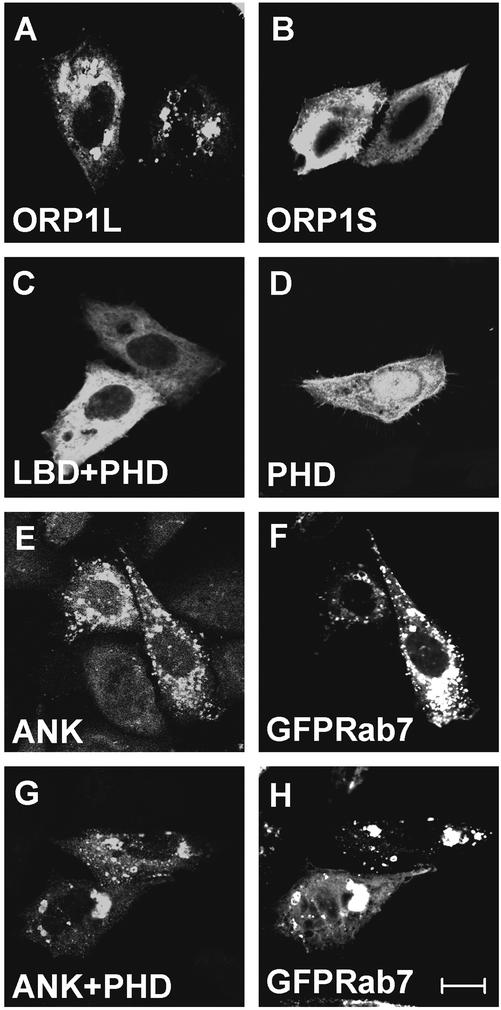
To identify which regions of the protein determine the intracellular localization of ORP1, a set of truncated cDNA constructs was generated (Figure 7). The constructs were expressed using pcDNA4HisMax, which allows detection of the proteins using the N-terminal Xpress epitope tag encoded by the vector. In addition to the full-length ORP1S and ORP1L, this set included constructs encoding the N-terminal ankyrin repeat region only (ANK; aa 1–237), the PHD only (PHD; aa 211–345); the ANK and PHD regions together (ANK + PHD; aa 1–408), and a construct lacking the N-terminal ANK region (PHD + ligand binding domain [LBD]; aa 211–950). Immunofluorescence microscopic localization of the proteins revealed that the epitope-tagged ORP1S and L showed localization identical to that of the untagged proteins (Figure 8, A and B; see also Figure 5, A and C). The PHD + LBD construct displayed an even staining throughout the cells, similar to ORP1S (Figure 8C). The PHD construct showed, in addition to cytosolic-like staining, prominent nuclear localization and was often seen concentrated in the plasma membrane region (Figure 8D). However, the ANK repeat fragment was observed on the surface of vacuole-like organelles such as the full-length ORP1L (Figure 8, E–F). This staining colocalized with GFP-Rab7, demonstrating that the stained structures are late endosomes.
Figure 7.
Schematic presentation of the ORP1 protein fragments used in this study. The fragment names are shown on the left, and amino acid positions of functional domains are shown above each fragment. Ankyrin repeats are shown as gray boxes. The pleckstrin homology domain and the putative ligand binding domain are indicated as striped and black boxes, respectively.
Figure 8.
Localization of Xpress epitope-tagged ORP1 protein fragments in CHO-K1 cells. The cells were transfected with cDNA constructs in pcDNA4HisMax, fixed with paraformaldehyde, and immunostained with anti-Xpress monoclonal antibody. ORP1L localizes on the surface of vacuole-like structures (A), whereas ORP1S is largely cytosolic (B). The fragment consisting of the ligand binding domain and the pleckstrin homology domain shows a cytosolic-appearing staining pattern (C), whereas the PHD on its own is seen within the nucleus, in the cytosol, and often in association with the plasma membrane (D). The ankyrin repeat region alone (E–F) as well as together with the PHD colocalizes with the late endosomal marker EGFP-Rab7. Clustering of endosomal membranes is evident in cells expressing the ANK + PHD fragment (G–H). Bar, 10 μm.
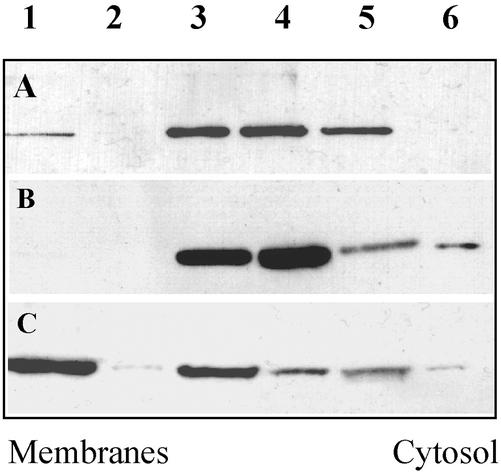
This finding was pursued further by separating total membrane and cytosolic fractions from the postnuclear supernatant of cells transfected with the ORP1S, ORP1L, or the ANK + PHD constructs, by ultracentrifugation in sucrose gradients. The distribution of the ORP1 proteins in the gradients was determined by SDS-PAGE and Western blotting (Figure 9). In this analysis, only a minor fraction of ORP1L was found to associate with the floated total membranes, whereas a majority was recovered in the bottom fractions containing cytosolic proteins. ORP1S was almost completely found in the cytosolic fractions. In contrast, a major portion of the ANK + PHD protein fragment associated with the total membrane fraction. It therefore seems that the ORP1L membrane association, or specifically its association with late endosomal membranes, is dictated by N-terminal sequences encompassing the ankyrin repeat region and the pleckstrin homology domain.
Figure 9.
Association of ORP1S, ORP1L, and the ankyrin repeat region-PHD fragment (Figure 7) with cellular membranes. Postnuclear supernatants of transiently transfected CHO-K1 cells were subjected to ultracentrifugation in a sucrose gradient after which fractions were collected and Western blotted. Fractions from the top, 1–5; pellet, 6. A minor fraction of ORP1L flotated with total membranes (A), whereas ORP1S was only recovered from the cytosolic fractions (B). The membrane association was most pronounced with the N-terminal fragment of ORP1L containing the ankyrin repeat region and the PHD (C).
Modulation of Liver X Receptor (LXR) α Transactivation Potential by ORP1L
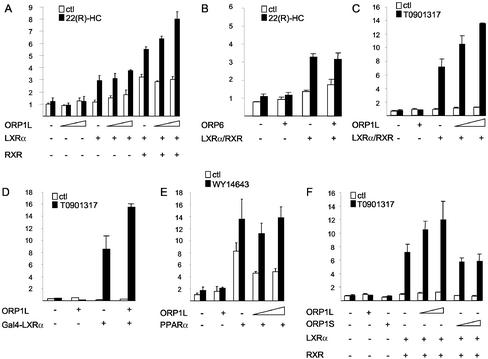
LXRs belong to the nuclear hormone receptor superfamily. They bind oxysterol ligands and function as obligate heterodimers with RXRs (Repa and Mangelsdorf, 2000; Schoonjans et al., 2000; Chawla et al., 2001). LXRα has been shown to control the expression of a number of genes with central roles in sterol metabolism. In macrophages, LXRα is an important regulator of genes whose products are instrumental in cholesterol efflux. The finding that ORP1L mRNA is strongly up-regulated during monocyte differentiation into macrophages prompted us to investigate whether ORP1 can modulate the LXRα-mediated transactivation of a reporter gene. We transfected COS cells with a luciferase reporter construct expressed under a TK promotor containing three copies of the consensus LXR response element (Willy et al., 1995), together with LXRα and RXR expression plasmids, in the presence or absence of an ORP1L expression construct (Figure 10). A plasmid encoding ORP6 (Lehto et al., 2001) was transfected into the cells for comparison. The transfected cells were incubated in the presence or absence of the LXR ligand 22(R)-HC, and reporter gene transactivation was determined using a luciferase assay. Transfection of cells with ORP1L in the absence of LXRα or RXR had no effect on reporter gene expression (Figure 10A). When coexpressed with LXRα, ORP1L seemed to mildly increase reporter activation but the change was not significant. However, upon ORP1L coexpression with both LXRα and RXR a clear and reproducible enhancement of reporter activation was detected in the presence of 22(R)-HC, whereas there was no effect in the absence of the ligand. The ORPlL-mediated enhancement was dependent on the amount of the ORP1L plasmid used and was maximally ∼45%. The induction was specific for ORP1L because ORP6 had no effect on expression of the reporter gene (Figure 10B).
Figure 10.
Effects of ORP1 expression on liver X receptor α-mediated transactivation of a luciferase reporter gene. (A) ORP1L potentiates LXR-RXR transactivation in a dose-dependent manner. COS-1 cells were transfected with a reporter plasmid containing a TK promotor with three copies of a consensus LXRE, LXRα, and/or RXR expression plasmids, and increasing quantities of pcDNAHisMax/ORP1L and incubated in the absence (white bars) or presence (black bars) of 10 μM 22(R)-hydroxycholesterol. (B) Another member of the ORP protein family, ORP6, has no effect on transactivation by LXRα-RXR. COS-1 cells were transfected with LXRα-RXR and/or ORP6 expression plasmids, and incubated in the absence or presence of 10 μM 22(R)-hydroxycholesterol. (C) ORP1L potentiation of LXRα-RXR transactivation is ligand dependent but not specific for the oxysterol ligand or the cell-type used. HEK 293 cells were transfected with LXRα-RXR and/or ORP1L expression plasmids and incubated in the absence or presence of the nonsterol synthetic LXR agonist T0901317 (0.1 μM). (D) ORP1L effect is not mediated through RXR and is independent of the specific interaction of LXRα/RXR with LXRE. HEK 293 cells were transfected with expression plasmids encoding a Gal4 DNA binding domain-LXRα ligand binding domain fusion (Gal4-LXRα) and/or ORP1L. The cells were incubated in the absence or presence of T0901317. (E) ORP1L does not influence transactivation by PPARα-RXR. COS-1 cells were transfected with a reporter plasmid containing six copies of a PPAR response element and the expression plasmids for PPARα, RXR, and/or ORP1L and incubated in the absence or presence of the PPARα agonist WY 14643 (50 μM). (F) ORP1S is not capable of enhancing reporter transactivation by LXRα. HEK 293 cells were transfected with LXRα, RXR, and/or ORP1L or ORP1S expression constructs and incubated in the absence or presence of T0901317.
To pursue this finding further in a different cell system, the effect of ORP1L on LXRα/RXR-induced activation of the luciferase reporter was studied in HEK 293 cells in the absence or presence of a very potent synthetic agonist of LXRα, T0901317 (Figure 10C). ORP1L alone had no significant effect on reporter activity either in the absence or the presence of the agonist. In the presence of LXRα/RXR the agonist T0901317 caused an approximately eightfold induction of the reporter, and this effect was potentiated in a dose-dependent manner by increasing amounts of the ORP1L expression plasmid. The maximal enhancement reached with ORP1L was 88%. To study whether the ORP1L enhancement of reporter expression was mediated by LXRα rather than through the RXR heterodimer partner, and whether the ORP1L enhancement is dependent on modulation of LXR/RXR DNA binding affinity for the LXRE, similar experiments were carried out in HepG2 cells using a Gal4 DNA binding domain-LXRα ligand binding domain fusion protein. In the absence of RXR, this fusion protein binds to a Gal4 promotor element present in five copies in the reporter construct and the expression of the luciferase reporter gene is induced upon LXR ligand binding (Figure 10D). In this assay, ORP1L enhanced reporter expression by 80% in the presence of the LXR agonist T0901317. These results suggest that the ORP1L effect is mediated in a ligand-dependent manner by LXRα, not RXR, and the enhancement is independent of the LXR-RXR heterodimer DNA binding. Furthermore, we wanted to determine whether the ORP1L enhancement of reporter transactivation was specific for LXRα, or whether this effect could also be seen with other nuclear receptors. We therefore carried out cotransfection experiments in HEK 293 cells with ORP1L and peroxisome proliferator-activated receptor α (Chinetti et al., 2000) expression plasmids. The luciferase reporter gene was under a TK promotor with six copies of a consensus peroxisome proliferator-activated receptor (PPAR) response element, and luciferase activity was assayed in the presence or absence of the PPARα agonist WY 14643 (Figure 10E). In these experiments, ORP1L had no significant effect on reporter transactivation by PPARα. Finally, we studied whether enhancement of LXRα-mediated transactivation could also be observed with ORP1S (Figure 10F). In an experimental set-up identical to that in Figure 10C, increasing amounts of either ORP1L or ORP1S expression plasmids were transfected into HEK 293 cells in the absence or presence of LXRα and RXR expression constructs, and cells were incubated in the absence or presence of the LXR agonist T0901317. Although ORP1L induced a clear agonist-dependent enhancement of reporter transactivation, ORP1S had no effect.
DISCUSSION
The family of OSBP-related proteins in different organisms consists of members that comprise merely a ligand binding domain with an OSBP fingerprint sequence or a ligand binding domain together with an N-terminal extension, including a pleckstrin homology domain (Beh et al., 2001; Jaworski et al., 2001; Lehto et al., 2001). The two variants of human ORP1 cDNA that have been described, ORP1S and ORP1L, encode predicted proteins belonging to these two classes. The proteins encoded from the same gene are markedly different in structure, and it is therefore likely that they are also functionally distinct. This is corroborated by the present finding that ORP1S and ORP1L display highly different tissue mRNA expression patterns. The difference in expression patterns may result from the use of two separate promotors (Jaworski et al., 2001), differential tissue-specific splicing, or tissue-specific modulation of mRNA stability. The ORP1S message was most abundant in skeletal muscle and heart, whereas ORP1L mRNA was predominant in macrophages, brain, and lung. Interestingly, the ORP1L mRNA underwent remarkable (>100-fold) up-regulation during differentiation of monocytes into macrophages in culture. Also the ORP1S mRNA was significantly up-regulated during this process. The gene up-regulation could also be verified at the protein level. A multitude of genes involved in a variety of cellular functions are induced during the maturation of monocytes into macrophage-type cells. These include genes involved in lipid metabolism, such as scavenger receptors (Tontonoz et al., 1998; Tomokiyo et al., 2002), peroxisome proliferator activated receptor-γ (Tontonoz et al., 1998; Moore et al., 2001), and ATP-binding cassette transporter A1 (Kielar et al., 2001), as well as regulators of inflammatory functions, the machinery of antigen presentation (Tontonoz et al., 1998; Angenieux et al., 2001; Lapteva et al., 2001; Moore et al., 2001), and factors controlling the coagulation process (Meisel et al., 2002). The present finding indicates that ORP1, particularly the ORP1L variant, has a specific function in macrophages.
ORP1S and ORP1L expressed from cDNA constructs were shown to display different intracellular localization in CHO cells. Although ORP1S localizes mainly to the cytosol with a substantial fraction found within the nucleus, ORP1L is distributed between the cytosol and late endosomes. It is thus evident that the N-terminal extension of ORP1L, which contains three ankyrin repeats and a PHD, carries determinants for intracellular localization. Experiments with fragments of ORP1L expressed in CHO cells indicate that the N-terminal ankyrin repeat region (aa 1–270) alone contains the minimal information for late endosomal targeting. The PHD alone did not localize to endosomes, but a fragment comprising the ANK and PHDs together did, and seemed to cause aggregation of these organelles in many cells. In sucrose gradient centrifugation analysis this protein fragment associated with cellular membranes more extensively than the full ORP1L. Therefore, we suggest that the targeting of ORP1L to late endosomes is due to the combined action of the ANK region and PHD. The affinity of the PHD toward its endosomal ligand(s) may not be sufficient for recruitment onto the endosomal membrane in the absence of the ANK domain, thus the PHD protein fragment may instead preferentially associate with the plasma membrane that has the highest concentration of polyphosphoinositide lipids (Lemmon and Ferguson, 2000). The C-terminal ligand binding domain could according to our data have a negative regulatory effect on the membrane association of ORP1. This situation resembles that reported for OSBP, the PHD of which associates with Golgi membranes constitutively when it is detached from the context of the C-terminal sterol binding domain (Ridgway et al., 1992; Levine and Munro, 1998). The ligands of the ORP1L ankyrin repeat region and PHD are still unknown, but identifying the components of late endosomal membranes that they associate with will be crucial in elucidating the function of the protein. The present electron microscopy findings suggest that stable overexpression of ORP1L induces changes in the morphology and function of late endosomal compartments, as indicated by the appearance of apparently enlarged endosomes that have an abnormal abundance of electron dense internal membranes. The clustering or aggregation of late endocytic compartments when ANK + PHD fragments are overexpressed is an interesting observation that further indicates a role of ORP1L in late endosomal membrane dynamics. It is possible that this protein fragment, when detached from the context of the C-terminal parts of ORP1L, tends to either recruit an abnormal density of tethering factors onto the membrane (Guo et al., 2000) or self-associate and thus cluster endosomes.
Association of OSBP with Golgi membranes is promoted by plasma membrane cholesterol depletion by sphingomyelinase treatment or upon incubation of cells with lipoprotein deficient serum. In contrast, OSBP detaches from the Golgi under conditions of ample low-density lipoprotein cholesterol supply in normal cells but not in fibroblasts of Niemann-Pick disease type C patients, in which trafficking of cholesterol from late endocytic compartments is disturbed (Ridgway et al., 1998; Storey et al., 1998; Mohammadi et al., 2001). These findings on OSBP have led to the proposal that OSBP responds to or senses altered cellular cholesterol status or transport. The localization of ORP1L on late endosomes may, by analogy, indicate that it could act as a sensor for the lipid status of late endocytic compartments and mediate information to yet unknown downstream machineries.
ORP1S showed significant affinity for the nucleus. This localization may be due to the presence of three putative nuclear localization signals: 510RKHR513, 531KKHR534, and 893KKRLEEKQRAARKNRSK909 in ORP1. The last two are located in the ORP1S region of the amino acid sequence. Nuclear localization of ORP1L was weaker than that of ORP1S, which may be due to the presence of functional domains that target the protein to endosomal membranes. The finding that ORP1L, but not ORP1S is capable of enhancing the LXRα transactivation potential is highly interesting. This enhancement is, according to the present data, specific for LXRs (similar results were obtained for LXRβ; our unpublished data) and dependent on the presence of LXR agonists. This could, in principle, indicate direct involvement of ORP1L in the ligand interactions of the LXRs. ORP1L could bind either LXR or its ligand facilitating their interaction. However, ORP1 has not been found to bind oxysterols but was suggested to bind phosphatidic acid and phosphoinositides (Xu et al., 2001). Furthermore, the effect of ORP1L was not only seen in the presence of 22(R)-HC but also of T091317, which is a synthetic nonsterol LXR agonist. It is therefore hard to imagine that, if the ORP1L effect were due to interaction with LXR agonists, it would be similar on both of them. The N-terminal region of ORP1L contains three LXXLL motifs, which are absent from ORP1S. These motifs are typically found in transcriptional coactivator proteins (Heery et al., 1997). Therefore, one possible explanation for the observed effect of ORP1L on LXR-mediated transcativation would be a coactivator-type function. However, we find this unlikely because 1) we were unable to detect a direct interaction between LXRα and the N-terminal ankyrin repeat region of ORP1L that includes the LXXLL motifs (our unpublished data); and 2) we detected only minor amounts of the protein within the nucleus. These findings, together with the fact that the ORP1L enhancement of LXR function is relatively weak, rather support an indirect effect on LXR, e.g., ORP1L mediated modulation of the cellular levels of LXR antagonists, such as polyunsaturated fatty acids (Ou et al., 2001) or oxidized cholesterol 3-sulfate derivatives (Song et al., 2001). The presence of ORP1S neither enhanced nor inhibited LXR transactivation. This, together with the differences in tissue distribution and intracellular localization, supports the view that ORP1S and L are functionally distinct.
ACKNOWLEDGMENTS
Seija Puomilahti and Pirjo Ranta are acknowledged for expert technical assistance. Drs. Marino Zerial and Angela Wandinger-Ness are thanked for kindly providing the EGFP-Rab5 and EGFP-Rab7 cDNA constructs, respectively. This study was supported by the Academy of Finland (grants 45817, 49987, 50641, and 54301 to V.M.O.; grant 51883 to M.L.), the Sigrid Juselius Foundation (to V.M.O.), and the Finnish Cultural Foundation (to V.M.O.). M.J. is a member of the Helsinki Graduate School of Biotechnology and Molecular Biology.
Abbreviations used:
- ANK
ankyrin repeat
- GFP
green fluorescent protein
- LBD
ligand binding domain
- LXR
liver X receptor
- ORP
OSBP-related protein
- OSBP
oxysterol binding protein
- PCR
polymerase chain reaction
- PHD
pleckstrin homology domain
- TRITC
tetramethylrhodamine B isothiocyanate
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0459. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0459.
REFERENCES
- Alphey L, Jimenez J, Glover D. A Drosophila homologue of oxysterol binding protein (OSBP): implications for the role of OSBP. Biochim Biophys Acta. 1998;1395:159–164. doi: 10.1016/s0167-4781(97)00159-0. [DOI] [PubMed] [Google Scholar]
- Angenieux C, Fricker D, Strub JM, Luche S, Bausinger H, Cazenave JP, Van Dorsselaer A, Hanau D, de la Salle H, Rabilloud T. Gene induction during differentiation of human monocytes into dendritic cells: an integrated study at the RNA and protein levels. Funct Integr Genomics. 2001;1:323–329. doi: 10.1007/s101420100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Jessup W. Oxysterols and atherosclerosis. Atherosclerosis. 1999;142:1–28. doi: 10.1016/s0021-9150(98)00196-8. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res. 2000;49:497–505. doi: 10.1007/s000110050622. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Van der Westhuyzen DR, Goldstein JL, Brown MS. Purification of oxysterol binding protein from hamster liver cytosol. J Biol Chem. 1989;264:9046–9052. [PubMed] [Google Scholar]
- Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- Guo W, Sacher M, Barrowman J, Ferro-Novick S, Novick P. Protein complexes in transport vesicle targeting. Trends Cell Biol. 2000;10:251–255. doi: 10.1016/s0962-8924(00)01754-2. [DOI] [PubMed] [Google Scholar]
- Hanley K, Ng DC, He SS, Lau P, Min K, Elias PM, Bikle DD, Mangelsdorf DJ, Williams ML, Feingold KR. Oxysterols induce differentiation in human keratinocytes and increase Ap-1-dependent involucrin transcription. J Invest Dermatol. 2000;114:545–553. doi: 10.1046/j.1523-1747.2000.00895.x. [DOI] [PubMed] [Google Scholar]
- Hayden JM, Brachova L, Higgins K, Obermiller L, Sevanian A, Khandrika S, Reaven PD. Induction of monocyte differentiation and foam cell formation in vitro by 7-ketocholesterol. J Lipid Res. 2002;43:26–35. [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Huang Z, Fasco MJ, Kaminsky LS. Optimization of DNase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. Biotechniques. 1996;20:1012–1014. doi: 10.2144/96206st02. , 1016, 1018–1020. [DOI] [PubMed] [Google Scholar]
- Jaworski CJ, Moreira E, Li A, Lee R, Rodriguez IR. A family of 12 human genes containing oxysterol-binding domains. Genomics. 2001;78:185–196. doi: 10.1006/geno.2001.6663. [DOI] [PubMed] [Google Scholar]
- Kolsch H, Lutjohann D, Tulke A, Bjorkhem I, Rao ML. The neurotoxic effect of 24-hydroxycholesterol on SH-SY5Y human neuroblastoma cells. Brain Res. 1999;818:171–175. doi: 10.1016/s0006-8993(98)01274-8. [DOI] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem J. 1997;326:205–213. doi: 10.1042/bj3260205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND. Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J Lipid Res. 1999;40:109–116. [PubMed] [Google Scholar]
- Laitinen S, Lehto M, Lehtonen S, Hyvarinen K, Heino S, Lehtonen E, Ehnholm C, Ikonen E, Olkkonen VM. ORP2, a homolog of oxysterol binding protein, regulates cellular cholesterol metabolism. J Lipid Res. 2002;43:245–255. [PubMed] [Google Scholar]
- Laitinen S, Olkkonen VM, Ehnholm C, Ikonen E. Family of human oxysterol binding protein (OSBP) homologues. A novel member implicated in brain sterol metabolism. J Lipid Res. 1999;40:2204–2211. [PubMed] [Google Scholar]
- Lapteva N, Ando Y, Nieda M, Hohjoh H, Okai M, Kikuchi A, Dymshits G, Ishikawa Y, Juji T, Tokunaga K. Profiling of genes expressed in human monocytes and monocyte-derived dendritic cells using cDNA expression array. Br J Hematol. 2001;114:191–197. doi: 10.1046/j.1365-2141.2001.02910.x. [DOI] [PubMed] [Google Scholar]
- Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM. The OSBP-related protein family in humans. J Lipid Res. 2001;42:1203–1213. [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognizes a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–78. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel SR, Xu XP, Edgington TS, Dimayuga P, Kaul S, Lee S, Fishbein MC, Cercek B, Shah PK. Differentiation of adherent human monocytes into macrophages markedly enhances tissue factor protein expression and procoagulant activity. Atherosclerosis. 2002;161:35–43. doi: 10.1016/s0021-9150(01)00616-5. [DOI] [PubMed] [Google Scholar]
- Mohammadi A, Perry RJ, Storey MK, Cook HW, Byers DM, Ridgway ND. Golgi localization and phosphorylation of oxysterol binding protein in Niemann-Pick C and U18666A-treated cells. J Lipid Res. 2001;42:1062–1071. [PubMed] [Google Scholar]
- Moore KJ, Rosen ED, Fitzgerald ML, Randow F, Andersson LP, Altshuler D, Milstone DS, Mortensen RM, Spiegelman BM, Freeman MW. The role of PPAR-γ in macrophage differentiation and cholesterol uptake. Nat Med. 2001;7:41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- Moreira EF, Jaworski C, Li A, Rodriguez IR. Molecular and biochemical characterization of a novel oxysterol-binding protein (OSBP2) highly expressed in retina. J Biol Chem. 2001;276:18570–18578. doi: 10.1074/jbc.M011259200. [DOI] [PubMed] [Google Scholar]
- Ou J, Tu H, Shan B, Luk A, DeBose-Boyd RA, Bashmakov Y, Goldstein JL, Brown MS. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc Natl Acad Sci USA. 2001;98:6027–6032. doi: 10.1073/pnas.111138698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panini SR, Sinensky MS. Mechanisms of oxysterol-induced apoptosis. Curr Opin Lipidol. 2001;12:529–533. doi: 10.1097/00041433-200110000-00008. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:E45–E45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway ND, Lagace TA, Cook HW, Byers DM. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J Biol Chem. 1998;273:31621–31628. doi: 10.1074/jbc.273.47.31621. [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Brendel C, Mangelsdorf D, Auwerx J. Sterols and gene expression: control of affluence. Biochim Biophys Acta. 2000;1529:114–125. doi: 10.1016/s1388-1981(00)00141-4. [DOI] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- Song C, Hiipakka RA, Liao S. Auto-oxidized cholesterol sulfates are antagonistic ligands of liver X receptors: implications for the development and treatment of atherosclerosis. Steroids. 2001;66:473–479. doi: 10.1016/s0039-128x(00)00239-7. [DOI] [PubMed] [Google Scholar]
- Storey MK, Byers DM, Cook HW, Ridgway ND. Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. Biochem J. 1998;336:247–256. doi: 10.1042/bj3360247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Morita K, Ueno N, Shibuya H. BIP, a BRAM-interacting protein involved in TGF-β signaling, regulates body length in Caenorhabditis elegans. Genes Cells. 2001;6:599–606. doi: 10.1046/j.1365-2443.2001.00444.x. [DOI] [PubMed] [Google Scholar]
- Taylor FR, Kandutsch AA. Oxysterol binding protein. Chem Phys Lipids. 1985;38:187–194. doi: 10.1016/0009-3084(85)90066-0. [DOI] [PubMed] [Google Scholar]
- Taylor FR, Saucier SE, Shown EP, Parish EJ, Kandutsch AA. Correlation between oxysterol binding to a cytosolic binding protein and potency in the repression of hydroxymethylglutaryl coenzyme A reductase. J Biol Chem. 1984;259:12382–12387. [PubMed] [Google Scholar]
- Tomokiyo R, Jinnouchi K, Honda M, Wada Y, Hanada N, Hiraoka T, Suzuki H, Kodama T, Takahashi K, Takeya M. Production, characterization, and interspecies reactivities of monoclonal antibodies against human class A macrophage scavenger receptors. Atherosclerosis. 2002;161:123–132. doi: 10.1016/s0021-9150(01)00624-4. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- Wang C, JeBailey L, Ridgway ND. Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol, and interacts with vimentin intermediate filaments. Biochem J. 2002;361:461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaya J, Aviram M, Mahmood S, Hayek T, Grenadir E, Hoffman A, Milo S. Selective distribution of oxysterols in atherosclerotic lesions and human plasma lipoproteins. Free Radic Res. 2001;34:485–497. doi: 10.1080/10715760100300431. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu Y, Ridgway ND, McMaster CR. Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicle transport. J Biol Chem. 2001;276:18407–18414. doi: 10.1074/jbc.M101204200. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Release of cellular cholesterol: molecular mechanism for cholesterol homeostasis in cells and in the body. Biochim Biophys Acta. 2000;1529:231–244. doi: 10.1016/s1388-1981(00)00152-9. [DOI] [PubMed] [Google Scholar]