Abstract
Pex3p is a peroxisomal integral membrane protein required early in peroxisome biogenesis, and Pex3p-deficient cells lack identifiable peroxisomes. Two temperature-sensitive pex3 mutant strains of the yeast Yarrowia lipolytica were made to investigate the role of Pex3p in the early stages of peroxisome biogenesis. In glucose medium at 16°C, these mutants underwent de novo peroxisome biogenesis and exhibited early matrix protein sequestration into peroxisome-like structures found at the endoplasmic reticulum-rich periphery of cells or sometimes associated with nuclei. The de novo peroxisome biogenesis seemed unsynchronized, with peroxisomes occurring at different stages of development both within cells and between cells. Cells with peripheral nascent peroxisomes and cells with structures morphologically distinct from peroxisomes, such as semi/circular tubular structures that immunostained with antibodies to peroxisomal matrix proteins and to the endoplasmic reticulum-resident protein Kar2p, and that surrounded lipid droplets, were observed during up-regulation of peroxisome biogenesis in cells incubated in oleic acid medium at 16°C. These structures were not detected in wild-type or Pex3p-deficient cells. Their role in peroxisome biogenesis remains unclear. Targeting of peroxisomal matrix proteins to these structures suggests that Pex3p directly or indirectly sequesters components of the peroxisome biogenesis machinery. Such a role is consistent with Pex3p overexpression producing cells with fewer, larger, and clustered peroxisomes.
INTRODUCTION
Peroxisomes are ubiquitous organelles that in electron micrographs look spherical with diameters between 0.1 μm and 1.0 μm, are surrounded by a single unit membrane, and contain a granular matrix and sometimes a paracrystalline core. Their number, size, and enzyme content vary in response to changing environmental and physiological conditions. Peroxisomes metabolize lipids, nitrogen bases, and carbohydrates. The importance of peroxisomes for human growth and development is underscored by the lethality of a group of genetic disorders known collectively as the peroxisome biogenesis disorders, including Zellweger syndrome, rhizomelic chondrodysplasia punctata, and neonatal adrenoleukodystrophy, in which peroxisome assembly is compromised (for reviews, see Lazarow and Fujiki, 1985; van den Bosch et al., 1992; Subramani, 1998; Purdue and Lazarow, 2001).
Proteins required for peroxisome biogenesis are termed peroxins, which are encoded by the PEX genes. Twenty-five PEX genes have been identified to date (Purdue and Lazarow, 2001; Titorenko and Rachubinski, 2001; Smith et al., 2002; Tam and Rachubinski, 2002). Protein targeting to peroxisomes is compromised in most mutants of the PEX genes. Because peroxisomes lack DNA, all peroxisomal proteins are encoded in the nucleus and imported after synthesis on cytoplasmic polysomes. Most soluble matrix proteins are targeted to peroxisomes by one of two peroxisome targeting signals (PTS). PTS1 is a carboxyl-terminal tripeptide of the sequence Ser-Lys-Leu (or conserved variants thereof), which is found in the majority of matrix proteins. PTS2 is a sometimes cleaved amino-terminal nonapeptide found in a smaller subset of matrix proteins. Membrane proteins are sorted to peroxisomes by targeting signals distinct from PTS1 or PTS2. Various peroxins have been shown to act as receptors for the different PTSs or as docking sites for these receptors (for reviews, see Subramani et al., 2000; Purdue and Lazarow, 2001; Titorenko and Rachubinski, 2001).
Peroxisomes can arise by budding/fission of preexisting peroxisomes and by de novo biogenesis. Two models have been proposed for the de novo biogenesis of peroxisomes (Gould and Valle, 2000; Titorenko and Rachubinski, 2001). One model proposes that small peroxisomal vesicles, distinct from mature peroxisomes, fuse to form larger vesicles, which in turn assemble the import machineries for peroxisomal membrane and matrix proteins (Titorenko et al., 2000; Titorenko and Rachubinski, 2000). In the second model, the early stages of peroxisome biogenesis do not involve vesicular fusion but instead nascent peroxisomes (preperoxisomes) are proposed to progressively import peroxisomal membrane proteins and matrix proteins, eventually maturing to peroxisomes (Gould and Valle, 2000).
The site of de novo peroxisome biogenesis remains an issue of intense debate. The peroxins Pex3p, Pex16p, and Pex19p have been implicated in the early events of peroxisome biogenesis in numerous organisms. Pex3p, an integral membrane protein, has been shown to be required for the formation of peroxisomal structures in all species so far investigated. Herein, we report the cloning and characterization of the Yarrowia lipolytica PEX3 gene and our observations of the early events of peroxisome biogenesis in cells lacking peroxisomes by using temperature-sensitive Pex3p mutants with slower than normal peroxisome biogenesis. We show that de novo peroxisome biogenesis in Y. lipolytica likely occurs in the endoplasmic reticulum (ER)-rich peripheral regions of the cell and in areas sometimes associated with the nucleus. We also demonstrate the presence of semi/circular tubular membrane structures that wrap around the surface of lipid droplets, contain peroxisomal matrix proteins, and may be involved in peroxisome biogenesis. In addition, we provide evidence that Y. lipolytica Pex3p might initiate peroxisome biogenesis by directly or indirectly sequestering components of the peroxisome biogenesis machinery.
MATERIALS AND METHODS
Strains and Culture Conditions
The Y. lipolytica strains used in this study are listed in Table 1. Media, growth conditions, and genetic techniques for Y. lipolytica have been described (Nuttley et al., 1993; Szilard et al., 1995). Media components were as follows: YPD, 1% yeast extract, 2% peptone, 2% glucose; YPA, 1% yeast extract, 2% peptone, 2% sodium acetate; YPBO, 0.3% yeast extract, 0.5% peptone, 0.5% K2HPO4, 0.5% KH2PO4, 1% Brij 35, 1% (wt/vol) oleic acid; YND, 1.34% yeast nitrogen base without amino acids, 2% glucose; YNO, 1.34% yeast nitrogen base without amino acids, 0.05% (wt/vol) Tween 40, 0.2% (wt/vol) oleic acid; and YNA, 1.34% yeast nitrogen base without amino acids, 2% sodium acetate. YNA, YND, and YNO media were supplemented with Complete Supplement Mixture (minus the appropriate amino acids) (Bio 101, Vista, CA) at twice the manufacturer's recommended concentration or with leucine, uracil, or lysine, each at 50 μg/ml, as required. Unless otherwise stated, all cells grown at 16°C were used in temperature shift experiments. For temperature shift experiments, stock cells were consistently grown on YNA plates at 30°C to keep them clear of peroxisomes. Starter cultures were made from the 30°C stock cells by growth of cells at 30°C in liquid culture before shifting the cells to 16°C to initiate peroxisome biogenesis.
Table 1.
Y. lipolytica strains used in this study
| Strain | Genotype |
|---|---|
| E122 | MATA, ura3-302, leu2-270, lys8-11 |
| 22301-3 | MATB, ura3-302, leu2-270, his1 |
| mut12-1 | MATA, ura3-302, leu2-270, lys8-11, mut12-1 |
| M12TR | MATA, ura3-302, leu2-270, lys8-11, p12(LEU2) |
| pex3KOA | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3 |
| pex3KOB | MATB, ura3-302, leu2-270, his1, pex3∷URA3 |
| D-WT | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1 |
| D3-1 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, mut12-1/+ |
| D3-2 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, +/pex3∷URA3 |
| D3-3 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex3∷URA3/+ |
| D3-4 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, mut12-1/pex3∷URA3 |
| D3-5 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex3∷URA3/pex3∷URA3 |
| Myc-3lnt | MATA, ura3-302, leu2-270, lys8-11, pex3∷Myc-PEX3 |
| PMyc-3 | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3(LEU2) |
| PMyc-3Q36P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3Q36P(LEU2) |
| PMyc-3R143P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3R143P(LEU2) |
| PMyc-3L162P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3L162P(LEU2) |
| PMyc-3L226P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3L226P(LEU2) |
| PMyc-3L320P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3L320P(LEU2) |
| PMyc-3E415Stop | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3E415Stop(LEU2) |
| PMyc-3Y424Stop | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3Y424Stop(LEU2) |
| PMyc-3TH-WT | MATA, ura3-302, leu2-270, lys8-11, pMyc-PEX3TH(URA3) |
| PMyc-3TH-12 | MATA, ura3-302, leu2-270, lys8-11, mut12-1, pMyc-PEX3TH(URA3) |
| PEYFP-SKL-WT | MATA, ura3-302, leu2-270, lys8-11, pEYFP-SKL (HygB) |
| PEYFP-SKL-PMyc-3R143P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3R143P(LEU2), pEYFP-SKL (HygB) |
| PEYFP-SKL-Pmyc-3L320P | MATA, ura3-302, leu2-270, lys8-11, pex3∷URA3, pMyc-PEX3L320P(LEU2), pEYFP-SKL (HygB) |
Cloning, Sequencing, and Integrative Disruption of the PEX3 Gene
The mut12-1 mutant strain was isolated from randomly mutagenized Y. lipolytica wild-type strain E122 by screening for the inability to use oleic acid as sole carbon source (Nuttley et al., 1993). The PEX3 gene was isolated by functional complementation of the mut12-1 strain by using a Y. lipolytica genomic DNA library in the autonomously replicating Escherichia coli shuttle vector pINA445 (Nuttley et al., 1993). Total DNA was recovered from a Leu+ transformant that recovered growth on YNO and was used to transform E. coli for plasmid recovery. Restriction fragments prepared from the genomic insert were subcloned and tested for their ability to functionally complement the mut12-1 strain. Dideoxynucleotide sequencing of the complementing Y. lipolytica genomic DNA was performed on both strands. The deduced Pex3p amino acid sequence was compared with other known protein sequences by using the GENEINFO (R) Blast Network Service (Blaster) of the National Center for Biotechnology Information (Bethesda, MD).
Targeted integrative deletion of the PEX3 gene was performed with the URA3 gene of Y. lipolytica. A 4.8-kbp SphI-HindIII DNA fragment containing the PEX3 gene and flanking regions was first subcloned into the vector pGEM7Zf (+) (Promega, Madison, WI). The PEX3 gene was removed by digestion with BspE1 and XbaI, and the resultant ends were made blunt with the Klenow fragment of DNA polymerase I. A SalI fragment containing the Y. lipolytica URA3 gene was made blunt with Klenow fragment and then ligated into the disrupted PEX3 locus. A fragment containing the URA3 gene flanked by ∼1.5 and ∼1.7 kbp of genomic sequence at the 5′ and 3′ ends of the insertion, respectively, was released by digestion with HindIII and SphI. This fragment was used to transform E122 cells to uracil prototrophy. Ura+ transformants were selected and screened for the inability to grow in the presence of oleic acid as carbon source. Integration into the correct locus was confirmed by Southern blot analysis (Ausubel et al., 1994). Deletion strains and the original mutant were crossed with wild-type strains and with each other, and the resultant diploids were checked for growth on YNO agar.
Tagging of Pex3p with the c-Myc Epitope
The c-Myc epitope (EQKLISEE) was inserted immediately after the initiator methionine of Pex3p by amplification of the PEX3 gene by the polymerase chain reaction (PCR) with primers 5′-ACACCACCATGGACGAACAAAAGCTGATATCTGAAGAAGACTTGTTCT-TCAGACGGCACCAG and 5′-AGGTGGAGCTCTCGAGAG. The 371-base pair amplified fragment was cleaved with NcoI and XhoI and inserted into the identically cleaved PEX3 gene. Positive clones were sequenced to verify the amplified sequence. The tagged PEX3 gene was cleaved with HindIII and SphI, and the linear DNA fragment was introduced into the pex3KOA strain by electroporation. Stable integrants were screened for growth on appropriately supplemented YNO medium.
Construction of the EYFP-SKL Reporter
Promoter and terminator regions of the gene encoding peroxisomal thiolase were amplified by PCR from the plasmid pTEC (Eitzen et al., 1997) with primers 5′-CTCAGATCTAACCTACCGGTTG and 5′-CAGAGATCTAGTCTGCCGGCGG. The amplified fragment was digested with BglII and inserted into the BamHI site of a modified pUB4 vector (Kerscher et al., 2001) with its EcoRI site destroyed to make the vector pOET. The pUB4 vector contains the hygromycin resistance gene as a selectable marker. The gene encoding enhanced yellow fluorescent protein (EYFP) was amplified by PCR from the plasmid pEYFP (BD Biosciences Clontech, Palo Alto, CA) with the primers 5′-CGCGAATTCCCCCGGGTACCGG and 5′-CCGGAATTCTTTACAGCTTGGACTTGTACAGCTCG to make a chimeric EYFP tagged at its carboxy terminus with the PTS1 tripeptide Ser-Lys-Leu (EYFP-SKL). The amplified product encoding EYFP-SKL was digested with EcoRI and inserted into the EcoRI site between the thiolase promoter and terminator regions of the pOET vector.
Site-directed Mutagenesis
Site-directed mutagenesis was performed with the QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) by using 50 ng of plasmid pINA445 containing the gene for the c-Myc tagged form of Pex3p and 125 ng of site-directed mutant primer. Site-directed mutations were confirmed by sequencing.
Antibodies
Antibodies to Pex3p were raised in guinea pig against a maltose-binding protein-Pex3p fusion containing the carboxyl 351 amino acids of Pex3p. The anti-Pex3p serum was partially purified by absorption against a lysate of pex3KOA cells or by affinity chromatography on immobilized maltose-binding protein-Pex3p fusion (Harlow and Lane, 1999).
A mouse monoclonal antibody to the human c-Myc epitope (9E10) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A rabbit polyclonal antibody to Saccharomyces cerevisiae carboxypeptidase Y was purchased from Nordic Immunological Laboratories (Tilburg, The Netherlands). Antibodies to the PTS1 tripeptide SKL, thiolase, acyl-CoA oxidase (AOX), isocitrate lyase (ICL), and Kar2p have been described previously (Aitchison et al., 1992; Eitzen et al., 1996; Titorenko et al., 1997; Wang et al., 1999). Fluorescein isothiocyanate-conjugated anti-rabbit IgG and rhodamine-conjugated anti-guinea pig IgG secondary antibodies were used to detect primary antibodies in immunofluorescence microscopy.
Light Microscopy
Cells were prepared for epifluorescence microscopy as described previously (Szilard et al., 1995). Formaldehyde-fixed cells were permeabilized by treatment with 0.2% Triton X-100 for 6 min and incubated with 10 μM BODIPY 493/503 (Molecular Probes, Eugene, OR) in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl for 15 min to label lipid droplets. Images were collected with a BX50 microscope (Olympus, Tokyo, Japan) mounted with a digital fluorescence camera (Spot Diagnostic Instruments, Sterling Heights, MI).
Electron Microscopy
Cells were prepared for transmission electron microscopy as described previously (Goodman et al., 1990). For immunogold labeling, cells were fixed with 3.0% paraformaldehyde/0.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.3, overnight at 4°C. Fixed cells were dehydrated in ethanol, embedded in LR White, and sectioned. Sections were incubated with guinea pig anti-thiolase serum at 1:250 dilution for 1 h at room temperature. Guinea pig primary antibodies were detected with 12-nm colloidal gold-conjugated donkey anti-guinea pig IgG secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA). Sections were observed with a Phillips EM 410 transmission electron microscope.
Subcellular Fractionation
Cells were grown for 16 h in glucose-containing YPD medium, transferred to oleic acid-containing YPBO medium, and incubated for 8 h. Fractionation of cells was performed as described previously (Szilard et al., 1995) and included the differential centrifugation of homogenized spheroplasts at 1000 × g for 10 min at 4°C in a JS13.1 rotor (Beckman Coulter, Fullerton, CA) to yield a postnuclear supernatant fraction. The postnuclear supernatant was subjected to further differential centrifugation at 20,000 × g for 30 min at 4°C in a JS13.1 rotor to yield a pellet (20KgP) fraction enriched for peroxisomes and mitochondria.
Subfractionation of Peroxisomes and Protease Protection Analysis
To subfractionate peroxisomes, ice-cold Ti8 buffer (10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and leupeptin, pepstatin, and aprotinin, each at 1 μg/ml) was added to the 20KgP fraction. The resuspended 20KgP fraction was incubated on ice for 15 min and then subjected to centrifugation at 200,000 × g at 4°C in a Beckman Coulter TLA 120.2 rotor to yield a pellet (Ti8P) enriched for membranes and a supernatant (Ti8S) enriched for soluble proteins. The Ti8P fraction was extracted separately with 1 M NaCl, 1 M urea, and 0.1 M Na2CO3, pH 11, by incubation on ice for 45 min. After extraction, membranes were pelleted by centrifugation as described above.
Protease protection analysis was performed on 20KgP fractions isolated in the absence of protease inhibitors, as described previously (Eitzen et al., 1997). Then 0, 10, 20, and 50 μg of trypsin was combined with 240 μg of protein of the 20KgP fraction in the absence or presence of 0.5% (wt/vol) Triton X-100. Reactions were incubated on ice for 40 min and terminated by precipitation with trichloroacetic acid. Proteins from equal fractions of each reaction were separated by SDS-PAGE, transferred to nitrocellulose, and subjected to immunoblotting.
Other Methods
Oligonucleotides were synthesized on an Oligo 1000 M DNA synthesizer (Beckman Coulter). Sequencing was performed with an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA). Preparation of yeast lysates, growth of E. coli, and manipulations of DNA were performed as described previously (Ausubel et al., 1994). Protein concentrations were determined with a commercially available kit (Bio-Rad, Mississauga, Ontario, Canada). SDS-PAGE was performed essentially as described in Laemmli (1970). Proteins were transferred to nitrocellulose for immunoblotting by using a semidry electrophoretic transfer system, and antigen-antibody complexes were detected by enhanced chemiluminescence.
RESULTS
Yarrowia lipolytica Cells Mutant for the Peroxisomal Integral Membrane Protein Pex3p Lack Peroxisomes and Mislocalize Peroxisomal Matrix Proteins
The mut12-1 strain (Table 1) was identified among randomly mutagenized wild-type Y. lipolytica E122 cells by its inability to grow on medium containing oleic acid as the carbon source (ole− phenotype). Morphological (Figure 1A) and biochemical (our unpublished data) analyses showed that this strain mislocalized peroxisomal matrix proteins targeted by PTS1 or PTS2 to the cytosol and that therefore this strain was affected in peroxisome assembly. Electron microscopical analysis showed that the mut12-1 mutant strain did not contain wild-type peroxisomes, but instead contained numerous small vesicles (our unpublished data).
Figure 1.
PTS1- and PTS2-containing matrix proteins are mistargeted to the cytosol in Y. lipolytica cells mutant for Pex3p. (A) Wild-type (WT) strain E122, original mutant strain mut12-1, and transformed strain M12TR were grown at 30°C in glucose-containing YND medium, transferred to oleic acid-containing YNO medium, and incubated at 30°C for an additional 8 h. Cells were processed for immunofluorescence microscopy with rabbit antibodies to the PTS1 tripeptide SKL and guinea pig antibodies to the PTS2-targeted enzyme thiolase (THI). Rabbit primary antibodies were detected with fluorescein-conjugated goat anti-rabbit immunoglobulin G secondary antibodies, and guinea pig primary antibodies were detected with rhodamine-conjugated donkey anti-guinea pig immunoglobulin G secondary antibodies. Bar, 5 μm. (B) Nucleotide sequence of the YlPEX3 gene and deduced amino acid sequence of YlPex3p. Translation was shown experimentally to occur from the indicated ATG rather than from an upstream, inframe ATG. These sequence data have been deposited in the DDBJ/EMBL/GenBank databases under accession number AF474003. (C) Sequence similarity between YlPex3p and Pex3 proteins of other organisms. Numeric scale at top indicates protein length in amino acids. Colorimetric scale of protein similarity is defined at right. The five blocks of amino acid sequence conserved among the different Pex3 proteins as defined by the program Macaw2, and the amino acid changes made to generate ts mutant forms of Pex3p, are presented at bottom. (D) Ultrastructure of wild-type and pex3KOA cells. The wild-type (WT) strain E122 and the pex3KOA mutant strain were grown in YPD medium and then incubated for 8 h at 30°C in YPBO medium. Cells were fixed in 1.5% KMnO4 and processed for electron microscopy. Peroxisomes (P) were observed in wild-type cells but not in pex3KOA cells, which instead contained numerous small vesicles (V). L, lipid droplet; M, mitochondrion, N, nucleus. Bar, 0.5 μm.
The MUT12 gene was isolated from a library of Y. lipolytica genomic DNA by functional complementation, i.e., restored growth on oleic acid-containing medium (the ole+ phenotype), of the mut12-1 strain. Functional complementation of the mut12-1 strain restored import into peroxisomes of proteins targeted by either PTS1 or PTS2 (Figure 1A). DNA was isolated from the complemented strain, and the complementing plasmid was recovered by transformation of E. coli. The complementing fragment was mapped by restriction endonuclease digestion, and various restriction fragments were subcloned and introduced by transformation into the mut12-1 strain to define the minimal complementing fragment. This fragment was sequenced in both directions and shown to contain two overlapping open reading frames, one coding for 482 amino acids and a second coding for 431 amino acids (Figure 1B). Insertion of a nucleotide between the first and second potential initiating codons, followed by transformation of the mut12-1 strain and determination of the ole+ phenotype, confirmed that initiation of translation originates at the second ATG codon (our unpublished data).
A search of databases with the use of the GENEINFO(R) BLAST Network Service of the National Center for Biotechnology Information indicated that the protein encoded by the open reading frame complementing the mut12-1 mutant strain shows significant sequence similarity (expected [E] values from 2e−54 to 2e−11) to Pex3 proteins from other species (Figure 1C). Therefore, the complementing gene was designated YlPEX3 and its encoded protein as YlPex3p. A comparison of YlPex3p with Pex3 proteins from eight other species by using the program Macaw2 (http://www.ncbi.nlm.nih.gov) identified five conserved blocks in these proteins (Figure 1C). The most conserved block occurs between residues 127–174 of YlPex3p (Figure 1C, block II) and is predicted to be hydrophobic in nature. None of the conserved blocks or variable intervening sequences contain any known functional motifs as determined by pattern and profile searching of YlPex3p against appropriate databases such as Prosite, Pfam, and Blocks (http://www.up.univ-mrs.fr/∼wabim/english/logligne.html). In addition, Position Specific Iterated-Blast searching (http://www.ncbi.nlm.nih.gov) did not reveal any evidence of distant relationships that could provide clues to the functions of the blocks.
The entire coding region of the PEX3 gene was replaced by targeted integration of the Y. lipolytica URA3 gene to make the strains pex3KOA and pex3KOB in the Y. lipolytica A (E122) and B (22301-3) mating types, respectively (Table 1). The strains deleted for PEX3 were unable to grow on oleic acid and like the mut12-1 strain did not contain wild-type peroxisomes, but instead contained numerous small vesicles (Figure 1D). Mating of the two wild-type strains to various pex3 mutant strains (Table 1) demonstrated that the authentic PEX3 gene had been cloned and that the ability to use oleic acid as the carbon source required at least one intact copy of the PEX3 gene (our unpublished data).
Pex3p was detected weakly in peroxisomes by immunofluorescence labeling of oleic acid-incubated wild-type E122 cells by using an anti-Pex3p polyclonal antibody (Figure 2A). Treatment of a peroxisome-enriched 20KgP subcellular fraction with various salts showed that a form of Pex3p tagged with the c-Myc epitope (MycPex3p), which can functionally complement the pex3KOA strain (our unpublished data), displays the characteristics of an integral membrane protein (Figure 2B). Pex3p is also exposed to the cytosol, as seen by the sensitivity of MycPex3p in a 20KgP fraction treated with exogenously added trypsin in the absence of detergent, in contrast to the matrix protein thiolase (Figure 2C).
Figure 2.
Pex3p is a peroxisomal integral membrane protein exposed to the cytosol. (A) Pex3p is localized to peroxisomes. Wild-type E122 cells incubated in oleic acid-containing YPBO medium for 8 h were processed for immunofluorescence microscopy with guinea pig primary antibodies to Pex3p and rabbit primary antibodies to the PTS1 tripeptide SKL. Primary antibodies were detected as described in the legend to Figure 1A. Bar, 2.5 μm. (B) Pex3p exhibits properties of an integral membrane protein. A functionally complementing form of Pex3p tagged with the c-Myc epitope (MycPex3p) resists extraction from a 20KgP subcellular fraction by treatment with 1.0 M NaCl, 1.0 M urea, and 0.1 M Na2CO3, pH 11. (C) Pex3p is exposed to the cytosol. MycPex3p in a 20KgP subcellular fraction is sensitive to exogenously added trypsin in both the absence and the presence of the detergent Triton X-100. In contrast, the peroxisomal matrix protein thiolase (THI) is resistant to exogenously added trypsin in the absence of detergent and sensitive in the presence of detergent.
During De Novo Peroxisome Biogenesis, Matrix Protein Sequestration Is First Detected at Single or Multiple Sites in ER-rich Regions of Pex3p Temperature-sensitive Mutant Cells
Temperature-sensitive (ts) mutants of Pex3p were made to investigate the early stages of peroxisome biogenesis. These ts mutants were constructed by site-directed mutagenesis of the regions of the PEX3 gene coding for each of the five amino acid blocks conserved in the known Pex3 proteins. Because ts mutations are generally thought to be missense mutations that affect protein structure (Pringle, 1975), amino acid residues to be mutated were chosen on their ability to change local secondary structure when mutated and on their degree of conservation among the Pex3 proteins (Figure 1C).
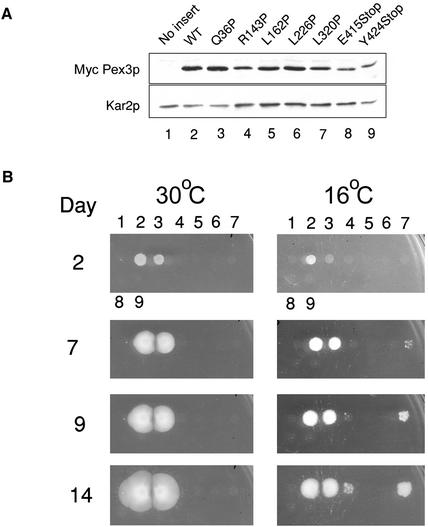
Genes encoding mutant forms of MycPex3p were expressed under control of the native PEX3 promoter in the plasmid pINA445 in the peroxisome-deficient pex3KOA mutant strain background. Immunoblot analysis showed that the levels of the various mutant forms of MycPex3p were similar to that of MycPex3p in the wild-type strain after incubation of the strains in oleic acid-containing medium (Figure 3A). Therefore, phenotypic differences between the strains could not be attributed to differences in the levels of the various forms of MycPex3p. The functionality of the individual mutant MycPex3 proteins was determined by testing their ability to restore growth of the pex3KOA strain on YNO agar medium containing oleic acid as the carbon source. At 30°C, only one mutant MycPex3p, Q36P, restored growth of the pex3KOA strain on oleic acid agar (Figure 3B). The six MycPex3p mutants that were unable to complement the pex3KOA strain at 30°C were checked for temperature sensitivity by incubation of their pex3KOA hosts on oleic acid agar plates at 16°C. At 16°C, the strain expressing MycPex3p-Q36P grew more slowly than the strain expressing MycPex3p, suggesting that this mutant strain is cold sensitive. Cells expressing the L320P and R143P forms of MycPex3p were able to complement the pex3KOA mutant strain at 16°C. Although they exhibited temperature sensitivity at this temperature, they grew more slowly than did cells expressing MycPex3p. In addition, at 16°C the strain expressing MycPex3p-R143P grew more slowly than the strain expressing MycPex3p-L320P (Figure 3B). The slower growing nature of the MycPex3p-R143P mutant strain is consistent with the R143 residue being more highly conserved across the different Pex3 proteins than the L320 residue. The spotted nature of the mutant colonies at 16°C (Figure 3B) suggested that the correct folding of Pex3p may be somewhat stochastic, resulting in the ts phenotype not being fully penetrant. To verify this, the genes encoding the ts forms of MycPex3p were again transformed into the pex3KOA strain, and the phenotypes observed were identical to those observed previously, indicating that the phenotypes did not result from spontaneous mutation elsewhere in the genome but were the direct result of the transformed genes.
Figure 3.
Temperature-sensitive growth of Pex3p variants. (A) Levels of the wild-type and mutant c-Myc-tagged Pex3 proteins expressed in transformed pex3KOA cells are comparable. “No insert” refers to the pex3KOA strain containing the vector pINA445 alone. Lysates of the various strains were subjected to immunoblotting with the 9E10 antibody against the c-Myc epitope. The immunodetection of Kar2p was used as a loading control. (B) Mutant MycPex3 proteins R143P and L320P confer ts growth on oleic acid-containing agar to the pex3KOA strain. The days of incubation at 30°C or 16°C are indicated. Numbering of strains is as in A.
Immunofluorescence staining of cells grown in glucose-containing YND medium at 30°C and then incubated in oleic acid-containing YNO medium at 30°C showed that the peroxisomal matrix protein thiolase was targeted to peroxisomes in cells able to grow in oleic acid (wild-type and MycPex3p-Q36P) but was mislocalized to the cytosol of cells unable to grow in this medium at 30°C (MycPex3p-R143P, -L162P, -L226P, -L320P, -E415Stop, and -Y424stop) (Figure 4A). Immunostaining of cells grown at 30°C and switched to glucose-containing YND medium at 16°C for 48 h without subsequent incubation in oleic acid-containing medium showed that thiolase was correctly targeted to peroxisomes in both the wild-type strain and in the strain expressing MycPex3p-Q36P (Figure 4B). Targeting to peroxisomes appeared less efficient in the strain expressing MycPex3p-Q36P, consistent with its cold sensitivity. In the mutants that were unable to grow in oleic acid-containing medium at 30°C, thiolase was generally dispersed throughout the cytosol of cells grown in glucose-containing medium at 16°C (Figure 4B), although a small percentage (much <1%) of the cells expressing MycPex3p-L226, -E415Stop, and –Y424Stop showed evidence of single peroxisome-like structures. These structures were probably nonfunctional, given the inability of these strains to grow on oleic acid-containing medium (Figure 3B). However, peroxisome biogenesis was observed in the ts mutant cells expressing MycPex3p-R143P and MycPex3p-L320P when grown in glucose-containing medium, because some thiolase was targeted to peroxisomes in these cells (Figure 4B). Peroxisome biogenesis in media containing glucose is not surprising, given that peroxisomes are multifunctional organelles involved in metabolic processes other than lipid catabolism (reviewed in Titorenko and Rachubinski, 2001). Because thiolase is present at low levels under conditions of glucose growth, peroxisomes could be detected at early stages of biogenesis in cells expressing MycPex3p-R143P and MycPex3p-L320P by immunostaining (Figure 4B). These peroxisomes might not be detected in the same cells incubated in oleic acid-containing medium due to the presence of very high levels of thiolase levels in the cytosol, which could mask their presence (Figure 4A). After 8 d of growth in glucose-containing YND medium at 16°C, 22% of MycPex3p-R143P and 30% of MycPex3p-L320P–expressing cells contained one or more peroxisomes.
Figure 4.
Peroxisome biogenesis occurs in cells grown in glucose-containing medium at 16°C. (A) Cells of strains able to grow on oleic acid-containing YNO agar plates at 30°C (wild-type [WT] and MycPex3p-Q36P) localized thiolase to peroxisomes by immunofluorescence, whereas cells of strains unable to grow on YNO agar plates at 30°C (MycPex3p-R143P, -L162P, -L226P, -L320P, -E415Stop, and -Y424Stop) showed a predominantly cytosolic localization for thiolase, upon incubation in oleic acid-containing YNO medium at 30°C. (B) At 16°C in glucose-containing YND medium, thiolase was targeted to peroxisomes of the WT strain and of the strain expressing MycPex3p-Q36P; however, targeting was less efficient in the latter strain. Strains expressing MycPex3p-L162P, -L226P, -E415Stop, and -Y424Stop showed a cytosolic localization for thiolase, whereas thiolase sequestration was first detected in MycPex3p-R143P and MycPex3p-L320P at one site, or sometimes at multiple sites, in the peripheral regions of cells.
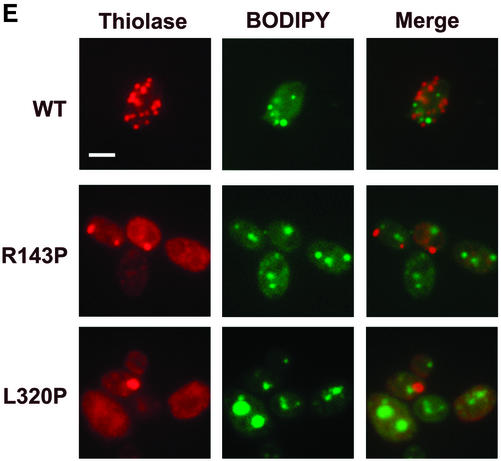
The first evidence of thiolase sequestration in the ts mutants MycPex3p-R143P and MycPex3p-L320P grown in glucose-containing medium at 16°C was in individual peroxisomes located close to the cell periphery. These individual peroxisomes varied in size and were frequently positioned at the tip of a cell, or close to it, although they were also found along the cell edge (Figure 4B). Acyl-CoA oxidase was sometimes detected in these peroxisomes, while thiolase was completely or mostly in the cytosol, indicating that these structures are nascent peroxisomes and not mature peroxisomes (Figure 5A). In addition to cells containing a single peroxisome at their periphery, some cells contained at their periphery multiple, small peroxisomes that were well separated from one another (Figure 4B). The same results were observed for a strain in which the MycPex3p-L320P allele had been integrated into the pex3KOA genome, indicating that these observations were not an artifact of plasmid expression (our unpublished data). Double labeling of cells with antibodies to thiolase and to other matrix proteins indicated that the nascent peroxisomes were functional in importing both PTS1- and PTS2-containing proteins (Figure 5B), consistent with the ability of the cells to grow on oleic acid plates. Because the ER has been suggested to play a role in peroxisome biogenesis, cells were doubly labeled with antibodies to thiolase and to the ER resident protein Kar2p (Figure 5C). The periphery of cells was often immunostained with the antibody to Kar2p, indicating it to be rich in ER elements. Nascent peroxisomes were closely associated with the ER in the peripheral regions of the cells and sometimes with a region that seemed to be the nuclear envelope (Figure 5C). To confirm that this region was indeed the nuclear envelope, cells were doubly labeled with antibodies to thiolase and the nuclear stain 4,6-diamidino-2-phenylindole (DAPI) (Figure 5D). Approximately 50% of the individual, peripherally located nascent peroxisomes were positioned close to nuclei, suggesting a possible role for the nuclear envelope in peroxisome biogenesis. Double labeling of cells with antibodies to thiolase and to the neutral lipid-specific stain BODIPY 493/503 showed that nascent peroxisomes were randomly distributed with respect to lipid droplets within the glucose-grown cells (Figure 5E).
Figure 5.
De novo peroxisome biogenesis occurs in cells grown in glucose-containing medium in regions associated with ER elements. (A) Immunofluorescence analysis demonstrating that the peroxisome-like structures found at the periphery of glucose-grown MycPex3p-R143p– or MycPex3p-L320P–expressing cells contain the matrix enzyme AOX but not the matrix enzyme thiolase, suggesting that these structures are nascent peroxisomes undergoing biogenesis. Bar, 3 μm. (B) Peripherally located peroxisomes contain different matrix proteins. Y. lipolytica cells were analyzed by immunofluorescence with antibodies to thiolase, the PTS1 SKL, AOX, and ICL. Bar, 3 μm. (C) Nascent, peripherally located peroxisomes are found in ER-rich regions of the cell and sometimes associated with the nuclear envelope. Cells were processed for immunofluorescence and doubly labeled with antibodies to thiolase and to the ER resident protein Kar2p. Bar, 3 μm. (D) Individual, peripherally located peroxisomes are often positioned close to nuclei. Cells were labeled with antibodies to thiolase and with the nuclear stain, DAPI. Bar, 3 μm. (E) Peroxisomes are randomly positioned with respect to lipid droplets in glucose-grown cells. Cells were labeled with antibodies to thiolase and with the neutral lipid stain BODIPY 493/503. Bar, 3 μm. (F) Peroxisome-like structures (P) are observed by electron microscopy closely associated with ER elements (E) in cells undergoing de novo peroxisome biogenesis. Cells were grown in glucose-containing YND medium at 16°C for 48 h.
Analysis by electron microscopy of cells grown in glucose-containing YND medium at 16°C showed that the ts mutants MycPex3p-R143P and MycPex3p-L320P contained peroxisome-like structures at the cell periphery in proximity to ER elements. These peroxisome-like structures were often positioned at, or close to, the tips of the ER elements (Figure 5F).
The various ts mutant cells were next grown at 16°C in glucose-containing YND medium for 8 d and then incubated in oleic acid-containing YNO medium for an additional 8 h at 16°C to study the events of peroxisome biogenesis under conditions of peroxisome proliferation. Under these conditions, 32% of MycPex3p-R143P and 50% of MycPex3p-L320P–expressing cells were observed to contain one or more peroxisomal structures, indicating that peroxisome biogenesis was up-regulated when the cells were incubated in oleic acid-containing medium compared with growth in medium containing glucose (see above). The populations of the MycPex3p-R143P– and MycPex3p-L320P–expressing ts mutants were comprised not only of cells with peripheral, often individual, peroxisomes but also of cells with morphologically distinct semicircular to circular (hereafter, for convenience, termed semi/circular) tubular structures that stained with antibodies to thiolase and which were clearly distinguishable from clusters of peroxisomes (Figure 6A). The immunostained semi/circular tubular structures often showed “bulges” of more intense staining. The cold-sensitive MycPex3p-Q36P–expressing strain also contained a small percentage of cells with these structures. When the ts MycPex3p-L320P allele was integrated into the genome of the pex3KOA strain, the same semi/circular tubular structures were observed, albeit at a lower frequency, indicating that the structures were not an artifact of plasmid expression (our unpublished data). The semi/circular tubular structures were never observed in cells of the wild-type strain or the pex3KOA strain.
Figure 6.
Peroxisomal matrix proteins are present in semi/circular tubular structures in Pex3p ts mutants undergoing peroxisome proliferation in oleic acid-containing medium. Cells were grown in glucose-containing YND medium for 48 h at 16°C and then incubated in oleic acid-containing YNO medium for 8 h at 16°C. (A) Immunofluorescence analysis with antibodies to thiolase showing that MycPex3p-R143P– and MycPex3p-L320P–expressing cells incubated in oleic acid-containing (YNO) medium at 16°C contain both peripherally located peroxisomes and distinct semi/circular tubular structures that immunostain for thiolase. (B) The semi/circular tubular structures immunopositive for thiolase do not surround nuclei. Nuclei were detected by staining with DAPI. (C) Semi/circular tubular structures immunopositive for thiolase surround lipid droplets. Lipid droplets were detected by staining with BODIPY 493/503. (D) Different peroxisomal matrix proteins are present in the semi/circular tubular structures. Immunofluorescence analysis showing colocalization of signals for antibodies to the PTS1 SKL (i), AOX (ii), and ICL (iii) with the signal for antibodies to thiolase in semi/circular tubular structures. (E) Immunofluorescence analysis showing the ER resident protein Kar2p partially colocalizes with thiolase in semi/circular tubular structures. (F) Immunofluorescence analysis shows no colocalization of thiolase and the vacuole resident protein carboxypeptidase Y (CPY), indicating that the semi/circular tubular structures are not vacuoles in the process of degrading peroxisomes. Bars, 3 μm.
To determine whether the semi/circular tubular structures surround nuclei, cells were doubly labeled with antibodies to thiolase and with DAPI (Figure 6B). Semi/circular tubular structures immunostained for thiolase rarely, if ever, surrounded nuclei. In addition, the unstained centers of these structures varied greatly in size. Given the circular nature of the immunostaining structures, the variable sizes of their unstained cores, and the fact that peroxisomes catabolize lipids, the lipid droplets of cells incubated in oleic acid-containing medium at 16°C were detected with BODIPY 493/503, and the cells were immunostained with anti-thiolase antibodies (Figure 6C). The semi/circular tubular structures that immunostained for thiolase were closely apposed to the surface of lipid droplets, sometimes completely surrounding them. Some cells with multiple lipid droplets contained just one droplet with an associated thiolase-immunostaining semi/circular tubular structure. Labeling of cells with antibody to thiolase and with another lipid droplet stain, Nile red, produced comparable results (our unpublished data).
To determine whether other peroxisomal matrix proteins were localized to these semi/circular tubular structures, cells were doubly labeled with antibodies to thiolase, which is targeted to peroxisomes by a PTS2, and antibodies to the PTS1, Ser-Lys-Leu (SKL). Structures stained by the anti-SKL antibodies often overlapped with the semi/circular tubular structures stained by the anti-thiolase antibodies, indicating that peroxisomal matrix proteins detected by anti-SKL antibodies were also localized to these structures (Figure 6D, i). The peroxisomal matrix proteins AOX and ICL similarly colocalized with thiolase in the semi/circular tubular structures (Figure 6D, ii and iii). Cells doubly labeled with antibodies to thiolase and to the ER chaperone Kar2p showed a strong thiolase immunosignal partially overlapping with a weaker Kar2p immunosignal in cells expressing MycPex3p-R143P or MycPex3p-L320P but not in wild-type cells (Figure 6E).
Vacuoles are amorphous structures that sometimes surround lipid droplets and exhibit characteristics similar to those of semi/circular tubular structures. To determine whether the semi/circular tubular structures were vacuoles in the process of degrading peroxisomes, cells were doubly labeled with antibodies to thiolase and to the vacuolar protein carboxypeptidase Y. The semi/circular tubular structures were distinct from vacuolar elements, although the two were often closely associated (Figure 6F).
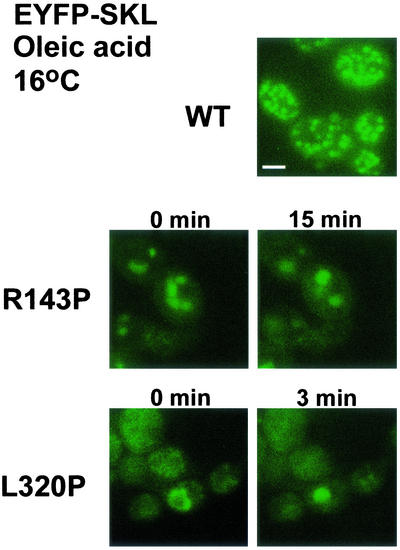
To investigate whether peroxisomes could arise from these semi/circular tubular structures, a gene encoding EYFP tagged at its carboxy terminus with the PTS1 Ser-Lys-Leu (EYFP-SKL) was expressed together with either MycPex3p-R143P or MycPex3p-L320P in pex3KOA cells incubated in oleic acid-containing medium. In both cases, in living cells, large spherical structures formed with time from the semi/circular tubular structures, consistent with a role for these structures in peroxisome biogenesis (Figure 7).
Figure 7.
Semi/circular tubular structures give rise to peroxisome-like structures in living cells. Cells expressing the chimeric protein EYFP-SKL and either MycPex3p-R143P or MycPex3p-L320P were grown at 16°C for 48 h in glucose-containing YND medium and then incubated for 8 h in oleic acid-containing YNO medium. EYFP-SKL is imported into peroxisomes in wild-type (WT) cells. In cells coexpressing MycPex3p-R143P or MycPex3p-L320P, the EYFP-SKL protein is targeted to the semi/circular tubular compartment at 0 min, which give rises to peroxisome-like structures by 15 min. Bar, 3 μm.
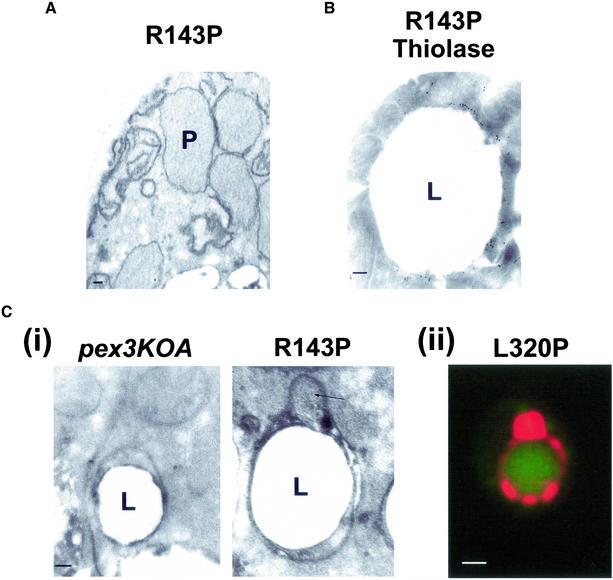
Given the unsynchronized nature of peroxisome biogenesis in pex3KOA cells expressing MycPex3p-R143P or MycPex3p-L320P at 16°C, and the fact that <15% of cells of these strains incubated in oleic acid-containing medium at 16°C contain thiolase immunostaining semi/circular tubular structures, we deemed it inappropriate, at least for the present, to attempt subfractionation and biochemical characterization of these strains as a way to analyze these structures. We instead focused on an electron microscopic analysis of pex3KO cells expressing MycPex3p-R143P, because these cells exhibit a higher frequency of thiolase immunostaining semi/circular tubular structures. Peroxisomes were observed in some MycPex3p-R143P–expressing cells incubated in oleic acid-containing YNO medium at 16°C (Figure 8A). Lipid droplets of varying size were also observed in the MycPex3p-R143P–expressing cells. Each droplet showed as a neutral lipid core surrounded by a phospholipid monolayer. We analyzed the surface of the lipid droplets as a possible site of peroxisome biogenesis. Although some lipid droplets apparently had no structures associated with them, others were associated with distinct bilayered membrane structures that closely wrapped around the surface of the lipid droplet. These structures varied in size from short segments of membrane to extensive membrane networks completely surrounding lipid droplets. Immunoelectron microscopy showed the structures to be labeled with antibodies to thiolase (Figure 8B). Similar membrane structures on the surface of lipid droplets were observed by electron microscopy of pex3KOA cells incubated in oleic acid-containing medium at 16°C (Figure 8C, i), suggesting that Pex3p is not required for their formation. No peroxisome-like structures were seen associated with these structures in pex3KOA cells. In contrast, bulges, peroxisome-like in appearance, were sometimes observed associated with the surface lipid droplet structures of cells expressing MycPex3p-R143P (Figure 8C, i, arrow). The bulges appeared within the membrane sheets of the surface lipid droplet structures. Morphologically, the membranous structures on the surface of lipid droplets observed by electron microscopy are the only structures that resemble the fluorescently labeled semi/circular tubular structures observed by epifluorescence microscopy (Figure 8C, ii), and they do not resemble morphologically peroxisomes undergoing microautophagy, as reported by Sakai et al. (1998).
Figure 8.
Microscopic analysis of peroxisomes in cells expressing MycPex3p-R143P and of structures surrounding lipid droplets. Cells were grown for 48 h in glucose-containing YND medium at 16°C, transferred to oleic acid-containing YNO medium, and incubated for a further 8 h at 16°C. (A) Electron micrograph of peroxisomes (P) in MycPex3p-R143P–expressing cells. (B) Electron micrograph of the structure surrounding a lipid droplet (L) in cells expressing MycPex3p-R143P. The structure shows immunogold labeling with antibody to thiolase. (C, i) Electron micrographs of structures surrounding lipid droplets (L) in the pex3KOA strain (left) and the pex3KOA strain expressing MycPex3p-R143P (right). The structures are distinct from the lipid droplets themselves, with their phospholipid bilayer membranes seen to be wrapping around the phospholipid monolayer of the lipid droplets. The structure around the lipid droplet of the strain expressing MycPex3p-R143P contains a bulge (arrow), which is peroxisome-like in appearance. (ii) A semi/circular tubular structure immunostained for thiolase and surrounding a lipid droplet detected with BODIPY 493/503 resembles the structure surrounding a lipid droplet observed by electron microscopy of a MycPex3p-R143P-expressing cell seen in Figure 8B. Bar, 1.0 μm. Bars in electron micrographs, 0.1 μm.
Overexpression of Pex3p Results in Enlarged Peroxisomes That Cluster
To determine whether Pex3p, normally a protein of low abundance, was targeted to the putative sites of peroxisome biogenesis, we overexpressed MycPEX3 from the oleic acid-inducible thiolase promoter in cells of both the wild-type E122 strain and the original mut12-1 mutant strain. We also used overexpression of MycPEX3 to analyze the ability of Pex3p to sequester peroxisomal factors. Cells were incubated for 7 h to 48 h at 30°C in oleic acid-containing YNO medium. Expression from the thiolase promoter led to enhanced levels of MycPex3p vis-à-vis expression from the native PEX3 promoter (Figure 9A).
Figure 9.
Overexpression of Pex3p produces a reduced number of enlarged and clustered peroxisomes. (A) Immunoblot with 9E10 antibody of MycPex3p expressed from the native PEX3 promoter in the plasmid pINA445 (pINA445Pex3p) and overexpressed from the thiolase promoter (TC3Pex3p) in the wild-type E122 strain. Immunodetection of glucose-6-phosphate dehydrogenase (G6PDH) was done as a loading control. (B) Overexpressed MycPex3p is detected in the perinuclear/juxtanuclear region (i), as well as in semicircular structures on the surface of lipid droplets (ii). Wild-type cells overexpressing MycPex3p were analyzed by immunofluorescence with an affinity-purified antibody to Pex3p and with antibodies to the ER resident protein Kar2p (i). Nuclei were detected by staining with DAPI (i), and lipid droplets were detected by staining with BODIPY 493/503 (ii). Bars, 3 μm. (C) Wild-type E122 and original mut12-1 mutant cells overexpressing MycPex3p have reduced numbers of enlarged peroxisomes that exhibit clustering. Peroxisomes were detected by immunofluorescence with anti-thiolase antibodies. Nuclei were detected by staining with DAPI. Bar, 3 μm. (D) Electron micrographs of the wild-type (WT) E122 strain (left) and of the wild-type (middle) and the original mut12-1 (right) strains overexpressing MycPex3p. Overexpression of MycPex3p produces a reduced number of enlarged peroxisomes that cluster. L, lipid droplet; N, nucleus; P, peroxisome. Bar, 0.5 μm.
Immunolabeling of MycPEX3–overexpressing cells incubated for 7 h in YNO medium with affinity-purified, anti-Pex3p antibodies showed Pex3p to be localized to a reticular system containing the ER resident protein Kar2p (Figure 9B). An intense perinuclear to juxtanuclear Pex3p immunosignal was detected in cells of both the wild-type and mut12-1 backgrounds, and the signal overlapped with the immunosignal for Kar2p (Figure 9B, i). A strong, semicircular Pex3p immunosignal was also observed around the surface of lipid droplets stained with BODIPY 493/503 (Figure 9B, ii), whereas a weaker Pex3p immunosignal was detected in the ER-rich periphery of cells.
Immunofluorescence labeling with antibody to thiolase showed that cells overexpressing MycPex3p incubated for 48 h in oleic acid-containing YNO medium contained reduced numbers of peroxisomes that were larger in size and more clustered in appearance than peroxisomes of wild-type cells (Figure 9C). Several of the cells overexpressing MycPex3p contained a single large peroxisome, whereas others contained peroxisomes that exhibited clustering and were larger than those of wild-type cells (Figure 9C). Peroxisomes were sometimes observed closely associated with nuclei (Figure 9C). Electron microscopy confirmed the peroxisomes of cells overexpressing MycPex3p to be larger and more clustered than peroxisomes of wild-type cells (Figure 9D). Enlarged and clustered peroxisomes are consistent with an ability of Pex3p to sequester factors required for peroxisome biogenesis, producing larger peroxisomes by promoting membrane fusion or by inhibiting peroxisomal division.
DISCUSSION
Pex3p has been shown to be essential for peroxisome biogenesis in all eukaryotic organisms investigated so far. In addition, it is the only reported integral membrane peroxin whose null phenotype in all organisms so far investigated results in cells lacking peroxisomal structures. Thus, Pex3p is an important candidate protein for initiating peroxisome biogenesis.
We have made ts mutants of Pex3p and have overexpressed Pex3p in the yeast Y. lipolytica to better understand the early stages of peroxisome biogenesis, as well as to identify a possible role for Pex3p in this process. The strains expressing the ts forms of Pex3p were able to assemble functional peroxisomes only at the lower temperature of 16°C, as shown by the ability of these strains to grow on oleic acid-containing medium at 16°C but not at 30°C. More importantly, however, these ts strains exhibited retarded peroxisome biogenesis, as indicated by their slower growth at 16°C compared with cells expressing the wild-type form of Pex3p. Immunofluorescence and electron microscopy studies support the concept of retarded peroxisome biogenesis in the ts strains, because these methodologies readily revealed apparent intermediate structures in the peroxisome biogenesis pathway. These intermediates may arise because Pex3p plays a central role in a rate-limiting step of peroxisome biogenesis, and this step is sufficiently retarded in the ts mutant strains to allow visualization of the intermediates. That these intermediates were observed in a percentage of the cells of the ts strains may result from a lack of synchronization in the peroxisome biogenesis process and/or an incomplete penetrance of the mutant phenotype.
The presence of individual, peripherally located peroxisomes strongly immunoreactive for different peroxisomal matrix proteins in the ts strains at 16°C in glucose-containing medium, and the absence of these structures in a strain deleted for the PEX3 gene under identical conditions, suggest that Pex3p is involved in the initiation of peroxisome assembly by sequestering directly or indirectly the components of peroxisome biogenesis, such as functional PTS1 and PTS2 import machineries. The enlarged and clustered peroxisomes observed in cells overexpressing Pex3p is also consistent with Pex3p sequestering peroxisome biogenesis factors. Excess Pex3p in the membrane of one peroxisome could lead to the aggregation of peroxisomes through its interaction with Pex3p or other proteins in the membranes of adjacent peroxisomes. Enlarged peroxisomes may result from an inhibition of peroxisome division, perhaps through interaction or complex formation with peroxins such as Pex11p and Pex25p or with enzymes of peroxisomal β-oxidation, both of which have been implicated in the regulation of peroxisome division (Erdmann and Blobel, 1995; Marshall et al., 1995; Schrader et al., 1998; Chang et al., 1999; Smith et al., 2000, 2002). Also interesting are the requirement for Pex3p for the proper localization and stability of peroxisomal membrane proteins in S. cerevisiae (Hettema et al., 2000), and the interaction of Pex3p with Pex19p (Götte et al., 1998; Soukupova et al., 1999), which acts in the stabilization of proteins within the peroxisomal membrane (Snyder et al., 2000). Both of these observations are consistent with a role for Pex3p in sequestering components of the peroxisome biogenesis machinery. How Pex3p achieves this sequestration remains unknown.
The site of de novo peroxisome biogenesis is an issue of intense debate. This work suggests that de novo peroxisome biogenesis, or at least that part involving matrix protein sequestration, can occur in regions of the cell rich in ER elements. Peroxisomes located at the periphery of cells and in association with ER elements were the first site at which matrix protein sequestration was observed. In addition, the fact that only some of these peripherally located peroxisomes contained acyl-CoA oxidase but not thiolase is consistent with these structures being nascent peroxisomes undergoing biogenesis and not mature peroxisomes. The occurrence of multiple, well separated peroxisomes at the cell periphery in some ts mutant cells suggests that peroxisome biogenesis can occur at multiple sites in the cell. In addition, the presence of cells with a single peripheral peroxisome suggests that peroxisome biogenesis within a cell may not be synchronized and that when one peroxisome forms early, it may have a selective advantage by sequestering available biogenesis components. Alternatively, the presence of individual peroxisomes in cells undergoing biogenesis may suggest the existence of a preferential site for peroxisome biogenesis at the cell periphery.
It remains a formal possibility that the nascent peroxisomes could have arisen from unidentified preperoxisomal/peroxisomal ghosts (Santos et al., 1988). The small, abundant vesicles observed in some of the oleic acid-incubated mut12-1 and pex3KO cells (Figure 1D) do not seem to be ghosts, because they were not detected at the early stages of peroxisome biogenesis in pex3KO cells expressing MycPex3p-R143P or MycPex3p-L320P at 16°C in glucose-containing medium (Figure 5F). The vesicles could have arisen in the mut12-1 and pex3KO cells because of starvation of the cells during their incubation for 8 h in medium containing oleic acid as the carbon source, which the cells were unable to metabolize. Similarly, the bilayered membrane structures surrounding lipid droplets (Figure 8C) do not seem to preperoxisomal/peroxisomal ghosts, because they contain the ER-resident protein Kar2p. ER-like structures have been reported to surround lipid droplets (Prattes et al., 2000).
Targeting of overexpressed Pex3p to membrane elements that contain Kar2p (Figure 9B) suggests a role for the ER in de novo peroxisome biogenesis. It is not unexpected that the ER could be a site of peroxisome biogenesis, given the intimate association between these organelles, and previous studies have suggested an essential role for the ER in peroxisome biogenesis in Y. lipolytica (Titorenko and Rachubinski, 1998). The proximity between peroxisomes and nuclei in approximately half of the ts mutant cells suggests that the nuclear envelope may also play a role in peroxisome biogenesis, and, indeed the outer membrane of the nuclear envelope is continuous with the ER system. The overexpressed Pex3p could be detected in the nuclear envelope (Figure 9B, i), and expression of the amino-terminal 50 amino acids of Pex3p in the methylotrophic yeast Hansenula polymorpha has been reported to produce Pex14p-containing vesicles thought to be derived from the nuclear envelope (Faber et al., 2002). Interestingly, however, South et al., (2001) have shown that Sec61p or its homolog Ssh1p, which are involved in protein targeting to the ER, is not required for transport of Pex3p to the peroxisome. This observation can be explained if targeting of Pex3p to the ER does not require the “usual” proteins required for translocation across, or integration into, the ER membrane. Indeed, some membrane proteins have been shown to insert posttranslationally into the ER (Kutay et al., 1993). South et al. (2000) have also reported that inhibitors of COPI and COPII do not block Pex3p-mediated peroxisome biogenesis. This observation is consistent with the results of our electron microscope investigations of cells expressing MycPex3p-R143P or MycPex3p-L120p (Figure 5F), in which we failed to identify numerous small vesicles but instead observed microperoxisomes in very close association with the ER.
What role semi/circular tubular structures play in the cell is still unclear. They seem unlikely to be involved in peroxisome degradation, or at least are not vacuoles in the process of degrading peroxisomes, given their intense immunostaining with antibodies to various peroxisomal matrix proteins and their distinct immunostaining pattern compared with the immunostaining pattern of vacuoles labeled with antibodies to carboxypeptidase Y (Figure 6F). Although it cannot be ruled out that the semi/circular tubular structures are simply oddly shaped peroxisomes, the presence of bulges of more intense immunostaining to peroxisomal matrix proteins within the structures suggests that this is not the case. Electron microscopy has also shown that the semi/circular tubular structures do not resemble mature peroxisomes. The presence of overexpressed MycPex3p in semi/circular tubular structures is consistent with them playing a role in peroxisome biogenesis, as are their association with lipid droplets and the demonstration of a partial localization of the ER-resident protein Kar2p within them. Lipid droplets are reported to associate closely with the ER, and several enzymes are shared between the two organelles (e.g., Erg1p). Indeed, lipid droplets are speculated to arise between the two leaflets of the ER membrane (for reviews, see Murphy and Vance, 1999; Zweytick et al., 2000; Brown, 2001). The association between peroxisome biogenesis and lipid bodies is not unprecedented. During postgerminative seedling growth of cotton (Gossypium hirsutum), enlarged glyoxysome (peroxisome) membranes were shown to derive their lipids not from the ER but from lipid bodies (Chapman and Trelease, 1991). In addition, microperoxisomes have been identified by catalase immunostaining to be closely associated with lipid droplets in 3T3-L1 adipocytes (Blanchette-Mackie et al., 1995). Furthermore, cholesterol-rich, ER-like structures have been associated with the surface of lipid bodies in murine 3T3-F442A adipocytes (Prattes et al., 2000). Besides containing the ER resident proteins BiP and calnexin, they have been shown to be a primary storage site for unesterified cholesterol. These ER-like structures may be equivalent to the semi/circular tubular structures described herein. Alternatively, the semi/circular tubular structure may represent a preperoxisomal/peroxisomal reticulum. The morphological changes of semi/circular tubular structures observed in living cells expressing EYFP-SKL and MycPex3p-R143P or MycPex3p-L320P is more consistent with these structures being a dynamic preperoxisomal/peroxisomal reticulum rather than static ER. If the surface lipid droplet structures do turn out to be involved in peroxisome biogenesis, then it would suggest a difference in peroxisome biogenesis between cells grown in glucose-containing medium and cells grown in oleic acid-containing medium, with peroxisome biogenesis occurring preferentially at the major “worksite ” of peroxisomes in oleic acid-grown cells.
In conclusion, our results show that the process of de novo peroxisome biogenesis, or at least some aspect of it, occurs in an unsynchronized manner in close association with elements of the ER, such as those found at the cell periphery, near the nucleus, and possibly associated with lipid droplets. Our data are consistent with Pex3p playing a role in the early stages of peroxisome biogenesis by directly or indirectly sequestering components of the peroxisome biogenesis machinery, such as the PTS1 and PTS2 import machineries. Finally, our work demonstrates that temperature-sensitive Pex3p mutants can be used to define the early stages of peroxisome biogenesis.
ACKNOWLEDGMENTS
This work was supported by grant MOP-15131 from the Canadian Institutes of Health Research (to R.A.R.). R.A.R. is a Canadian Institutes of Health Research Senior Investigator, Canada Research Chair in Cell Biology, and an International Research Scholar of the Howard Hughes Medical Institute. R.A.B. is the recipient of postdoctoral fellowships from the Canadian Institutes of Health Research and from the Alberta Heritage Foundation for Medical Research.
Abbreviations used:
- AOX
acyl-CoA oxidase
- ER
endoplasmic reticulum
- EYFP
enhanced yellow fluorescent protein
- ICL
isocitrate lyase
- PCR
polymerase chain reaction
- PTS
peroxisome targeting signal
- ts
temperature-sensitive
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0633. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–10–0633.
REFERENCES
- Aitchison JD, Szilard RK, Nuttley WM, Rachubinski RA. Antibodies directed against a yeast carboxyl-terminal peroxisomal targeting signal specifically recognize peroxisomal proteins from various yeasts. Yeast. 1992;8:721–734. doi: 10.1002/yea.320080905. [DOI] [PubMed] [Google Scholar]
- Ausubel FJ, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- Brown DA. Lipid droplets: proteins floating on a pool of fat. Curr Biol. 2001;11:R446–R449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- Chang CC, South S, Warren D, Jones J, Moser AB, Moser HW, Gould SJ. Metabolic control of peroxisome abundance. J Cell Sci. 1999;112:1579–1590. doi: 10.1242/jcs.112.10.1579. [DOI] [PubMed] [Google Scholar]
- Chapman KD, Trelease RN. Acquisition of membrane lipids by differentiating glyoxysomes: role of lipid bodies. J Cell Biol. 1991;115:995–1007. doi: 10.1083/jcb.115.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Titorenko VI, Smith JJ, Veenhuis M, Szilard RK, Rachubinski RA. The Yarrowia lipolytica gene PAY5 encodes a peroxisomal integral membrane protein homologous to the mammalian peroxisome assembly factor PAF-1. J Biol Chem. 1996;271:20300–20306. doi: 10.1074/jbc.271.34.20300. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Haan GJ, Baerends RJ, Kram AM, Veenhuis M. Normal peroxisome development from vesicles induced by truncated Hansenula polymorpha Pex3p. J Biol Chem. 2002;277:11026–11033. doi: 10.1074/jbc.M112347200. [DOI] [PubMed] [Google Scholar]
- Goodman JM, Trapp SB, Hwang H, Veenhuis M. Peroxisomes induced in Candida boidinii by methanol, oleic acid and d-alanine vary in metabolic function but share common integral membrane proteins. J Cell Sci. 1990;97:193–204. doi: 10.1242/jcs.97.1.193. [DOI] [PubMed] [Google Scholar]
- Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau W-H, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Valle D. Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 2000;16:340–345. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Hettema EH, Girzalsky W, van den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher SJ, Eschemann A, Okun PM, Brandt U. External alternative NADH:ubiquinone oxidoreductase redirected to the internal face of the mitochondrial inner membrane rescues complex I deficiency in Yarrow lipolytica. J Cell Sci. 2001;114:3915–3921. doi: 10.1242/jcs.114.21.3915. [DOI] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM. Pmp27 promotes peroxisomal proliferation. J Cell Biol. 1995;129:345–355. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- Nuttley WM, Brade AM, Gaillardin C, Eitzen GA, Glover JR, Aitchison JD, Rachubinski RA. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- Prattes S, Horl G, Hammer A, Blaschitz A, Graier WF, Sattler W, Zechner R, Steyrer E. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J Cell Sci. 2000;113:2977–2989. doi: 10.1242/jcs.113.17.2977. [DOI] [PubMed] [Google Scholar]
- Pringle JR. Induction, selection, and experimental uses of temperature-sensitive and other conditional mutants of yeast. Methods Cell Biol. 1975;12:233–272. doi: 10.1016/s0091-679x(08)60959-0. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MJ, Imanaka T, Shio H, Small GM, Lazarow PB. Peroxisomal membrane ghosts in Zellweger syndrome—aberrant organelle assembly. Science. 1988;239:1536–1538. doi: 10.1126/science.3281254. [DOI] [PubMed] [Google Scholar]
- Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ. Expression of PEX11β mediates peroxisome proliferation in the absence of external stimuli. J Biol Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Brown TW, Eitzen GA, Rachubinski RA. Regulation of peroxisome size and number by fatty acid β-oxidation in the yeast Yarrowia lipolytica. J Biol Chem. 2000;275:20168–20178. doi: 10.1074/jbc.M909285199. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA, Aitchison JD. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder WB, Koller A, Choy AJ, Subramani S. The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J Cell Biol. 2000;149:1171–1177. doi: 10.1083/jcb.149.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukupova M, Sprenger C, Gorgas K, Kunau W-H, Dodt G. Identification and characterization of the human peroxin PEX3. Eur J Cell Biol. 1999;78:357–374. doi: 10.1016/S0171-9335(99)80078-8. [DOI] [PubMed] [Google Scholar]
- South ST, Baumgart E, Gould SJ. Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc Natl Acad Sci USA. 2001;98:12027–12031. doi: 10.1073/pnas.221289498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Sacksteder KA, Li X, Liu Y, Gould SJ. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol. 2000;149:1345–1360. doi: 10.1083/jcb.149.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Subramani S, Antonius K, Synder WB. Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem. 2000;69:399–418. doi: 10.1146/annurev.biochem.69.1.399. [DOI] [PubMed] [Google Scholar]
- Szilard RK, Titorenko VI, Veenhuis M, Rachubinski RA. Pay32p of the yeast Yarrowia lipolytica is an intraperoxisomal component of the matrix protein translocation machinery. J Cell Biol. 1995;131:1453–1469. doi: 10.1083/jcb.131.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YYC, Rachubinski RA. Yarrowia lipolytica cells mutant for the PEX24 gene encoding a peroxisomal membrane peroxin mislocalize peroxisomal proteins and accumulate membrane structures containing both peroxisomal matrix and membrane proteins. Mol Biol Cell. 2002;13:2681–2691. doi: 10.1091/mbc.E02-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol. 2000;150:881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. The life cycle of the peroxisome. Nature Rev Mol Cell Biol. 2001;2:357–368. doi: 10.1038/35073063. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Chan H, Rachubinski RA. Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J Cell Biol. 2000;148:29–43. doi: 10.1083/jcb.148.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol. 1998;18:2789–2803. doi: 10.1128/mcb.18.5.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Ogrydziak DM, Rachubinski RA. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol. 1997;17:5210–5226. doi: 10.1128/mcb.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RBH, Wanders RJA, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Le Dall M-T, Waché Y, Laroche C, Belin J-M, Gaillardin C, Nicaud J-M. Evaluation of acyl coenzyme A oxidase (Aox) isoenzyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol. 1999;181:5140–5148. doi: 10.1128/jb.181.17.5140-5148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta. 2000;1469:101–120. doi: 10.1016/s0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]