Abstract
To understand the many roles of the Krebs tricarboxylic acid (TCA) cycle in cell function, we used DNA microarrays to examine gene expression in response to TCA cycle dysfunction. mRNA was analyzed from yeast strains harboring defects in each of 15 genes that encode subunits of the eight TCA cycle enzymes. The expression of >400 genes changed at least threefold in response to TCA cycle dysfunction. Many genes displayed a common response to TCA cycle dysfunction indicative of a shift away from oxidative metabolism. Another set of genes displayed a pairwise, alternating pattern of expression in response to contiguous TCA cycle enzyme defects: expression was elevated in aconitase and isocitrate dehydrogenase mutants, diminished in α-ketoglutarate dehydrogenase and succinyl-CoA ligase mutants, elevated again in succinate dehydrogenase and fumarase mutants, and diminished again in malate dehydrogenase and citrate synthase mutants. This pattern correlated with previously defined TCA cycle growth–enhancing mutations and suggested a novel metabolic signaling pathway monitoring TCA cycle function. Expression of hypoxic/anaerobic genes was elevated in α-ketoglutarate dehydrogenase mutants, whereas expression of oxidative genes was diminished, consistent with a heme signaling defect caused by inadequate levels of the heme precursor, succinyl-CoA. These studies have revealed extensive responses to changes in TCA cycle function and have uncovered new and unexpected metabolic networks that are wired into the TCA cycle.
INTRODUCTION
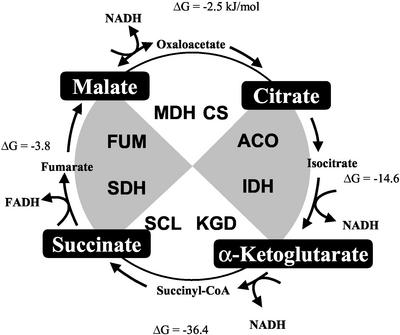
The Krebs tricarboxylic acid (TCA1) cycle is a central pathway of metabolism. Its main catalytic function is to provide reducing equivalents to the respiratory complexes through the oxidative decarboxylation of acetyl-CoA (see Figure 1). However, the TCA cycle also functions in a biosynthetic capacity, primarily in the synthesis of amino acids, heme, and glucose. Glutamate and aspartate are synthesized from α-ketoglutarate and oxaloacetate, respectively, via transamination. Succinyl-CoA and glycine are condensed in the committed step of heme synthesis. Assimilation of two-carbon molecules, such as ethanol and acetate, through the glyoxylate cycle for the synthesis of carbohydrates depends on part of the TCA cycle. Indeed, every TCA cycle intermediate, with the exception of aconitate, is commonly used by other metabolic reactions in a wide variety of cell types. Anaplerotic enzymes, such as pyruvate carboxylase, replenish TCA cycle intermediates that are utilized elsewhere. These metabolic networks extend throughout the cell from this central pathway.
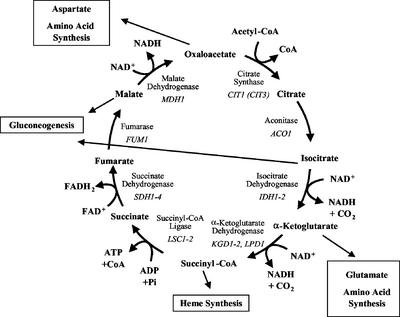
Figure 1.
The yeast TCA cycle. The eight enzymes of the TCA cycle are encoded by at least 15 nuclear genes in S. cerevisiae. Although CIT3 may encode an enzyme with citrate synthase activity, it may actually be a methylcitrate synthase and is not formally included as a TCA cycle gene. The TCA cycle contributes to gluconeogenesis in conjunction with the glyoxylate cycle, which is not active in higher animals.
The eight enzymes of the classic TCA cycle are encoded by 15 nuclear genes in Saccharomyces cerevisiae (McAlister-Henn and Small, 1997; Przybyla-Zawislak et al., 1999). Four of the enzymes: citrate synthase, aconitase, fumarase, and malate dehydrogenase, are encoded by single genes: CIT1, ACO1, FUM1, and MDH1, respectively. The other four enzymes are composed of subunits encoded by distinct genes: IDH1 and IDH2 for the NAD+-dependent isocitrate dehydrogenase, KGD1, KGD2, and LPD1 for the α-ketoglutarate (2-oxoglutarate) dehydrogenase complex, LSC1 and LSC2 for succinyl-CoA ligase (synthetase), and SDH1–4 for succinate dehydrogenase. Other genes encode isozymes of the TCA cycle enzymes that function in other pathways that are often localized to other cellular compartments, such as the cytosol or peroxisomes (McAlister-Henn and Small, 1997).
TCA cycle flux appears to be constricted at two steps on the basis of the limited availability of the substrates oxaloacetate and α-ketoglutarate. This has led to a model dividing the TCA cycle into two minicycles that are interconnected by these substrates and their transamination products, glutamate and aspartate (Yudkoff et al., 1994; Rustin et al., 1997). This model is consistent with the unique regulation of the first three enzymes of the TCA cycle in yeast. In cells with reduced or compromised mitochondrial function, the RTG genes regulate expression of genes encoding the first three steps of the TCA cycle to maintain cellular glutamate levels (Liu and Butow, 1999). The RTG system appears to integrate necessary components of the TCA cycle with nitrogen metabolism in yeast (Hardwick et al., 1999; Komeili et al., 2000; Epstein et al., 2001a). The genes encoding TCA cycle proteins are regulated by several other factors. Starvation for any one of several amino acids induces multiple genes for enzymes involved in the biosynthesis of amino acids. This has been called the general amino acid response and is regulated by the transcription factor Gcn4p (Hinnebusch and Natarajan, 2002). TCA cycle genes, such as LPD1, ACO1, and IDH1–2, that are involved in the synthesis of amino acids are induced by Gcn4p in response to amino acid starvation (Natarajan et al., 2002). The CAATT-binding Hap2/3/4/5p transcription complex controls expression in response to glucose availability (Forsburg and Guarente, 1989; De Winde and Grivell, 1993). Depletion of glucose results in a 3-fold to 10-fold increase in TCA cycle mRNAs (DeRisi et al., 1997). This appears to be the major mechanism for the increased expression of TCA cycle enzymes that is necessary for oxidative metabolism. Other factors and binding sites have been described for some TCA cycle genes, but a complete picture of TCA cycle gene regulation is lacking.
Adding to the complexity of understanding TCA cycle function is the growing appreciation that many TCA cycle proteins may play additional cellular roles beyond their catalytic roles within this pathway (Jeffery, 1999). Several TCA cycle proteins have recently been identified in nucleoids with mitochondrial DNA (mtDNA) and have been reported to play an active role in stabilizing mtDNA (Kaufman et al., 2000). The iron response binding protein (IRP-1) is a cytosolic form of aconitase that regulates iron metabolism in mammals (Hentze, 1994). The IRP-1 binds to mRNAs of iron metabolic proteins, such as ferritin subunits and the transferrin receptor, and regulates expression in response to available cellular iron levels (Paraskeva and Hentze, 1996). Yeast isocitrate dehydrogenase binds to mitochondrially encoded mRNAs and appears to regulate their translation (Elzinga et al., 1993). RNA binding also affects isocitrate dehydrogenase catalytic activity, suggesting a mechanism for coordinating expression of mitochondrial respiratory complexes and TCA cycle oxidative metabolism (Anderson et al., 2000).
TCA cycle genetic defects in humans are extremely rare, and the most severe are probably embryonic lethal events (Rustin et al., 1997). Recent studies have revealed that fumarase and succinate dehydrogenase genes act as tumor suppressors in humans. Fumarase defects were associated with dominantly inherited uterine fibroids, skin leiomyomata, and renal cell cancer (Tomlinson et al., 2002), whereas two types of brain tumors were found to be caused by mutations in three of the genes encoding subunits of succinate dehydrogenase (Baysal et al., 2000; Niemann and Muller, 2000; Astuti et al., 2001). In yeast, none of the TCA cycle genes are essential, but all display growth defects on nonfermentable carbon sources (McAlister-Henn and Small, 1997; Przybyla-Zawislak et al., 1999). Interestingly, these growth phenotypes vary with the enzyme defect. Although cells lacking aconitase, α-ketoglutarate dehydrogenase, succinate dehydrogenase, or fumarase are essentially unable to grow on any nonfermentable carbon source, strains lacking the other TCA cycle enzymes are able to grow to varying degrees on ethanol, glycerol, lactate, or acetate (Przybyla-Zawislak et al., 1999). This indicates that not all TCA cycle mutations are the same and implies that there are differential responses to distinct blocks within the pathway. One specific and unique response to TCA cycle dysfunction occurs on the loss of isocitrate dehydrogenase. Yeast strains lacking isocitrate dehydrogenase grow poorly on glycerol, and they accumulate extragenic nuclear suppressor mutations that enhance growth on this nonfermentable carbon source (McCammon, 1996; Gadde and McCammon, 1997; Przybyla-Zawislak et al., 1999). It is not clear whether isocitrate dehydrogenase defects induce genetic instability or whether the secondary mutations occur randomly and overtake the parental strain by clonal expansion. Both types of mutation accumulation occur in cancer cell lines (Lengauer et al., 1998).
To define metabolic and regulatory networks responsive to TCA cycle function, we used transcriptional profiling on a collection of yeast mutants defective in each of the 15 genes that encode TCA cycle proteins. We have observed both global responses to any of these TCA cycle defects and specific responses to single enzyme defects. We have uncovered a new and unique pattern of gene expression that alternates between elevated and diminished levels in response to contiguous pairs of TCA cycle enzyme defects. These studies provide a metabolic framework to understand the cellular signaling responsive to TCA cycle function.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Saccharomyces cerevisiae strains were routinely maintained on 1% yeast extract, 2% peptone, and 2% glucose (YPD) with 2% agar. For gene disruptions, Ura+ prototrophs were selected on 0.67% yeast nitrogen base, 2% glucose, and 20 mg/l each histidine and adenine, 50 mg/l leucine, and 20 mg/l tryptophan (HALT). Media for growth on nonfermentable carbon sources (ethanol, lactate, pyruvate, glycerol, and acetate) have been described previously (Przybyla-Zawislak et al., 1999). All yeast strains were derived from MMYO11 (MATα ade2–1 can1-100 his3-11,15 leu2-3112 trp1-1, Ole+), a derivative of W303-1B (McCammon et al., 1990). TCA cycle gene mutations were constructed by disruption of the gene of interest by insertion of the URA3 gene. Disruption of ACO1, KGD1, LPD1, LSC2, SDH2, and MDH1 were previously constructed (Przybyla-Zawislak et al., 1999). CIT1, IDH1, IDH2, KGD2, LSC1, SDH1, SDH3, SDH4, and FUM1 were disrupted by PCR amplification of URA3 using hybrid primers containing ∼40–45 nucleotides homologous to the TCA cycle gene on the 5′ ends and 22 nucleotides with homology to the URA3 gene at the 3′ end. A 1.2-kb HindIII fragment containing URA3 was used as a template. The gel-purified DNA was transformed into MMYO11 (Gietz et al., 1992). A wild-type reference strain was constructed by converting the ura3-1 locus of MMYO11 to URA3 by “knock-in” transformation with the functional URA3 gene that was purified from plasmid DNA. Ura+ transformants were screened for disruption of the locus of interest in several ways. Chromosomal DNA was prepared from the transformants (Hoffman and Winston, 1987), and positive mutations were detected by PCR amplification and by Southern blotting. These mutants displayed growth phenotypes on nonfermentable carbon sources that were identical to those previously reported, and they would not complement previously defined TCA cycle mutants (Przybyla-Zawislak et al., 1999). Finally, microarray analysis confirmed almost complete loss of mRNA from the deleted gene.
Strains from the Saccharomyces genome deletion project (Winzeler et al., 1999) harboring disruptions of USV1 (YPL230W) and YGR067C were obtained from the American Type Culture Collection (Manassas, VA). The usv1::KAN and ygr067c::KAN disruption cassettes were PCR-amplified from chromosomal DNA and transformed into MMYO11 and a strain disrupted for IDH2. Positive disruptants were confirmed by PCR using flanking oligonucleotides. The mutant genes displayed normal growth phenotypes on nonfermentable carbon sources, except for the growth enhancement on YPG plates by Δygr067c in the Δidh2 strain.
Petite mutations were routinely analyzed as the ratio of smaller white (petite) to larger red (grande) colonies after growth on YPD plates. The red pigment was produced as a result of the ade2 mutation in these strains. For aco1 mutants, the red color of the grande colonies did not develop, and so the petite frequency was calculated from the frequency of small smooth colonies (petites) to larger rough colonies that harbored petite sectors. Petite frequencies were also confirmed by use of tetrazolium red (Ogur and John, 1956). However, color development in the overlay was also not observed for the aco1 strain.
Microarray Analysis
From YPD plus HALT precultures (OD600 ∼5), strains were diluted 500- to 1000-fold into 200 ml of YPGal (1% yeast extract, 2% peptone, 2% galactose plus HALT). Cultures were grown for 5 or 6 generations and harvested at approximately OD600 = 0.8. Aliquots were plated onto YPD to assay the petite frequency of each culture as described above. Cells were collected and mRNA samples prepared as previously described (Epstein et al., 2001a). RNA from three independent cultures of the wild-type strain was pooled and used as the reference against which mRNAs from the mutants were compared. Cy3- and Cy5-labeled cDNAs were prepared and hybridized to a microarray containing 6219 yeast genes. Replica experiments were performed using independent liquid mutant cultures and with the opposite configuration of Cy3 and Cy5. Methods for background subtraction, low value rejection, and normalization were described previously (Epstein et al., 2001a,b). Array data for each TCA cycle mutation represent the average of two independent microarray hybridizations with reversal of Cy labels as described above. The numerical data may be found at http://www.molbiolcell.org. The averaged and normalized log10 expression ratios were analyzed by cluster analysis (Eisen et al., 1998) and included all genes showing a threefold change in at least one TCA cycle mutant. For the initial dataset (see Figure 3), arrays were clustered using uncentered correlation as the similarity metric for average linkage clustering. Arrays were weighted to account for the number of genes coding for a particular TCA cycle enzyme (e.g., Eweight = 1.0 for CIT1, 0.5 for IDH1 and IDH2, and 0.25 for SDH1–4), whereas responsive genes were unweighted. Genes were arranged manually for other figures. The TreeView program (Eisen et al., 1998) was used to display all data.
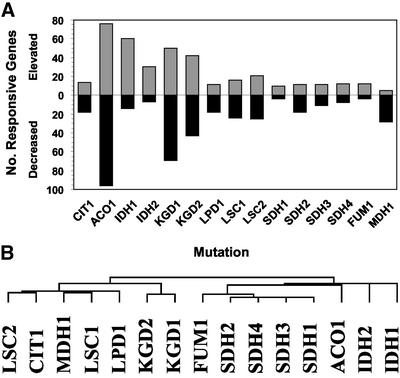
Figure 3.
Genes responsive to TCA cycle dysfunction. Microarray analysis was performed in duplicate for each of the TCA cycle mutations, and the data were normalized and averaged as described in the MATERIALS AND METHODS section. Expression of 406 genes was altered threefold or greater by at least one TCA cycle mutation. (A) Summation of genes responding to TCA cycle gene dysfunction. The numbers of genes responding to each TCA cycle mutation are shown. Genes with elevated expression are shown by gray bars; genes with diminished expression are shown by black bars. Total responsive genes represent the sum of gray and black bars. (B) Clustering of TCA cycle mutant arrays. Array datasets for each TCA cycle mutation are clustered (Eisen et al., 1998). Similarities among arrays are indicated by the dendogram.
Aneuploidy
The microarray datasets were investigated to test for chromosomal aneuploidy in the TCA cycle mutant strains (Hughes et al., 2000). Gene expression ratios from individual microarray experiments were sorted by chromosome, and the average expression for each chromosome was calculated to test for chromosome-wide expression bias indicative of chromosomal aneuploidy. For most of the chromosomes, the average from the log10 expression ratios of wild-type to mutant was between +0.1 and −0.1. Average expression ratios greater than +0.1 or less than −0.01 were observed in a handful of chromosomes. However, when the expression ratios of individual genes from these chromosomes were compared with a “nonextreme” strain, no systematic expression bias indicative of localized or whole-chromosome aneuploidy was observed. It is concluded that none of the strains used for these studies contain an aberrant chromosome number or segment.
Protein Methods
Whole-cell lysates were prepared as described previously (Gadde and McCammon, 1997) from cultures grown on YPGal, and protein was quantified using a Bio-Rad Protein assay reagent with bovine gamma globulin as a standard. Proteins were separated on a 12% polyacrylamide gel and electrophoretically transferred to polyvinylidine difluoride membranes, and the Idp1p, Idp2p, and Idh1p proteins were immunodetected by chemiluminescence (Amersham Pharmacia). The rabbit antiserum that recognizes both Idp1p and Idp2p has been described previously (Haselbeck and McAlister-Henn, 1991). The Idh1p-specific antiserum was prepared against urea-solubilized pentahistidine-tagged Idh1p (Zhao and McAlister-Henn, 1997; Gadde et al., 1998). Idh1p was further purified by PAGE, electroeluted from the gel, and injected into rabbits for antibody production.
RESULTS
Construction of TCA Cycle Mutant Collection and Microarray Analysis
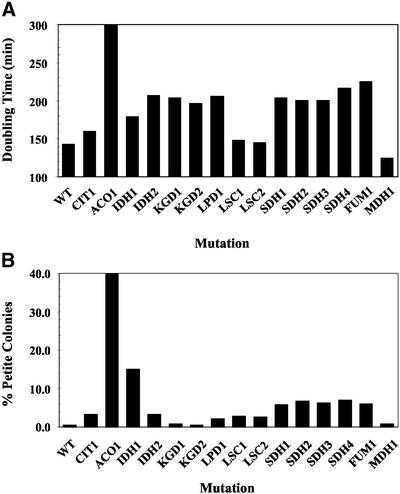
Transcriptional profiling was performed on yeast strains harboring mutations in each of the 15 genes encoding polypeptides comprising the eight TCA cycle enzymes (Figure 1). Galactose was chosen as a carbon source for growth of these strains, for several reasons. First, all of the TCA cycle mutant strains displayed growth defects on nonfermentable carbon sources (Przybyla-Zawislak et al., 1999), so a fermentable carbon source had to be used. Growth on the galactose medium was reproducible for each mutant strain and was consistent for strains harboring defects in the same enzyme (e.g., SDH1–4 strains; Figure 2A). Second, galactose was preferred over glucose, because carbon catabolite repression is partially relieved by galactose. Raffinose was tested as an alternative to galactose and had the benefit of less carbon catabolite repression. However, the mutant strain deleted for the ACO1 gene encoding aconitase was unable to grow with raffinose as a carbon source. In addition, the frequency of spontaneous petite mutations was significantly elevated in many TCA cycle mutant strains during growth on raffinose (>70% of all colonies for some mutations). In contrast, the frequency of petite mutations was only slightly elevated (by ∼5%) when strains were cultured on galactose compared with glucose. For most of the TCA cycle mutant strains, the frequency of petite mutations was <10% (Figure 2B). The most obvious exception was the Δaco1 strain, for which 40–60% of the plated colonies were petite. As reported recently, mutations of the mitochondrial genome can have a significant effect on the expression of nuclear genes (Epstein et al., 2001a; Traven et al., 2001). Therefore, the expression profile of the aconitase-deficient strain probably represents a primary effect resulting from the loss of aconitase activity and a secondary effect resulting from the loss of mtDNA expression. For the other TCA cycle mutations, with the possible exception of IDH1, the expression profile should reflect only the primary effect of the nuclear mutation and not the complication caused by a significant subpopulation of cells harboring both a nuclear mutation and a mtDNA defect.
Figure 2.
Growth characteristics of TCA cycle mutants. (A) Doubling time of TCA cycle mutants in minutes in a rich medium with galactose as a carbon source. Values represent the average of at least two independent cultures. WT, wild-type strain. (B) Petite frequencies of TCA cycle mutants. Cultures were grown as described above and plated onto YPD plates at the time of harvest. After 3–4 d at 30°C, the percentage of small white colonies (i.e., petites) were counted. Values represent the average of at least two independent cultures for each mutation.
Expression changes in response to TCA cycle defects of threefold or greater were observed for 406 genes. On average, ∼50 genes were responsive per TCA cycle mutation. However, the number of responsive genes was quite variable. Although ∼170 and 120 genes responded to ACO1 and KGD1 defects, respectively, only 15–20 genes were responsive to FUM1, SDH1, SDH3, or SDH4 defects (Figure 3A). This wide variation in the responses to blocks in discrete steps of the TCA cycle suggests that genes are not simply responding in a general way to the loss of TCA cycle function (but see below). Approximately 25% of these responsive genes are of unknown function. We allowed clustering software to group the expression profiles by their similarities and independent of the TCA cycle enzyme affected. For the multimeric enzymes encoded by multiple genes, the expression profiles were grouped together in the same clusters, indicating that these defects tended to elicit similar responses (Figure 3B). The two main branches of array data are derived from four TCA cycle enzyme defects each. As shown below, this grouping of TCA cycle enzyme arrays is probably brought about by the expression pattern of a small group of genes. In one branch, the SDH1–4 arrays were closely grouped and displayed similarity to the FUM1 and ACO1 arrays. The IDH1–2 arrays were outgroups of this cluster. In the other branch, two main subgroups were represented by the KGD1–2 arrays and a larger group composed of LSC2, CIT1, MDH1, LSC1, and LPD1. LPD1 encodes the lipoamide dehydrogenase subunit of α-ketoglutarate dehydrogenase complex, which is also a subunit of pyruvate dehydrogenase, glycine decarboxylase, and the branched-chain amino acid dehydrogenase (Sinclair and Dawes, 1995; Pronk et al., 1996). Accordingly, genes involved in amino acid metabolism appeared to be specifically responsive to defects in LPD1. BAT1, which encodes a branched-chain amino acid transferase, was diminished 3.4-fold, and the LEU1 and LEU2 genes of leucine biosynthesis were diminished 7-fold. YLR089C, which encodes a potential alanine aminotransferase, was induced 3.8-fold. These differences may explain in part the outgrouping of LPD1 from the KGD1–2 arrays.
TCA Cycle Gene Expression in TCA Cycle Mutants
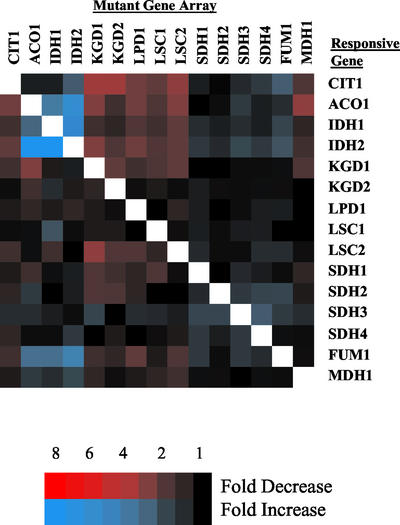
Of the 15 TCA cycle genes, the expression of only 6 was altered in response to other TCA cycle mutations (Figure 4). CIT1 expression was diminished by mutations in KGD1, KGD2, and LSC2. ACO1 expression was elevated by mutations in IDH1–2 and was diminished by mutations in KGD1 and MDH1. LSC2 expression was diminished by a mutation in a KGD1. FUM1 expression was elevated by mutations in IDH1–2 and ACO1. Finally, expression of the IDH1 and IDH2 genes was elevated by mutations in ACO1 and by mutations in their partner IDH gene. It was somewhat surprising that for three of the hetero-oligomeric TCA cycle enzymes, expression of their encoding genes was largely unresponsive to the loss of constituent subunits. On one hand, expression of genes encoding subunits of the α-ketoglutarate dehydrogenase complex, succinyl-CoA ligase and succinate dehydrogenase, did not significantly change on the loss of an essential subunit. This was consistent with the observation that expression of these genes was generally unresponsive to other defects within the TCA cycle as well. On the other hand, the two genes encoding the NAD+-dependent isocitrate dehydrogenase were highly responsive to the loss of IDH function. Expression of IDH1 increased 6-fold and IDH2 increased 10-fold on the loss of each partner IDH gene. Isocitrate dehydrogenase and aconitase are necessary for the synthesis of α-ketoglutarate, which is used for the synthesis of glutamate. Glutamate can repress the synthesis of these genes through the RTG system (Liu and Butow, 1999; Epstein et al., 2001a; Liu et al., 2001), and this may be a mechanism through which the expression of these genes was affected. However, the response may be more complex, because a similar expression increase is not observed for CIT1, which is also involved in the synthesis of α-ketoglutarate and is similarly regulated by the RTG system (Liu and Butow, 1999).
Figure 4.
Expression of TCA cycle genes in TCA cycle mutant arrays. The expression of TCA cycle genes in each of the 15 sets of TCA mutant array sets are displayed relative to a wild-type strain with a complete TCA cycle. Results are visualized by TreeView program (Eisen et al., 1998). Missing or deleted data are displayed in white. The expression of individual TCA cycle genes in its own array (e.g., ACO1 gene expression in the ACO1 mutant array) was deleted and displayed in white in this and all subsequent array figures. Genes with an elevated expression are displayed in blue, and genes with decreased expression are displayed in red. Expression differences are indicated by the color intensity scale for this and subsequent figures.
Genes Affected by Multiple TCA Cycle Defects
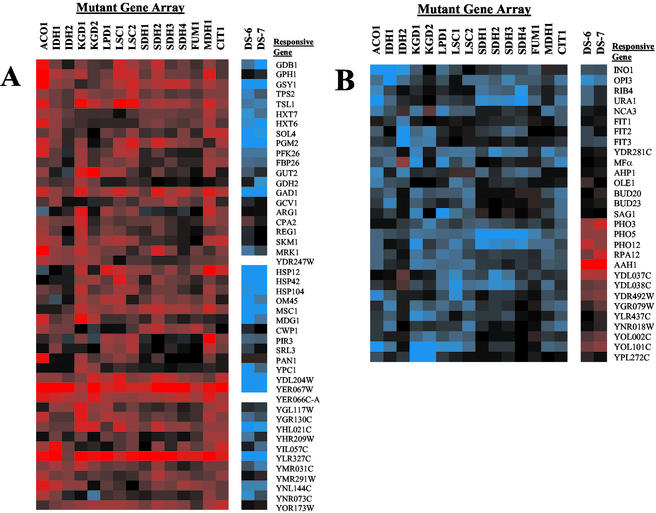
The expression of ∼100 genes was affected by multiple defects within the TCA cycle. We used the diauxic shift data of DeRisi et al. (1997) to identify genes whose expression changed after glucose was exhausted and metabolism was completely oxidative (i.e., the last two time points). Three major expression patterns could be discerned. For 46 genes, mRNA levels decreased with essentially any TCA cycle mutation (Figure 5A). Responsive genes encoded enzymes of glycogen and trehalose metabolism (e.g., GSY1, TSL1), hexose metabolism (e.g., HXT6), glycolysis (e.g., PGM2), amino acid metabolism (e.g., GAD1), heat shock (e.g., HSP104), and cell signaling pathways (e.g., MDG1). Most of these genes were highly induced after the diauxic shift when nonfermentable carbon sources, such as ethanol, were being used. Genes in this group are not properly induced in cells with a dysfunctional TCA cycle, suggesting that oxidative metabolism may be slowed. This fairly uniform change in expression is probably not brought about by a change in the growth rate of the mutant strains, because, as noted above, growth rates were quite variable with different mutations, and strains with some defects (e.g., in MDH1, or LSC1) had little if any discernable difference in growth compared with the wild-type strain (Figure 2A).
Figure 5.
Genes responsive to multiple TCA cycle defects. Common expression patterns in TCA cycle mutant strains grouped by pattern. For comparison, the last two time points from the diauxic shift microarray experiment of DeRisi et al. (1997) are also shown (DS-6 and DS-7). (A) Genes with decreased expression in TCA cycle mutants. (B) Genes with elevated expression in TCA cycle mutants.
For a second group of 29 genes, mRNA levels were elevated in response to TCA cycle gene mutations (Figure 5B). Affected genes encode enzymes of phosphate (e.g., PHO3), lipid (e.g., INO1, OPI3), nucleotide (e.g., AAH1), and iron metabolism (e.g., FIT1–3). However, unlike the previous set of genes, these genes did not display the same expression pattern during the diauxic shift. Some genes, such as OPI3, were also induced in the postdiauxic shift dataset. However, for a larger group, including AAH1 and the phosphate genes PHO3 and PHO12, expression was decreased in the postdiauxic shift dataset. This suggests that these genes may not be involved in oxidative metabolism, because they are induced with a dysfunctional TCA cycle but are not normally highly expressed during oxidative metabolism. Alternatively, they may represent genes that are induced to supply metabolites that are normally produced by a functional TCA cycle.
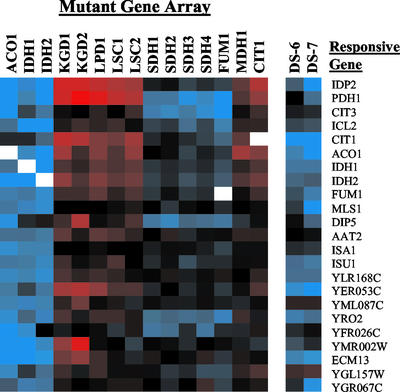
The most interesting group consisted of 23 genes whose expression was elevated in aconitase, isocitrate dehydrogenase, succinate dehydrogenase, and fumarase mutants but was decreased in citrate synthase, α-ketoglutarate dehydrogenase, succinyl-CoA ligase, and malate dehydrogenase mutants (Figure 6). This resulted in an alternating expression pattern that was either elevated or diminished in response to defects in pairs of contiguous TCA cycle enzymes as one moves around the TCA cycle. In Figures 5 and 6, we have placed the array dataset from the CIT1 mutant adjacent to MDH1 to aid in displaying this pattern. This pairing of enzyme arrays was probably responsible for the array-clustering pattern described earlier (Figure 3B). The expression patterns of these genes were most similar between defects in enzymes catalyzing adjacent reactions. For instance, the expression patterns were very similar among aconitase and isocitrate dehydrogenase defects or among succinate dehydrogenase and fumarase defects, but there were slight differences in the expression patterns between these two pairs. Similar differences could be observed between the enzyme pairs in the other branch.
Figure 6.
Alternating expression of genes in response to different TCA cycle enzyme defects. Abrupt changes in gene expression are observed between defects in adjacent pairs of TCA cycle enzymes.
The genes that display this alternating expression pattern in response to TCA cycle dysfunction are induced in postdiauxic shift cells, and most of the encoded proteins are involved in oxidative metabolism. The three distinct genes of the methylcitrate cycle, CIT3, PDH1, and ICL2, were among the most prominent genes displaying this profile. Three genes involved in mitochondrial protein assembly were also among this group. YLR168C is involved in mitochondrial protein sorting, and ISA1 and ISU1 are involved in the maturation of mitochondrial iron sulfur proteins, such as aconitase and succinate dehydrogenase. Other genes encoding metabolic enzymes in the group include IDP2, which encodes a cytosolic NADP+-dependent isocitrate dehydrogenase; MLS1, which encodes the glyoxylate cycle malate synthase; and DIP5, which encodes a plasma membrane dicarboxylic acid permease. Finally, five genes of the TCA cycle: CIT1, ACO1, IDH1–2, and FUM1, also displayed this profile. These TCA cycle genes are the most responsive to TCA cycle defects, as mentioned previously, whereas the other TCA cycle genes were unresponsive.
The methylcitrate cycle is used for the assimilation of propionate via propionyl-CoA, which is generated from the oxidation of odd-chain fatty acids or amino acids, such as threonine or methionine (Luttik et al., 2000). The methylcitrate cycle genes are induced by propionate (Epstein et al., 2001a) and are not expressed under anaerobic conditions in a glucose-limited chemostat culture (Ter Linde et al., 1999). The function and species distribution of the methylcitrate cycle are still poorly understood, but this pathway is intimately linked to the TCA cycle. The three distinct enzymes of this mitochondrial pathway: methylcitrate synthase, methylaconitate dehydratase, and methylisocitrate lyase, appear to be encoded by the CIT3, PDH1, and ICL2 genes, respectively (Luttik et al., 2000; Horswill and Escalante-Semerena, 2001). Expression of PDH1 was altered in 13 of the 15 mutant strains, and CIT3 and ICL2 were also highly responsive to TCA cycle defects. The CIT3 gene encodes a protein with citrate synthase activity (Jia et al., 1997), and its methylcitrate synthase activity has not been assayed. However, on the basis of its regulation (Epstein et al., 2001a), it appears to be responsive to propionate metabolism. By analogy to Salmonella enterica, aconitase is also required in this pathway to convert 2-methylaconitate to 2-methylisocitrate (Horswill and Escalante-Semerena, 2001). Methylisocitrate lyase cleaves methylisocitrate into pyruvate and succinate, and succinate dehydrogenase and other TCA cycle enzymes are necessary to further metabolize the succinate generated during propionate metabolism.
The expression pattern of IDP2 was similar to that of the methylcitrate cycle genes. All of these genes are highly expressed in aerobic cultures and poorly expressed in anaerobic cultures (Ter Linde et al., 1999). mRNA levels were elevated fourfold to eightfold in aconitase- or isocitrate dehydrogenase–deficient strains and were diminished in strains deficient in citrate synthase, α-ketoglutarate dehydrogenase, and succinyl-CoA ligase. IDP2 encodes a cytosolic form of NADP+-dependent isocitrate dehydrogenase (Idp2p). This enzyme is induced during the diauxic shift and is regulated by the Cat8p transcription factor that controls expression of glyoxylate cycle and gluconeogenic and related proteins (Bojunga and Entian, 1999; Haurie et al., 2001). In addition to providing cytosolic α-ketoglutarate, Idp2p also appears to be important for the generation of cytosolic NADPH, which is used for cellular antioxidant functions (Minard and McAlister-Henn, 2001).
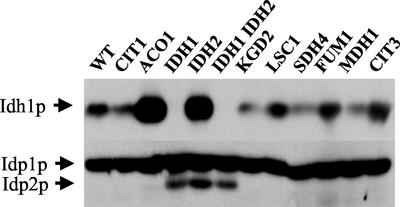
To test these alterations, we investigated the steady-state levels of several isocitrate dehydrogenase polypeptides in TCA cycle mutants. Lysates from TCA cycle mutants representing each enzyme were probed with antisera to detect Idh1p, Idp2p, and Idp1p, a mitochondrial NADP+-isocitrate dehydrogenase (Haselbeck and McAlister-Henn, 1991). Protein levels of Idh1p were elevated in strains deleted for ACO1 and IDH2 (Figure 7) and appeared to be diminished in several other strains deleted for CIT1, KGD2, and MDH1. Idp2p was detected only in strains lacking the NAD+-isocitrate dehydrogenase genes IDH1 or IDH2. By contrast, Idp1p was largely unresponsive to all TCA cycle defects. IDP1 mRNA levels were largely unaffected by the TCA cycle mutations in the microarray dataset. These results are consistent with previous studies in which Idp1p or IDP1 mRNA levels are essentially constant over a variety of growth conditions in several mutant backgrounds (Minard et al., 1998). Thus, in these examples, protein levels correlated with the microarray results.
Figure 7.
Expression pattern of Idh1p, Idp1p, and Idp2p in TCA cycle mutants. Strains harboring defects in TCA cycle genes were cultured in YPGal and analyzed as in the MATERIALS AND METHODS section. Protein levels of Idh1p, Idp1p, and Idp2p were detected by Western blotting.
Alternating Gene Expression Pattern and Suppressor Mutations
Another interesting aspect of the alternating expression pattern was its correlation with suppressor mutations of isocitrate dehydrogenase defects. Strains with defects in isocitrate dehydrogenase grow poorly on media containing glycerol and accumulate extragenic suppressor mutations that enhance growth on this nonfermentable carbon source (McCammon, 1996; Gadde and McCammon, 1997; Przybyla-Zawislak et al., 1999). Mutations in CIT1 were the first and most abundant class of suppressor mutations identified. A systematic search of TCA cycle genes that could function as suppressors when mutated divided the TCA cycle genes into two sets. Mutations in CIT1, KGD1, KGD2, LPD1, LSC1, LSC2, and MDH1 were capable of enhancing growth of IDH-inactivated strains on glycerol medium, whereas mutations in ACO1, IDH1, IDH2, SDH1, SDH2, SDH3, SDH4, and FUM1 were not (Przybyla-Zawislak et al., 1999). Remarkably, the same two sets of mutant genes give rise to the differential expression pattern displayed in Figure 6. The suppressor defects elicit the diminished mRNA expression pattern, whereas the TCA cycle defects that cannot function as suppressors elicit the elevated expression pattern. Thus, transcriptional profiling has independently identified a metabolic network that is in concordance with a genetically defined set of growth-enhancing mutations. This obviously suggests a relationship between the glycerol suppressor mutations of isocitrate dehydrogenase defects and the alternating expression pattern in response to TCA cycle dysfunction.
To examine this relationship further, the mRNA expression pattern of a double mutant deleted for both CIT1 and IDH2 was investigated using DNA microarrays. We focused on the genes displaying the alternating expression pattern by comparing their mRNA levels in the double-mutant and the corresponding single-mutant strains. Expression of these genes was essentially intermediate in the double-mutant strain compared with each of the single-mutant strains (Table 1). For instance, in the strain in which CIT1 was inactivated, the mRNA levels of PDH1 and ICL2 were down-regulated relative to the wild-type strain, whereas when IDH2 was inactivated, the mRNA levels of these genes were elevated ∼12-fold. However, mRNA levels were elevated only ∼2.5-fold in the double mutant. Similar changes in relative mRNA levels of threefold to fourfold (up-regulated relative to the CIT1 inactivated strain and down-regulated relative to the IDH2 inactivated strain) were observed in the double mutant for most of the genes in this group. This resulted in a slight net increase (1.5- to 2.5-fold) of these mRNAs in the double mutant relative to the wild-type. Hence, the CIT1 suppressor mutation decreased the expression of this set of genes and minimized their expression differences between normal and isocitrate dehydrogenase–deficient strains. We have observed previously that aconitase and fumarase levels were elevated in a strain lacking isocitrate dehydrogenase but that the levels of these enzymes were similar between the wild-type and a strain lacking both citrate synthase and isocitrate dehydrogenase (Gadde and McCammon, 1997). We have also observed elevated levels of Idh1p in strains lacking IDH2 and a decreased level of Idh1p in a strain that is also deficient in citrate synthase (Gadde et al., 1998). These results indicate that the expression changes revealed from the microarray analysis translate into altered protein levels and confirm the effect of suppressor mutations, such as those in CIT1.
Table 1.
Effect of mutations in IDH2 and CIT1on alternating gene expression
| Responsive gene | Fold expression (mutant/WT)
|
||
|---|---|---|---|
| Δidh2 | Δidh2 Δcit1 | Δcit1 | |
| IDP2 | 8.9 | 2.2 | −3.6 |
| PDH1 | 12.0 | 2.6 | −2.7 |
| CIT3 | 3.2 | 1.0 | −1.6 |
| ICL2 | 12.0 | 2.8 | −1.4 |
| ACO1 | 6.1 | 2.0 | −2.5 |
| IDH1 | 5.7 | 2.2 | −1.7 |
| FUM1 | 4.6 | 2.9 | −1.6 |
| MLS1 | 4.8 | 1.1 | −1.1 |
| DIP5 | −1.3 | 2.0 | 1.2 |
| AAT2 | 3.5 | 1.5 | −1.4 |
| ISA1 | 3.9 | 2.2 | 1.1 |
| ISU1 | 4.4 | 2.6 | −1.2 |
| YLR168C | 4.9 | 1.5 | −1.4 |
| YER053C | 3.1 | 1.3 | −1.8 |
| YML087C | 3.8 | 2.4 | 1.1 |
| YRO2 | 4.0 | 1.8 | 1.2 |
| YFR026C | 1.0 | 1.0 | −1.2 |
| YMR002W | 4.3 | 1.9 | 1.6 |
| ECM13 | 5.6 | 3.3 | 1.1 |
| YGL157W | 3.0 | 1.0 | −1.7 |
| YGR067C | 3.2 | 1.2 | −1.3 |
Expression of alternating genes (Figure 6) is displayed from strains deleted for IDH2, IDH2 plus CIT1, and CIT1. Fold expression of mRNA in mutant strains relative to the same pool of wild-type mRNA is shown. Data for CIT1 and IDH2 mRNAs have been removed for greater clarity.
How might these positively correlated patterns of alternating gene expression and genetic suppression result in the altered growth of strains deficient in isocitrate dehydrogenase? It is possible that the overexpression of one or more of these proteins in the isocitrate dehydrogenase–deficient strain is detrimental to growth. Inactivation of CIT1 somehow signals for a decrease in expression of those genes with an alternating expression profile, which in essence serves to cancel out the expression increase brought about by the isocitrate dehydrogenase defect. The end result is an increased rate of growth, even though the cells have accumulated a mutation in CIT1 that also results in a slightly diminished ability to grow on glycerol. The implication is that the absence of citrate synthase may be less deleterious than the overexpression of one of these target genes. This would apply only to genes displayed in Figure 6 and not those in Figure 5, because these latter genes respond in a similar manner to both CIT1 and IDH defects.
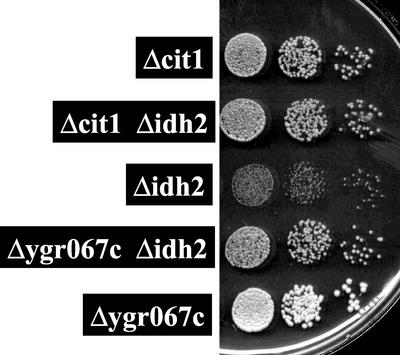
Although this model is speculative, it is readily testable. One approach is to delete a candidate gene and to look at its effect on the glycerol growth in a strain lacking functional isocitrate dehydrogenase. Several of the genes with an alternating expression profile have already been tested because they encode TCA cycle genes (CIT1, ACO1, IDH1, and FUM1) or a homologue (CIT3) (Przybyla-Zawislak et al., 1999). Of these, only CIT1 is able to enhance growth on glycerol when inactivated. However, the other genes, except for CIT3, display severe growth defects on nonfermentable carbon sources when inactivated. To extend these studies, we deleted the gene YGR067C, the expression of which is elevated in isocitrate dehydrogenase–deficient strains (Figure 6, Table 1). YGR067C is predicted to encode a zinc finger transcription factor similar to Adr1p, Mig1p, and Cat8p (Bohm et al., 1997). Like these other transcription factors, YGR067C is induced during the diauxic shift at a time when the cell shifts to oxidative metabolism. However, little else is known about this open reading frame. A haploid strain in which YGR067C was deleted displayed no obvious growth defect on nonfermentable carbon sources. However, similar to a mutation in CIT1, a defect in YGR067C serves as a growth enhancer for an isocitrate dehydrogenase–deficient strain on glycerol (Figure 8). Another gene encoding a potential zinc-finger transcription factor, USV1 (YPL230W), was also present in the TCA cycle defect microarray dataset but does not display an alternating pattern of expression. Like YGR067C, very little is known about USV1, except that it is induced in post–diauxic shift cells. However, deletion of USV1 does not enhance growth on glycerol in a Δidh2 background (our unpublished observations), indicating that this gene does not serve as a suppressor mutation. Because YGR067C may encode a transcription factor, it is possible that it may regulate genes whose expression is relevant to TCA cycle function or to isocitrate dehydrogenase dysfunction.
Figure 8.
CIT1 and YGR067c defects are growth enhancers of isocitrate dehydrogenase dysfunctional strains. Strains with the genotypes indicated were cultured overnight in YPD; an aliquot was washed once in water and resuspended to an initial concentration of 2 OD600/ml water. This was diluted in a 10-fold series, and 10 μl of each dilution was spotted onto a YPG plate that was incubated 4 d at 30°C. Larger colony sizes of Δcit1 Δidh2 and Δygr067c Δidh2 strains relative to Δidh2 strain indicate that the Δcit1 and Δygr067c are growth enhancers of Δidh2 defect (Gadde and McCammon, 1997; Przybyla-Zawislak et al., 1999). Growth of the wild-type and Δusv1 strains were indistinguishable from that of the Δygr067c strain.
Hypoxic and Aerobic Genes Responsive to α-Ketoglutarate Dehydrogenase and Aconitase Defects
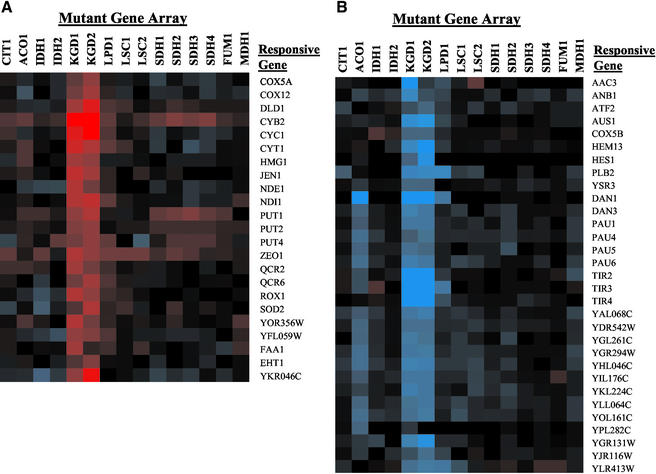
The expression of 54 genes was affected by defects in aconitase and the α-ketoglutarate dehydrogenase complex. These genes fell into two predominant categories (Figure 9): expression of aerobic genes, predominantly of the mitochondrial respiratory complexes, was diminished by defects in KGD1, KGD2, and LPD1 and to a lesser extent by a defect in ACO1, whereas expression of anaerobic and hypoxic genes was elevated with these defects. Aerobic genes (Figure 9A) with diminished expression include the major isoform of cytochrome c (CYC1), cytochromes c1 (CYT1) and b2 (CYB2), isoforms of NADH dehydrogenase (NDE1 and NDI1), subunits of cytochrome c oxidase (COX5A and COX12), ubiquinol cytochrome c reductase (QCR2 and QCR6), and ROX1, a transcriptional repressor of hypoxic genes. The hypoxic genes (Figure 9B) include AAC3, which encodes an isoform of the ADP/ATP carrier protein COX5B and HEM13, which also encodes isoforms of proteins that are expressed under hypoxic conditions; and 16 of the 31 members of the seripauperin family, a group of genes involved in cell wall synthesis (Ter Linde et al., 1999; Rachidi et al., 2000). The hypoxic induction of the seripauperin genes is for the most part independent of Rox1p and dependent on Upc2p (Mox4p) (Rachidi et al., 2000; Kwast et al., 2002).
Figure 9.
Oxidative and hypoxic genes in α-ketoglutarate dehydrogenase and aconitase mutants. (A) Oxidative genes. (B) Hypoxic/anaerobic genes.
The affected genes correlate well with genes that are regulated by cellular heme levels (Kwast et al., 1998; Zhang and Hach, 1999). Heme regulates gene expression in yeast in an oxygen-dependent manner, because it requires molecular oxygen for its synthesis. Heme is a cofactor of the Hap1p transcription factor that regulates expression of respiratory complex protein genes, such as CYC1 and CYB2. Heme also seems to stimulate the function of the Hap2/3/4/5p complex that regulates expression of other respiratory complex proteins, such as COX4 and QCR8. Heme appears to block the binding of the HDS binding factor, allowing the expression of other genes, such as SOD2 (Kwast et al., 1998). Hypoxic genes, however, are negatively regulated by heme. Several transcriptional repressors of hypoxic genes are induced under oxidative conditions when heme levels in the cell are high. These include ROX1, which represses CYC7 and ANB1 (Zitomer et al., 1997). Other heme-dependent factors, such as Upc2, negatively regulate expression of the seripauperin family, such as DAN1 and other hypoxic genes (Kwast et al., 1998; Rachidi et al., 2000).
DISCUSSION
We have performed transcription profiling using DNA microarrays on a collection of mutants defective in each of the 15 genes that encode subunits of TCA cycle proteins. This analysis revealed >400 genes that were highly responsive to TCA cycle defects, suggesting that nuclear gene signaling is responsive to TCA cycle function. In this report, we have concentrated on two sets of genes that appear to be responding to distinct metabolic signals resulting from TCA cycle dysfunction. The first signaling pathway appears to monitor the general state of the TCA cycle, because defects throughout the cycle elicit a response in nuclear gene expression. The second pathway represents signaling resulting from a single enzyme defect in the α-ketoglutarate dehydrogenase complex and appears to be the result of aberrant heme-dependent signaling. To the best of our knowledge, these responses to TCA cycle dysfunction have not been reported previously.
Four of the eight TCA cycle enzymes that we inactivated are encoded from a single gene, whereas the other four enzymes are encoded by multiple (2–4) genes. By comparing expression profiles in response to defects in genes encoding different subunits of hetero-oligomeric TCA cycle proteins, it was possible to analyze how the cell responds to different defects in the same enzyme. Although isocitrate dehydrogenase and succinyl-CoA ligase require both subunits for activity, subcomplexes composed of only some subunits of α-ketoglutarate dehydrogenase complex and succinate dehydrogenase can be detected (Repetto and Tzagoloff, 1991; Scheffler, 1998). However, cluster analysis suggested that the responses to defects in genes encoding different subunits of the hetero-oligomeric TCA cycle enzymes were, for the most part, very similar (Figure 3). This was most apparent for the SDH1–4 mutant arrays, which were clustered into the same branch. Some differences among arrays of genes encoding subunits of the same enzyme could also be explained. For instance, although KGD1 and KGD2 arrays were observed in the same branch, the LPD1 array was clustered in a close outgroup. Because the lipoamide dehydrogenase encoded by LPD1 is also a subunit in three other proteins, the slightly different response to an LPD1 defect probably reflects an aggregate response to loss of all four enzyme complexes. Because the responses to different mutations encoding distinct subunits of the same enzyme produced similar expression patterns, it is possible that one mutation per enzyme is sufficient to establish a reliable expression profile.
A specific response to defects in the KGD1–2 and LPD1 genes was the elevated expression of hypoxic genes and a diminished expression of oxidative genes. This appeared to be a specific response to defects in the genes encoding the α-ketoglutarate dehydrogenase complex and was not apparent with other TCA cycle defects (Figure 9), although some hypoxic genes were elevated in aconitase-deficient cells. The most likely rationale for the observed changes in oxidative and hypoxic gene expression is that heme levels are diminished in the α-ketoglutarate dehydrogenase mutants. Succinyl-CoA is produced by the α-ketoglutarate dehydrogenase complex, and this metabolite is used directly for heme biosynthesis by δ-aminolevulinate synthase (Figure 1). An α-ketoglutarate dehydrogenase defect should result in lowered succinyl-CoA and therefore diminished cellular heme. The heme deficiency would result in the diminished expression of oxidative genes that are regulated through the Hap1p, Hap2/3/4/5, and HDS complexes. Diminished heme levels also result in lower levels of active Rox1p and other factors that repress hypoxic genes, leading to the elevated expression of hypoxic genes, such as CYC7, ANB1, and DAN1. The magnitude of the expression changes in the α-ketoglutarate dehydrogenase mutants appears to be close to but less than the maximal changes in expression reported in heme auxotrophs or by mutations in HAP1 or ROX1 (Kwast et al., 2002; Ter Linde and Steensma, 2002). Because the α-ketoglutarate dehydrogenase mutants are not heme auxotrophs, there must be other routes for the synthesis of succinyl-CoA. One such enzyme might be succinyl-CoA ligase, which is a reversible enzyme that can synthesize succinyl-CoA from succinate (Przybyla et al., 1998). However, on the basis of the magnitude of the expression changes, α-ketoglutarate dehydrogenase may be a significant source of succinyl-CoA under these conditions.
It is not entirely clear why the changes in heme-dependent genes are also observed with aconitase deficiency (Figure 9). Aconitase deficiency should result in the accumulation of citrate and aconitate and a deficiency of isocitrate and α-ketoglutarate. Sources of glutamate should be able to compensate for the α-ketoglutarate deficiency; however, under the culture conditions used in these experiments, this may not be occurring. The seripauperin family genes were the most responsive to the aconitase defect, suggesting that the response by these genes may not be entirely because of cellular heme levels (Cohen et al., 2001). Mutants in ACO1 display the most severe growth defects on both fermentable and nonfermentable carbon sources. They are glutamate auxotrophs and are unable to grow on some fermentable carbon sources that do not repress oxidative gene expression, such as raffinose, suggesting that oxidative functions may be severely compromised. Many aconitase-deficient cells are also petites (Figure 2B), and such mtDNA mutations can have profound effects on nuclear gene expression (Epstein et al., 2001b; Traven et al., 2001). Hence, the expression profile displayed by ACO1 defects is expected to be complex and to result from many different factors, such as altered heme levels, mtDNA defects, and a slower growth rate.
Many genes responded in a similar manner to generalized defects within the TCA cycle. Whereas some genes did not appear to be properly induced (Figure 5A), other genes were hyperinduced (Figure 5B), perhaps in an attempt to overcome the absence of this critical metabolic pathway. Of particular interest was a set of genes that responded in a similar manner to TCA cycle dysfunction but whose response pattern varied with the enzyme defect (Figure 6). Expression was elevated in response to aconitase and isocitrate dehydrogenase deficiencies, diminished in response to α-ketoglutarate dehydrogenase complex and succinyl-CoA ligase deficiencies, elevated again in response to succinate dehydrogenase and fumarase deficiencies, and diminished again in response to malate dehydrogenase and citrate synthase deficiencies. Although it is not immediately apparent why this alternating pattern of expression occurs in response to defects in contiguous pairs of TCA cycle enzymes, it is presumably a reflection of changes in metabolic signals. As an approach to understanding this pattern, we have reduced the TCA cycle into four steps by combining the adjacent enzymatic reactions that yielded similar expression patterns (Figure 10). From this framework, we looked for similarities and differences among these four enzyme sets. Each of these enzyme pairs contains one reaction that generates reduced nucleotides (NADH or FADH2) during TCA cycle function, suggesting that changes in redox state are not primarily responsible for generating this response. The net ΔG values for each pair are negative, indicating an overall exergonic reaction (Matthews et al. 2000). In addition, each set contains one reaction that is highly reversible and one reaction that is essentially irreversible when assayed individually (except for the succinate dehydrogenase–fumarase pair, in which both reactions are reversible). Four carboxylic acids separate these enzyme pairs: citrate, α-ketoglutarate, succinate, and malate. It is possible that changes in one or more of these metabolites may be critical in establishing this expression pattern as a result of TCA cycle dysfunction. For instance, an increase in succinate caused by a succinate dehydrogenase or fumarase defect may signal for increased transcription of these genes, whereas a decrease in succinate formation caused by a succinyl-CoA ligase or an α-ketoglutarate dehydrogenase defect might signal for a decrease in gene expression. Although changes in these TCA cycle metabolites within the mitochondrial matrix may initiate a signaling pathway that results in altered nuclear gene expression, it is not certain whether they serve directly as the signaling molecules. For instance, glutamate and glutamine are derived from α-ketoglutarate through successive transamination reactions, and both amino acids have been demonstrated to signal changes in the metabolic state of the cell that regulate nuclear gene expression (Butow, 2002; Crespo et al., 2002).
Figure 10.
A sectored model of the TCA cycle to explain the alternating gene expression pattern in response to TCA cycle dysfunction. TCA cycle enzymes are paired on the basis of the alternating gene expression pattern in response to enzyme dysfunction. Enzyme defects in white resulted in decreased expression, and enzyme defects in gray resulted in elevated gene expression. ΔG values (kJ/mol) for each pair are calculated as the sum of the individual enzymes (Matthews et al., 2000). Potential signaling metabolites at the inflection points between these pairs are shown in black boxes. CS, citrate synthase; ACO, aconitase; IDH, isocitrate dehydrogenase; KGD, α-ketoglutarate dehydrogenase; SCL, succinyl-CoA ligase; SDH, succinate dehydrogenase; FUM, fumarase; MDH, malate dehydrogenase.
One of the more interesting aspects of the alternating gene expression pattern is its correlation with a previously identified set of TCA cycle gene defects that were identified as growth-enhancing mutations of isocitrate dehydrogenase–dysfunctional cells. Mutations in the CIT1, KGD1–2, LPD1, LSC1–2, and MDH1 genes can serve as growth enhancers of isocitrate dehydrogenase dysfunction, and these same defects result in the diminished expression response (Figure 6). This suggests a link between glycerol growth enhancement of isocitrate dehydrogenase dysfunction and the oscillating gene expression pattern reported here.
An extensive analysis of the glycerol suppressor accumulation phenotype associated with isocitrate dehydrogenase dysfunction has been reported (Przybyla-Zawislak et al., 1999). A collection of mutations in each of the 15 genes encoding TCA cycle polypeptides was screened for this phenotype. In addition, two complementary approaches were taken to determine the identities of the suppressor mutations. First, on the basis of the previous characterization of defects in CIT1 as the most abundant class of suppressors (Gadde and McCammon, 1997), mutations in genes encoding TCA cycle proteins were tested for their ability to enhance growth of isocitrate dehydrogenase–dysfunctional cells on glycerol. Second, a collection of spontaneous suppressor mutations was characterized. Several conclusions were drawn from these studies. First, the glycerol suppressor accumulation phenotype is a unique phenotype associated with the loss of isocitrate dehydrogenase polypeptides. Second, defects in genes (CIT1, KGD1–2, LPD1, LSC1–2, and MDH1) encoding half of the TCA cycle enzymes could function as growth enhancers; partial function alleles of KGD1–2 and LPD1 that can grow on glycerol were capable of growth enhancement, whereas deletion mutations could not. Third, neither deletion mutations nor partial function glycerol+ alleles of ACO1, IDH1, SDH1–4, and FUM1 could function as growth enhancers. Fourth, only defects in MDH and CIT genes encoding the TCA cycle isozymes were capable of suppression. Finally, eight other genes involved in oxidative metabolism were identified as growth enhancers, indicating that not all defects in oxidative metabolism were capable of suppression. These results divided the TCA cycle genes between suppressing genes and nonsuppressing genes and established limits to the number and types of genes that are capable of growth enhancement. However, the physiological basis for the growth enhancement remained undefined.
To investigate the relationship between the alternating gene expression pattern and the growth enhancing mutations, microarray analysis was used to determine the effects of an Δidh2 Δcit1 double mutation. Inactivation of either CIT1 or IDH2 resulted in diminished (CIT1) or elevated (IDH2) patterns of gene expression. With defects in both genes, the expression pattern was largely corrected and was very similar to wild-type levels (Table 1). These results led to the hypothesis that overexpression of one or several of these responsive genes is deleterious to growth and that the suppressor mutations function to correct this altered expression. We have begun to test this idea by assaying the effect of inactivation of genes showing the alternating pattern of expression on glycerol growth. To date, we have found that two of six genes tested can serve as growth enhancing mutations in isocitrate dehydrogenase–deficient strains, CIT1 and YGR067C (Figure 8). CIT1 is the only TCA cycle suppressor gene that also displays the alternating gene expression pattern in response to TCA cycle dysfunction. YGR067C appears to encode a transcription factor and may therefore regulate the expression of a number of other genes.
Eight other suppressor genes were also identified that do not encode TCA cycle proteins (Przybyla-Zawislak et al., 1999). While the identities of these genes have not been determined, the mutations display growth phenotypes on nonfermentable carbon sources, suggesting that the encoded proteins are involved in oxidative metabolism. It will be interesting to determine how these defects affect the alternating gene expression pattern of the genes reported here. For instance, do these and other suppressor mutations affect the alternating genes in a similar manner? In addition, is it possible the suppressor mutations enhance growth by decreasing the expression of one or more of the genes displaying an alternating expression pattern that may be deleterious when overexpressed? There are several reasons why the suppressor mutations may be specifically detected with isocitrate dehydrogenase defects and not with other TCA cycle mutations that result in the same pattern of alternating gene expression. First, strains in which IDH1 or IDH2 are inactivated are able to grow on certain nonfermentable carbon sources whereas the other related TCA cycle gene defects (i.e., in ACO1, SDH1–4, FUM1) cannot (Przybyla-Zawislak et al., 1999). Second, many of the responsive genes display their highest expression defect in strains deleted for IDH1 and IDH2 (Figure 6), and, therefore, the levels of the potentially deleterious proteins may be highest in the isocitrate dehydrogenase dysfunctional strains. Finally, these potentially deleterious genes may be particularly sensitive to metabolic signals resulting from isocitrate dehydrogenase dysfunction.
Isocitrate dehydrogenase dysfunction results in two types of DNA instability. As described above, second site nuclear mutations arise that enhance growth of cells lacking isocitrate dehydrogenase on glycerol. Second, isocitrate dehydrogenase dysfunction results in mtDNA instability, and strains lacking this enzyme have a high frequency of petite [ρ−] mutations with large deletions in mtDNA (Elzinga et al., 1993; Lin et al., 2001). mtDNA instability is a phenotype associated with other TCA cycle defects and with a number of genes encoding proteins in mitochondrial oxidative phosphorylation and biogenesis (Contamine and Picard, 2000). Many of these latter proteins are bifunctional and appear to play additional roles in the translation and/or assembly of mitochondrially encoded proteins. Isocitrate dehydrogenase is a bifunctional protein since it binds to mitochondrially encoded mRNA and appears to regulate translation of these transcripts (Elzinga et al., 1993; de Jong et al., 2000). However, it is not clear whether the mtDNA instability associated with isocitrate dehydrogenase dysfunction results from the loss of catalytic activity or from the aberrant expression and turnover of mitochondrial respiratory complexes (de Jong et al., 2000; Lin et al., 2001). These functions may not be distinct, because mRNA binding by isocitrate dehydrogenase inhibits its catalytic activity (Anderson and McAlister-Henn, 2000). These observations suggest a mechanism whereby TCA cycle metabolic flux and the synthesis of respiratory complexes are coordinately regulated (Anderson and McAlister-Henn, 2000). Mutations in CIT1 also suppress the mtDNA instability of isocitrate dehydrogenase dysfunctional cells (our unpublished results), indicating that these two properties are functionally linked.
Recent studies have revealed that fumarase and succinate dehydrogenase genes act as tumor suppressors in humans. Fumarase defects were associated with dominantly inherited uterine fibroids, skin leiomyomata, and renal cell cancer (Tomlinson et al., 2002), whereas two types of brain tumors were found to be caused by mutations in genes encoding succinate dehydrogenase subunits (Baysal et al., 2000; Niemann and Muller, 2000; Astuti et al., 2001). Our studies in yeast have revealed that defects in either succinate dehydrogenase or fumarase produce similar responses in gene expression, and aconitase and isocitrate dehydrogenase defects produce similar response patterns. The correlation of this pattern with genetic suppressor defects of isocitrate dehydrogenase dysfunction suggests that genetic instability may be a consequence of TCA cycle dysfunction. Thus, the genetic instability caused by isocitrate dehydrogenase dysfunction may provide clues to aberrant growth properties of tumor cells, and this yeast model may prove useful in understanding how metabolic signaling and changes in TCA cycle function affect cell function and genomic stability. Given the ubiquity of the TCA cycle, especially in eukaryotes, it is predicted that similar signaling pathways between the TCA cycle and the nucleus are operative in higher organisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grant MCB9604225 (M.M.C.), National Institutes of Health grants GM-51265 and AG-17477 (L.M.-H.), GM-22525, and CA-77811, and grant I-0642 from the Robert A. Welch Foundation (R.A.B.).
Footnotes
Online version of this article contains supplementary dataset material. The online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0422. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0422.
REFERENCES
- Anderson SL, Minard KI, McAlister-Henn L. Allosteric inhibition of NAD+-specific isocitrate dehydrogenase by a mitochondrial mRNA. Biochemistry. 2000;39:5623–5629. doi: 10.1021/bi000272e. [DOI] [PubMed] [Google Scholar]
- Astuti D, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Bohm S, Frishman D, Mewes HW. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 1997;25:2464–2469. doi: 10.1093/nar/25.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojunga N, Entian K-D. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p) Mol Gen Genet. 1999;262:869–785. doi: 10.1007/s004380051152. [DOI] [PubMed] [Google Scholar]
- Butow RA. Cellular responses to mitochondrial dysfunction: it's not always downhill. Cell Death Differ. 2002;9:1043–1045. doi: 10.1038/sj.cdd.4401083. [DOI] [PubMed] [Google Scholar]
- Cohen BD, Sertil O, Abramova NE, Davies KJA, Lowry CV. Induction, and repression of DAN1, and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 2001;29:799–808. doi: 10.1093/nar/29.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Bio Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong L, Elzinga SD, McCammon MT, Grivell LA, van der Spek H. Increased synthesis and decreased stability of mitochondrial translation products in yeast as a result of loss of mitochondrial NAD(+)-dependent isocitrate dehydrogenase. FEBS Lett. 2000;483:62–66. doi: 10.1016/s0014-5793(00)02086-x. [DOI] [PubMed] [Google Scholar]
- De Winde JH, Grivell LA. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog Nucleic Acids Res. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga SDJ, Bednarz AL, van Oosterum K, Dekker PJT, Grivell LA. Yeast mitochondrial NAD+-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res. 1993;21:5328–5331. doi: 10.1093/nar/21.23.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Hale W, IV, Butow RA. Numerical methods for handling uncertainty in microarray data: an example analyzing perturbed mitochondrial function in yeast. Methods Cell Biol. 2001b;65:439–482. doi: 10.1016/s0091-679x(01)65026-x. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Waddle JA, Hale W, IV, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001a;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1989;5:153–180. doi: 10.1146/annurev.cb.05.110189.001101. [DOI] [PubMed] [Google Scholar]
- Gadde DM, McCammon MT. Mutations in the IDH2 gene encoding the catalytic subunit of the yeast NAD+-dependent isocitrate dehydrogenase can be suppressed by mutations in the CIT1 gene encoding citrate synthase and other genes of oxidative metabolism. Arch Biochem Biophys. 1997;344:139–149. doi: 10.1006/abbi.1997.0191. [DOI] [PubMed] [Google Scholar]
- Gadde DM, Yang E, McCammon MT. An unassembled subunit of NAD+-dependent isocitrate dehydrogenase is insoluble and covalently modified. Arch Biochem Biophys. 1998;354:102–110. doi: 10.1006/abbi.1998.0677. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JS, Kurvilla FG, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbeck RJ, McAlister-Henn L. Isolation, nucleotide sequence, and disruption of the Saccharomyces cerevisiae gene encoding mitochondrial NADP(H)-specific isocitrate dehydrogenase. J Biol Chem. 1991;266:2339–2345. [PubMed] [Google Scholar]
- Haurie V, Perrot M, Mini T, Jeno P, Sagliocco F, Boucherie H. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2001;276:76–85. doi: 10.1074/jbc.M008752200. [DOI] [PubMed] [Google Scholar]
- Hentze MW. Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends Biochem Sci. 1994;24:101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryotic Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Horswill AR, Escalante-Semerena JC. In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry. 2001;40:4703–4713. doi: 10.1021/bi015503b. [DOI] [PubMed] [Google Scholar]
- Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Jia Y-K, Becam A-M, Herbert CJ. The CIT3 gene of Saccharomyces cerevisiae encodes a second mitochondrial isoform of citrate synthase. Mol Microbiol. 1997;24:53–59. doi: 10.1046/j.1365-2958.1997.3011669.x. [DOI] [PubMed] [Google Scholar]
- Kaufman BA, Newman SN, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc Natl Acad Sci USA. 2000;96:14866–14870. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Wedaman KP, O'Shea EK, Powers T. Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Poyton RO. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- Kwast KE, Lai LC, Menda N, James DT, III, Aref S, Burke PV. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol. 2002;184:250–265. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lin AP, McCammon MT, McAlister-Henn L. Kinetic and physiological effects of alterations in homologous isocitrate-binding sites of yeast NAD(+)-specific isocitrate dehydrogenase. Biochem. 2001;40:14291–14301. doi: 10.1021/bi0111707. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in expression of tricarboxylic acid cycle genes in response to a reduction or loss of mitochondrial function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Epstein CB, Butow RA. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 2001;17:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik MA, Kotter P, Salomons FA, van der Klei IJ, van Dijken JP, Pronk JT. The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme A metabolism. J Bacteriol. 2000;182:7007–7013. doi: 10.1128/jb.182.24.7007-7013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews CK, van Holde KE, Ahern KG. Biochemistry. 3rd ed. San Francisco: Addison Wesley Longman; 2000. [Google Scholar]
- McAlister-Henn L, Small WC. Molecular genetics of yeast TCA cycle isozymes. Prog Nucleic Acids Res Mol Biol. 1997;57:317–339. doi: 10.1016/s0079-6603(08)60285-8. [DOI] [PubMed] [Google Scholar]
- McCammon MT, Veenhuis M, Trapp SB, Goodman JM. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J Bacteriol. 1990;172:5816–5827. doi: 10.1128/jb.172.10.5816-5827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon MT. Yeast mutants of acetate metabolism: isolation and characterization of Acn− mutants. Genetics. 1996;144:57–69. doi: 10.1093/genetics/144.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard KI, Jennings GT, Loftus TM, Xuan D, McAlister-Henn L. Sources of NADPH and expression of mammalian NADP+-specific isocitrate dehydrogenases in Saccharomyces cerevisiae. J Biol Chem. 1998;273:31486–31493. doi: 10.1074/jbc.273.47.31486. [DOI] [PubMed] [Google Scholar]
- Minard KI, McAlister-Henn L. Antioxidant function of cytosolic sources of NADPH in yeast. Free Radic Biol Med. 2001;31:832–843. doi: 10.1016/s0891-5849(01)00666-9. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2002;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- Ogur M, John RS. A differential and diagnostic plating method for population studies of respiration deficiency in yeast. J Bacteriol. 1956;72:500. doi: 10.1128/jb.72.4.500-504.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva E, Hentze MW. Iron-sulfur clusters as genetic regulatory switches: the bifunctional iron regulatory protein-1. FEBS Lett. 1996;389:40–43. doi: 10.1016/0014-5793(96)00574-1. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Steensma HY, Van Dijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Przybyla-Zawislak B, Dennis RA, Zakharkin SO, McCammon MT. Genes of the tricarboxylic acid cycle succinyl-CoA ligase from Saccharomyces cerevisiae. Eur J Biochem. 1998;248:736–743. doi: 10.1046/j.1432-1327.1998.2580736.x. [DOI] [PubMed] [Google Scholar]
- Przybyla-Zawislak B, Gadde DM, Ducharme K, McCammon MT. Genetic and biochemical interactions involving tricarboxylic acid (TCA) cycle function using a collection of mutants defective in all TCA cycle genes. Genetics. 1999;152:153–166. doi: 10.1093/genetics/152.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi N, Martinez M-J, Barre P, Blondin B. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol Microbiol. 2000;35:1421–1430. doi: 10.1046/j.1365-2958.2000.01807.x. [DOI] [PubMed] [Google Scholar]
- Repetto B, Tzagoloff A. In vivo assembly of yeast mitochondrial alpha-ketoglutarate dehydrogenase complex. Mol Cell Biol. 1991;11:3931–3939. doi: 10.1128/mcb.11.8.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P, Bourgeron T, Parfait B, Chretien D, Munnich A, Rotig A. Inborn errors of the Krebs cycle: a group of unusual mitochondrial diseases in human. Biochim Biophys Acta. 1997;1361:185–197. doi: 10.1016/s0925-4439(97)00035-5. [DOI] [PubMed] [Google Scholar]
- Scheffler IE. Molecular genetics of succinate:quinone oxidoreductases in eukaryotes. Prog Nucl Acid Res Mol Biol. 1998;60:267–315. doi: 10.1016/s0079-6603(08)60895-8. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Dawes IW. Genetics of the synthesis of serine from glycine and the utilization of glycine as sole nitrogen source by Saccharomyces cerevisiae. Genetics. 1995;140:1213–1222. doi: 10.1093/genetics/140.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Linde JJM, Liang H, Davis RW, Steensma HY, van Dijken JP, Pronk JT. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J Bacteriol. 1999;181:7409–7413. doi: 10.1128/jb.181.24.7409-7413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Linde JJM, Steensma HY. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast. 2002;19:833–848. doi: 10.1002/yea.879. [DOI] [PubMed] [Google Scholar]
- Tomlinson IPM, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication: altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J Biol Chem. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yudkoff M, Nelson D, Daikhin Y, Erecinska M. Tricarboxylic acid cycle in rat brain synaptosomes: fluxes and interactions with aspartate aminotransferase and malate/aspartate shuttle. J Biol Chem. 1994;269:27414–27420. [PubMed] [Google Scholar]
- Zhang L, Hach A. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell Mol Life Sci. 1999;56:415–426. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W-N, McAlister-Henn L. Affinity purification and kinetic analysis of mutant forms of yeast NAD+-specific isocitrate dehydrogenase. J Biol Chem. 1997;272:21811–21817. doi: 10.1074/jbc.272.35.21811. [DOI] [PubMed] [Google Scholar]
- Zitomer RS, Limbach MP, Rodriguez-Torres AM, Balasubramanian B, Deckert J, Snow PM. Approaches to the study of Rox1 repression of the hypoxic genes in the yeast Saccharomyces cerevisiae. Methods. 1997;11:279–288. doi: 10.1006/meth.1996.0422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.