Abstract
In eukaryotic cells, RNA polymerase II (RNA pol II) transcription and pre-mRNA processing are coordinated events. We have addressed how individual components of the transcription and pre-mRNA processing machinery are organized during mitosis and subsequently recruited into the newly formed daughter nuclei. Interestingly, localization studies of numerous RNA pol II transcription and pre-mRNA processing factors revealed a nonrandom and sequential entry of these factors into daughter nuclei after nuclear envelope/lamina formation. The initiation competent form of RNA pol II and general transcription factors appeared in the daughter nuclei simultaneously, but prior to pre-mRNA processing factors, whereas the elongation competent form of RNA pol II was detected even later. The differential entry of these factors rules out the possibility that they are transported as a unitary complex. Telophase nuclei were competent for transcription and pre-mRNA splicing concomitant with the initial entry of the respective factors. In addition, our results revealed a low turnover rate of transcription and pre-mRNA splicing factors during mitosis. We provide evidence to support a model in which the entry of the RNA pol II gene expression machinery into newly forming daughter nuclei is a staged and ordered process.
INTRODUCTION
In eukaryotic cells, RNA polymerase II (RNA pol II) transcription and pre-mRNA processing are coordinated events that require finely tuned interactions among a large number of proteins (Misteli and Spector, 1999; Maniatis and Reed, 2002; Orphanides and Reinberg, 2002; Proudfoot et al., 2002). One fundamental feature of mammalian cell nuclei is that many components of the RNA synthesis and processing machinery are organized into compartments (Spector, 1993; Lamond and Earnshaw, 1998; Misteli, 2000; Spector, 2001; Hernandez-Verdun et al., 2002; Huang, 2002). The best characterized nuclear compartment is the nucleolus where rRNA synthesis and processing as well as preribosome assembly takes place (Scheer and Hock, 1999; Hernandez-Verdun et al., 2002; Huang, 2002). The nucleus is further divided into other nonmembrane-enclosed compartments, including but not limited to chromosome territories, nuclear speckles, Cajal bodies, promyelocytic leukemia bodies, etc. (Spector, 1993, 2001; Lamond and Earnshaw, 1998; Gall, 2000). Active transcription sites can be visualized by bromouridine triphosphate (bromo-UTP) incorporation as several thousand foci scattered throughout the nucleus that colocalize with transcription factors, heterogeneous nuclear RNA-associated proteins (hnRNPs), and other RNA processing factors (Iborra et al., 1996; Pombo et al., 2000). However, these sites are not generally coincident with the larger and less abundant “nuclear speckles” where splicing factors are enriched. Mammalian interphase nuclei typically contain 20–50 nuclear speckles. By electron microscopy, the speckled pattern corresponds to interchromatin granule clusters (IGCs) and perichromatin fibrils (Spector et al., 1983, 1991; Fakan et al., 1984; Fakan, 1994). Each IG cluster is composed of granules measuring 20–25 nm in diameter (Fakan and Puvion, 1980; Spector, 1993). Biochemical purification of IGCs from mouse liver nuclei revealed that IGCs contain ∼136 proteins (Mintz et al., 1999), a large number of which are pre-mRNA processing factors. IGCs also contain transcription factors (Larsson et al., 1995), a hyperphosphorylated form of RNA pol II (Bregman et al., 1995), and 3′-processing factors (Schul et al., 1998; Calado and Carmo-Fonseca, 2000). Observations that splicing factors are recruited from IGCs to transcription sites support the possibility that one function of IGCs is the assembly/modification of spliceosomal components (Jiménez-Garcia and Spector, 1993; Spector, 1993; Misteli et al., 1997; Lamond and Earnshaw, 1998). Recent results further point to the involvement of IGCs in coordinating transcription and pre-mRNA splicing (Misteli and Spector, 1999; Sacco-Bubulya and Spector, 2002).
On transcriptional elongation, pre-mRNA processing events are typically initiated immediately, at the site of transcription (Beyer and Osheim, 1988; Bauren and Wieslander, 1994; Proudfoot et al., 2002). The efficient processing of nascent pre-mRNA is crucial for mRNA biogenesis and is also a prerequisite for the export of processed mRNA to the cytoplasm (Custodio et al., 1999; Reed and Hurt, 2002). Numerous in vivo and in vitro studies indicate that RNA pol II couples transcription and pre-mRNA splicing (Hirose and Manley, 2000; Maniatis and Reed, 2002; Proudfoot et al., 2002). Various biochemical studies have established that the largest subunit of RNA pol II shows differential hyperphosphorylation on serine residues within heptad repeats in its carboxyl-terminal domain (CTD). These modifications regulate its ability to initiate and elongate transcripts (Bensaude et al., 1999; Hirose and Manley, 2000; Komarnitsky et al., 2000) and to interact with specific components of the RNA processing machinery such as capping enzyme, various splicing factors, and 3′-end cleavage factors (Kim et al., 1997; McCracken et al., 1997; Ho and Shuman, 1999; Misteli and Spector, 1999; Komarnitsky et al., 2000; Ryan et al., 2002).
In higher eukaryotes, mitosis is accompanied by dramatic transformations in the structural organization of both the cytoplasm and nucleus. The onset of mitosis is accompanied by chromatin condensation, breakdown of the nuclear envelope (John and Workman, 1998), and cessation of bulk cellular transcription (Prescott and Bender, 1962; Johnson and Holland, 1965; Gottesfeld and Forbes, 1997). The constituents of many nuclear domains, such as the pre-mRNA processing factors in nuclear speckles, become distributed diffusely throughout the cytoplasm (Reuter et al., 1985; Spector and Smith, 1986; Spector et al., 1991; Ferreira et al., 1994). During metaphase, pre-mRNA processing factors begin to reassemble into discrete structures called mitotic interchromatin granules (MIGs) (Verheijen et al., 1986; Leser et al., 1989; Ferreira et al., 1994). MIGs seem to be structurally analogous to IGCs (Leser et al., 1989; Thiry, 1993, 1995a), and the number and size of MIGs increase progressively from metaphase to telophase. However, the function of MIGs has not been addressed.
The gene expression machinery must be rapidly reactivated when cells exit from mitosis. Because the onset of transcription in newly formed daughter nuclei relies on the presence of transcription and RNA processing factors, it is important to determine when the bulk of these factors reenter daughter nuclei and whether factors are recycled or are newly translated at the end of mitosis. Herein, we report on the localization and stability of transcription and pre-mRNA processing factors during the mitosis/G1 transition. We found that RNA pol II and its transcription and pre-mRNA processing factors entered daughter nuclei in a sequential and nonrandom manner after nuclear envelope/lamina formation. Bromo-UTP incorporation was temporally correlated with the presence of the initiation-competent form of RNA pol II in daughter nuclei, and it was further enhanced with increased accumulation of the elongation-competent form of RNA pol II. The pre-mRNA processing machinery appeared in daughter nuclei after the entry of the general transcription factors but just before the appearance of the elongation competent form of RNA pol II. Interestingly, transcription and pre-mRNA processing factors had a low turnover rate during mitosis and were recycled from the cytoplasm into daughter nuclei. Our data demonstrate that nuclear entry of the RNA pol II gene expression machinery after mitosis is a staged process whereby the transcription apparatus enters in a separate and reproducible window of time prior to the entry of the pre-mRNA processing machinery.
MATERIALS AND METHODS
cDNA Constructs
Polymerase chain reaction generated a restriction site at the stop codon of human SC35 cDNA for convenient subcloning into pEYFP-N1 (BD Biosciences Clontech, Palo Alto, CA). SF2/ASF was subcloned from GFP-SF2/ASF vector (Misteli et al., 1997) into the corresponding pEYFP vector.
Cell Culture and Transfection
HeLa cells were grown in DMEM containing low glucose (Invitrogen, Carlsbad, CA) supplemented with penicillin-streptomycin and 10% fetal bovine serum (Hyclone Laboratories, Logan, UT). Electroporation was performed on trypsinized cells resuspended in 250 μl of growth medium and transferred to cuvettes containing 2 μg of fusion protein plasmid plus 20 μg of salmon sperm DNA. Cells were seeded onto acid-washed coverslips and processed for immunofluorescence localization of nuclear speckle proteins 2 d after transfection.
Cell Synchronization and Drug Treatments
To obtain large numbers of mitotic cells for protein turnover studies (Figure 9), HeLa cells were arrested at prometaphase with 50 ng/ml nocodazole for 14–18 h. Cells were treated with or without cycloheximide 50 μg/ml during the final 30 min of nocodazole treatment. Prometaphase cells were collected and were continuously incubated in medium either with or without (control) cycloheximide during release from mitosis. Cells were incubated with [35S]methionine (10 mCi/ml) in cold methionine-free medium for 30 min before collection. Aliquots of cells were collected at the indicated times (as in text) and the efficacy of the protein synthesis block was analyzed by measuring incorporated [35S]methionine by autoradiography. Parallel samples were also analyzed by immunoblotting, fluorescence-activated cell sorting (FACS), and immunofluorescence.
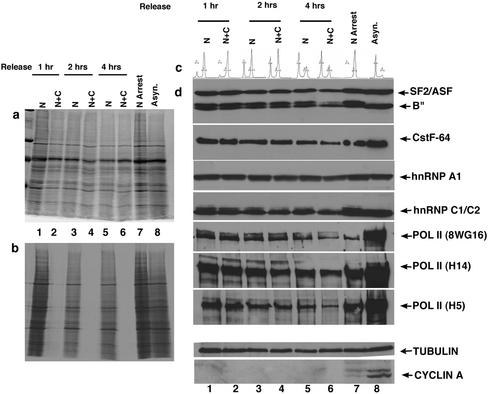
Figure 9.
RNA processing factors exhibit a low turnover rate during mitosis. Cells blocked in prometaphase by nocodazole treatment were incubated with (N + C) or without (N) cycloheximide and pulsed with [35S]methionine during release from the mitotic block. Aliquots of cells were collected at indicated times and efficacy of the protein synthesis block was analyzed by measuring incorporated [35S]methionine by autoradiography (b) (see MATERIALS AND METHODS). The corresponding Coomassie-stained gel is shown in a. Lanes 1, 3, and 5 are cells treated with nocodazole for 14–18 h and collected at 1, 2, and 4 h postrelease, respectively. Lanes 2, 4, and 6 represent cells treated with nocodazole and cycloheximide at indicated time points. Lanes 7 and 8 represents nocodazole-arrested cells and an asynchronous population, respectively. DNA content of the respective cell populations indicated in d, 1–8, was determined by FACS analysis and is shown in c. Cells treated with or without cycloheximide were collected at indicated time points and extracted for immunoblot analysis of components of the gene expression machinery (d). The results show a low turnover of splicing factors (SF2/ASF and B"), CstF-64, hnRNP A1, hnRNPC1/C2, and various forms of RNA pol II during mitosis. Tubulin and cyclin A are shown as internal controls for protein loading and cell cycle progression, respectively.
Immunofluorescence
Cells were rinsed briefly in phosphate-buffered saline (PBS) and then fixed for 15 min in 1.7% formaldehyde in PBS (pH 7.4) or for 5 min in cold methanol (−20°C) for optimal penetration of IgM antibodies into nuclei. Cells were permeabilized in PBS containing 0.5% Triton X-100 and 1% goat serum for 7–10 min on ice, and primary antibodies were added for 1 h at room temperature: anti-RNA pol II (8WG16 [1:200], H14 [1:20], H5 [1:20], hnRNP A1 [1:1500], hnRNP C1/C2 [1:2500], p56 [TFIIE subunit] [1:50], TFIIB [1:300], TFIIF [RAP74; 1:300], TFIIH [ERCC2, 1:200; ERCC3, 1:100], DRIP130 [1:500], DRIP150 [1:300], TATA binding protein [TBP] [1:50], CstF-64 [1:50], SC35 [1:200], 4G3 anti-B" [1:100], mAb103 anti-SF2/ASF [1:50], SF3a-60 [1:800], lamin B1 [1:500], lamin A/C [1:500], p62 [nuclear pore protein] [1:100], poly (A) binding protein II [PABP II] [1:200], m3G anti-snRNA [1:50], ubiquitin [1:1500]). Cells were rinsed in PBS containing 1% goat serum and then secondary anti-species–specific antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) were added for 30 min at room temperature: goat anti-mouse (GAM) IgG1-Texas Red (1:1000), GAM IgG H+L Texas Red (1:500), GAM IgM-Cy5 (1:1000), goat anti-rat IgG-fluorescein (1:1200). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Cells were mounted in medium containing 90% glycerol, 10% PBS pH 8.0 plus 1 mg/ml paraphenylenediamine. Cells were examined using an Axioplan 2i fluorescence microscope (Carl Zeiss, Thornwood, NY) equipped with Chroma filters (Chroma Technology, Brattleboro, VT). OpenLab software (Improvision, Boston, MA) was used to collect digital images from a ORCA cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ).
Bromo-UTP Incorporation
HeLa cells were rinsed briefly in glycerol buffer (20 mM Tris-HCl pH 7.4, 5.0 mM MgCl2, 25% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5 mM EGTA) followed by permeabilization for 3 min in glycerol buffer supplemented with 2 μg/ml digitonin. Transcription buffer (100 mM KCl, 50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 1.0 mM phenylmethylsulfonyl fluoride, 2.0 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 0.2 mM 5-bromo-UTP, and 1.0 μg/ml RNAsin) was added for 5 min at 37°C. Colocalization of bromo-UTP (rat anti-bromo, 1:30) and splicing factors was performed as described above.
RNA Fluorescence In Situ Hybridization
To detect splicing of β-tropomyosin minigene pre-mRNA, a 24-mer oligodeoxynucleotide conjugated with a single Texas Red molecule at the 5′ end (Invitrogen) was designed to hybridize to the splice junction between exons 5 and 6 (5′-tgcctggctcggctctcagccacc-3′). Control 12-mer oligos were designed to hybridize to the 3′ end of exon 5 (5′-tgcctggctcgg-3′) or the 5′ end of exon 6 (5′-ctctcagccacc-3′). Cells were extracted in CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 10 mM PIPES pH 6.8) supplemented with 0.5% Triton X-100 and 2 mM vanadyl ribonucleoside complex. After immunolabeling as described above, cells were again fixed for 15 min in 2% formaldehyde. Hybridization of oligo probes (1 ng/μl) was performed in 35% deionized formamide, 10% dextran sulfate, 1 mg/ml yeast tRNA, and 2× SSC for 3 h at 37°C. Cells were washed for 30 min in 25% formamide/2× SSC at 37°C and then 30 min in 2× SSC. Cells were mounted and images obtained using an Axioplan 2i-fluorescence microscope (Carl Zeiss) equipped with Chroma filters (Chroma Technology) as described above.
Live Cell Microscopy
HeLa cells were transiently transfected with 2 μg of YFP-SF2/ASF or SC35-YFP and live-cell observations were initiated 2 d posttransfection to ensure that fusion protein would be detectable in mitotic cells. The cells were transferred to an FCS2 live-cell chamber (Bioptechs, Butler, PA) mounted onto the stage of an IX70 inverted fluorescence microscope (Olympus, Melville, NY) and kept at 37°C in L-15 medium (minus phenol red) containing 10% fetal bovine serum. Time-lapse images acquired with a 100× 1.4 numerical aperture heated objective lens were captured with a Peltier-cooled IMAGO charge-coupled device camera by using an SVGA interline chip (1280 × 1024) with a pixel size of 6.7 μm (Till Photonics, Munich, Germany).
Online Supplemental Materials
Video 1.
A HeLa cell in metaphase exhibits a diffuse localization of YFP-SF2/ASF with a few small MIGs near the metaphase plate. On entry into anaphase, the number and size of MIGs increases. As nuclear entry of YFP-SF2/ASF begins, there is a concomitant decrease in YFP-SF2/ASF signal in MIGs, consistent with recycling of splicing factors into newly forming daughter nuclei. Image sequences were acquired using TillVision software (Till Photonics) and animated using QuickTime software. The video is comprised of a sequence of 260 images. Exposures (20 ms) were collected every 15 s for 65 min.
RESULTS
Components of Transcription and pre-mRNA Processing Machinery Enter Daughter Nuclei after Nuclear Envelope/Lamina Formation
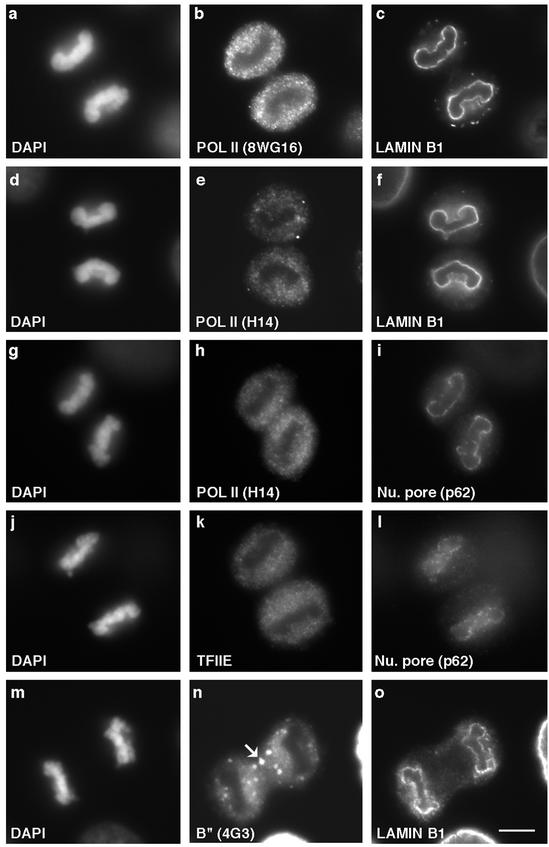
One of the earliest events in establishing daughter nuclei after mitosis is deposition of the nuclear envelope/lamina around segregated chromosomes (Haraguchi et al., 2000; Moir et al., 2000; Gerlich et al., 2001; Goldman et al., 2002). To determine whether the bulk of RNA pol II transcription and pre-mRNA processing factors associate with daughter nuclei before or after nuclear envelope/lamina formation, we performed coimmunolocalization experiments with antibodies against various transcription or RNA processing factors and members of the nuclear lamina/nuclear pore complex in asynchronous HeLa cells. The largest subunit of RNA pol II in its hypo- (Figure 1b) as well as hyperphosphorylated (Figure 1, e and h) forms was predominantly present in the cytoplasm when lamin B1 (Figure 1, c and f) or nuclear pore complex protein p62 (Figure 1i) had already localized around the periphery of newly forming daughter nuclei. Similarly, the largest subunit of transcription factor TFIIE, p56 (Figure 1k), was not yet detectable in daughter nuclei at a stage when p62 (Figure 1l) or lamin B1 (our unpublished data) had already begun to localize at the nuclear periphery. Lamins A/C also showed peripheral nuclear staining before the entry of RNA pol II and general transcription factors (data not shown). Next, we examined the localization of the B" protein, an integral component of the U2 small nuclear (sn)RNP complex (Habets et al., 1989). In early telophase cells, B" was diffusely distributed in the cytoplasm as well as enriched in MIGs (Figure 1n, arrow). Similar to RNA pol II and transcription factors, B" remained in the cytoplasm (Figure 1n), whereas the nuclear lamina and envelope was assembled (Figure 1o). Similar observations were made with other pre-mRNA splicing factors, including SF2/ASF and SF3a-60 (our unpublished data). An antibody that recognizes the trimethyl guanosine cap of snRNAs (m3G) also showed positive nuclear staining only after nuclear envelope/lamina formation occurred (our unpublished data). These results clearly indicated that nuclear envelope/lamina formation occurs before nuclear entry of various components of the transcription and pre-mRNA processing machinery.
Figure 1.
Nuclear envelope/lamina is assembled before the entry of components of the gene expression machinery into daughter nuclei. Both the hypo- (b) and ser-5–phosphorylated form of RNA pol II (e) decorate the cytoplasm after the nuclear lamina formation (c and f). Similarly, colocalization studies with the ser-5–phosphorylated form of RNA pol II (h) and nuclear pore protein p62 (i) showed that the nuclear envelope was established before the nuclear entry of H14. Transcription factor TFIIE (k) and splicing factor B" (n) also remained in the cytoplasm until after nuclear envelope (l) and lamina (o) were assembled. Chromosomes were stained with DAPI (a, d, g, j, and m). Bar 5 μm.
Initiation-Competent Form of RNA pol II and Its Associated Transcription Factors Appear in Daughter Nuclei prior to Nuclear Entry of pre-mRNA prior to Processing Machinery
Next, we evaluated the sequence of nuclear import of various components of the gene expression machinery. First, we wanted to establish the timing of RNA pol II entry with regard to entry of transcription and pre-mRNA processing factors. Antibody 8WG16 (Thompson et al., 1989) predominantly detects the hypophosphorylated form of the large subunit of RNA pol II. Antibody H14 (Bregman et al., 1995) recognizes the hyperphosphorylated ser-5 moiety in the CTD repeats, and preferentially recognizes the form of RNA pol II that is competent for the initiation of transcription (Komarnitsky et al., 2000). Both the 8WG16 and H14 epitopes were simultaneously detectable in midtelophase daughter nuclei (Figure 2, b and c). However, although there was a negligible amount of cytoplasmic H14 staining (Figure 2b), there was a significant level of cytoplasmic 8WG16 staining that persisted until late telophase/early G1 (Figure 2c). TFIIE entered daughter nuclei at the same time as RNA pol II (H14; Figure 2, e and f). Similar observations were made with other general transcription factors, including TFIIB, TFIIF (RAP 74) and TFIIH (ERCC2, ERCC3) as well as components of mediator complex (DRIP 130, 150) (Figure 4). This differs from the behavior of TBP, a member of the TFIID complex that was found on the chromosomes throughout mitosis (our unpublished data) (Chen et al., 2002). Thus, RNA pol II and multiple components of the transcription machinery entered daughter nuclei simultaneously during a narrow temporal window in telophase.
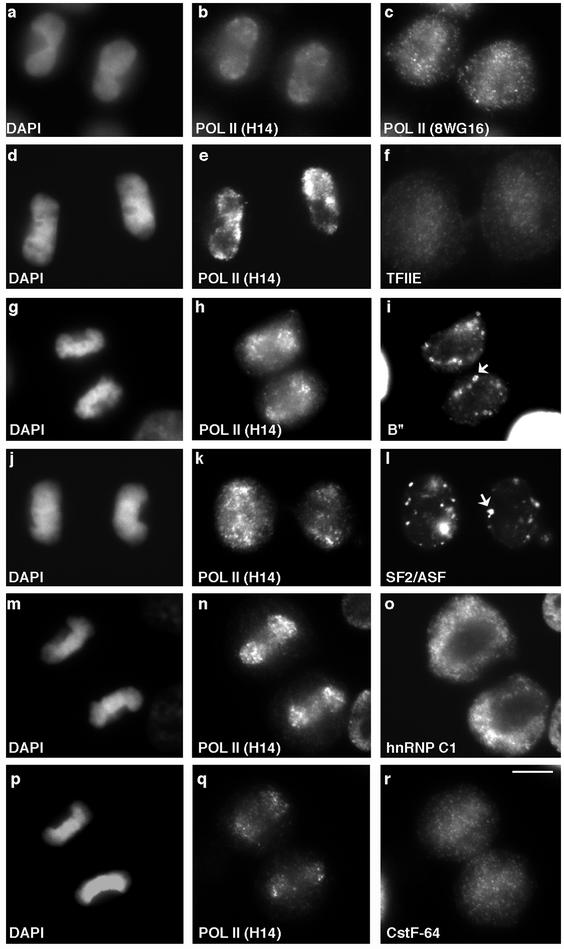
Figure 2.
Appearance of the transcription initiation-competent form of RNA pol II and associated transcription factors in postmitotic nuclei precedes pre-mRNA processing factors. The initiation-competent ser-5 phosphorylated form of RNA pol II (b and e), hypophosphorylated form of RNA pol II (c) and TFIIE (f) appeared in daughter nuclei almost simultaneously. Splicing factors B" (i) and SF2/ASF (l) were largely localized to the cytoplasmic MIGs (i and l, arrows) when H14 labeling was already apparent in daughter nuclei (h and k). During this same time period, the hnRNP C1/C2 proteins (o) were predominantly present in the cytoplasm, whereas H14 had already entered nuclei (n). However, CstF-64 (r) and H14 (q) appeared in daughter nuclei simultaneously. DNA was stained with DAPI (a, d, g, j, m, and p). Bar 5 μm.
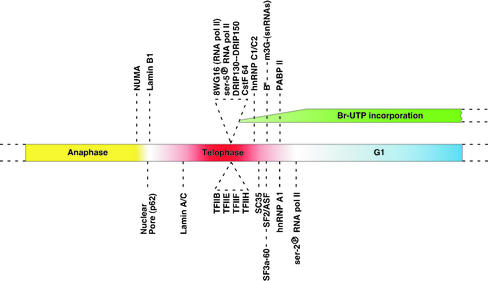
Figure 4.
Schematic representation of the order of events in the recruitment of the gene expression machinery into daughter nuclei. A consolidated overview of sequential entry of transcription and pre-mRNA processing factors into newly formed daughter nuclei.
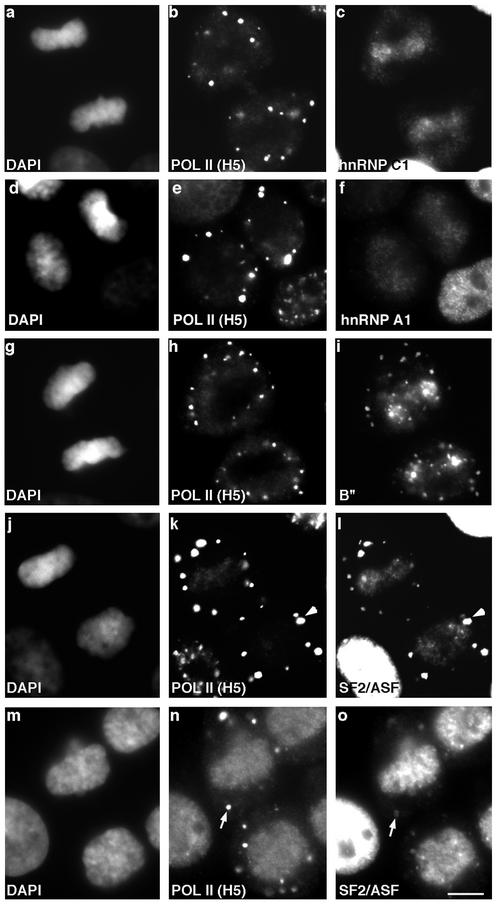
We then compared the import of various pre-mRNA processing factors with respect to entry of the transcription machinery. Interestingly, the bulk of B" (Figure 2i) and SR splicing factor SF2/ASF (Figure 2l) remained localized in the cytoplasm of telophase cells, intensely concentrated in the MIGs, even as the ser-5 phosphorylated RNA pol II appeared in the nucleus (Figure 2, h and k). Similarly, splicing factor SF3a-60 (Schmidt-Zachmann et al., 1998) and the snRNAs, as well as hnRNP C1/C2 (Figure 2o) (Pinol-Roma and Dreyfuss, 1991), hnRNP A1, and PABP II entered the daughter nuclei after RNA pol II and transcription factors (see Figure 4 for details). In contrast, the cleavage stimulation factor CstF-64 entered the daughter nuclei at the same time as RNA pol II and transcription factors (Figure 2, q and r). In summary, the hypo- and ser-5–hyperphosphorylated forms of the large subunit of RNA pol II and its associated factors, as well as CstF-64 entered the daughter nuclei almost simultaneously, prior to entry of the bulk of other pre-mRNA processing factors.
Elongation-Competent Form of RNA pol II Appears in Daughter Nuclei after Import of RNA Processing Machinery
Next, we examined the presence in daughter nuclei of the serine-2 phosphorylated form of RNA pol II, which plays an essential role in transcriptional elongation (Komarnitsky et al., 2000) and which is recognized by the H5 antibody (Bregman et al., 1995). Ser-2 phosphorylated RNA pol II colocalized with splicing factors in speckles in interphase nuclei and in the cytoplasmic MIGs in mitotic cells (Bregman et al., 1994, 1995). This form of RNA pol II remained in the cytoplasm (Figure 3, b and e) when hnRNP C1/C2 (Figure 3c) and hnRNP A1 (Figure 3f) had already appeared in the daughter nuclei. Splicing factors B" (Figure 3i), SF2/ASF (Figure 3l), and SF3a-60 and m3G-containing snRNPs (our unpublished data) also appeared in daughter nuclei before the ser-2 modified form of RNA pol II (Figure 3, h and k). This result implies that the ser-5 phosphorylated form of RNA pol II that accumulates in daughter nuclei during midtelophase is not hyperphosphorylated at the ser-2 moiety until after the pre-mRNA processing machinery has entered the nuclei (see DISCUSSION).
Figure 3.
Transcription elongation competent RNA pol II appears in daughter nuclei subsequent to the pre-mRNA processing machinery. Double-label immunofluorescence by using antibodies against the ser-2 phosphorylated, elongation-competent form of RNA pol II (H5) (b, e, h, k, and n) and hnRNP C1/C2 (c), hnRNP A1 (f), B" (i), and SF2/ASF (l and o), respectively, demonstrated that entry of the pre-mRNA processing machinery preceded the appearance of H5 labeling in daughter nuclei. Splicing factors (i, l, and o) colocalized with H5 in MIGs (k and l, see arrowhead). SF2/ASF was nearly absent from MIGs in G1 cells (o, arrow), whereas H5 labeling was largely retained in MIGs through G1 (n, arrow). DNA was stained with DAPI (a, d, g, j, and m). Bar, 5 μm.
We extended our analysis to monitor the timing of nuclear entry of splicing factors vs. other pre-mRNA processing factors. A schematic diagram of the complete analysis summarizing the data is shown in Figure 4. It was previously shown that hnRNP C proteins are transported into the daughter nuclei before hnRNP A1 (Pinol-Roma and Dreyfuss, 1991). In addition, our results showed that splicing factors (B", SF2/ASF and SF3a) and snRNPs entered daughter nuclei after hnRNP C1/C2, but prior to hnRNP A1. However, PABP II and hnRNP A1 entered almost simultaneously.
Transcription Machinery Is Active Immediately upon Nuclear Entry
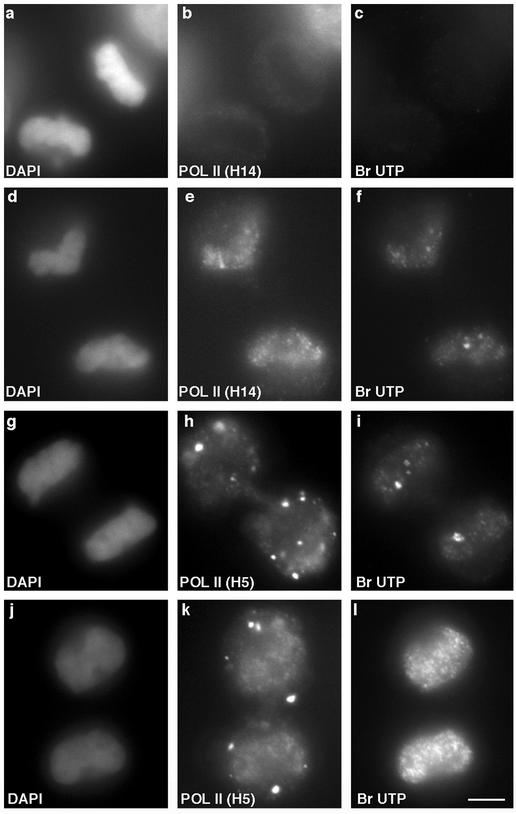
We were interested in determining whether RNA pol II and its associated factors immediately begin transcribing RNAs upon entry into daughter nuclei. To visualize the earliest active transcription sites in daughter nuclei, HeLa cells were permeabilized with digitonin and incubated in transcription buffer containing bromo-UTP for 5 min at 37°C. Before appearance of RNA pol II in daughter nuclei (H14 epitope; Figure 5b), transcription was not detectable (Figure 5c). When RNA pol II (H14 epitope) was first detectable in daughter nuclei during telophase (Figure 5e), global transcription was detected by bromo-UTP incorporation as faint, punctate foci throughout the nuclei (Figure 5f), which likely result from initiation of transcription by ser-5 phosphorylated RNA pol II. Daughter nuclei with low levels of elongation-competent RNA pol II (H5; Figure 5h) showed faint bromo-UTP incorporation in the nucleoplasm, as well as some bright foci (Figure 5i), the latter of which likely correspond to RNA pol I transcription sites in the nucleolar organizing regions. With increased amounts of H5 epitope in daughter nuclei (Figure 5k), bromo-UTP incorporation increased (Figure 5l), consistent with additional initiation and transcript elongation at this stage. The increase in nuclear H5 signal in late telophase nuclei is probably a result of the change in hyperphosphorylation (ser-5 to ser-2) of the CTD of RNA pol II already situated at transcription sites.
Figure 5.
Global transcription increases with accumulation of the ser-2 phosphorylated form of RNA pol II in daughter nuclei. Double-label immunofluorescence by using antibodies against ser-5 phosphorylated (H14) (b and e) or ser-2 phosphorylated RNA pol II (H5) (h and k) and bromo-UTP (c, f, i, and l) shows that there is no transcription in daughter nuclei prior to entry of RNA pol II (b and c). When ser-5 phoshorylated RNA pol II begins to enter daughter nuclei there is only a low level of nucleoplasmic bromo-UTP incorporation (e and f), similar to the presence of low levels of ser-2 phosphorylated RNA pol II (h and i). With further accumulation of the ser-2 phosphorylated RNA pol II, bromo-UTP incorporation dramatically increased (k and l). DNA was stained with DAPI (a, d, g, and j). Bar, 5 μm.
The pre-mRNA Splicing Machinery Is Recruited to Transcription Sites and Is Functional Immediately upon Nuclear Entry
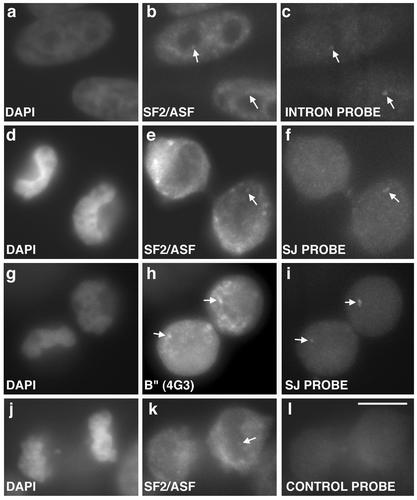
A HeLa cell line with a stably integrated β-tropomyosin minigene reporter was used to determine whether splicing factors are competent for pre-mRNA splicing during late telophase in daughter nuclei. Using an intron-specific oligonucleotide probe to β-tropomyosin pre-mRNA, the site of transcription was localized in interphase nuclei by RNA fluorescent in situ hybridization (Figure 6c, arrows). Splicing factors such as SF2/ASF are recruited to this transcription site during interphase (Figure 6b, arrows). We found that the β-tropomyosin pre-mRNA is also transcribed during late telophase. To determine whether this newly transcribed pre-mRNA was being spliced at the transcription site we used a 24-mer splice junction oligonucleotide probe that hybridizes only to spliced message by virtue of its complementarity to 12 nucleotides at the 3′ end of exon 5 and 12 nucleotides at the 5′ end of exon 6. Hybridization of this oligonucleotide in mitotic cells indicated that splicing factors are competent for intron excision as early as telophase (Figure 6, f and i, arrows). Furthermore, we could detect splicing factors such as SF2/ASF (Figure 6e) and B" (Figure 6h) at the transcription site immediately upon their entry into daughter nuclei, before their significant accumulation in the nucleoplasm and prior to the formation of nuclear speckles. Importantly, we excluded the possibility that the splice junction probe partially hybridizes to pre-mRNA, via either half of this probe, because we did not detect hybridization of control 12-mer oligonucleotides (Figure 6l) in telophase cells in regions where SF2/ASF has accumulated (Figure 6k, arrow) at what likely corresponds to the β-tropomyosin transcription site.
Figure 6.
Splicing factors are functionally competent upon entry into daughter nuclei. RNA fluorescence in situ hybridization was performed on HeLa cells stably expressing a β-tropomyosin minigene. The locus is detected during interphase as a single dot (c, arrow) that recruits splicing factors such as SF2/ASF (b, arrow). During telophase, SF2/ASF (e) and B" (h) are recruited to the locus, which is detected by a splice junction probe (SJ) (f and i), demonstrating that splicing occurs at this stage. Control hybridization of 12-mer oligonucleotides (l) shows no hybridization signal at loci decorated with SF2/ASF (k, arrow indicates probable transcription site). DNA was stained with DAPI (a, d, g, and j). Bar, 5 μm.
Release of Splicing Factors from MIGs during Telophase Is Concurrent with Their Nuclear Entry
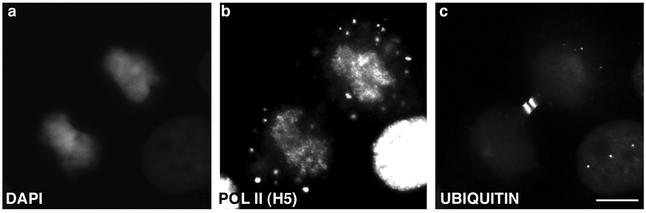
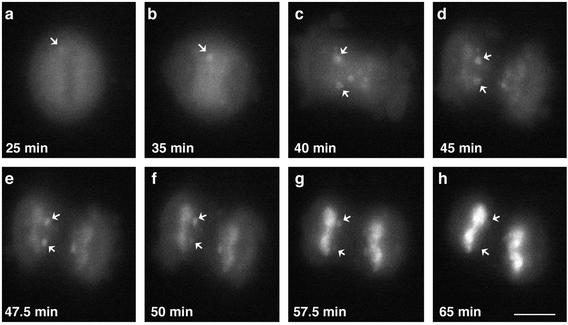
The number and size of cytoplasmic MIGs increases when cells transit from metaphase to telophase. SF2/ASF in MIGs decreased by late telophase, the time when SF2/ASF entered the daughter nuclei (compare Figure 3, l and o). Surprisingly, in the same telophase cells, the ser-2 phosphorylated form of RNA pol II (labeled by H5) persisted in the MIGs (compare Figure 3, n and o). Other splicing factors such as U2-snRNP (detected by B") and m3G-containing snRNPs behaved in a similar manner as SF2/ASF (our unpublished data). Surprisingly, immunostaining with an antibody against essential splicing factor SC35 (Fu and Maniatis, 1990) revealed that it remains in the cytoplasmic MIGs of telophase cells after all modified forms of RNA pol II (detected by 8WG16, H14, and H5) and other factors of the gene expression machinery entered daughter nuclei (our unpublished data). However, transient transfection of an SC35 cDNA fused to yellow fluorescent protein (YFP) showed that SC35-YFP entered daughter nuclei at the same time as other splicing factors (our unpublished data). The difference in nuclear entry detected using SC35 antibody vs. SC35-YFP fusion protein is likely due to the fact that the SC35 antibody recognizes a subpopulation of SC35 protein, most likely a specific modified (i.e., phosphorylated) form that enters at a time point later than other splicing factors. To confirm that MIGs are not merely protein degradation sites, cells were immunolabeled with an antibody against ubiquitinated proteins and no positive labeling of MIGs was observed (Figure 7). Taken together, these results indicate that all MIGs have a similar composition at a given stage of mitosis, and raises the possibility that different factors are released from MIGs at different time points and enter daughter nuclei sequentially. To investigate this further, HeLa cells were transiently transfected with either YFP-SF2/ASF or SC35-YFP and localization of the fusion protein was monitored in living mitotic cells by time-lapse microscopy (Figure 8; see video, Mitosis.mov). During metaphase, YFP-SF2/ASF was localized in a diffuse cytoplasmic pattern and in addition one to two small MIGs were routinely observed (Figure 8a). As mitosis progressed from late-anaphase to early telophase, the MIGs increased in size and number (Figure 8, b and c). During midtelophase, YFP-SF2/ASF began to enter daughter nuclei (Figure 8d) and the MIG signal decreased simultaneously (Figure 8, e-h) (see supplementary movie). The majority of YFP-SF2/ASF protein entered daughter nuclei within 10 min; however, some faint signal was still detectable in MIGs. Similar results were seen with SC35-YFP (our unpublished data). These results raised the possibility that during telophase, splicing factors are recycled from the cytoplasm (MIGs) into daughter nuclei.
Figure 7.
MIGs are not protein degradation sites. Immunostaining with an antibody against ubiquitinated proteins (c) did not reveal any positive labeling of MIGs but predominantly labeled the midbody between daughter cells. MIGs were positively stained with the elongation competent form of RNA pol II, H5 (b). DNA was stained with DAPI (a). Bar, 5 μm.
Figure 8.
Live cell observations of entry of SF2/ASF into daughter cell nuclei. HeLa cells were transiently transfected with YFP-SF2/ASF and live-cell observations were initiated 2 d posttransfection. Metaphase cells exhibit one to two MIGs near the metaphase plate (a, arrow). As the cell enters anaphase, MIGs become more abundant (b and c). SF2/ASF begins to enter daughter nuclei at telophase (d). Nuclear entry of YFP-SF2/ASF is nearly complete in ∼20 min (h). Time is indicated in min from the initiation of imaging the metaphase cell (see video, Mitosis.mov).
RNA Processing Factors Have a Low Turnover Rate during Mitosis
As observed in fixed and living cells, the disappearance of splicing factors from MIGs was coincident with their appearance in daughter nuclei, suggesting that cycling was occurring. We pursued this possibility further by addressing whether the transcription and RNA processing factors found in the newly formed daughter nuclei were synthesized just after mitosis, or whether they were recycled from the preexisting cytoplasmic population. We arrested HeLa cells at prometaphase with the microtubule disrupting drug nocodazole, and then subsequently released the cells from mitotic arrest either in the presence or absence of the protein synthesis inhibitor cycloheximide (see MATERIALS AND METHODS). Cells were collected over a time course and total cellular proteins were subjected to SDS-PAGE. The Coomassie Blue staining (Figure 9a) showed the level of proteins in each lane and the corresponding [35S]methionine incorporation (Figure 9b) revealed a complete inhibition of protein synthesis in the cells incubated with cycloheximide (Figure 9b, lanes 2, 4, and 6). Cell cycle progression was monitored by assaying the DNA content of propidium iodide stained cells by FACS (Figure 9c). It was clear that in the absence of protein synthesis, the majority of cells progressed normally through the cell cycle and entered G1, as determined by FACS and phase contrast microscopy (data not shown). Immunoblot analysis was carried out with the above-mentioned samples by using antibodies against various splicing factors (SF2/ASF and B"), CstF-64, hnRNP A1, hnRNP C1/C2, and different forms of RNA pol II (8WG16, H14, and H5) (Figure 9d). These results showed that there is not a significant turnover of the components of the pre-mRNA splicing and RNA processing machinery during mitosis (Figure 9d, lanes 1–4). A similar low turnover rate was observed for the RNA pol II transcription factors TFIIB and TBP (our unpublished data). Consistent with this finding, the recruitment of factors into daughter nuclei as observed by immunofluorescence was similar in the presence or absence of cycloheximide (data not shown). Note that as the cells progressed into G1, there was a fresh round of protein synthesis as seen by the quantitative difference in protein levels as detected by immunostaining after 4 h of release from nocodazole arrest with or without cycloheximide (Figure 9d, compare SF2/ASF, B", CstF-64, and hnRNP C1/C2 between lanes 5 and 6). α-Tubulin and cyclin A are shown as internal controls for equal protein loading and cell cycle progression, respectively. These results conclusively demonstrate that transcription factors and splicing factors have a low rate of turnover during mitosis, and it is the preexisting population of proteins that is recycled into daughter nuclei during the transition from mitosis to G1.
DISCUSSION
A physical coupling between transcription and pre-mRNA processing components is now thought to be instrumental for efficient gene expression (Maniatis and Reed, 2002). During mitosis, there is a global shut down of gene expression, due at least in part to the hyperphosphorylation of a large number of proteins involved in this process and resulting in the disassembly of many cellular structures (Gottesfeld and Forbes, 1997). A problem confronted by the mitotic cell is the establishment of the gene expression machinery in daughter nuclei so that these cells become competent to undergo transcription/RNA processing as they exit from mitosis. To investigate whether components of the gene expression machinery enter the postmitotic nuclei individually or as a unitary complex, we used immunofluorescence and live cell imaging to monitor the dynamics of these factors from mitosis to G1. We have demonstrated that transcription and pre-mRNA splicing factors enter the nuclei sequentially, not as a unitary complex, after nuclear envelope/lamina formation. This ordered entry of transcription factors prior to pre-mRNA splicing factors seems to be a general phenomenon because we have observed similar results in transformed cells (HeLa) as well as in cells of defined passage number (IMR90). Furthermore, we have shown that telophase nuclei are competent for transcription and pre-mRNA splicing immediately upon reentry of the gene expression machinery. The present study also established a low turnover of transcription and pre-mRNA splicing factors during mitosis demonstrating that the preexisting population of proteins recycled into daughter nuclei as the cells progress through telophase.
The nuclear envelope/lamina of higher eukaryotes breaks down during prophase and is reconstituted around chromosomes during late anaphase to telophase, reestablishing the boundary of the interphase nucleus (Haraguchi et al., 2000; Goldman et al., 2002). Our observations that import of transcription and pre-mRNA processing factors into daughter nuclei occurs after the nuclear envelope and lamina are assembled suggests that import occurs through nuclear pore complexes by active transport, because many of the presently studied proteins are above the size exclusion for transit by diffusion (Adam, 1999). Although all the members of the RNA pol II gene expression machinery do not enter newly formed daughter nuclei as a unitary complex, we cannot exclude the possibility that groups of some of these proteins enter as heterotypic complexes. Although the nuclear entry of these factors coincides with activation of transcription in daughter nuclei, transcription inhibition does not interfere with entry of most of these proteins (Ferreira et al., 1994), with the exception of hnRNP A1, which is known to be retained in the cytoplasm of transcriptionally inhibited postmitotic cells (Pinol-Roma and Dreyfuss, 1991).
Immunolabeling with specific antibodies that detect the largest subunit of RNA pol II clearly shows that hypo- and ser-5–phosphorylated RNA pol II appear in daughter nuclei before the ser-2 phosphorylated form of RNA pol II. CTD phosphorylation at ser-2 and ser-5 represents an important regulatory mechanism for coordinating RNA pol II activity (West and Corden, 1995; Bensaude et al., 1999). Phosphorylation of the CTD occurs soon after initiation of transcription and is necessary for interaction of RNA pol II with other RNA processing factors (Hirose and Manley, 2000). Studies from various laboratories suggest that RNA pol II with a hypophosphorylated CTD is preferentially included in the transcription preinitiation complex formed at the promoter. Once bound to the promoter, the CTD is phosphorylated at ser-5 and becomes initiation competent (Komarnitsky et al., 2000; Cho et al., 2001). Chromatin immunoprecipitation studies showed that the phosphorylated ser-5 (H14) epitope persists until RNA pol II transcribes up to 200 nucleotides downstream of the promoter, at which time the ser-5 phosphate is either removed or the CTD is further modified in a way that blocks the H14 epitope (Komarnitsky et al., 2000). On the other hand, the ser-2 (H5) epitope was not detected on RNA pol II at the promoter, but was detected on the polymerase in the coding regions of the genes studied leading to the interpretation that it is the elongation competent form of the polymerase (Komarnitsky et al., 2000). Our study revealed that the initiation competent form of RNA pol II appeared in daughter nuclei almost simultaneously with other transcription factors but before splicing factors. Dual immunostaining studies with 8WG16 and H14 revealed the concomitant presence of these two forms of RNA pol II in the daughter nuclei. However, we have not been able to determine whether appearance of the ser-5 (H14) phosphorylated form of RNA pol II in daughter nuclei is a consequence of entry of this modified form from the cytoplasmic population or whether the unphosphorylated form enters the nucleus and is subsequently phosphorylated. Nocodazole block and release in the presence or absence of cycloheximide showed that the level of all forms of RNA pol II was constant from M phase to G1. Continuous oscillation of phosphorylation/dephosphorylation of RNA pol II makes it difficult to address the issue of recycling of the ser-5 modified form. However, phosphorylation at ser-5 may occur in daughter nuclei because it is the hypophosphorylated form of RNA pol II that is preferentially detected at the preinitiation complex at the promoter (Hirose and Manley, 2000). Consistent with this, we observed that a subpopulation of postmitotic nuclei positive for the ser-5 phosphorylated form of RNA pol II and other transcription factors showed weak bromo-UTP incorporation, suggesting that only initiation of transcription was occurring in these cells at this time point. In support of this possibility, those cells that showed weak bromo-UTP incorporation also showed weak to no H5 immunolabeling, leading us to speculate that the RNA pol II present in these nuclei is not the elongation competent form. Furthermore, we observed that the level of bromo-UTP incorporation increased dramatically in cells showing intense H5 labeling.
An important question concerning the organization of the gene expression machinery during cell division concerns the timing of import of various transcription and RNA processing factors into daughter nuclei with respect to activation of transcription and splicing in these cells. Our data demonstrate that the transcription apparatus reproducibly enters daughter nuclei independently of pre-mRNA splicing factors, which are recruited into daughter nuclei after transcription is initiated. Therefore, a mechanism must exist to coordinate transcription and pre-mRNA splicing during the transition from telophase to the establishment of the interphase nucleus. Nuclear speckles have been suggested to play a role in coupling transcription and pre-mRNA splicing in mammalian interphase nuclei (Misteli and Spector, 1999; Sacco-Bubulya and Spector, 2002). Although MIGs have been proposed to be the mitotic equivalent of nuclear speckles, their function in mitotic cells has not been addressed (Leser et al., 1989; Thiry, 1995a,b). In telophase cells, some MIGs were found to be in proximity to the newly formed nuclear envelope. This proximity of MIGs to the nuclear periphery and the disappearance of MIGs in late telophase cells with the appearance of IGCs in daughter nuclei have suggested that the MIGs might be directly transported into the nuclei (Leser et al., 1989; Thiry, 1995a,b). However, colocalization of SF2/ASF and RNA pol II (H5) in MIGs of late telophase cells (Figure 3, j–o) clearly shows that various components of MIGs are differentially released for subsequent entry into the nucleus, whereas some factors such as modified forms of SC35 and RNA pol II (H5) still maintain a subpopulation in MIGs until G1. This result raises the possibility that MIGs might be playing important roles either in modification of the components of the splicing machinery before their nuclear entry, or as enriched populations of these factors allowing for protein–protein interactions to occur between subsets of proteins before their nuclear entry.
Our data demonstrating that splicing factors are competent for pre-mRNA splicing immediately upon entry into daughter nuclei supports the possibility that MIGs may be responsible for splicing factor modification, allowing for immediate targeting of modified RNA processing complexes to transcription sites in telophase nuclei. Because daughter nuclei late in telophase have not yet assembled nuclear speckles, cytoplasmic MIGs are likely to function as their counterparts to provide competent pre-mRNA splicing factors to the initial sites of transcription in newly formed nuclei. Perhaps splicing factors are released from MIGs via hyperphosphorylation as has been demonstrated for their release from nuclear speckles in interphase nuclei. Of particular interest in this regard are two kinase families, SR protein specific kinases, including SRPK1, 2 and cdc2-like kinases (Clk/STY 1–4) that have been shown to specifically phosphorylate the RS domains of SR splicing factors (Howell et al., 1991; Colwill et al., 1996a,b; Wang et al., 1998a,b; Yeakley et al., 1999). Although Clk/STY has been shown to modulate the localization of SR proteins in interphase nuclei (Colwill et al., 1996a,b; Sacco-Bubulya and Spector, 2002), SRPK family has been implicated in the reorganization of splicing factors associated with mitosis (Gui et al., 1994a,b). Future studies will address the role of SRPK or Clk/STY family members in the release of splicing factors from MIGs, thereby making them available to enter daughter nuclei.
Our study provides two lines of evidence to support recycling of MIG constituents (splicing factors) during mitosis. First, observation of YFP-tagged splicing factors in living cells demonstrates a decrease in the MIG population of splicing factors concomitant with an increase in the nuclear population of these factors. Second, we demonstrate that there is a low turnover rate of protein constituents of the splicing machinery during mitosis, suggesting recycling of a preexisting population into newly formed daughter nuclei. Based upon these observations, we propose a direct role for MIGs in recycling of splicing factors during mitosis. Our data are consistent with the possibility that proteins and/or protein complexes are released from MIGs for subsequent entry into daughter nuclei. Future studies will examine this possibility in living cells.
Our results demonstrating that the entry of RNA pol II, transcription and pre-mRNA processing factors into postmitotic daughter nuclei is a staged process that occurs after the deposition of nuclear envelope/lamina, suggests the existence of a possible regulatory mechanism, at the level of nuclear import for each class of these factors, as the cells exit mitosis. Facilitated transport of proteins into the nucleus requires the presence of signal motifs, nuclear localization signals (NLSs), which are recognized by specific soluble shuttling receptors of the importin/karyopherin family (Mattaj and Englmeier, 1998; Kuersten et al., 2001). Nuclear import of proteins with a classical basic NLS is mediated by the dimeric complex of importin α/β, of which the importin-α subunit binds to the NLS directly and serves as the adaptor to importin-β (Jans et al., 2000). The importin-β receptor facilitates nuclear translocation by binding to nuclear pore proteins and Ran GTPase in conjunction with other proteins leading to nuclear import. To date, many nonprototypical NLSs have been reported (Nakielny and Dreyfuss, 1999). The well-characterized nonclassical NLS is the M9 sequence of hnRNP A1 (Siomi and Dreyfuss, 1995). M9-dependent import is mediated by transportin 1, which is related to importin-β. Similarly, the majority of the SR proteins are imported into the nuclei with the help of two different importins-β, transportin SR1 (TRN-SR1) and transportin SR2 (TRN-SR2) (Kataoka et al., 1999; Lai et al., 2000). TRN-SR2 mediates the nuclear import of phosphorylated SR proteins (Lai et al., 2001; Allemand et al., 2002), suggesting that differentially modified proteins could be imported by independent mechanisms by different importin receptors (Nakielny and Dreyfuss, 1999). It is tempting to speculate that the differential entry of RNA pol II transcription and pre-mRNA processing factors at the end of mitosis could be the result of regulated interactions with various importin superfamily members. This possibility will be pursued in future studies.
The coupling of the transcription and pre-mRNA processing machinery is clearly an important aspect of accurate gene expression. Our data provides important insight with regard to how the gene expression machinery is recruited into daughter nuclei at the end of mitosis. To our surprise, the gene expression machinery did not enter daughter nuclei in a stochastic manner but instead components entered in a reproducible sequence. It is possible that this sequential recruitment of proteins into daughter nuclei establishes favorable cues for transcription initiation within the context of the decondensing chromosomes. The formidable challenge will be to identify the critical signals that trigger this sequential recruitment.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Spector laboratory for insightful discussions and suggestions and for critical review of the manuscript. We acknowledge D. Bregman, L. Davis, G. Dreyfuss, L. Freedman, A. Krainer, A. Krämer, D. Reinberg, S. Warren, X.-D. Fu, J. Manley, N. Hernandez, and W. van Venrooij for providing antibodies. We thank Jim Duffy for artwork and photography. D.L.S. is supported by National Institutes of Health/National Institute of General Medical Sciences 42694.
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0669. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–10–0669.
REFERENCES
- Adam SA. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- Allemand E, Dokudovskaya S, Bordonne R, Tazi J. A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1. Mol Biol Cell. 2002;13:2436–2447. doi: 10.1091/mbc.E02-02-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Bensaude O, Bonnet F, Casse C, Dubois MF, Nguyen VT, Palancade B. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD) Biochem Cell Biol. 1999;77:249–255. [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, Li Y, Ribisi S, Warren SL. Cytostellin distributes to nuclear regions enriched with splicing factors. J Cell Sci. 1994;107:387–396. doi: 10.1242/jcs.107.3.387. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Carmo-Fonseca M. Localization of poly(A)-binding protein 2 (PABP2) in nuclear speckles is independent of import into the nucleus and requires binding to poly(A) RNA. J Cell Sci. 2000;113:2309–2318. doi: 10.1242/jcs.113.12.2309. [DOI] [PubMed] [Google Scholar]
- Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996a;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996b;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fakan S, Leser G, Martin TE. Ultrastructural distribution of nuclear ribonucleoproteins as visualized by immunocytochemistry on thin sections. J Cell Biol. 1984;98:358–363. doi: 10.1083/jcb.98.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Gebhard M, Ellenberg J, Eils R. Four-dimensional imaging and quantitative reconstruction to analyze complex spatiotemporal processes in live cells. Nat Cell Biol. 2001;3:852–855. doi: 10.1038/ncb0901-852. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994a;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994b;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets WJ, Hoet MH, De Jong BA, Van der Kemp A, Van Venrooij WJ. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci. 2000;113:779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Gebrane-Younes J. Emerging concepts of nucleolar assembly. J Cell Sci. 2002;115:2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Howell BW, Afar DE, Lew J, Douville EM, Icely PL, Gray DA, Bell JC. STY, a tyrosine-phosphorylating enzyme with sequence homology to serine/threonine kinases. Mol Cell Biol. 1991;11:568–572. doi: 10.1128/mcb.11.1.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Building an efficient factory: where is pre-rRNA synthesized in the nucleolus? J Cell Biol. 2002;157:739–741. doi: 10.1083/jcb.200204159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Jiménez-García LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- John S, Workman JL. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998;20:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Johnson LH, Holland JJ. Ribonucleic acid and protein synthesis in mitotic HeLa cells. J Cell Biol. 1965;27:565–574. doi: 10.1083/jcb.27.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Ohno M, Mattaj IW. Nucleocytoplasmic transport. Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-β family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci USA. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- Leser GP, Fakan S, Martin TE. Ultrastructural distribution of ribonucleoprotein complexes during mitosis. snRNP antigens are contained in mitotic granule clusters. Eur J Cell Biol. 1989;50:376–389. [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- Pombo A, Jones E, Iborra FJ, Kimura H, Sugaya K, Cook PR, Jackson DA. Specialized transcription factories within mammalian nuclei. Crit Rev Eukaryot Gene Expr. 2000;10:21–29. [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Reuter R, Appel B, Rinke J, Luhrmann R. Localization and structure of snRNPs during mitosis. Immunofluorescent and biochemical studies. Exp Cell Res. 1985;159:63–79. doi: 10.1016/s0014-4827(85)80038-0. [DOI] [PubMed] [Google Scholar]
- Ryan K, Murthy KG, Kaneko S, Manley JL. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol Cell Biol. 2002;22:1684–1692. doi: 10.1128/MCB.22.6.1684-1692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya P, Spector DL. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J Cell Biol. 2002;156:425–436. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Knecht S, Kramer A. Molecular characterization of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp Cell Res. 1998;238:1–12. doi: 10.1006/excr.1997.3808. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Spector DL, Smith HC. Redistribution of U-snRNPs during mitosis. Exp Cell Res. 1986;163:87–94. doi: 10.1016/0014-4827(86)90560-4. [DOI] [PubMed] [Google Scholar]
- Thiry M. Differential location of nucleic acids within interchromatin granule clusters. Eur J Cell Biol. 1993;62:259–269. [PubMed] [Google Scholar]
- Thiry M. Behavior of interchromatin granules during the cell cycle. Eur J Cell Biol. 1995a;68:14–24. [PubMed] [Google Scholar]
- Thiry M. The interchromatin granules. Histol Histopathol. 1995b;10:1035–1045. [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Verheijen R, Kuijpers H, Vooijs P, van Venrooij W, Ramaekers F. Protein composition of nuclear matrix preparations from HeLa cells: an immunochemical approach. J Cell Sci. 1986;80:103–122. doi: 10.1242/jcs.80.1.103. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998a;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xiao SH, Manley JL. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998b;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Tronchere H, Olesen J, Dyck JA, Wang HY, Fu XD. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.