Abstract
hic-5 was originally isolated as an H2O2-inducible cDNA clone whose product was normally found at focal adhesions. In this study, we found that Hic-5 accumulated in the nucleus in response to oxidants such as H2O2. Other focal adhesion proteins including paxillin, the most homologous to Hic-5, remained in the cytoplasm. Mutation analyses revealed that the C- and N-terminal halves of Hic-5 contributed to its nuclear localization in a positive and negative manner, respectively. After the finding that leptomycin B (LMB), an inhibitor of nuclear export signal (NES), caused Hic-5 to be retained in the nucleus, Hic-5 was demonstrated to harbor NES in the N-terminal, which was sensitive to oxidants, thereby regulating the nuclear accumulation of Hic-5. NES consisted of a leucine-rich stretch and two cysteines with a limited similarity to Yap/Pap-type NES. In the nucleus, Hic-5 was suggested to participate in the gene expression of c-fos. Using dominant negative mutants, we found that Hic-5 was actually involved in endogenous c-fos gene expression upon H2O2 treatment. Hic-5 was thus proposed as a focal adhesion protein with the novel aspect of shuttling between focal adhesions and the nucleus through an oxidant-sensitive NES, mediating the redox signaling directly to the nucleus.
INTRODUCTION
The hic (hydrogen peroxide-inducible clone)-5 gene was originally isolated as one of the TGFβ1- and H2O2-inducible cDNA clones from mouse osteoblastic cells in an effort to understand the biological relevance of the finding that TGFβ1-stimulated cells to release H2O2 (Ohba et al., 1994; Shibanuma et al., 1994). Since its discovery, its biological functions have been studied, and its ectopic expression has been found to affect cellular growth and differentiation; in immortalized human fibroblasts, it suppressed cellular growth and induced a morphological change similar to that of senescent cells (Shibanuma et al., 1997). Rat osteoblast clones overexpressing Hic-5 exhibited a slowed rate of growth and more differentiated phenotypes in response to retinoic acid (Shibanuma and Nose, 1998). A Drosophila ortholog of Hic-5 was proposed to be a switch to block the differentiation process and to induce cell death in muscle development (Hu et al., 1999), whereas we found that mammalian Hic-5 could be a positive regulator of myogenic differentiation of an early stage (Shibanuma et al., 2002). However, the precise mechanisms underlying these biological effects of Hic-5 as well as their relation to H2O2 and TGFβ1 signals remained undefined.
hic-5 encodes a LIM protein that is most homologous to the focal adhesion protein, paxillin (reviewed by Turner, 2000). Together with two other newly identified LIM proteins, leupaxin and paxB (Lipsky et al., 1998), paxillin and Hic-5 are suggested to constitute a new family of LIM proteins. In addition to having four contiguous LIM domains in their C-terminal half, the members have common LD motifs in the N-terminal half (Brown et al., 1998). Both the LD motifs and LIM domains potentially serve as an interface for protein–protein interactions (Dawid et al., 1995). The members of this family are localized at specialized structures at the cell surface, so called focal adhesion sites or complexes in fibroblasts. These structures not only mediate the physical link between the extracellular matrix (ECM) and intracellular actin cytoskeleton but also coordinate and transmit the information from the cell surface to the nucleus (reviewed e.g., by Giancotti and Ruoslahti, 1999). Paxillin has been demonstrated to interact with a host of signaling molecules at focal adhesion sites and is proposed to function as an adapter under integrin (reviewed by Turner, 2000). As predicted from the extensive similarity in structural features, Hic-5 shares some of the interacting proteins such as FAK/FRNK, PYK2, Csk, vinculin, and PTP-PEST with paxillin (Fujita et al., 1998; Matsuya et al., 1998; Nishiya et al., 1999; Thomas et al., 1999). Like paxillin, Hic-5, thus, has been expected to serve as an adapter protein at focal adhesions either competitively or complementarily to paxillin (Fujita et al., 1998; Nishiya et al., 2001).
In a previous study, we occasionally observed that Hic-5, although only in small amounts, was localized in the nucleus of certain cells (Shibanuma et al., 1997). Recently, we proposed that Hic-5 accumulated in the nucleus under oxidative stress in mouse osteoblastic cells (Shibanuma et al., 2001), inconsistent with the idea that Hic-5 functions only as an adapter at focal adhesions. Identification of Hic-5 as a coactivator of steroid receptors also challenged the previous notion (Fujimoto et al., 1999; Yang et al., 2000). Besides, Yang et al. (2000) reported that a fraction of Hic-5 was associated with the nuclear matrix. These apparently incompatible findings tempted us to hypothesize that Hic-5 could be distributed both at focal adhesions and in the nucleus depending on cellular conditions. Recently, members of another LIM protein family, zyxin, LPP, and Trip6, have been demonstrated to shuttle between focal adhesions and the nucleus in a Crm1/exportin-dependent manner (Nix and Beckerle, 1997; Petit et al., 2000; Nix et al., 2001; Wang and Gilmore, 2001). Most recently, paxillin was added to the list (Woods et al., 2002). In zyxin, LPP and Trip6, nuclear export signal (NES)s have been identified which were leucine-rich and conserved. These findings led us to the idea that members of these LIM protein families including Hic-5 were localized both at focal adhesions and the nucleus with dual functions, thereby integrating signals from outside of cells into nuclear activity directly.
In the present study, we attempted to substantiate the finding that Hic-5 is accumulated in the nucleus in response to oxidants such as H2O2 and clarify the molecular mechanisms underlying the nuclear accumulation. Furthermore, we addressed the pertinent function of Hic-5 in the nucleus.
MATERIALS AND METHODS
Cell Culture and Chemicals
Mouse MC3T3 osteoblastic cells, C3H10T1/2 fibroblastic cells, and human TIG-7 normal diploid fibroblasts were grown in Dulbeco's modified MEM (MC3T3 and TIG-7) and MEM (C3H10T1/2) supplemented with 10% fetal calf serum, respectively, as reported previously (Ohba et al., 1994). C2C12 mouse myoblastic cells were maintained as described previously (Nishiya et al., 1999). LMB was kindly provided by Dr. M. Yoshida (Tokyo University). Diethyl maleate was obtained from Aldrich Chemical Company, Inc. (Milwaukee, WI). TGFβ1 was obtained from Genzyme/Techne (Minneapolis, MN). Other reagents used in this study were purchased from Sigma Chemical Co. (St. Louis, MO).
The MEF#43/Tet-Off/LD1 mhic-5 cell line was established from the MEF/3T3 Tet-Off cell line by stably introducing pTRE-LD1 mhic-5, a pTRE-based tetracycline-responsive expression plasmid for HA-tagged LD1mhic, which was constructed by integrating the LD1 mhic-5 cDNA (Nishiya et al., 2001) into the cloning site of pTRE. This cell line was maintained in MEM supplemented with 10% fetal calf serum and 2 ng/ml doxycycline. Both pTRE and the Tet-Off cell line were purchased from Clontech Laboratories, Inc. (Palo Alto, CA), and the above procedure was carried out according to the manufacturer's manual. Immortalized mouse fibroblasts including the parental MEF/3T3 Tet-Off cell line showed lower expression levels of Hic-5 than normal and mortal mouse fibroblasts (Ishino et al., 2000). In MEF#43/Tet-Off/LD1 mhic-5, HA-tagged Hic-5 was induced and the total amount of Hic-5 protein reached almost the same level as in normal and mortal mouse fibroblasts 24 h after the removal of doxycycline (−Tet) (our unpublished results) and used for the experiments.
Construction of Expression Plasmids
Mammalian expression plasmids, pCG-LD1 mhic-5, -hhic-5, and pcDNA3.1A-hhic-5, for HA- or Myc-tagged wild-type mouse (m) or human (h) Hic-5, respectively, or pCG-pax for paxillin (HA-tagged) were as previously described (Nishiya et al., 2001). pCG-mhic-5 was the alternative form of pCG-LD1 mhic-5, including the insert of the PCR-amplified mouse original, the LD1-null form of hic-5 cDNA (Shibanuma et al., 1994; designated as mhic-5). Flag-mhic-5/pcDNA3 was generated enzymatically by excising the mhic-5 cDNA fragments from the pCG series and inserting into the cloning vector of pcDNA3 (Invitrogen, San Diego, CA) including Flag tag.
To construct expression plasmids for the chimeric proteins, pCG-hic/pax and pCG-pax/hic, an amino acid portion (228–461) of the human hic-5 cDNA fragment in pCG-hhic-5 was substituted with residues 324–557 of paxillin in pCG-hic/pax, and 1–227 of Hic-5 with 1–322 of paxillin in pCG-pax/hic, respectively. In both cases, the cDNA fragments for paxillin were generated by PCR. The 5′ ScaI or 3′ StuI sites incorporated at the ends of the primers for PCR were used to join each paxillin fragment to the Hic-5 fragment in-frame at the StuI site of Hic-5 codon 226 to yield pCG-hic/pax or pCG-pax/hic, respectively.
pCG-paxLIM was for the LIM-only domain of paxillin and was constructed by PCR amplification of the sequence encoding amino acids 307–557 of paxillin. For the expression plasmid of the LIM-only domain of Hic-5, the human hic-5 cDNA fragment encoding amino acids 220–461 was amplified. A Kozak consensus ATG start codon was incorporated into the 5′ primer immediately in front of codon 220. The insert was subcloned into the vectors, pcDNA3.1(−)/Myc-HisA (Invitrogen) and pCG-N-BL (Hirai et al., 1994), to construct pcDNA3.1A-hhicLIM and pCG-hhicLIM, respectively.
pCG-LD1 mhic/mL1,/mL2,/mL3 were created with restriction enzymes by replacing the wild-type portion on pCG-LD1 mhic with the corresponding portion containing the LIM disrupted by site-directed mutagenesis as described before (Nishiya et al., 1999).
A set of C-terminal deletion mutants of LD1 mhic-5 was created on the basis of pCG-LD1 mhic-5. The regions that involved the selected LIM domains were deleted using a pair of restriction enzymes followed by blunting when necessary. The deleted regions and the flanking enzymes used were as follows; pCG-LD1 mhic/delL4 and/ml3-delL4, 5′ PmaCI to 3′ EcoRV (from amino acids 408–461);/delL1,2, 5′ PstI to 3′ PstI (212 to339);/delL2,3,4, 5′ BstP1–3′ EcoRV (301–461).
A set of N-terminal deletion mutants of hhic-5 was created on the basis of pCG-hhic-5. pCG-delLD3–4hhic-5 and pCG-delLD3hhic-5 were described previously (Nishiya et al., 2001). pCG-delLD1–2hhic-5 was constructed by inserting the PCR-amplified cDNA fragment containing the coding region of human hic-5 (146–461) with the first additional methionine into pCG-N-BL vector.
The site-directed mutagenesis for the NES functional analysis was carried out on pKF 18k (Takara Shuzoh Co., Kyoto, Japan) with Takara's Mutan-Super Express Km Kit according to the manufacturer's instructions. The amino acid substitutions in each mutant, which were based on pCG-mhic-5, were as follows: pCG-mLD2 mhic-5; L73A/L76A, mLD3; L144A, mCf/N; C47N, mCl/S; C74S, mCfl/NS; C47N and C74S. The mutant of LD4 was described elsewhere (Shibanuma et al., 2001). The mutated fragments in pKF 18k were transferred to pCG-mhic-5 by restriction enzymes.
The fusion protein for the analysis of NES activity was expressed from a plasmid constructed as follows: parts of the coding sequence, LD3; amino acids 131–154, CC-LD3; 41–154 of mhic-5, were inserted into pCG-paxLIM, so as to be expressed as (LD3)LIM and (CC-LD3)LIM in which the LD3 and CC-LD3 portions were placed at the N-terminal of the LIM domains of paxillin. (Hic(N))LIM was equivalent to the chimeric protein expressed from pCG-hic/pax described above.
The nuclear targeted version of the proteins used as effectors in the luciferase assay was engineered by inserting a PCR-amplified nuclear localization signal (NLS) from SV40 large T antigen into the plasmids to express the fusion proteins carrying NLS at their N-terminals.
All PCR amplifications were carried out with pfu grade polymerase, and the products including those with the mutations introduced were verified by DNA sequencing.
Immunocytochemistry, Immunoblotting, and Antibodies
The expression plasmids were introduced into cells using a conventional calcium phosphate precipitation method. In the case of TIG-7, TransIT-LT1 reagent purchased from PanVera (Madison, WI) was used. After being incubated in complete medium for 24 h, the cells were processed for immunocytochemistry or lysed for immunoblotting.
Immunofluorescence labeling and immunoblotting were performed as described previously (Ishino et al., 2000). Fluorescence microscopy was carried out using an Axioskope microscope (Zeiss, Tokyo, Japan) equipped with a high speed cooled digital CCD camera fluorescence imaging system (Argus HiSCA, Hamamatsu Photonics, Hamamatsu, Japan) or a confocal system composed of a multi-pinhole confocal scanner (CSU10Z, Yokogawa Electric, Tokyo, Japan).
The anti-mouse Hic-5 polyclonal antibody (number 1024) and the monoclonal anti-HA antibody (12CA5) were as described previously (Nishiya et al., 1999; Ishino et al., 2000). The monoclonal antivinculin antibody was purchased from Sigma Chemical Co. (St. Louis, MO) and antimyc tag antibodies from Invitrogen, respectively.
Luciferase Assay
The transient transfection of plasmids was performed by a conventional calcium phosphate precipitation method. The cells used here were C2C12 cells in which the nuclear localization of endogenous Hic-5 was suppressed for some unknown reason (unpublished data). For construction of the c-fos luciferase reporter, a 5′ upstream fragment of human c-fos (−2.2 kb) including the TATA and Cap site was subcloned into the pGL3 basic luciferase reporter plasmid (Promega Corporation, Madison, WI). The transfected plasmid mixture included 1 μg each of the reporter and effector plasmids together with 0.02 μg of an internal control plasmid, pRL/CMV (Promega Corporation) per assay. Total DNA was kept constant by addition of an empty vector. Luciferase activities were quantified 24 h after the transfection using a Dual Luciferase Assay kit (Promega). Each assay was done in duplicate and repeated at least three times, and values were normalized with the renilla luciferase activity expressed from pRL/CMV.
RESULTS
Nuclear Accumulation of Hic-5 in Response to Oxidants
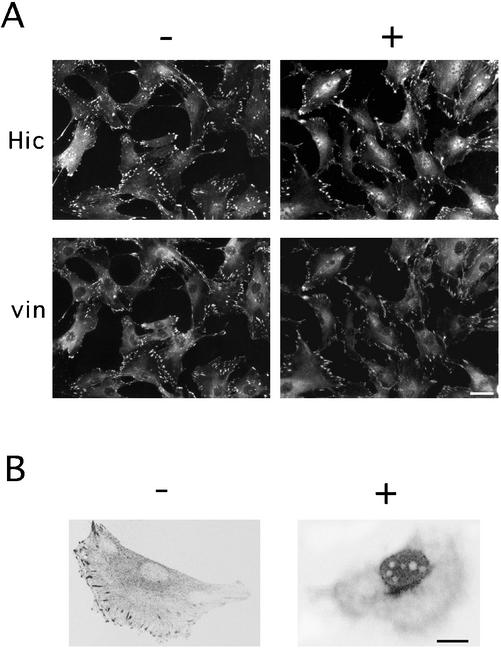
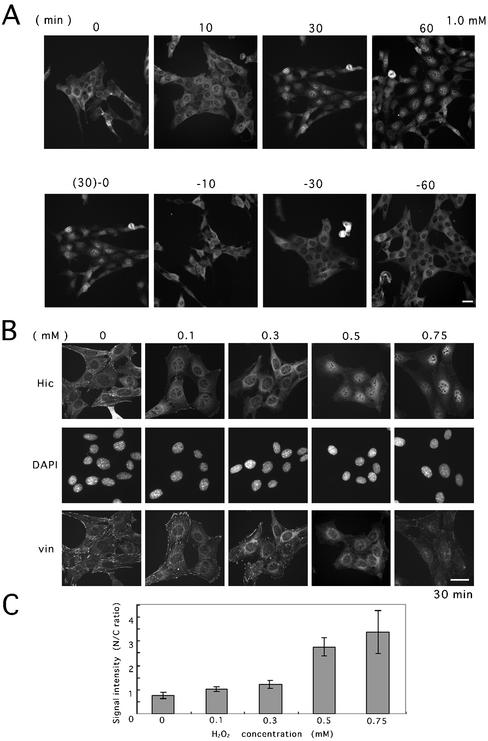
Among a variety of agents including growth factors (serum, TGFβ1, TNF-α, PDGF), protein kinase and phosphatase inhibitors, and genotoxic substances (bleomycin, adriamycin), a set of chemicals including H2O2 that modify the cellular redox state induced a marked change in the subcellular localization of Hic-5 in several cell lines such as murine fibroblasts; a significant fraction of Hic-5 was localized in the nucleus by H2O2 with the rest remaining at the focal adhesions 60 min after the treatment (Figure 1A, Hic-5, +). Although a faint signal was detected in the nucleus even under the unstimulated condition, depending on the cell lines as mentioned above and in our previous work (Shibanuma et al., 1997), the intensity was significantly increased by H2O2. In contrast to Hic-5, a focal adhesion marker, vinculin, was retained at the focal adhesions or in the cytoplasm, being apparently excluded from the nucleus (Figure 1A, vin, +). The localization of Hic-5 within the nucleus was proved by confocal laser scanning microscopy; as shown in Figure 1B, +, an optical section passing through the center of the nucleus clearly showed Hic-5 present within the nucleus except the nucleoli after the treatment. The nuclear localization was discerned from 10 min and clearly evident 30 min after the treatment as shown in the upper row of Figure 2A. Of note, upon removal of H2O2, the signal intensity promptly shifted from the nucleus to the cytoplasm and relocalized to the cytoplasm within 30 min as shown in the lower row of Figure 2A. On the contrary, Hic-5 stayed in the nucleus as long as H2O2 was present in the medium. The pretreatment with cycloheximide did not inhibit the nuclear localization induced by H2O2 (our unpublished results), suggesting that Hic-5 localized at focal adhesions was redistributed into the nucleus. In Figure 2B, the nuclear accumulation observed at several H2O2 doses is shown. Above 0.5 mM of H2O2, most of the signal was detected in the nucleus, suggesting that considerable amount of Hic-5 was localized in the nucleus. Through image analysis, the increase in the signal intensity in the nucleus was quantitatively shown in Figure 2C. At higher doses, the signal of vinculin became faint and diffusely distributed in the cytoplasm surrounding the nucleus, suggesting that the focal adhesion structure was disturbed to some extent. However, the disruption of the focal adhesion structure was considered to play only a minor role, if any, in inducing the nuclear localization of Hic-5 as noted below. The survival rate of the cells was retained above 80% for 15 h under these conditions (our unpublished results).
Figure 1.
Nuclear accumulation of endogenous Hic-5 in the presence of H2O2. C3H10T1/2 (A) and MC3T3 (B) cells were treated with (+) or without (−) 1.5 mM (A) or 0.5 mM (B) H2O2 for 60 min. After fixation, the cells were processed for coimmunostaining with the antibody to Hic-5 and vinculin. The localization of the proteins was examined using a microscope (A) or a laser scanning confocal microscope (B). Bars, 20 (A) and 10 (B) μm.
Figure 2.
Time- and dose-dependent nuclear accumulation of Hic-5 in response to H2O2. (A) MEF#43/Tet-Off/LD1 mhic-5 induced to express the HA-tagged Hic-5 by removal of doxycycline (−Tet) was exposed to H2O2 (1.0 mM) for the period indicated (upper row). In the lower row, after 30 min exposure to H2O2, the medium was replaced with the conditioned medium without H2O2, and the cells were further incubated for 0, 10, 30, and 60 min. The cells were then fixed and immunolabeled with the antibody to HA tag. (B) Cells were exposed to H2O2 at the indicated doses for 30 min, and then coimmunstained with the antibody to HA tag (Hic) and vinculin (vin). DNA was stained with DAPI for localizing the nuclei. Bars, 20 μm. (C) The extent of the nuclear localization was evaluated on the image of (B, Hic) by measuring the cytoplasmic and nuclear signal intensity of individual cells with AquaCosmos image acquisition and analysis system (Hamamatsu Photonics; K. K. Hamamatsu, Japan). The values were mean ± SD obtained from at least six cells.
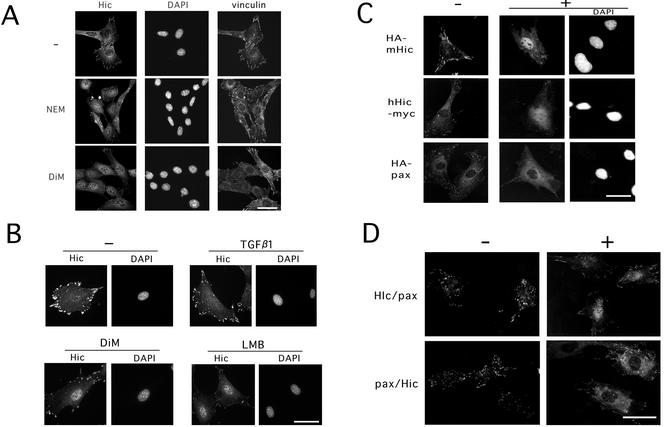
The oxidants, N-ethyl maleimide and diethyl maleate (DiM), both of which are known as sulfhydryl reactive reagents were also tested as to whether they affected the localization of Hic-5 and vinculin. Both agents appeared as effective as H2O2 in inducing the nuclear accumulation of Hic-5, but almost unaffected the localization of vinculin at focal adhesions (Figure 3A). Therefore, disruption of the focal adhesions was not necessarily required for the nuclear localization of Hic-5. In the previous work, we observed that Hic-5 mutated in LIM3 did not accumulate in the nucleus in spite of the release from focal adhesions (Nishiya et al., 1999). On the basis of these observations, we argued against the significance of collapse of focal adhesions in the induction of the nuclear localization. Neither heat nor osmotic stress significantly affected the localization of Hic-5 (unpublished data). These observations suggested that the relocalization of Hic-5 from focal adhesions or cytoplasm to the nucleus was not merely a consequence of cellular damage but was a specific response to the oxidative state in cells.
Figure 3.
Nuclear accumulation of Hic-5 induced by sulfhydryl reagents and subcellular localization of Hic-5, paxillin, and their chimeric proteins in the presence or absence of H2O2. (A) MEF#43/Tet-Off/LD1 mhic-5 (−Tet) was exposed to 20 μM N-ethylmaleimide (NEM) or 2.0 mM diethyl maleate (DiM) for 30 min and then processed as in Figure 2. (B) MC3T3 cells were untreated (−) or treated with TGFβ1 (5 ng/ml for 90 min) as described previously (Ohba et al., 1994), DiM (2.0 mM for 90 min), and LMB (10 ng/ml for 6h), and processed for immunostaining with the antibody to Hic-5 as in Figure 1. (C) C3H10T1/2 cells were transfected with the plasmids, pCG-mhic-5, pcDNA3.1A-hhic-5 and pCG-pax, for expressing the indicated tagged-proteins, HA-mHic-5, hHic-5-myc, and HA-pax, respectively. After 24 h, the cells were treated with (+) or without (−) 1.5 mM H2O2 for 60 min, and then the expressed proteins were visualized with the antibodies to the tags. The nuclei were located by staining the DNA with DAPI (DAPI). (D) The expression plasmids for the chimeric proteins, pCG-hic/pax (Hic/pax) and pCG-pax/hic (pax/Hic), were introduced into C3H10T1/2 cells, and immunostaining with the antibody to HA tag was performed as in B. Bars, 20 μm.
Previously, we found that TGFβ1 stimulated MC3T3 osteoblastic cells to accumulate H2O2 within the cells with a peak at 90 min after the treatment (Ohba et al., 1994). Therefore, we tested as to whether TGFβ1 induced the nuclear localization of Hic-5. The Hic-5 localization was not changed noticeably by TGFβ1 from 90 min to 6 h after the treatment, whereas DiM and LMB (see below) clearly induced the nuclear localization of Hic-5 in this cell line as well as other cell lines (Figure 3B).
The nuclear localization was further confirmed with tagged proteins expressed exogenously in the cells, and the Hic-5 proteins were mostly detected in the nucleus, like endogenous Hic-5, by the antibody to the tag after the treatment with H2O2 (Figure 3C). This behavior was conserved between human and mouse Hic-5 (Figure 3C, hHic and mHic) or between two alternative forms with or without the LD1 domain (our unpublished results) and was observed in the cells of mouse osteoblastic MC3T3 (Figure 1B and as in previous reports), MEF embryonic fibroblasts (Figures 2 and 3) and human fibroblastic TIG-7 (Figure 9A) as well as mouse fibroblastic C3H10T1/2 (Figures 1A and 3C). No processing of the protein was predicted to accompany the nuclear entry, because the nuclear localization was similarly visualized with the antibodies to the tags in HA-mHic and hHic-myc, whose tags were placed at the N- and C-terminal ends of the proteins, respectively. In contrast to Hic-5, however, paxillin was detected in the cytoplasm, being excluded from the nucleus, under the same conditions (Figure 3C, HA-pax). Even at higher doses of H2O2, no sign of the nuclear localization of paxillin was observed. Endogenous paxillin apparently behaved similar to HA-paxillin (our unpublished results), whereas an antibody against paxillin showed a cross-reactivity to Hic-5 to some extent, and the result was not so unambiguous as that of HA-paxillin.
Figure 9.
Alignments of the NES sequences. (A) The NESs with cysteine residues were aligned. (B) (C) The NESs found among the LIM protein family were aligned and compared with NES of Rev. The consensus proposed as the Rev-type NES is shown at the bottom. The amino acids emphasized by open letters and filled boxes were those whose importance was demonstrated experimentally. The large group hydrophobic amino acids are shaded.
Distinctive Contributions of C- and N-Terminal Halves to Regulation of the Nuclear Localization of Hic-5
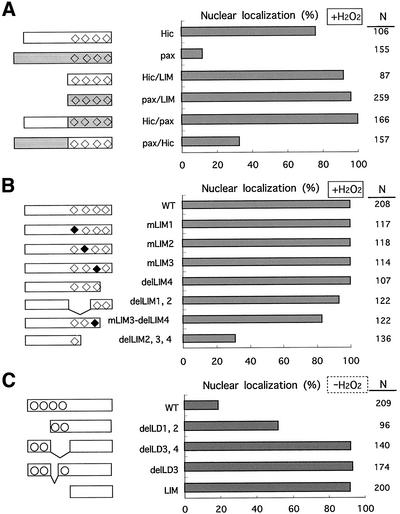
As mentioned above, the C- and N-terminal halves of Hic-5 and paxillin exhibit the unique structural features; most of the C-terminal half is composed of four LIM domains, the N-terminal comprising four (Hic-5) or five (paxillin) LD motifs along with proline-rich regions. To determine the role of each half of the protein in the regulation of nuclear localization, we constructed a series of plasmids expressing HA-tagged variants of Hic-5, including those expressing LIM-only regions or chimeric proteins in which the N- and C-terminal halves of Hic-5 and paxillin were mutually interchanged. The variants studied here are schematically shown in the left side of Figure 4. After transfection of the plasmids into the cells, the expressed proteins were visualized with the antibody to HA tag as in Figure 3C, and their nuclear localization was quantitatively examined in the presence (Figure 4, A and B) or absence (Figure 4C) of H2O2. The subcellular localization of the chimeric proteins under unstimulated conditions was at focal adhesions as were wild-types of the parental proteins (Figure 3D, −). The following possibilities were deduced from this series of experiments: First, the LIM region of Hic-5 and paxillin intrinsically carried the capacity to localize in the nucleus as well as that of other proteins (Thomas et al., 1999; Petit et al., 2000; Wang and Gilmore, 2001), and their nuclear localization was unaffected by H2O2 (Figure 4C, LIM; Figure 4A, Hic/LIM and pax/LIM; and Figure 6C, LIM, HA). Their residual distribution to focal adhesions was possibly due to the focal adhesion targeting capacity identified previously in LIM 3 (Nishiya et al., 1999). Second, under normal conditions, the N-terminal halves of both proteins negatively regulated their nuclear localization; in contrast to the LIM-only proteins, the full-length proteins were excluded from the nucleus and mostly recruited to focal adhesions as hitherto described (Figures 3C, −, and 4C, WT vs. LIM). Finally, the N-terminal half of Hic-5 lost its negative effect on the nuclear localization in response to H2O2 but not that of paxillin, resulting in a distinctive localization of the two proteins under the treatment; when exposed to H2O2, the N-terminal half of Hic-5 turned permissive and as a consequence, wild-type Hic-5 and Hic/pax, whose N-terminal halves were derived from Hic-5, were detected in the nucleus of ∼80% of the cells expressing them (Figures 4A and 3D, +). In contrast, the N-terminal of paxillin was consistently nonpermissive; wild-type paxillin and pax/Hic whose N-terminals were derived from paxillin were excluded from the nucleus in the presence and absence of H2O2 (Figures 4A and 3D, +). In summary, the nuclear localization of Hic-5 was speculated to be controlled primarily by both the driving force to the nucleus, which was provided by the LIM region, and the capacity to sequester a protein from the nucleus, which was displayed by the N-terminal region and undermined by the oxidants. Because some of the mutants studied here suffered the rigorous deletions, more detailed studies were performed as below.
Figure 4.
The nuclear localization of Hic-5 variants either in the presence or absence of H2O2. (A) Wild-type, LIM-only regions or the chimeric proteins of Hic-5 and paxillin were expressed from the expression plasmids, pcDNA3.1A-hhic-5 (Hic), pCG-pax (pax), pcDNA3.1A-hhicLIM (Hic/LIM), pCG-paxLIM (pax/LIM), pCG-hic/pax (Hic/pax), and pCG-pax/hic (pax/Hic), respectively, in C3H10T1/2 cells. Their localization was examined by immunostaining with antibodies to the tags as in Figure 5 after treatment with 1.0 mM H2O2 for 60 min. The nuclear localization was scored as positive and shown as a percentage when the signal in the nucleus was more intense than that in the cytoplasm or the nucleus and the cytoplasm were evenly labeled. N was the total number of cells scored in three independent transfections. (B) A series of LIM mutants of Hic-5 was expressed, and their localization was examined as in A. The plasmids introduced into the cells were pCG-LD1 mhic (WT),/mL1 (mLIM1),/mL2 (mLIM2),/mL3 (mLIM3),/delL4 (delLIM4),/delL1,2 (delLIM1,2),/ml3-delL4 (mLIM3-delLIM4), and/delL2,3,4 (delLIM2,3,4). The squares indicate LIM domains; open squares for the wild-type and closed squares for mutated versions. (C) The N-terminal deletion mutants of Hic-5 were expressed, and their nuclear localization was examined as in A and B but without the treatment with H2O2. The expression plasmids used here were pCG-hhic-5 (WT), pCG-delLD1–2hhic-5 (delLD1,2), pCG-delLD3–4hhic-5 (delLD3,4), pCG-delLD3hhic-5 (delLD3), and pCG-hhicLIM (LIM). The open circles represent a LD motif.
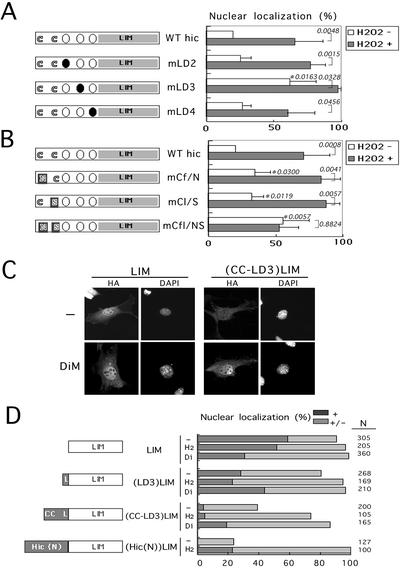
Figure 6.
Identification and characterization of oxidant-sensitive NES containing leucine and cysteine residues in the N-terminal region of Hic-5. (A and B) The variants of mouse LD1-null form of Hic-5 in which leucine or cysteine residues were substituted as shown in Figure 5B were expressed in C3H10T1/2 cells from pCG-mhic-5 (WT hic) and its derivatives (mLD2, mLD3, mLD4, mCf/N, mCl/S, and mCfl/NS), and then visualized as in Figure 3C after treatment with (H2O2+) or without (H2O2 −) 1.0 mM H2O2 for 60 min. The nuclear localization of the variant proteins was evaluated as in Figure 4 and graphed (mean ± SD derived from a series of experiments repeated more than five times, in each of which around one hundred cells were examined). The significance of the differences was assessed by t test. P values with no mark were for each treatment and those marked with asterisks were for each mutation. The cartoon outlined the protein structure examined, in which open and closed circles indicate wild-type and mutant LD motifs, and the substitutions of the cysteines are also indicated. (C) The LIM and (CC-LD3)LIM were expressed from pCG-paxLIM and its derivatives (MATERIALS AND METHODS), and the cells were exposed to 2.0 mM DiM for 60 min. The control (−) and DiM-exposed cells (DiM) were then immunolabeled with the antibody to HA tag (HA). The nuclei were located by staining the DNA with DAPI (DAPI). Bar, 20 μm. (D) The fusion proteins as illustrated were expressed from respective expression plasmids based on pCG-paxLIM (MATERIALS AND METHODS), and then visualized as in C under untreated conditions (−), or after exposure to 1.0 mM H2O2 (H2) or 2.0 mM DiM (Di) for 60 min. The nuclear localization was scored as nuclear (+) and nuclear and cytoplasmic (+/−). LIM; LIM region of paxillin, L or LD3; amino acids 131–154 of mouse LD1-null Hic including the LD3 motif, CC-LD3; residues 41–154 of the Hic-5 including the two cysteines and the LD3 motif, Hic-5(N); 1–227 of human Hic-5.
Nuclear Targeting Capacity of LIM Domains of Hic-5
We here tried to characterize the nuclear targeting capacity of Hic-5 found in its C-terminal region by disrupting the four LIM domains individually or in combinations, and effects on the Hic-5 nuclear localization were examined in the presence of H2O2. No obvious nuclear localization signal (NLS) was found in the region. Individual mutations of each of the LIM domains did not interfere with the nuclear localization, whereas simultaneous removal of two or more LIMs severely impeded it (Figure 4B). None of the four LIM domains were thus critical for the nuclear targeting function of the C-terminal region; rather it was most likely that all four domains cooperated as an unconventional NLS. This type of NLS was previously reported in zyxin and Trip6, both of which are LIM proteins related to Hic-5; similar to Hic-5, they had no classical NLS and their nuclear targeting capacity was found in the plural LIM domains in their C-terminals (Nix et al., 2001; Wang and Gilmore, 2001).
NES Module Identified in the N-Terminal Half of Hic-5
In light of the negative role of the N-terminal half of Hic-5 in controling the nuclear localization of the protein, the four LD motifs (three in the alternative form) were noteworthy, because their sequences resembled the Rev-type or leucine-rich nuclear export signal (NES) found in an increasing number of proteins including aforementioned Hic-5-related zyxin, LPP and Tirp6 (Figure 5B; see also Figure 9B; Nix and Beckerle, 1997; Petit et al., 2000; Wang and Gilmore, 2001). Then, we first examined the effect of LMB, a specific inhibitor of NES function, on the subcellular localization of Hic-5 in murine fibroblasts and found that it caused a marked accumulation of Hic-5 in the nucleus within 2 h (Figure 5A). A similar result was obtained in mouse osteoblastic cells previously (Shibanuma et al., 2001). Theses results suggested the presence of the NES in Hic-5. Among a series of deletions of the N-terminal region of Hic-5, the deletion of 24 amino acids from 131 to 154 including the LD3 motif in mutants such as del-LD3 and del-LD3, 4, significantly promoted the nuclear localization of the proteins to almost the same extent as the LIM-only protein, implying that the LD3 motif is an essential region for the NES (Figure 4C). The deletion of the LD1 and 2 motifs (del-LD1, 2) increased the nuclear localization to some extent, suggesting another NES or negative regulatory activity on nuclear entry around the LD1 or 2 motifs, although less significant than that around the LD3 motif.
Figure 5.
Nuclear accumulation induced by LMB and the sequences of LD motifs as the assumptive NES of Hic-5. (A) MEF#43/Tet-Off/LD1 mhic-5 (−Tet) was exposed to LMB (10 ng/ml) for 2 h, and then processed as in Figure 2. Bar, 20 μm. (B) The amino acid sequences of the LD motifs (LD2, 3, 4) of mouse and human Hic-5 are shown along with the corresponding LD of paxillin. The sequences around LD2 are also shown, extended to contain the two cysteines marked by asterisks. The leucine residues in each LD motif are overlined. The sequences under the wild-type were those of the mutants in which leucine or cysteine residues were replaced as indicated.
We focused in a further study on the characterization of the assumptive NES around the LD3 motif at the amino acid level in the naturally occurring LD1-null form of Hic-5 (Mashimo et al., 2000), because the involvement of LD1 in the NES activity was less likely as mentioned below. A growing body of evidence has established the importance of leucine residues in the NES (reviewed by Nigg, 1997). Accordingly, we introduced point mutations into the motifs, LD2, 3, and 4, of Hic-5 as illustrated in Figure 5B, disrupting the assumptive NES. The mutants were expressed in the cells, and the nuclear localization in the presence or absence of H2O2 was quantitatively evaluated as above (Figure 6A). The disruption of the LD3 motif by the substitution of the leucine residue with an alanine resulted in an increase in the nuclear localization of Hic-5 from 22 to 69% in the absence of H2O2 (Figure 6A, hic vs. mLD3; H2O2 −), suggesting that the NES activity is carried by the LD3 motif or its leucine residue. Unlike for mLD3, similar mutations of the LD2 (mLD2 and mCl/S-mLD2) and LD4 motifs (mLD4) did not affect the subcellular distribution of the Hic-5 protein, displaying essentially the same behavior as the wild-type irrespective of the treatment with H2O2 (Figure 6A). Then, the oxidants might disable the LD3 motif from exerting its function, thereby inducing the nuclear accumulation of Hic-5. In mLD3, however, its nuclear localization was further augmented by H2O2 to the maximum level of 100% (Figure 6A, mLD3 H2O2 − vs. H2O2 +), suggesting another element outside the LD3 motif sensitizing the nuclear-cytoplasmic transport and/or anchoring system of Hic-5 to oxidants.
Woods et al. (2002) recently reported the nuclear accumulation of paxillin induced by LMB, suggesting the presence of the NES in paxillin, whose LD motifs are closely homologous to those of Hic-5. However, unlike Hic-5, paxillin did not accumulate in the nucleus on treatment with H2O2 (Figures 3C and 4A). Concerning the difference in amino acids between the two proteins, we noticed that the two cysteine residues around the LD2 motif of Hic-5 were replaced by asparagine and serine at the corresponding positions in paxillin (Figure 5B). We thus examined the effect of the mutations, converting these cysteine residues into asparagine and serine, respectively, as shown in Figure 5B, and found that the mutations promoted the nuclear localization of Hic-5 from 22 to 62% under normal conditions (Figure 6B, hic vs. mCfl/NS, H2O2 −), for which effect each mutation was insufficient (Figure 6B, mCf/N and mCl/S, H2O2 −), suggesting a redundant function of the two cysteines. Importantly, compared with mLD3, the responsiveness to H2O2 was completely lost in mCfl/NS (Figure 6B, mCfl/NS H2O2 − vs. H2O2 +). In conclusion, the mutation analysis highlighted the two elements involved in the negative control of the nuclear localization of Hic-5; one was the leucine around the LD3 motif, and the other was the two cysteine residues around the LD2. It was also noted that the enhancement of nuclear localization observed by mutations of these elements was comparable to that induced by oxidants in the wild-type (Figure 6, A and B), suggesting that a majority of the oxidative-sensitive NES activity of Hic-5 was directed by these two elements. In conclusion, based on the results of the deletion and mutation analysis, we hypothesized that the NES module of Hic-5 consisted of the LD3 motif and the cysteines playing an important role particularly in sensing the cellular redox state. The role of the LD3 motif in sensing redox was ambiguous at this stage, because H2O2 enhanced the nuclear localization of mLD3 up to the maximum level as described above, thereby covering a possible further increase.
To characterize the above NES module further and substantiate its NES activity, we fused the regions of the hypothesized module with the heterologous protein of the C-terminal LIM region of paxillin. The region designated as CC-L3 from amino acids 41–154 contained both of the cysteines and the LD3 motif, and that of LD3 from amino acids 131–154 contained only the LD3 motif. Although the LIM region was localized in the nucleus regardless of oxidative states as described above (Figure 6, C and D), the considerable amount of protein relocalized from the nucleus to the cytoplasm when fused with CC-LD3 (Figure 6, C and D, (CC-LD3)LIM, −). On exposure to the oxidants, the fusion protein accumulated in the nucleus again (Figure 6, C and D, (CC-LD3)LIM, H2 and Di). Notably, its NES activity and sensitivity to the oxidants appeared almost equivalent to those shown by the entire N-terminal half of Hic-5 (Figure 6D, (CC-LD3)LIM vs. (Hic(N))LIM), revealing the CC-LD3 region to be the major functional NES module of Hic-5 sensitive to the oxidants. This observation excluded the involvement of LD1, which was included in (Hic(N)) but not in (CC-LD3), in the NES activity. In contrast to CC-LD3, the leucine-rich stretch of LD3 alone could not change the subcellular distribution of the fused protein (Figure 6D, (LD3)LIM), presenting a marked contrast to the similar leucine-rich stretches of zyxin and Trip6, which were active by themselves.
Contribution of Hic-5 to c-fos Induction by H2O2
We here tried to clarify the pertinent function of Hic-5 in the nucleus. In our previous study, the forced expression of Hic-5 in certain cells was demonstrated to lead to changes in the expression of several genes. They included the extracellular matrix-related genes (Shibanuma et al., 1997) and c-fos (our unpublished results). Among them, the c-fos gene is well known as being transcriptionally induced in response to various stimuli including oxidants (Crawford et al., 1988; Shibanuma et al., 1988; Nose et al., 1991). These facts led us to speculate that Hic-5 localized in the nucleus under the normal condition or accumulated in response to the oxidants participates in the transcriptional up-regulation of the gene.
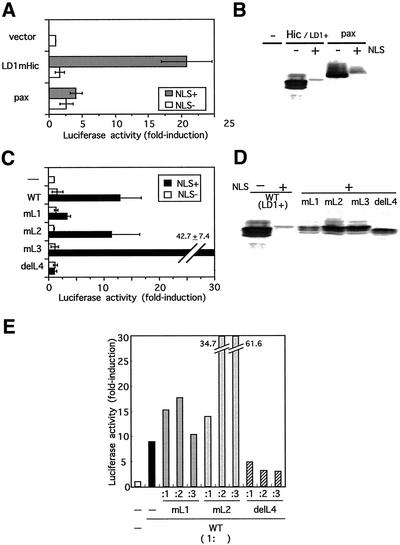
First we tested whether nuclear localized Hic-5 had the potential to affect gene expression. To do this, we engineered +NLS forms of Hic-5 and its variants, which carried NLS derived from SV40 large T antigen, and used them as effectors in experiments together with −NLS forms as a control. When expressed in the cells, being exclusively localized in the nucleus (our unpublished results), NLS+Hic-5 was found to transactivate the reporter construct of c-fos containing the −2.2-kb upstream region of the gene in a transient reporter assay (Figure 7A, LD1mHic, NLS+). The wild-type (−NLS form) had almost no effect on the activity at any concentrations studied including lower ones (Figure 7A, LD1mHic, NLS−). In contrast, paxillin affected only marginally the reporter activity even in NLS+ form (Figure 7A, pax, NLS+), suggesting that the transactivation by NLS+Hic-5 was not merely a consequence of overexpression of the exogenous protein in the nucleus but was the inherent effect of Hic-5. Immunoblotting examining the expression levels of these effectors showed that the NLS+ forms of paxillin and Hic-5 were almost equally expressed, whereas their levels were remarkably lower than those of wild types (−NLS forms) for an unknown reason (Figure 7B).
Figure 7.
The ability of the nuclear localized Hic-5 to transactivate the c-fos gene, for which LIM1 and 4 were required. (A) The reporter plasmid of c-fos and the effector plasmid expressing Hic-5 or paxillin were transiently cotransfected into C2C12 cells together with an internal control plasmid, pRL/CMV. The effector plasmids used here were pCG-LD1 mhic-5 (LD1mHic) and pCG-pax (pax), respectively, which were either the wild-type (NLS−) or nuclear-targeted (NLS+). Luciferase activities were determined as outlined in MATERIALS AND METHODS and shown as a ratio to the controls transfected with the pCG vector (vector) after normalization with the internal control. Bars represent the mean ± SD from at least three independent experiments. (B) pCG vector (−), pCG-LD1 mhic-5 (Hic/LD1+) and pCG-pax (pax) with (+) or without (−) NLS were transfected into C2C12, and 24 h later the cells were lysed and processed for Western blotting. The expressed proteins were detected with the antibody to HA tag. (C) The wild-type and the LIM mutants of Hic-5 were expressed as effectors from pCG-LD1 mhic (WT),/mL1–3 (mL1–3),/delL4 (delL4) with (NLS+) or without NLS (NLS−) in the cells with the c-fos reporter, and a luciferase assay was performed as in A. (D) The expression plasmids of the LIM mutants used in the above assay of C were transfected into C2C12 cells, and 24 h later, Western blotting was carried out using the antibody to HA tag. (E) The c-fos reporter was introduced into the cells with combinations of plasmids at various ratios of the wild-type (1:) and each LIM mutant (:1–3) as indicated, and a luciferase assay was carried out as in A and C. Each assay was done in duplicate. The experiments were repeated three times and similar results were obtained.
Our previous study suggested that Hic-5 bound to DNA through the LIM domains (Nishiya et al., 1998). Thus, the effect of disrupting the four LIM domains was examined on the transactivating ability of NLS+Hic-5. Distinctive contributions of each LIM domain were revealed; particularly noteworthy was the elimination of LIM4, which almost completely deprived NLS+Hic-5 of the ability to transactivate the reporter (Figure 7C, delL4). The mutagenesis of LIM1 also resulted in loss of the transactivating ability, although less completely than in delL4 (Figure 7C, mL1). Immunoblot analysis indicated that the expression levels of the mutants (NLS+forms) used here were almost the same or even higher than the level of NLS+ wild-type Hic-5 (Figure 7D), suggesting that the inability of LIM1,4 mutants to transactivate the reporter was unrelated to their expression levels. The finding that mL1 and delL4 were nonfunctional types of Hic-5 led us to assume that they were dominant-negative against the transactivating function of wild-type Hic-5. The results in Figure 7E supported this idea. In Figure 7E, the transactivation by NLS+wild-type Hic-5 was assayed under the coexpression of NLS+mL1, 2 or delL4 at the ratio indicated. When delL4 was coexpressed, the transactivation of the c-fos reporter by the wild-type Hic-5 was inhibited in a dose-dependent manner not completely but to less than half of the control.
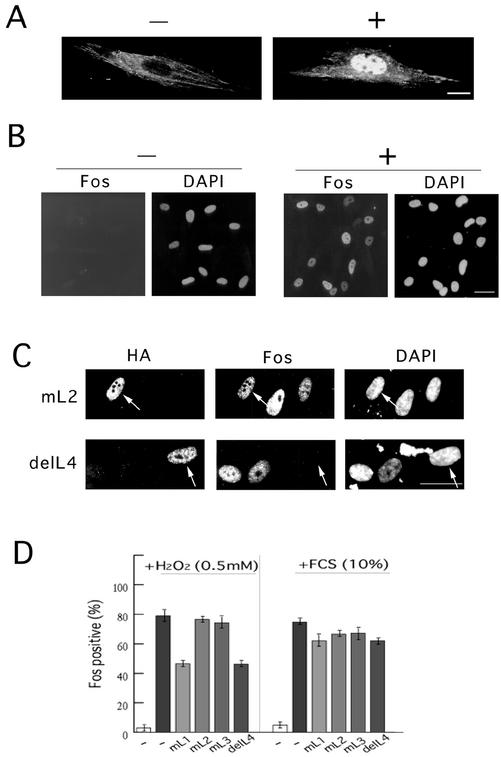
After these findings, we next examined whether the endogenous Hic-5 that accumulated in the nucleus in response to H2O2 actually participated in the transcriptional control of c-fos under the conditions. The cell line we chose was TIG-7, normal human fibroblasts. In mortal fibroblasts such as TIG-7, the endogenous expression level of Hic-5 was relatively high when compared with that in immortalized cell lines (Ishino et al. 2000), and a clear nuclear localization of Hic-5 was observed on H2O2 treatment (Figure 8A). Besides, after the same treatment with H2O2, c-fos was remarkably induced in this cell line in comparison with other cell lines such as mouse C3H10T1/2 fibroblasts, human KMST6 fibroblasts and HaCaT keratinocytes that were immortalized, and the induction was long-lasting (Figure 8B). In parallel with the endogenous gene, the luciferase reporter of c-fos responded by around fivefold to the stimulus in a transient assay (our unpublished results). To test the involvement of Hic-5 in c-fos induction, we first introduced the NLS+LIM mutants of Hic-5 including delL4 into the cells and then exposed the cells to H2O2. The introduction of delL4 but not mL2 before the treatment frequently resulted in the loss of c-fos expression in the presence of H2O2 (Figure 8C). Quantitative evaluation revealed a considerable decrease in c-fos expression caused by NLS+delL4 or mL1 expression. The decrease was modest but highly reproducible. The experiment was repeated three times, and for each mutant, ∼100 cells were examined in total. This attenuation was evident when c-fos expression was induced by H2O2 but not by 10% fetal calf serum (Figure 8D). These observations suggested a role for Hic-5 in the c-fos expression induced by H2O2. The response of the luciferase reporter was also inhibited in the presence of NLS+delL4 and mL1 (our unpublished results). The incompleteness of this inhibition might be due to the expression levels of NLS+delL4 and mL1 being insufficient to compete with the endogenous wild-type protein or alternatively, due to a residual ability of the mutants to transactivate the gene.
Figure 8.
Involvement of Hic-5 in endogenous c-fos induction by H2O2. (A and B) After serum deprivation for >24 h, TIG-7 cells were treated with (+) or without (−) 0.4 mM H2O2 for 5 h, and endogenous Hic-5 (A) or c-Fos (B) was immunolabeled with specific antibodies. DAPI was used to stain DNA. Bars, 10 (A) and 20 (B) μm. (C) (D) The same expression plasmids for the nuclear localized type of LIM mutants used in Figure 7C were first introduced into TIG-7 cells, and then, the cells were serum-deprived and exposed to the stimuli for 5h. Subsequent to fixation, coimmunostaining was performed with the antibodies to HA tag and Fos, along with DAPI. The immunolabels with antibody to HA identify the cells expressing the mutants. Among the cells expressing the indicated LIM mutants, those expressing Fos were enumerated microscopically, and the counts are shown as percentages in D. The experiment was repeated three times, and for each mutant, around one hundred cells were examined in total. In C, representative results of the immunostaining are shown in the cases of mL2 and delL4 expression in the cells treated with 0.5 mM H2O2. Bar, 20 μm.
DISCUSSION
Nuclear Localization of a Focal Adhesion Protein Hic-5 in Response to the Oxidative State of Cells
One of the ultimate goals of cellular signaling is the control of gene expression in the nucleus. Several signal transducers, including MAP and abl kinases, regulatory molecules of transcription such as JAK/STAT, Smads, and JAB1, have been shown to translocate from the cytoplasm to nucleus and thereby transmit signals into the nucleus. Besides such canonical signal transducers, certain components constituting cell-ECM or cell–cell adhesion complexes have been found to translocate into the nucleus and potentially transduce signals from sources to the nucleus directly. The candidates for such structural signal transducers are β-catenin, ZO-1, ajuba, zyxin, and others (reviewed by Lelièvre and Bissel, 1998; Alpin and Juliano, 2001).
In the present study, we presented findings that Hic-5, a focal adhesion protein, shuttled between focal adhesions and the nucleus through an oxidant-sensitive NES, the consequence of which was the nuclear accumulation of Hic-5 under the oxidative state of cells. Neither such NES nor the accumulation in the nucleus in response to the redox state have been reported in other focal adhesion proteins. Among the members of the LIM protein family, zyxin family members and paxillin were recently demonstrated to shuttle between focal adhesions and the nucleus in a Crm1/exportin-dependent manner (Nix and Beckerle, 1997; Petit et al., 2000; Nix et al., 2001; Wang and Gilmore, 2001; Woods et al., 2002). In these cases, however, no conditions or signals have been defined that could induce the bulk redistribution of the proteins from focal adhesions to the nucleus, and the biological relevance of the nuclear localization is unclear. Thus, to our knowledge, Hic-5 is the first example of a focal adhesion protein of the LIM family that accumulates in the nucleus in response to distinct stimuli with a pertinent function there.
One of the most surprising findings in this study was that despite its high homology and many common characteristics including nuclear-cytoplasmic shuttling, paxillin did not accumulate in the nucleus under the conditions in which Hic-5 accumulated in the nucleus. The difference emphasizes the specificity of Hic-5 for nuclear localization, implying a biological significance. However, the possibility has not been excluded that paxillin accumulates in the nucleus under certain circumstances and participates in some nuclear event. Regarding the biological significance of the nuclear-cytoplasmic shuttling of paxillin, paxillin was recently implicated in the transport of mRNA from the nucleus to sites of protein synthesis at the endoplasmic reticulum and the leading lamella during cell migration through binding to poly (A)-binding protein 1 (Woods et al., 2002). Several other differences between the two proteins, particularly their distinctive expression patterns, have been noted in processes including immortalization of murine fibroblasts, maturation of megakaryocytes to platelets, and in several tissues in vivo (Ishino et al., 2000; Hagmann et al., 1998; Jia et al., 2001). Thus, not simply complementary but distinctive biological functions are predicted for paxillin and Hic-5.
Mechanistic View of the Nuclear Localization of Hic-5
The N-terminal region of Hic-5 has been demonstrated to serve as the interface for interactions with several signaling molecules at focal adhesions as mentioned above. In particular, LD3 was shown to be important for interaction with FAK and for inhibiting cell spreading (Nishiya et al., 2001). In the present study, we assigned additional role to the region containing the LD3 motif and the two cysteines around the LD2 motif in controlling the nuclear export of the protein in an oxidant-sensitive manner, suggesting that the region was developed to detect, integrate adhesion and ROS signals, and couple the signals to transcriptional control in the nucleus. It is an open question how the two functions of LD3 as NES and as an interface for a signaling complex are influenced by each other. Recently, Aoto et al. (2002) reported that a mutant of CAKβ/PYK2 that lost a PXXP motif accumulated in the nucleus, and then Hic-5 also accumulated in the nucleus. Thus, PYK2 might be involved in the nuclear localization of Hic-5 in the particular condition, but its relation to the cellular oxidative state was unclear. The involvement of FAK also appeared to be unlikely, because FAK remained in the cytoplasm in the same cells in which Hic-5 accumulated in the nucleus under the oxidative condition (our unpublished results).
In Figure 9B, the amino acid sequence of the LD3 motif is aligned with the sequences of NESs identified in LIM proteins. The hydrophobic amino acids including the leucine residues, which was proven to be critical for the NESs, are well conserved, whereas, unlike Hic-5, there were no conserved cysteine residues found around the stretches of zyxin and Trip6. The alignment in Figure 9A shows the similarity and difference between the sequence surrounding the cysteine residues in the NES module of Hic-5 and the NESs of Yap1 and Pap1. Yap/Pap family members are transcription factors involved in oxidative stress response in yeast, accumulating in the nucleus upon exposure to oxidants and thereby regulating gene transcription (Kuge et al., 1997; Toone et al., 1998). A redox-sensitive functional NES was characterized in Pap1 and classified as a novel type of leucine-rich NES containing two or three cysteine residues besides a leucine-rich stretch (Kudo et al., 1999; Figure 9A). In the NES of Yap1/Pap1, the cysteines are placed immediately before the leucine-rich stretch, whereas in the NES module of Hic-5, instead of the stretch of the LD2 motif corresponding to that of Yap1/Pap1, the LD3 motif residing ∼50 amino acids from it was assigned as the NES component. The NES module of Hic-5 might be a hybrid evolved from the Yap/Pap-type and zyxin family-type NESs, in which the leucine-rich stretch was insufficient by itself for functioning as a NES, requiring coexistence of the cysteines. The prominent involvement of the cysteine residues in the NES activity, sensitizing it to the oxidants, does not have other precedents among mammalian proteins. It is these unique characteristics of NES that enables Hic-5 to be a novel type of mediator of redox signaling, shuttling between focal adhesions and the nucleus, coupling the signal directly to the transcriptional activity in the nucleus. Overall, even among NESs sharing some characteristics, it appeared that each NES has a distinctive property originating from amino acid disposition or protein-context, providing a basis for the regulation of the subcellular distribution of individual proteins and eventual biological roles within cells.
Biological Significance of Nuclear Accumulation of Hic-5
Among Hic-5–related LIM proteins, Trip6 and LPP have been proposed to be involved in transcriptional activity within the nucleus under some experimental conditions. However, their exact target genes have not been identified yet (Petit et al., 2000; Wang and Gilmore, 2001). In this article, we presented evidence that Hic-5 functions as a transcriptional regulator of a certain gene in the nucleus. Hypothetically, Hic-5 at the focal adhesions could modify integrin signaling, thereby affecting gene expression indirectly in the nucleus. In fact, our recent work showed that Hic-5 had a profound effect on integrin signaling (Nishiya et al., 2001). However, once localized in the nucleus, Hic-5 was expected to have a role distinct from that at the focal adhesions. By placing the NLS signal at the N-terminal and thereby expressing the exogenous Hic-5 almost exclusively in the nucleus, we could demonstrate that Hic-5 had the potential to transactivate c-fos. The complete dependency of the transactivation on the presence of NLS confirmed that Hic-5 in the nucleus was directly involved in the transactivation. We also found the distinctive roles of each LIM domain in the activation, implying that the stimulation of the gene transcription by the nuclear localized Hic-5 appeared not to be promiscuously observed but relevantly based on some molecular mechanisms to which each LIM domain contributes in its own way. Interestingly, a close correlation was revealed between the ability of Hic-5 to activate gene expression and its ability to bind to DNA. In both cases, LIM4 was essentially required, whereas LIM3 had a negative effect (Nishiya et al., 1998 and the present study). Alternatively, it is possible that LIM3 and 4 interact with negative and positive regulators for the transcription, respectively.
We previously reported that the inducibility of c-fos in response to phorbol ester decreased in Hic-5 high-expressing cells (Shibanuma et al., 1997), which apparently contradicts the observation in the present study that Hic-5 is involved in the upregulation of c-fos by H2O2. In a preliminary study, we outlined the region responsive to Hic-5 over the c-fos gene encompassing from −1.4 to −1.2 kb, which was overlapped with that negatively affected the basal level. At this stage, it has been unclear whether this region is involved in the responsiveness to either stimuli of H2O2 or phorbol ester. A further study on the precise mechanisms by which Hic-5 affects the c-fos expression would help to answer the above issue.
Recently, Hic-5 was reported to bind to steroid receptors and act as their coactivator in a transient reporter assay (Fujimoto et al., 1999; Yang et al., 2000). Similarly, Trip6 was reported to interact with thyroid hormone receptor (Lee et al., 1995). However, the biological relevance of their interactions with steroid receptors within cells has not been fully elucidated. In the present study, the transcriptional activation of c-fos by Hic-5 was observed in the absence of the ligands, suggesting that this transactivation was unassociated with the function as a coactivator of steroid receptors. Moreover, the coactivator function was reportedly observed in the transient reporter assay, whereas the involvement of Hic-5 in the c-fos induction was elucidated in the endogenous gene. It is thus likely that Hic-5 is not a coactivator specific to steroid receptors but implicated more generally in gene transcriptional activity in the nucleus in association with the redox state of cells.
Considerable work remains to be done to achieve a full understanding of the biological role of Hic-5 in cells. For example, the precise functions of Hic-5 at focal adhesions and in the nucleus together with the physiological conditions inducing its redistribution should be elucidated, which would give insight not only into the biological role of Hic-5 but also the relevance of change in the cellular redox state as biological signaling. Our previous work suggested that H2O2 accumulated in the cells upon the TGFβ1 treatment and was involved in the TGFβ1 signaling (Ohba et al., 1994). In the present study, TGFβ1 failed to induce the nuclear localization of Hic-5, suggesting that the nuclear localization of Hic-5 might be a stress response that was caused by the higher doses of H2O2 than that accumulated upon TGFβ1 treatment. Alternatively, a signaling pathway that was activated by TGFβ1 along with the production of H2O2 would negatively regulate the nuclear localization of Hic-5. It is also possible that retention time of Hic-5 within the nucleus was longer under the TGFβ1 treatment without observable increase in the population of Hic-5 in the nucleus, leaving the conclusion obsecure. In the recent work, the change in redox state in cells was intimately related to that in the adhesion state of the cells, resulting in the induction of a gene through NFκB (Kheradmand et al., 1998). Hic-5 may be another entity that regulates and coordinates gene expression, depending on the cell adhesion state through ROS signal. Clarifying these issues would hopefully provide an understanding of the dynamics of gene expression as the basis for the plasticity of cells in response to microenvironmental change represented by cell adhesion and/or cellular redox status.
ACKNOWLEDGMENTS
We thank Dr. M. Yoshida (Tokyo University) for generously donating LMB. We also thank M. Asakawa for contributing to this work as the theme for his bachelor's degree. This work was supported in part by Grants-in-Aid for Scientific Research, a Grant-in-Aid for Cancer Research, and the High-Technology Research Center Project from the ministry for Education, Science, Sports, and Culture of Japan, and a Grant-in-Aid from the Takeda Science Foundation.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–06–0099. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–06–0099.
REFERENCES
- Alpin A, Juliano RL. Regulation of nucleocytoplasmic trafficking by cell adhesion receptors and he cytoskeleton. J Cell Biol. 2001;155:187–191. doi: 10.1083/jcb.200107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto H, Sasaki H, Ishino M, Sasaki T. Nuclear translocation of cell adhesion kinase beta/praline-rich tyrosine kinase 2. Cell Struct Funct. 2002;27:47–61. doi: 10.1247/csf.27.47. [DOI] [PubMed] [Google Scholar]
- Brown MC, Curtis MS, Turner CE. Paxillin LD motifs may define a new family of protein recognition domains. Nature Struct Biol. 1998;5:677–678. doi: 10.1038/1370. [DOI] [PubMed] [Google Scholar]
- Crawford D, Zbinden I, Amstad P, Cerutti P. Oxidant stress induces the protooncogene c-fos and c-myc in mouse epidermal cells. Oncogene. 1988;3:27–32. [PubMed] [Google Scholar]
- Dawid IB, Toyama R, Taira M. LIM domain proteins. CR Acad Sci Iii. 1995;318:295–306. [PubMed] [Google Scholar]
- Fujimoto N, Yeh S, Kang H-Y, Inui S, Chang H-C, Mizokami A, Chang C. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J Biol Chem. 1999;274:8316–8321. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. Interaction of Hic-5, a senescence-related protein, with focal adhesion kinase. J Biol Chem. 1998;273:26516–26521. doi: 10.1074/jbc.273.41.26516. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- Hagmann J, Grob M, Welman A, van Willigen G, Burger MM. Recruitment of the LIM protein hic-5 to focal contacts of human platelets. J Cell Sci. 1998;111:2181–2188. doi: 10.1242/jcs.111.15.2181. [DOI] [PubMed] [Google Scholar]
- Hirai H, Suzuki T, Fujisawa J, Inoue J, Yoshida M. Tax protein of human T-cell virus type I binds to the ankyrin motifs of inhibitory factor kappa B and induces nuclear translocation of transcription factor NF-kappa B proteins for transcriptional activation. Proc Natl Acad Sci USA. 1994;91:3584–3588. doi: 10.1073/pnas.91.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Cascone PJ, Cheng L, Sun D, Nambu JR, Schwartz LM. Lepidopteran DALP, and its mammalian ortholog HIC-5, function as negative regulators of muscle differentiation. Proc Natl Acad Sci USA. 1999;96:10218–10223. doi: 10.1073/pnas.96.18.10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino K, Kaneyama JK, Shibanuma M, Nose K. Specific decrease in the level of Hic-5, a focal adhesion protein, during immortalization of mouse embryonic fibroblasts, and its association with focal adhesion kinase. J Cell Biochem. 2000;76:411–419. doi: 10.1002/(sici)1097-4644(20000301)76:3<411::aid-jcb9>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Jia Y, Ransom RF, Shibanuma M, Liu C, Welch MJ, Smoyer WE. Identification and characterization of hic-5/ARA55 as an hsp27 binding protein. J Biol Chem. 2001;276:39911–39918. doi: 10.1074/jbc.M103510200. [DOI] [PubMed] [Google Scholar]
- Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J Biol Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- Lelièvre SA, Bissell MJ. Communication between the cell membrane and the nucleus: Role of protein compartmentalization. J Cell Biochem Suppls. 1998;30/31:250–263. [PMC free article] [PubMed] [Google Scholar]
- Lipsky BP, Beals CR, Staunton DE. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J Biol Chem. 1998;273:11709–11713. doi: 10.1074/jbc.273.19.11709. [DOI] [PubMed] [Google Scholar]
- Mashimo J-I, Shibanuma M, Satoh H, Chida K, Nose K. Genomic structure and chromosomal mapping of the mouse hic-5 gene that encodes a focal adhesion protein. Gene. 2000;249:99–103. doi: 10.1016/s0378-1119(00)00163-3. [DOI] [PubMed] [Google Scholar]
- Matsuya M, Sasaki H, Aoto H, Mitaka T, Nagura K, Ohba T, Ishino M, Takahashi S, Suzuki R, Sasaki T. Cell adhesion kinase β forms a complex with a new member, Hic-5, of proteins localized at focal adhesions. J Biol Chem. 1998;273:1003–1014. doi: 10.1074/jbc.273.2.1003. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Nishiya N, Sabe H, Nose K, Shibanuma M. The LIM domains of hic-5 protein recognize specific DNA fragments in a zinc-dependent manner in vitro. Nucleic Acids Res. 1998;26:4267–4273. doi: 10.1093/nar/26.18.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya N, Iwabuchi Y, Shibanuma M, Cote JF, Tremblay ML, Nose K. Hic-5, a paxillin homologue, binds to the protein-tyrosine phosphatase PEST (PTP-PEST) through its LIM 3 domain. J Biol Chem. 1999;274:9847–9853. doi: 10.1074/jbc.274.14.9847. [DOI] [PubMed] [Google Scholar]
- Nishiya N, Tachibana K, Shibanuma M, Mashimo J-I, Nose K. Hic-5-reduced cell spreading on fibronectin: Competitive Effects between paxillin and Hic-5 through interaction with focal adhesion kinase. Mol Cell Biol. 2001;21:5332–5345. doi: 10.1128/MCB.21.16.5332-5345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of Zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- Nose K, Shibanuma M, Kikuchi K, Kageyama H, Sakiyama S, Kuroki T. Transcriptional activation of early-response genes by hydrogen peroxide in a mouse osteoblastic cell line. Eur J Biochem. 1991;201:99–106. doi: 10.1111/j.1432-1033.1991.tb16261.x. [DOI] [PubMed] [Google Scholar]
- Ohba M, Shibanuma M, Kuroki T, Nose K. Production of hydrogen peroxide by transforming growth factor-β1 and its involvement in induction of egr-1 in mouse osteoblastic cells. J Cell Biol. 1994;126:1079–1088. doi: 10.1083/jcb.126.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MMR, Fradelizi J, Golsteyn RM, Ayoubi TAY, Menichi B, Louvard D, Van de Ven WJM, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbor a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–129. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibanuma M, Kuroki T, Nose K. Induction of DNA replication and expression of proto-oncogene c-myc and c-fos in quiescent Balb/3T3 cells by xanthin/xanthin oxidase. Oncogene. 1988;3:17–21. [Google Scholar]
- Shibanuma M, Mashimo J, Kuroki T, Nose K. Characterization of the TGFβ1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J Biol Chem. 1994;269:26767–26774. [PubMed] [Google Scholar]
- Shibanuma M, Mochizuki E, Maniwa R, Mashimo J, Nishiya N, Imai S, Takano S, Oshimura M, Nose K. Induction of senescence-like phenotypes by forced expression of hic-5, which encodes a novel LIM motif protein, in immortalized human fibroblasts. Mol Cell Biol. 1997;17:1224–1235. doi: 10.1128/mcb.17.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibanuma M, Nose K. Forced expression of HIC-5, a senescence-related gene, potentiates a differentiation process of RCT-1 cells induced by retinoic acid. Int J Biochem Cell Biol. 1998;30:39–45. doi: 10.1016/s1357-2725(97)00155-6. [DOI] [PubMed] [Google Scholar]
- Shibanuma M, Ishino K, Sakamoto N, Nose K. Accumulation of focal adhesion protein Hic-5 in the nucleus by hydrogen peroxide. Acta Histochem Cytochem. 2001;34:259–264. [Google Scholar]
- Shibanuma M, Iwabuchi Y, Nose K. Possible involvement of Hic-5, a focal adhesion protein, in the differentiation of C2C12 myoblasts. Cell Struct Funct. 2002;27:21–27. doi: 10.1247/csf.27.21. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Hagel M, Turner CE. Characterization of a focal adhesion protein, Hic-5, that shares extensive homology with paxillin. J Cell Sci. 1999;112:181–190. doi: 10.1242/jcs.112.2.181. [DOI] [PubMed] [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated MAP kinase. StyI/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CE. Paxillin and focal adhesion signaling. Nat Cell Biol. 2000;2:E231–E236. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gilmore TD. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim Biophys Acta. 2001;1538:260–272. doi: 10.1016/s0167-4889(01)00077-5. [DOI] [PubMed] [Google Scholar]
- Woods AJ, Roberts MS, Choudhary J, Barry ST, Mazaki Y, Sabe H, Morley SJ, Critchley DR, Norman JC. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and leading edge of migrating cells. J Biol Chem. 2002;277:6428–6437. doi: 10.1074/jbc.M109446200. [DOI] [PubMed] [Google Scholar]
- Yang L, Guerrero J, Hong H, DeFranco DB, Stallcup MR. Interaction of the τ2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol Biol Cell. 2000;11:2007–2018. doi: 10.1091/mbc.11.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]