Abstract
Sis1 and Ydj1, functionally distinct heat shock protein (Hsp)40 molecular chaperones of the yeast cytosol, are homologs of Hdj1 and Hdj2 of mammalian cells, respectively. Sis1 is necessary for propagation of the Saccharomyces cerevisiae prion [RNQ+]; Ydj1 is not. The ability to function in [RNQ+] maintenance has been conserved, because Hdj1 can function to maintain Rnq1 in an aggregated form in place of Sis1, but Hdj2 cannot. An extended glycine-rich region of Sis1, composed of a region rich in phenylalanine residues (G/F) and another rich in methionine residues (G/M), is critical for prion maintenance. Single amino acid alterations in a short stretch of amino acids of the G/F region of Sis1 that are absent in the otherwise highly conserved G/F region of Ydj1 cause defects in prion maintenance. However, there is some functional redundancy within the glycine-rich regions of Sis1, because a deletion of the adjacent glycine/methionine (G/M) region was somewhat defective in propagation of [RNQ+] as well. These results are consistent with a model in which the glycine-rich regions of Hsp40s contain specific determinants of function manifested through interaction with Hsp70s.
INTRODUCTION
Several prion proteins have been identified in Saccharomyces cerevisiae. Such proteins have the unusual ability to exist in distinct and heritable states (Wickner and Masison, 1996; Lindquist, 1997). The stability of these states allows for the faithful transmission of the prions during cell division. Genomic searches for Gln/Asn-rich domains, a domain critical for prion propagation, led to the identification of another prion protein, Rnq1 (Sondheimer and Lindquist, 2000). Like other prions, Rnq1 exists in two heritable states, a soluble form, [rnq−], and an aggregated form, [RNQ+], also known as the epigenetic element [PIN+] (Derkatch et al., 2001).
Molecular chaperones, known to function in a variety of cellular processes involving changes in protein conformation (Hartl, 1996; Bukau and Horwich, 1998; Craig et al., 1999), have recently been implicated in the maintenance of the prion form of proteins. Heat shock protein (Hsp)104 is required for the maintenance of the yeast prion states [PSI+] and [RNQ+] (Chernoff et al., 1995; Patino et al., 1996; Sondheimer and Lindquist, 2000). The efficient propagation of [PSI+] also requires the Hsp70 Ssa1 (Jung et al., 2000). Moreover, it was demonstrated that the function of the Hsp40 Sis1 was required for [RNQ+] propagation (Sondheimer et al., 2001). However, little is understood about the mechanism by which one or another molecular chaperone affects prion propagation.
Hsp40s, J-type proteins that function as molecular chaperones with Hsp70s (Kelley, 1998), assist Hsp70s function in two ways: 1) stimulating the intrinsic ATPase activity of Hsp70, thus promoting a relative stable complex between Hsp70 and unfolded polypeptides; and 2) in various cases Hsp40s can bind unfolded polypeptides themselves and transfer them to Hsp70s (Hartl, 1996; Bukau and Horwich, 1998). Working together, Hsp40s and Hsp70s facilitate the refolding of denatured proteins in vivo and in vitro (Kim et al., 1998; Lu and Cyr, 1998a,b; Liu et al., 2001).
Hsp40s have been divided into two classes (Cheetham and Caplan, 1998). Members of both classes have a signature J domain of ∼70 amino acids that is critical for the interaction with Hsp70s. A glycine-rich region follows the J domain of both classes, which are often rich in phenylalanine residues and vary in length. For instance, the glycine-rich region of Sis1, the focus of this work, has an extended glycine-rich region with one segment rich in phenylalanine residues (G/F) and a second rich in methionine residues (G/M). Adjacent to the glycine-rich region, class I Hsp40s have a cysteine-rich region, followed by an unconserved C terminus, whereas class II Hsp40s lack a cysteine-rich region, but do have an unconserved C-terminal region. The unconserved C-terminal region of both classes contains the unfolded polypeptide binding region (Banecki et al., 1996; Szabo et al., 1996; Sha et al., 2000).
Two Hsp40s of the yeast cytosol are well characterized: class I Ydj1 and class II Sis1. In vivo analyses indicate that Ydj1 is involved in the translocation of proteins from the cytosol into organelles (Becker et al., 1996). Sis1 has been linked to the initiation of protein synthesis (Zhong and Arndt, 1993). However, it is likely that both of these Hsp40s play multiple roles in the cytosol. Both can stimulate the ATPase of the Ssa class of Hsp70s, and function with Ssa1 to facilitate the refolding of denatured proteins (Lu and Cyr, 1998b), suggesting that they function with the same Hsp70 in vivo.
The functional relationship between these two Hsp40s in vivo is complex. SIS1 is an essential gene (Luke et al., 1991); YDJ1 is not essential, but cells lacking Ydj1 grow very poorly (Caplan and Douglas, 1991). Overexpression of Sis1 can suppress the slow growth phenotype of a ydj1 strain (Caplan and Douglas, 1991), but overexpression of Ydj1 cannot rescue a sis1 strain (Luke et al., 1991). Therefore, Sis1 can carry out Ydj1 functions, but Sis1 performs an essential function in vivo that Ydj1 cannot.
Genetic analysis has implicated the G/F segment as the region defining Sis1-specific function. A Sis1 truncation having only the J domain and the G/F region is able to rescue growth of a Δsis1 strain (Yan and Craig, 1999). In this 121 amino acid fragment, the critical region for Sis1-specific function is the G/F region, as a chimeric gene encoding the J domain of Ydj1 and the G/F region of Sis1 allows growth of Δsis1 cells. The importance of the G/F region is also indicated by the fact that a Sis1 protein lacking only the G/F region is unable to maintain the prion form of Rnq1, although it still allows growth (Sondheimer et al., 2001). Because of the importance of the G/F region in Sis1 function, we pursued an analysis of its specificity.
MATERIALS AND METHODS
Yeast Strains and Growth Methods
The following W303 isogenic strains were used in this study: wild-type PJ51-3A (MAT a trp1-1 ura3-1 leu2-3112 his3-11,15 ade2-1 can1-100 GAL2+ met2-Δ1 lys2-Δ2) (James and Craig, unpublished data); WY26 (MAT α lys2-Δ2 Δsis1::LEU2) (Yan and Craig, 1999); JJ160 (MAT a ydj1::HIS3) (Johnson and Craig, 2000). 74D-694 Δsis1 [RPS+] (MAT a ade1-14 trp his leu ura sup35::RMC Δsis1::KAN) (Sondheimer et al., 2001) was used for analysis of the synthetic prion protein RMC. Yeast cells were grown on rich medium (YPD) or minimal medium (SD) where specified (Sherman et al., 1986).
For testing the ability of sis1 mutants to support in vivo functions, “plasmid shuffling” (Sikorski and Boeke, 1991) was used as described elsewhere (Yan and Craig, 1999) by using WY26 cells. The growth of sis1 mutants was evaluated on YPD and SD media with 5-fluororotic acid (5-FOA) after 3 d of incubation at 30°C. Plasmid shuffling was also used to test the ability of full-length sis1g/f mutants to support [RPS+] maintenance in 74-D694 Δsis1 [RPS+] cells. Growth was evaluated on YPD after 3 d of incubation at 30° and on SD media lacking adenine after 5 d at 30°C.
Plasmids
Plasmids used in this study: pYW65 (pRS314-SIS1), pYW116 (pRS314-sis1Δg/f), pYW66 (pRS314-sis1-171), pYW62 (pRS314-sis1-121), pYW70 (pRS314-YS-121), p316-RNQ1-GFP have been described elsewhere (Yan and Craig, 1999; Sondheimer and Lindquist, 2000). p314-sis1Δg/m was created by engineering an internal deletion on pYW65 that removes sequences encoding from amino acid 121–170. psis1-121-N108I and psis1-121-D110G were isolated from a genetic screen by using a plasmid library originated from polymerase chain reaction (PCR) fragments with random mutations cloned onto pYW62. psis1-121-D110N was a single mutation created on pYW62 based on an isolated full-length sis1 mutant (SIS1-D110N). psis1Δg/m-N108I and psis1Δg/m-D110G resulted from generating a single mutation in p314-sis1Δg/m. psis1-N108I and psis1-D110G were obtained by introducing the 3′-end sequence of SIS1 in psis1-121-N108I and psis1-121-D110G. p314-sis1-121Δ67-78 and p314-sis1-121Δ101-113 were internal deletions generated on pYW62 by PCR reactions. Construction of p314-sis1-Δg/mΔ67-78 and p314-sis1-Δg/mΔ101-113 were done by cloning sis1-121 from p314-sis1-121Δ67-78 or p314-sis1-121Δ101-113 into psis1Δg/m. p314-sis1Δ67-78 and p314-sis1Δ101-113 were made as described above for psis1-N108I and psis1-D110G.
Human HDJ1 was cloned from pET3a-hHsp40 (Minami et al., 1996) into pUC21 and subcloned into a pRS424 GPD driven expression vector (424GPD-HDJ1). To create HDJ1 mutant clones, a stop codon was engineered by PCR at amino acid position 147 amino acids (HDJ1-147) followed by its 3′-end untranslated region. Resulting PCR products were cloned into the pRS424 GPD vector. The Drosophila melanogaster Hsp40 DROJ1 was cloned from pYX232DroJ1 (Marchler and Wu, unpublished data) to pUC21 and subcloned to the pRS424 GPD vector. Construction of droj1 mutant clones was achieved by generating a stop codon by using PCR at amino acid position 143 (DROJ1-143) followed by its 3′-untranslated region. PCR fragments were cloned into pRS424 GPD-driven expression vector. HDJ2 was amplified by PCR from a plasmid (Toft, unpublished data) and cloned into a pRS424 GPD promoter vector.
A plasmid library of sis1-121g/f containing random mutations in the coding sequence of the G/F region was created from the plasmid pYW62 by using error prone PCR (Yan and Craig, 1999). PCR-amplified DNA was cloned into pYW62 to replace the wild-type sequence of the G/F region. About 9000 independent Escherichia coli transformants were obtained with 1% of the population containing clones lacking inserts.
A second mutant library was derived from the sis1-121g/f mutant library to identify the sequences within Sis1's G/F region required to support prion maintenance. BamHI-NotI DNA fragments were subcloned into the same sites of YCplac-SIS1, replacing the indicated sequences in this wild-type gene. Ligations were transformed into E. coli and resulted in ∼26,000 independent transformants with 20% of the population lacking inserts.
Protein Analysis
Immunoprecipitations and aggregation assays were carried out as described elsewhere (Sondheimer et al., 2001). In aggregation assays, cell lysates were fractionated by differential centrifugation at 280,000 × g. Protein quantifications with the wild-type strain PJ51-3A was performed as described elsewhere (Pfund et al., 1998) by using purified Sis1 and Rnq1 as standards.
Purified Rnq1 and its specific polyclonal antibodies were kindly provided by Neal Sondheimer and S. Lindquist (Sondheimer and Lindquist, 2000). In addition, Rnq1 antibodies were generated using as an immunogen the first 185 amino acids of Rnq1 fused to glutathione S-transferase (GST). This GST-Rnq1-185 fusion was expressed in E. coli and purified using glutathione beads, and then injected into rabbits to raise polyclonal antibodies. Purified Sis1 was obtained from WY26 cells expressing histidine-tagged Sis1 from the plasmid p424-GPD-HisB-SIS1 by using Nickel chelate affinity chromatography (His-Bind Resin from Novagen, Madison, WI).
Fluorescence Microscopy
For fluorescence microscopy, cells were transformed with a copy of RNQ1-GFP that is regulated by the CUP1 promoter (Sondheimer and Lindquist, 2000). Transformants in mid-log phase were induced by the addition of 50 μM CuSO4 and then incubated for 4 h at 30°C. After induction, cells were mixed with an equivalent amount of 1% SeaPlaque agarose (FMC Products, Rockland, ME) and visualized in a fluorescence microscope Axioplan 2 (Carl Zeiss, Thornwood, NY). Digital images were captured using the QED software (QED Imaging, Pittsburgh, PA).
RESULTS
Sis1 and Rnq1 Are Present in Equimolar Amounts in Immunoprecipitates of Prion [RNQ+]
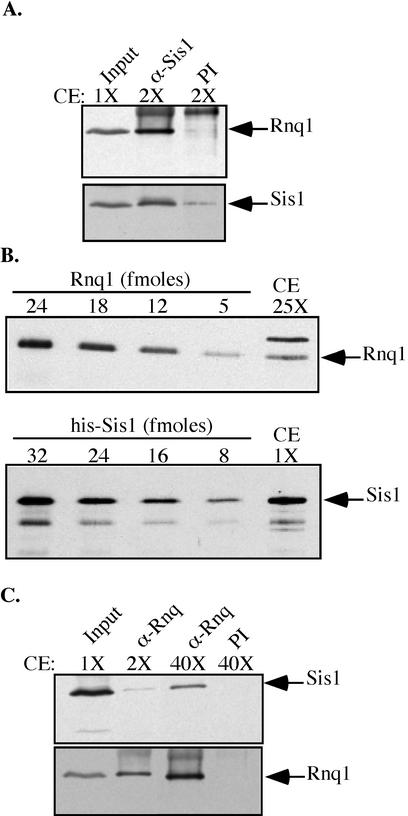
Sis1 can be coimmunoprecipitated with the prion, but not the soluble, form of Rnq1 (Sondheimer et al., 2001). To further characterize the Sis1:Rnq1 interaction found in [RNQ+] cells, we assessed the amounts of Sis1 and Rnq1 in these complexes by using Sis1-specific antibodies in immunoprecipitation assays. Nearly all of the Rnq1 in [RNQ+] lysates was associated with Sis1 (Figure 1A). However, Sis1 was determined to be ∼65 times more abundant in cell lysates than Rnq1 (Figure 1B). Therefore, the majority of Sis1 is available for other interactions, including those essential for cell viability.
Figure 1.
Sis1 coimmunoprecipitates with the prion form of Rnq1 in equimolar amounts. (A) Immunoprecipitation assays performed with lysates from an [RNQ+] W303 wild-type strain by using Sis1-specific antibodies. Relative amounts of cell extract (CE) loaded directly as input or used in immunoprecipitations are indicated as (1×) or two times (2×) with Sis1-specific antibodies (α-Sis1) or preimmune sera (PI). Relative amounts of proteins were determined by immunoblot analysis by using specific antibodies against Sis1 and Rnq1, followed by densitometry measurements. The efficiency of immunoprecipitation of Sis1 by α-Sis1 was ∼50%. (B) To determine the relative abundance of Rnq1 and Sis1 in extracts of [RNQ+] cells, dilution series of purified Rnq1, histidine-tagged Sis1 (his-Sis1) and CE were separated by SDS-PAGE. Numbers represent the femtomoles of purified proteins in each lane. Various dilutions of cell extract were loaded on gels but only those dilutions used for quantification are shown. The total amount of protein loaded for cell extract (CE1X) was 0.5 μg; in CE25X 12.5 μg. Proteins were resolve by electrophoresis and analyzed by immunoblot analysis by using antibodies against Rnq1 or Sis1. The amount of Rnq1 in 25X cell extract sample was determined to be 12 fmol, whereas 31 fmol Sis1 was in 1× cell extract sample. (C) Immunoprecipitation assays with Rnq1 specific antibodies (α-Rnq1) as described above. When 2× cell lysate was used the efficiency of immunoprecipitation by α-Rnq1 was ∼50%.
To determine the amount of Sis1 relative to Rnq1 in the Sis1:Rnq1 complexes, we carried out immunoprecipitation assays by using antibodies against Rnq1. Only ∼2% of the total amount of Sis1 was in association with Rnq1 (Figure 1C). Rnq1 and Sis1 are therefore found in approximately equimolar amounts in these complexes, consistent with an important role of Sis1 in prion maintenance.
Ydj1 Can Associate with the prion [RNQ+], but Is Dispensable for Its Propagation
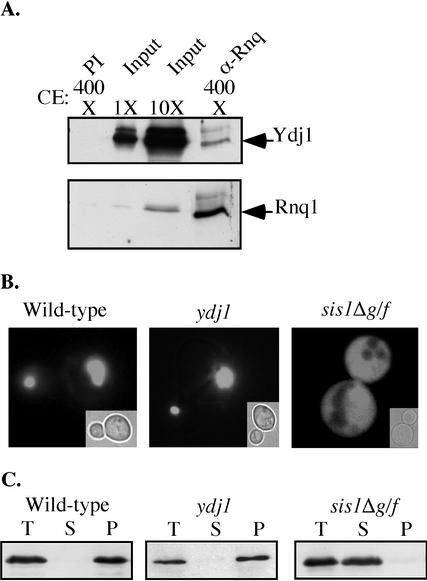
Another Hsp40, Ydj1, which is ∼10 times more abundant than Sis1 (Yan and Craig, 1999), is present in the yeast cytosol. To address whether Ydj1 is also involved in prion maintenance, immunoprecipitates isolated using Rnq1-specific antibodies were tested for the presence of Ydj1 by immunoblot analysis by using Ydj1-specific antibodies. Low amounts of Ydj1 were consistently coimmunoprecipitated with the prion [RNQ+] (Figure 2A).
Figure 2.
Ydj1 is dispensable for maintenance of the prion [RNQ+]. (A) Coimmunoprecipitation of Rnq1 and Ydj1. Immunoprecipitation assays performed with lysates from an [RNQ+] W303 wild-type strain by using Rnq1-specific antibodies (α -Rnq1) or preimmune serum (PI) as a control. We loaded ∼2.5 μg (1×) and 25 μg (10×) of total protein in lysate on a SDS-polyacrylamide gel together with the total immunoprecipitated material from 400 μl (400×) of lysate containing ∼1 mg of total protein. Samples were subjected to electrophoresis followed by immunoblot analysis by using specific antibodies against Ydj1 and Rnq1. (B) Fluorescence microscopy of Rnq1-GFP in wild-type and ydj1cells. The aggregation state of Rnq1-GFP was assessed by fluorescence microscopy after Rnq1-GFP expression was induced with 50 μM CuSO4for 4 h. Fluorescent images are representative of >60% of cells in the population. Insets, corresponding bright field images. (C) Aggregation assays for Rnq1 were carried out by fractionating crude lysates from a wild-type or ydj1 strain by using high-speed centrifugation. Equivalent aliquots of total (T) lysate, supernatant (S), and pellet (P) fractions were separated by SDS-PAGE, followed by immunoblot analysis by using Rnq1-specific antibodies.
To determine the biological significance of the Ydj1 association with [RNQ+], ydj1 cells were evaluated for the stability of [RNQ+]. The distribution of Rnq1 in living cells after transient overexpression of an Rnq1-green fluorescent protein fusion (Rnq1-GFP) was monitored. As previously shown, the fluorescence showed as a single focus in the majority of wild-type [RNQ+] cells (Sondheimer and Lindquist, 2000), whereas the remaining cells had multiple small foci. As typically seen in [rnq−] cells, the Rnq1-GFP fluorescence was diffuse in cells carrying Sis1 lacking the G/F region (Sis1ΔG/F) (Figure 2B; Sondheimer et al., 2001). Fluorescence microscopy of a ydj1null mutant expressing RNQ1-GFP was indistinguishable from a [RNQ+] wild-type strain (Figure 2B).
This in vivo approach was complemented by assaying cell lysates for the presence of the prion by high-speed centrifugation. Rnq1 from wild-type cells was all in the pellet fraction, whereas it was all of Rnq1 was in the supernatant fraction of sis1Δg/f lysates. As in analysis of lysates from a wild-type strain, all of the Rnq1 from the ydj1 cells was found in the pellet fractions, indicating that the [RNQ+] prion is maintained in the absence of Ydj1 (Figure 2C). Therefore, Ydj1 is dispensable for the propagation of the prion [RNQ+], whereas Sis1 plays a specific role in this process.
In Vivo Sis1 Functions Are Complemented by the Human and D. melanogaster Sis1 Homologs
Hsp40s related to Sis1 and Ydj1 have been conserved during evolution. For example, Hdj1 and Hdj2 are human homologs of Sis1 and Ydj1, respectively. In addition, it was recently found that the Drosophila homolog of Sis1 would rescue the viability of a Δsis1strain (Marchler and Wu, 2001). To test whether any of these heterologous proteins could function to maintain [RNQ+], a Δsis1 strain expressing SIS1 from a URA3-based centromeric plasmid was transformed with a TRP1-based plasmid containing either HDJ1, HDJ2, or DROJ1 driven by the yeast GPD1 promoter. Growth of selected transformants was evaluated on media containing 5-FOA, which selects for cells having lost the URA3-based plasmid (Sikorski and Boeke, 1991). Cells expressing either Hdj1 or Droj1 grew on media containing 5-FOA, but cells expressing Hdj2 did not (Figure 3A), indicating that both these class II Hsp40s can carry out the essential functions of Sis1, but the class I Hdj2 cannot. However, Hdj2 allowed wild-type growth of a ydj1 strain at 30°C, and thus is functional as a class I Hsp40 in yeast.
Figure 3.
J domain and G/F region of Drosophila and human Sis1 homologs can complement Sis1's roles in cell growth and [RNQ+] maintenance. (A) A 10-fold dilution series of cell suspensions from a Δsis1 (left) or ydj1 (right) strain carrying SIS1 or YDJ1, respectively, on a URA3-containing vector and second plasmid having no Hsp40 gene (−), SIS1, Drosophila DROJ1, human HDJ1, or human HDJ2. Cells were spotted on media containing 5-FOA and incubated for 3 d. (B–D) Top, fluorescence microscopy of Δsis1 cells coexpressing an Hsp40 gene and RNQ1-GFP. Cells were visualized 4 h after RNQ1-GFP induction as described above. Bottom, aggregation assays for the lysates of corresponding strains. After centrifugation, equivalent aliquots of total lysate T, supernatant fraction S, and pellet fraction P were subjected to immunoblot analysis by using Rnq1-specific antibody. (B) Full-length DROJ1 and HDJ1. (C) Truncation mutants of DROJ1 and HDJ1, droJ1-143 and hdj1-147, which encompass their corresponding J domain and glycine-rich sequences. (D) Sis1-121 truncation or a fusion that includes the J domain of Ydj1 fused to the G/F region of Sis1 (YS-121).
Because both Hdj1 and DroJ1 were able to maintain the viability of Δsis1cells, we asked whether cells having either of these heterologous proteins were able to maintain Rnq1 in the prion form. All of the Rnq1 protein was in the pellet fraction after sedimentation of lysates of Δsis1cells expressing either Hdj1 and DroJ1, indicating that Rnq1 was in the prion form (Figure 3B). In the in vivo assay with GFP-Rnq1, the majority of cells showed multiple small foci, with only a few cells showing a single center of fluorescence, most commonly seen in wild-type cells.
This pattern of multiple small foci resembled that observed in cells expressing Sis1-121, which contains only the J domain and G/F region of Sis1. Therefore, we proceeded to further dissect the sequences in Hdj1 and DroJ1 sufficient for supporting growth as well as prion maintenance. Truncation mutants of both HDJ1and DROJ1 that encoded only the J domain and glycine-rich region, Hdj1-147 and DroJ1-143, were constructed. Both were able to rescue the growth of a Δsis1strain and maintain the Rnq1 prion (Figure 3C; our unpublished data). Thus, the specificity of the Sis1-type (class II) Hsp40 has been conserved in evolution, and residues within the J domain:G/F region are sufficient for both Sis1's essential function and prion maintenance.
G/F Region Carries the Specificity for Prion Maintenance
The results described above demonstrate that the J domain and G/F region of Sis1 homologs from diverse organisms are sufficient to maintain [RNQ+]. Previously, the G/F region was shown to be required for Sis1's specific function(s) in maintaining cell viability, because the chimera between the J domain of Ydj1 and the G/F region of Sis1 (YS-121) was sufficient to allow growth of a Δsis1strain (Yan and Craig, 1999). Therefore, using this chimeric gene, we tested whether the Sis1 J domain was critical for prion maintenance. A strain containing YS-121 as the only protein having Sis1 G/F sequences was able to maintain Rnq1 in the prion form as indicated by both centrifugation and fluorescence assays (Figure 3D). Thus, Sis1's J domain is functionally exchangeable with that of Ydj1 but its G/F region contains sequences critical for maintenance of the Rnq1 prion.
A Unique Amino Acid Sequence (aa 101–113) in the G/F Region Determines Sis1's Functional Specificity
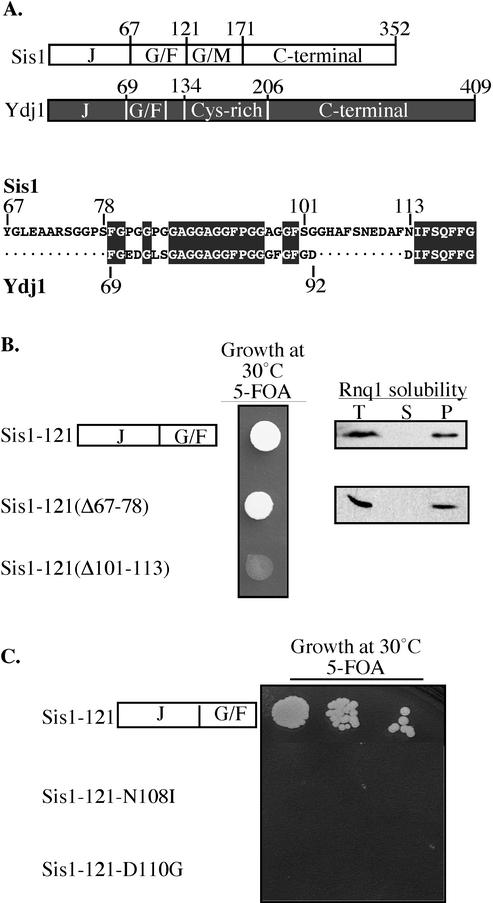
Sis1, but not Ydj1, is required for maintenance of the Rnq1 prion. Our observations indicate that the functional distinction between Sis1 and Ydj1 in prion maintenance is attributable to the G/F region. Thus, we compared the sequences within the G/F region between the two Hsp40s (Figure 4A). The G/F regions have extensive conservation in the glycine-rich segments, as well as in a small segment at the C terminus of this region that is devoid of glycine residues. However, the Sis1G/F region is longer than that of Ydj1 due to two unique stretches of amino acids (aa 67–78 and 101–113).
Figure 4.
Unique residues within the G/F region of Sis1-121 are required for in vivo function. (A) Top, comparison of overall structure between Sis1 and Ydj1: J domain (J), glycine/phenylalanine region (G/F), glycine/methionine region (G/M)-rich region, and cysteine-rich region (Cys-rich). Bottom, sequence comparison of the G/F regions of Sis1 (top) and Ydj1(bottom) by using the Jotun Hein algorithm of the MegAlign software of the DNAstar package (DNASTAR, Madison, WI). Identical residues are highlighted. Numbers refer to the position of the residues in the protein. (B) Left, cell suspensions of a Δsis1 strain carrying SIS1 on a URA3-containing plasmid and a second plasmid expressing sis1-121, sis1-121Δ67-78, or sis1-121Δ101-113 were spotted on medium containing 5-FOA and incubated for 2 d at 30°. Right, Rnq1 aggregation assays of lysates from Δsis1 cells expressing sis1-121 or sis1-121Δ67-78. After centrifugation, equivalent aliquots of total lysate T, supernatant fraction S, and pellet fraction P were subjected to immunoblot analysis by using Rnq1-specific antibody. (C) Dilution series (1/10) of cell suspensions from a Δsis1 strain carrying SIS1 on a URA3-based plasmid and a second plasmid expressing either Sis1-121 or Sis1-121 with two alterations (N108I or D110G) were spotted on media containing 5-FOA and incubated for 3 d at 30°C.
To determine the importance of these unique segments of the G/F region for function, internal deletion mutants of sis1-121 that encode proteins lacking one or the other of the unique segments of the Sis1 G/F region were created. Cells expressing sis1-121Δ67-78 were viable. In contrast, sis1-121Δ101-113 could not support growth (Figure 4B), even though protein levels similar to sis1-121 were produced (our unpublished data).
Because the N-terminal 121 amino acids of Sis1 (Sis1-121) are sufficient to maintain Rnq1 in the prion state, as well as support viability, we also tested the state of the Rnq1 prion in sis1-121Δ67-78 cells. All of Rnq1 was found in the pellet fractions after differential centrifugation (Figure 4B). Therefore, the unique stretch of amino acids between positions 67 and 78 is not important for either cell viability or prion maintenance.
Single Amino Acid Alterations within the aa 101–113 Segment of the G/F Region Alleviate the Ability of Sis1-121 to Support Cell Viability
Because the results described above suggest a critical role of the amino acids between positions 101–113 in the G/F region in maintaining the prion [RNQ+], we wanted to identify amino acids within this region important for this function. In the absence of a phenotype that distinguishes [RNQ+] from [rnq−] cells, we used an alternative genetic screen based on the ability of Sis1-121 to support cell viability (Yan and Craig, 1999). A Δsis1 strain expressing SIS1 from an URA3-based centromeric plasmid was transformed with a sis1-121 mutant library in which the G/F sequences were randomly mutagenized by error prone PCR. We screened 5000 transformants, resulting in two independent mutants that produced normal levels of Sis1-121 (our unpublished data), but did not support growth of Δsis1cells on 5-FOA–containing media (Figure 4C). The mutations caused single amino acid alterations: Asn to Ile at position 108 (N108I) and Asp to Gly at position 110 (D110G), demonstrating that single amino acid substitutions in this stretch of amino acids unique to Sis1 can disrupt its function. Thus, these amino acids found in the G/F region of Sis1, but not Ydj1, are not serving simply as a spacer between segments of the G/F, but are functionally important in their own right.
G/M Region of Sis1 Is Functionally Redundant with the Unique Stretch of Amino Acids in G/F Region
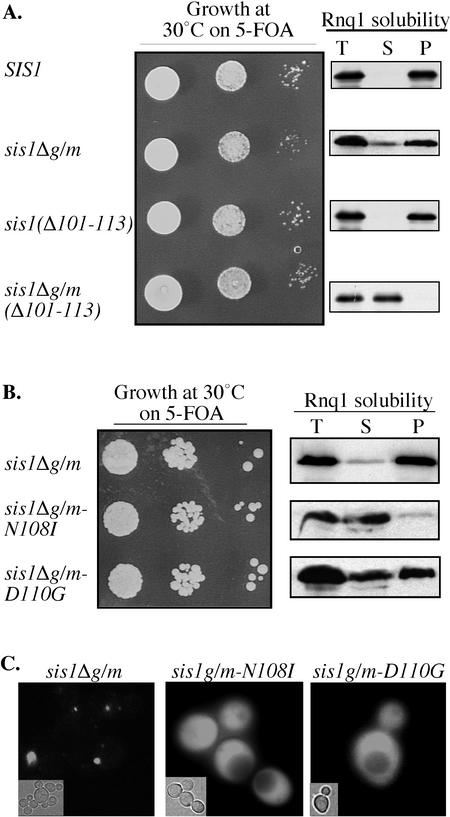
The results discussed above demonstrate that the G/F region contains sequences that can be critically important for prion propagation. However, because Sis1 contains a second glycine-rich region, the immediately adjacent G/M region, we wanted to test for any redundancy in function of these regions. First, we constructed a mutant containing a deletion of Sis1's G/M region to test its ability to complement functions. Cells only containing sis1Δg/m grew like wild-type cells. However, ∼5% of Rnq1 was consistently found in the soluble fraction in aggregation assays (Figure 5A). No Rnq1 was found in the soluble fraction upon analysis of wild-type cells, suggesting that the G/M region may play a role in prion maintenance.
Figure 5.
Unique residues within the G/F region of Sis1 are essential for the maintenance of [RNQ+] in the absence of the G/M region. (A and B) Left, 10-fold serial dilution series of cell suspensions from Δsis1 cells carrying SIS1 on a URA3-based plasmid, as well as a second plasmid expressing wild-type or the indicated mutant were grown on medium containing 5-FOA for 3 d at 30°C. Right, Rnq1 aggregation assays were performed using lysates from Δsis1 cells expressing the specified sis1 mutants. Equivalent amounts of total lysate (T), supernatant (S), and pellet (P) fractions from aggregation assays of lysates from the indicated strains were subjected to SDS-PAGE and analyzed for the presence of Rnq1 by immunoblot analysis. (C) Fluorescence microscopy of Rnq1-GFP in Δsis1 expressing sis1Δg/m, sis1Δg/m-N108I, or sis1Δg/m-D110G as described in Figure 2.
To further test this idea of functional redundancy, we combined alterations in the G/F region with deletion of the G/M region. First, a deletion of residues 101–113 in the context of otherwise full-length protein (Sis1Δ101-113) was constructed. Sis1Δ101-113 was able to maintain [RNQ+] in Δsis1 cells (Figure 5A), consistent with the idea that both glycine-rich regions play a role in prion maintenance. The construct sis1Δ101-113Δg/m sustained the growth of Δsis1 cells similar to a wild-type strain, however, [RNQ+] was not maintained (Figure 5A). Moreover, single alterations of residues 108 or 110 in combination with a deletion of the G/M region resulted in either complete or significant loss of the prion, respectively, without affecting cell growth (Figure 5B). In aggregation assays using Δsis1 lysates containing Sis1ΔG/M-N108I, Rnq1 was found only in supernatant fractions, whereas those containing Sis1ΔG/M-D110G resulted in 40% of the Rnq1 protein in supernatant fractions. These observations are consistent with analysis of fluorescence in cells expressing Rnq1-GFP. The fluorescence was dispersed in Δsis1 cells expressing either Sis1ΔG/M-N108I or Sis1ΔG/M-D110G (Figure 5C). These results suggest an important role for unique residues of Sis1 between positions 101 and 113 in prion maintenance. However, the G/M region complements the function of these residues.
Residues between Amino Acid 101 and 113 in Sis1 Are Important for Propagation of Synthetic Prion [RPS+]
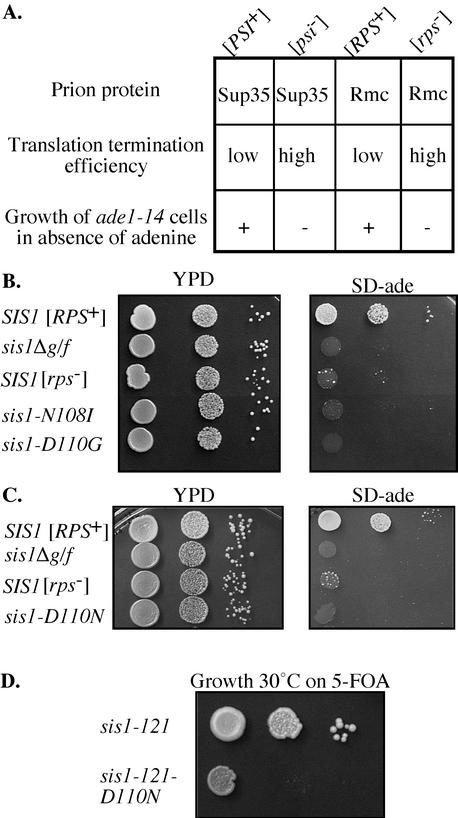
In the assays described above, no effect of the single amino acid alterations in the unique stretch between amino acids 101–113 on maintenance of [RNQ+] was found when in the context of the full-length protein, either in centrifugation assays (Figure 5A) or by observation of Rnq1-GFP fluorescence in vivo (our unpublished data). To test for more subtle effects on prion maintenance, we made use of an assay based on the ability of the prion conformation of Sup35, an essential yeast translation termination factor, to suppress the nonsense mutation ade1-14 (Patino, 1996; Figure 6A). When Sup35 is in the prion form, [PSI+], nonsense suppression occurs due to a reduced efficiency of translation termination. We made use of the synthetic prion protein, Rmc, which is composed of the Rnq1 prion domain and the catalytic region of Sup35 (Middle and C-terminal region) and can substitute for Sup35 in vivo (Sondheimer and Lindquist, 2000). In the prion form, Rmc mediates the nonsense suppression of ade1-14 cells, hence its designation as [RPS+], which stand for Rnq-PSI like-Suppression. This assay is very sensitive; lack of growth in the absence of adenine can result from minor changes in protein solubility that do not completely eliminate the prion conformation.
Figure 6.
Unique residues in the G/F region of Sis1 are important for maintenance of the prion [RPS+]. (A) Schematic representation of [RPS+]-based nonsense suppression. The translation terminator factor Sup35 is found in two protein states: prion state [PSI+] or the native state [psi−]. [PSI+] reduces the efficiency of translation termination resulting in nonsense suppression of the ade1-14 (UAG) allele. Rmc is composed of the Rnq1 prion domain fused to the Middle and C-terminal catalytic domain of Sup35. The resulting synthetic prion, [RPS+], can suppress the ade1-14 allele, hence its designation as Rnq1-PSI like-Suppression. (B and C) 74-D694 (Δsis1 Δsup35::RMC ade1-14 [RPS+]) cells expressing SIS1 or various full-length sis1 mutants. Serial dilutions (1/10) of cell suspensions of a Δsis1 strain containing plasmids containing the designated genes. Growth was evaluated after 3 d on YPD or 5 d on adenine depleted medium at 30°C. (D) Growth of Δsis1 W303 cells expressing either sis1-121 or the mutant sis1-121-D110N spotted onto YPD.
To validate the assay, 74-D694 Δsis1 [RPS+] cells expressing SIS1 or sis1Δg/f were constructed and tested for the ability to grow in the absence of adenine. As expected 74-D694 Δsis1 [RPS+] cells expressing SIS1 from a plasmid were able to grow in the absence of adenine (Figure 6B) due to read-through of the nonsense codon. However, cells expressing sis1Δg/f failed to grow in the absence of adenine, similar to [rps−] cells. Full-length Sis1 having the alterations N108I and D110G were tested for their ability to maintain [RPS+] in 74-D694 cells. sis1N108I and sis1D110Gdid not support the growth of 74-D694 strain in the absence of adenine (Figure 6B), establishing the importance of these residues in a Sis1 protein containing otherwise wild-type sequences for prion maintenance.
The essentiality of the unique G/F region sequences in [RPS+] maintenance allowed us to screen directly for alterations in the G/F region of the full-length gene that affected prion maintenance by screening for mutants that could not support growth on adenine. Efforts using this prion-based genetic screen identified a single mutation causing a substitution of Asn for Asp at position 110 (Figure 6C). Expression of sis1D110N allowed growth of 74-D694 cells in complete medium but resulted in impaired growth in medium lacking adenine. Because this mutation was located in the same codon as another alteration (D110G) that affected the ability of Sis1-121 to support cell growth, we also tested whether the alteration D110N affected the capacity of sis1-121 to support cell growth. A Δsis1 strain expressing sis1-121D110N grew poorly compared with cells expressing sis1-121 (Figure 6D). Therefore, based on our genetic analysis, we conclude that the unique sequences of Sis1's G/F region are important both for supporting cell growth and prion maintenance.
DISCUSSION
The glycine-rich regions of Hsp40 J-type proteins, which frequently have been considered simply linker regions between the J domain and the unfolded protein-binding domain, are critical for the protein's function. Herein, we report that the sequences in this region are important for the specific function of the Sis1/Hdj1 Hsp40s (class II), and distinguish them from the Ydj1/Hdj2 Hsp40s (class I). Class I and II J-type proteins often function with Hsp70s as soluble proteins within the same cellular compartment. Under such circumstances a single Hsp70 may function with more than one Hsp40, thus being a part of a complex network of chaperones. An example of such cross-function is the interaction of both Sis1 and Ydj1 with the Ssa class of Hsp70s.
The results presented above demonstrate that residues in the G/F region of Sis1 play important roles in Sis1-specific functions required for cell viability, as well as maintenance of [RNQ+]. Consistent with our hypothesis based on comparison of the G/F regions of Sis1 and Ydj1, a short sequence of amino acids found in Sis1, but not Ydj1, is required for both viability and prion maintenance. Therefore, this “insertion” in the G/F region of Sis1 is important for Sis1-specific function. However, this result does not mean that the conserved sequences in the G/F region do not play an important function in both Ydj1 and Sis1. The G/F region of Sis1 likely plays two roles, one generic and held in common with that of the G/F region of class I Hsp40s such as Ydj1, and the second specific for Sis1-type Hsp40s. This idea is consistent with the fact that overexpression of Sis1 can rescue growth defects of a ydj1 strain (Caplan and Douglas, 1991), but overexpression of Ydj1 cannot compensate for the absence of Sis1 (Luke et al., 1991).
Our observations demonstrate that Ydj1 does not complement Sis1's function in [RNQ+] maintenance. Nevertheless, it can interact with the prion form of Rnq1. Recently, it was reported that overexpression of Ydj1 can cure some variants of [RNQ+] (also known as [PIN+]) (Bradley et al., 2002), suggesting that Ydj1 antagonizes Sis1 in prion maintenance.
The sufficiency of the J domain and G/F region (Sis1-121) to support viability, as well as to maintain the prion form of Rnq1, facilitated dissection of the requirements for certain sequences in this truncated protein. The results of the analysis of full-length protein were more complex, revealing a redundancy in function within the glycine-rich region. The G/M portion of the glycine-rich region also plays a partially overlapping role with the G/F region. The single amino acid alterations in the G/F region that resulted in a nonfunctional Sis1-121 protein resulted in only mild defects when in the full-length protein. Similarly, deletion of only the G/M region had a modest effect on prion maintenance. However, in combination with the single amino alterations (N108I or D110G), the G/M deletion had dramatic effects on prion maintenance. Therefore, both sections of the glycine-rich region play a role in prion maintenance. Actually, the initial division of extended glycine-rich region into G/F and G/M segments was quite arbitrary. In fact, these two glycine-rich segments likely function together.
What does the glycine-rich region of Sis1 do? In vitro experiments indicate that deletion of the G/F region reduces the ability of an Hsp40 to enhance the interaction of an Hsp70 with a target protein (Wall et al., 1995) (Lopez, Johnson, and Craig, unpublished data). Because Sis1, but not Ydj1, is required for maintenance of the Rnq1 prion, and the sequences responsible are within the glycine-rich region, we propose that Sis1 type molecular chaperones uniquely have the ability to facilitate the productive interaction of Hsp70 with particular substrates, including Rnq1. More generally, the glycine-rich regions of an Hsp40 may determine the specificity of interaction of an Hsp70 with substrate proteins. This idea is an expansion of the hypothesis of Rapoport and coworkers developed from the results of experiments in which the presence of only the J domain greatly enhanced the affinity of an Hsp70 for a variety of protein/peptide substrates (Misselwitz et al., 1998). According to our expanded hypothesis, the J domain is required for this enhanced interaction with substrates, but the G/F region has evolved to determine the specificity of such interactions. In the case analyzed in our laboratory, the G/F region of the Sis1 family of Hsp40 can specifically enhance the productive interaction of Rnq1 with Hsp70.
However, it should be pointed out that glycine-rich regions are not required for function of all J-type proteins because members of class III, by definition, have no obvious region rich in glycine residues. Several members of that class have been found to function with Hsp70s at particular sites within organelles and some are important for targeting the Hsp70 to the site of function. Sec63 is localized to the ER membrane with a J domain located in the lumen targets Kar2/BiP to the translocation pore (Lyman and Schekman, 1995). Auxilin targets Hsp70 of mammalian cells to clathrin-coated vesicles where it functions in vesicle uncoating (Ungewickell et al., 1995). Zuo1 functions with Ssb Hsp70 on yeast ribosomes as a chaperone for nascent chains (Yan et al., 1998). G/F regions that may function to modulate the affinity of Hsp70 for particular substrates may not be as critical in cases where the Hsp70 is tethered to the location of the polypeptide substrate, thus dramatically increasing the local concentration. On the other hand, it is also possible that sequences, not recognizable to us because they are not rich in glycine residues, play a similar role.
An important goal of this study was to assess whether the role of Sis1 in prion maintenance was functionally different from its role in other cellular functions. This question was raised by the observation that Sis1 lacking the G/F region resulted in complete loss of the prion, but had little effect on cell viability. However, the analysis of mutants reported herein suggests that the sequence requirements are very similar, but that prion maintenance is particularly sensitive to slight alterations in Sis1 function. Therefore, we propose that biochemically Sis1 functions in prion maintenance as it does in other cellular processes, although its exact role in prion maintenance remains to be elucidated.
The ability of the Sis1 homologs from both Drosophila and humans to not only carry out the essential function of Sis1 but also maintain the Rnq1 prion was surprising. Neither of these Hsp40s have any apparent sequence similarity with Sis1 in the glycine-rich regions, other than the general preponderance of glycine and phenylalanine residues that are also found in yeast Ydj1 and human Hdj2. Therefore, divergent sequences are able to perform the function carried out by amino acids 101–113 of Sis1. This divergence suggests that a certain structural conformation that can be manifested through a variety of interactions is required for class II Hsp40-specific function, rather than a requirement for specific amino acid contacts between the G/F region and other domains. Current research to determine whether the G/F sequences affect the structure of the adjacent J domain or directly interact with Hsp70 and alter its function is in progress.
ACKNOWLEDGMENTS
We thank N. Sondheimer and S. Lindquist for purified Rnq1 and its specific antibodies; U. Hartl, D. Toft, and R. Morimoto for plasmids; P. Focke for technical assistance; and J. Johnson and R. Seiser for helpful comments on the manuscript. This work was supported by the National Institutes of Health grants GM-31107 (to E.A.C.) and 5 F31 GM-18507-05 (to N.L.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0593. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0593.
REFERENCES
- Banecki B, Liberek K, Wall D, Wawrzynow A, Georgopoulos C, Bertoli E, Tanfani F, Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J Biol Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic Hsp70 and DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions, and prion. “strains” in yeast. Proc Natl Acad Sci USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Douglas MG. Characterization of YDJ1: a yeast homologue of the bacterial DnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaption of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Craig E, Yan W, James P. Genetic dissection of the Hsp70 chaperone system of yeast. In: Bukau B, editor. Molecular Chaperones and Folding Catalysts: Regulation, Cellular Function, and Mechanisms. Amsterdam, the Netherlands: Harwood Academic Publishers; 1999. pp. 139–162. [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Prions affect the appearance of other prions. The story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Craig EA. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol Cell Biol. 2000;20:3027–3036. doi: 10.1128/mcb.20.9.3027-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- Kim S, Schilke B, Craig E, Horwich A. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc Natl Acad Sci USA. 1998;95:12860–12865. doi: 10.1073/pnas.95.22.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- Liu Q, Krzewska J, Liberek K, Craig EA. Mitochondrial Hsp70 Ssc1. role in protein folding. J Biol Chem. 2001;276:6112–6118. doi: 10.1074/jbc.M009519200. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998a;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998b;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Luke M, Suttin A, Arndt K. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial DnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler G, Wu C. Modulation of Drosophila heat shock transcription factor activity by the molecular chaperone DROJ1. EMBO J. 2001;20:499–509. doi: 10.1093/emboj/20.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Hohfeld J, Ohtsuka K, Hartl F-U. Regulation of the Heat-shock Protein 70 Reaction Cycle by the Mammalian DnaJ Homolog, Hsp40. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Rapoport T. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone SSB from S. cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure Fold Des. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sikorski RS, Boeke JO. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. In: Guthrie C, Fink GR, editors. Methods in Enzymology. Vol. 194. 1991. pp. 302–318. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1. an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SEH, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C. The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J Biol Chem. 1995;270:2139–2144. doi: 10.1074/jbc.270.5.2139. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Masison DC. Evidence for two prions in yeast: [URE3] and [PSI] Curr Top Microbiol Immunol. 1996;207:147–160. doi: 10.1007/978-3-642-60983-1_10. [DOI] [PubMed] [Google Scholar]
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong T, Arndt KT. The yeast SIS1 protein, a DnaJ homolog, is required for initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]