Abstract
Tor1p and Tor2p kinases, targets of the immune-suppressive antibiotic rapamycin, are components of a highly conserved signaling network that couples nutrient availability and cell growth. To gain insight into the molecular basis underlying Tor-dependent signaling, we used cell fractionation and immunoaffinity chromatography to examine the physical environment of Tor2p. We found that the majority of Tor2p associates with a membrane-bound compartment along with at least four other proteins, Avo1p-Avo3p and Lst8p. Using immunogold electron microscopy, we observed that Tor2p, as well as Tor1p, localizes in punctate clusters to regions adjacent to the plasma membrane and within the cell interior, often in association with characteristic membranous tracks. Cell fractionation, coimmunoprecipitation, and immunogold electron microscopy experiments confirmed that Lst8 associates with both Tor2p as well as Tor1p at these membranous sites. In contrast, we find that Kog1, the yeast homologue of the mammalian Tor regulatory protein Raptor, interacts preferentially with Tor1p. These findings provide evidence for the existence of Tor signaling complexes that contain distinct as well as overlapping components. That these complexes colocalize to a membrane-bound compartment suggests an intimate relationship between membrane-mediated signaling and Tor activity.
INTRODUCTION
Understanding how cell growth is controlled in response to environmental signals remains an outstanding biological problem. It has become clear in recent years that the Tor kinases act within an intracellular regulatory network used by eukaryotic cells to regulate their growth according to nutrient availability (reviewed by Dennis et al., 1999; Schmelzle and Hall, 2000; Raught et al., 2001; Rohde et al., 2001). This network regulates multiple aspects of gene expression, including transcription, translation, and intracellular protein trafficking, making the Tor pathway an important global regulator of cellular activity (Dennis et al., 1999; Schmelzle and Hall, 2000; Raught et al., 2001; Rohde et al., 2001). Two highly homologous Tor kinases exist in Saccharomyces cerevisiae, encoded by the TOR1 and TOR2 genes (Heitman et al., 1991; Helliwell et al., 1994; Zheng et al., 1995). Both Tor1p and Tor2p are targets of the immune-suppressive antibiotic rapamycin, which, in combination with the small immunophilin FKBP, inhibits the activity of these kinases that is required for nutrient-related growth control (Dennis et al., 1999; Schmelzle and Hall, 2000; Raught et al., 2001; Rohde et al., 2001). In addition to this Tor1p and Tor2p shared function, Tor2p is also required for proper actin cytoskeleton dynamics and polarized cell growth (Schmidt et al., 1996, 1997; Schmelzle and Hall, 2000). This Tor2p-specific function is essential for cell viability and is not inhibited by rapamycin.
In recent years, studies by many laboratories have been devoted to elaborating the circuitry of each of the major downstream branches of the Tor1p/Tor2p-shared as well as the Tor2p-specific functions in yeast (reviewed in Dennis et al., 1999; Schmelzle and Hall, 2000; Raught et al., 2001; Rohde et al., 2001). These studies have revealed many important details regarding the overall architecture, in terms of the molecular components involved, of the Tor signaling network. Moreover, they have revealed that this network intersects many other regulatory pathways that control cell growth and polarity. One example of this intersection is the observation that Tor regulates a concise set of metabolic genes, termed RTG target genes, that encode mitochondrial and peroxisomal enzymes required for de novo biosynthesis of glutamate and glutamine (Komeili et al., 2000; Shamji et al., 2000). Butow and coworkers originally identified these genes as targets of a mitochondria-to-nucleus signaling pathway, or retrograde response pathway, that adjusts their transcription in response to the respiratory state of the cell (Liao et al., 1991; Liao and Butow, 1993; Liu and Butow, 1999). Control of these genes by both pathways involves regulated nucleocytoplasmic transport of the heterodimeric bHLH/Zip transcription factors Rtg1p and Rtg3p, as well as depends on the cytoplasmic protein Rtg2p (Komeili et al., 2000; Sekito et al., 2000). These results highlight how Tor signaling is integrated with fundamental aspects of cellular metabolism.
Despite this progress, very little is presently understood regarding how the Tor kinases are themselves regulated. These are extremely large proteins, >2000 amino acids in length, and contain several predicted protein-interacting domains, including a large N-terminal domain consisting of multiple HEAT repeats (Andrade and Bork, 1995; Schmelzle and Hall, 2000). There have been reports that these repeats are important for interactions with other proteins, as well as membranes, although the precise nature of these interactions or their potential role in Tor signaling have not been well characterized (Sabatini et al., 1999; Bertram et al., 2000; Kunz et al., 2000; Wu et al., 2002). Given that control of Tor1p and Tor2p activity is likely to be complex, it would not be surprising if these proteins turn out to be involved in interactions with multiple partners. Indeed, this prediction has been born out by very recent studies of mammalian Tor (mTor), where several essential regulatory factors that interact directly with this protein have been identified (Gao et al., 2002; Hara et al., 2002; Inoki et al., 2002; Kim et al., 2002). One of these proteins, termed Raptor, associates with mTor in a nutrient-regulated manner (Hara et al., 2002; Kim et al., 2002).
It is also remains unclear precisely where within the cell the Tor kinases reside in yeast. Early immunofluorescence evidence suggested that Tor2p was associated with the vacuole (Cardenas and Heitman, 1995). In contrast, results from a more recent study suggest that both Tor2p as well as Tor1p localize primarily to the plasma membrane (Kunz et al., 2000). In studies of mammalian cells, mTOR is also primarily membrane associated and has been localized to punctate structures within the cell interior, possibly associated with membranes of exocytic and/or endocytic origin (Sabatini et al., 1999; Kim and Chen, 2000; Zhang et al., 2002). It has also been reported that mTOR shuttles between the cytoplasm and the nucleus or, alternatively, is localized constitutively to the nucleus in certain malignant cells (Kim and Chen, 2000; Zhang et al., 2002). Given that their function and regulation are likely to be intimately tied to their site(s) of action, a more complete understanding of intracellular location(s) of both Tor1p as well as Tor2p is necessary.
Herein, we describe studies that address many of these issues, through the biochemical identification of proteins that copurify with Tor2p. We characterize in detail the interaction between Tor2p and one of these proteins, Lst8p, which has recently been identified as a component of the retrograde response pathway (Liu et al., 2001). We demonstrate that Lst8p also interacts with Tor1p and that all three proteins localize to an intracellular membranous structure that seems distinct from the plasma membrane. Finally, we find that the yeast homologue of Raptor, Kog1p, associates specifically with Tor1p.
MATERIALS AND METHODS
Strains, Media, and General Methods
All strains were in the W303a background (leu2-3112 ura3-52 can1-100 ade2-1 his3-11 trp1-11 MATa). Culture medium used was YPD (2% yeast extract, 1% peptone, and 2% dextrose). Yeast cultures were grown at 30°C for all experiments. DNA ligations were performed using the Rapid DNA ligation kit (Roche Diagnostics, Indianapolis, IN). Plasmid DNA constructs were transformed into Escherichia coli DH5α and grown at 37°C. Yeast transformations were performed using a lithium acetate procedure (Geitz and Woods, 2002). Synthetic complete dextrose medium (0.8% yeast nitrogen base without amino acids, pH 5.5, 2% dextrose) was supplemented with amino acids as described previously (Sherman, 1991). Rapamycin (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) and added to a final concentration of 0.2 μg/ml. Anti-hemagglutinin (HA) (HA.11) and anti-Myc (9E10) monoclonal antibodies were purchased from Covance (Berkeley, CA). Anti-Tor1p polyclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmid Construction
Plasmid pFA6a-HisMX6-PTOR2-ATG-3HA (Figure 1A), used for amino terminal tagging of TOR2 with three copies of the HA epitope (HA3) by integration into the genome of W303a, was constructed in several steps. First, plasmid pFA6a-His3MX6-PGAL1–3HA (Brachmann et al., 1998; Longtine et al., 1998) was digested with BglII and PacI to remove the PGAL1 promoter region. The promoter region of TOR2 (600 base pairs) directly upstream of the start was polymerase chain reaction (PCR) amplified by using primers that contained the BglII or PacI sites and genomic DNA from strain S288c as template. The PCR product of the TOR2 promoter was digested and ligated into the above-mentioned plasmid to create pFA6a-His3MX6-PTOR2–3HA. Because removal of the PGAL1 promoter also removed the start site, a new start site was generated before the beginning of the 3HA tag. For this, plasmid pFA6a-His3MX6-PGAL1–3HA was used as the template in a PCR reaction where the forward primer included a PacI site as well as a start codon within its sequence and the reverse primer included the Asc I site at its 3′ end. The amplified PCR product and plasmid pFA6a-His3MX6-PTOR2–3HA were then digested with PacI and Asc I and ligated together. This final plasmid contained a new HA3 tag and a start codon and is termed pFA6a-HisMX6-PTOR2-ATG-3HA.
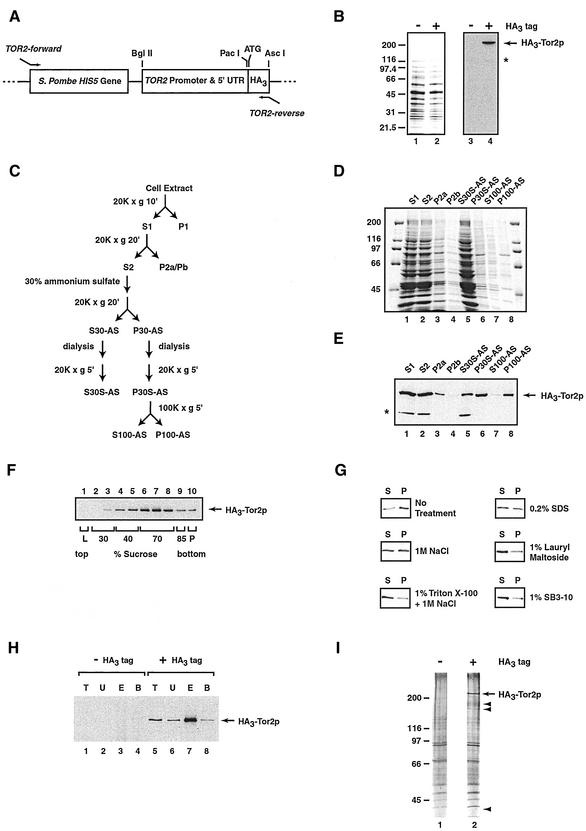
Figure 1.
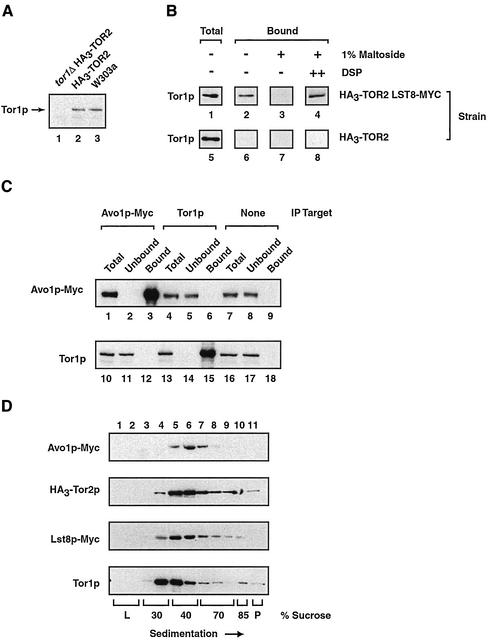
Strategy used to isolate Tor2p copurifying proteins. (A) Relevant portion of plasmid pFA6a-HisMX6-PTOR2-ATG-3HA, a derivative of pFA6a-His3MX6-PGAL1–3HA (Longtine et al., 1998), used to construct strain HA3-TOR2, as described in MATERIALS AND METHODS. Indicated are relevant restriction endonuclease sites, the translational start site, and primers used for PCR-mediated integration of the diagrammed cassette. (B) Characterization of HA3-Tor2. Western blot analysis of extracts prepared from HA3-TOR2 or W303a, indicated by + or −-HA3 tag, respectively (right). For comparison, a Coomassie-stained gel of each extract is also shown (left). (C) Scheme used to prepare HA3-Tor2p–enriched fraction P100-AS. Indicated are the relevant ultracentrifugation, and ammonium sulfate precipitation and dialysis steps, as described in detail in the text. (D) Coomassie-stained gel of total proteins in fractions, corresponding to steps outlined in C prepared during preparation of P100-AS. (E) Western blot analysis detecting HA3-Tor2p in fractions corresponding to those in D. (F) Equilibrium density centrifugation of the P100-AS fraction on a sucrose step gradient. After centrifugation, fractions were taken and analyzed by Western blotting to detect HA3-Tor2p. (G) Treating the P100-AS fraction with agents that disrupt membranes and membrane-protein interactions converts HA3-Tor2p into a soluble form. Samples were treated as described in MATERIALS AND METHODS and analyzed by Western blotting to detect HA3-Tor2p. (H) Testing interactions between HA3-Tor2p in the P100-AS fraction and immobilized anti-HA polyclonal antibody. Indicated is the amount of HA3-Tor2p present in the initial extract (T), the amount that does not bind to the antibody (U), as well as the amount that is either specifically eluted by competing HA dipeptide (E) or that remains bound (B). The amount of material in T and U fractions represents 10% of the material present in E and B fractions. Samples were prepared from strain W303a (−HA3 tag) or strain HA3-TOR2 (+HA3 tag), respectively. (I) Silver-stained SDS-PAGE gel of proteins in the P100-AS fraction that elute from immobilized anti-HA antibody, prepared from either W303a (lane 1) or HA3-TOR2 (lane 2). Arrows point to peptide species whose presence is unique to the tagged strain. In B and E, the * denotes a protein present in the W303a background that cross-reacts with anti-HA antibody. Note that this protein does not copurify with HA3-Tor2p in the P100-AS fraction.
Construction of a plasmid used for the HA3 amino terminal tagging of TOR1 was the same as described above except for the following: the pFA6a-HisMX6-PTOR2-start-3HA plasmid was digested with BglII and PacI to remove the TOR2 promoter. The promoter regions of TOR1 (240 base pairs) directly upstream of the start codon was amplified using genomic DNA from strain S288c as a template. Herein, the forward primer included a BglII site and the reverse primer included a PacI site. This PCR product was digested with BglII/PacI and ligated to create pFA6a-HisMX6-PTOR1-start-3HA.
Construction of Yeast Strains
To fuse the HA3 tag amino-terminally to TOR2 within the S. cerevisiae genome, PCR was performed using pFA6a-HisMX6-PTOR2-start-3HA and two TOR2-specific gene primers, TOR2-forward and TOR2-reverse (Figure 1A). The products from several PCR reactions were concentrated and transformed into W303a by using a lithium acetate procedure (Geitz and Woods, 2002) and grown at 30°C for 2–3 d on synthetic complete dextrose minus Histidine agar plates to select for integrants. Candidate colonies were screened by PCR, by using several primer pairs that identified putative correct integration events. Genomic DNA was prepared and DNA sequencing of the junctions was performed to confirm that the HA3 tag was properly integrated.
A similar strategy was used to produce amino-terminally tagged HA3-TOR1 except that for HA3-TOR1, the plasmid pFA6a-HisMX6-PTOR1-start-3HA was used as the template in a PCR reaction. This reaction also used specific TOR1-forward and TOR1-reverse primers. Transformation and identification of integrants proceeded in the same way as that for HA3-TOR2, as described above.
A double-tagged strain derived from strain HA3-TOR2 that produced a version of Lst8p tagged at its carboxy terminus with multiple copies of the Myc epitope was constructed using the PCR-based gene tagging method and plasmid pFA6a-13Myc-TRP1 (Brachmann et al., 1998; Longtine et al., 1998). LST8-specific primers targeted the region directly upstream and downstream of the stop codon. An identical approach was taken to construct versions of Kog1 and Avo1 that were tagged at their carboxy termini with multiple copies of the Myc epitope.
The TOR1 gene was disrupted in the HA3-TOR2 strain by using a similar PCR-based method and plasmid pFA6a-TRP1 as template (Longtine et al., 1998). Primers used corresponded to the very 5′ and 3′ ends of the TOR1 open reading frame.
To verify that strain HA3-TOR2 produced full-length HA3-Tor2p protein, Western blot analysis was performed. Briefly, 5 OD of cells from strain HA3-TOR2 W303a was pelleted and resuspended in 0.5 ml of trichloroacetic acid (TCA) buffer plus protease and phosphatase inhibitors. Resuspended cells were combined with 0.5 mg of glass beads and 0.5 ml of cold 30% TCA and were vortexed for four 30-s intervals with cooling on ice during the interim. Supernatants were centrifuged at 20,000 × g for 5 min at 4°C. Pellets were washed with 1 ml of cold acetone and centrifuged at 20,000 × g for 5 min at 4°C. Acetone was completely removed and pellets were air dried for 10 min. Pellets were resuspended in sample buffer, incubated for 5–10 min at 65°C, and loaded onto 7.5% SDS-PAGE gels followed by staining with Coomassie Blue or transfer to nitrocellulose for analysis for Western blotting. Anti-HA monoclonal antibody was used to detect HA3-Tor2p. A similar approach was used to confirm the sizes of the other epitope-tagged proteins described above.
Antibody Production
Anti-HA polyclonal antibodies were raised by immunizing rabbits with a peptide sequence from the influenza virus HA epitope (peptide sequence CYPYDVPDYA) conjugated to keyhole limpet hemocyanin. HA antibodies were affinity purified from serum by using a purified glutathione S-transferase-HA fusion protein coupled to Affigel 10 (Bio-Rad, Hercules, CA) as described previously (Kellogg and Alberts, 1992).
Immunoaffinity Purification and Mass Spectrometry Analysis of Tor2 Complex
The following protocol is a modified form of that published by Kellogg and coworkers (Mortensen et al., 2002). Cells producing HA3-Tor2p as well as untagged control cells were grown overnight at 30°C to 0.5 OD600/ml in YPD. Cells were pelleted, washed in H2O, pelleted again, and resuspended in yeast extract buffer (YEB; 50 mM HEPES-KOH, pH 7.1, 100 mM β-glycerol phosphate, 50 mM NaF, 5 mM EGTA, 5 mM EDTA, 10% glycerol, 0.25% Tween 20, and 150 mM KCl). The pellet was resuspended and transferred to a 50-ml conical tube, pelleted again, resuspended 1:1 (w/vol) in YEB containing protease inhibitors (cocktail tablet; Roche Diagnostics), 2 mM dithiothreitol, and 2 mM phenylmethylsulfonyl fluoride, and frozen dropwise by transfer pipet into liquid nitrogen. The cell pellet was then transferred to a prechilled mortar and pestle containing liquid nitrogen and ground into a fine powder (∼150 strokes followed by addition of liquid nitrogen repeated three times). After liquid nitrogen had boiled off, the powder that remained was transferred to 1.5-ml microfuge tubes, thawed, and centrifuged at 20,000 × g for 20 min at 4°C. The supernatants (S1) were pooled and centrifuged again as described above. Supernatants (S2) were pooled after removal of the lipid layer at the surface. Then 490 μl of S2 was combined with 210 μl of saturated ammonium sulfate (30% final) and stirred for 10 min at 4°C. Samples were centrifuged at 20,000 × g for 20 min. Supernatant was removed and held on ice and the pellet was resuspended in cold YEB buffer to 700 μl. Both the supernatant (S30-AS) and resuspended pellet (P30-AS) were dialyzed three times against excess YEB buffer. A clarifying spin of 20,000 × g for 5 min at 4°C followed and the supernatants (S30S-AS and P30S-AS) were removed and frozen in liquid nitrogen. Samples were thawed, placed in Beckman Coulter polyallomer tubes and centrifuged in a TLA45 rotor in a Beckman Coulter TL100 tabletop ultracentrifuge at 100,000 × g for 1 h at 4°C. Supernatant (S100-AS) was removed and pellet (P100-AS) was resuspended in YEB and passed ∼20 times through the needle of a Hamilton syringe.
The P100-AS extract was precleared by addition of 25 μl of protein A-Sepharose beads (Amersham Biosciences, Piscataway, NJ) and 25 μl of YEB in 1.5-ml Microfuge tubes to remove material that bound nonspecifically to protein A. The cleared P100-AS extract was then combined with polyclonal HA antibody bound to fresh protein A beads for 2 h at 4°C. The beads were then pelleted at 10,000 × g for 2 min at 4°C and washed 5× with 1 ml of YEB. After the last wash, the supernatant was removed completely using a Hamilton syringe. HA3Tor2p was released from the antibody by addition of 75 μl of elution buffer (YEB with 0.35 mg/ml of HA dipeptide [sequence CYPYDVPDYAGYPYDVPDYAG; Genemed Synthesis, South San Francisco, CA]), followed by incubation for 15 min at room temperature. At this point, the eluted material was either analyzed by SDS-PAGE and silver stained (Silver Stain Plus kit; Bio-Rad) or by mass spectrometry. For mass spectrometric analysis, 4 liters of starting material was used and the final eluted volume was ∼2 ml. Eluted material was TCA precipitated overnight at 4°C and the pellets were acetone washed and dried. A portion of the material was analyzed by SDS-PAGE and the remaining material was analyzed by mass spectrometry as described previously (Carroll et al., 1998; Link et al., 1999).
Coimmunoprecipitation Experiments
For coimmunoprecipitation experiments, 400 μl of P100-AS extract was prepared from an appropriate strain and incubated with anti-HA or anti-myc mAb overnight at 4°C. Then 25 μl of protein G beads (Amersham Biosciences) was added and incubated for 2 h at 4°C followed by five washes in YEB. After the final wash, the beads were completely cleared of the supernatant and 30 μl of sample buffer was added to the beads. They were then heated at 65°C for 5 min, 95°C for 5 min and loaded onto a 7.5% SDS-PAGE gel. The amount of material loaded for the total and unbound fractions corresponded to 5% of the material loaded for the bound fractions.
Detergent Treatment
To detergent treat material in the P100-AS extract, a P30S-AS extract was thawed and centrifuged at 100,000 × g for 1 h at 4°C and the pellets were resuspended in YEB. An equal volume of YEB that contained an appropriate concentration of detergent and/or salt was added such that the final concentrations were as follows: 1 M NaCl, 1% Triton-X 100/1 M NaCl, 0.2% SDS, 1% lauryl maltoside, or 1% SB3-10. Samples were incubated on ice for 30 min during which time they were passed through a Hamilton syringe five times approximately every 7 min. Samples were again centrifuged at 100,000 × g for 1 h 4°C. Supernatants and resuspended pellets were either analyzed directly by SDS-PAGE and Western blotting, applied to sucrose gradients, or used in immunoprecipitation experiments.
Dithiobis(succinimidylproionate) (DSP) Cross-linking
Proteins present in the P100-AS extract were cross-linked by addition of 18 mM DSP (Pierce Chemical, Rockford, IL) in DMSO to a final concentration of 1 mM or 1.8 mM for 2 h on ice. Cross-linked samples were adjusted to a final concentration of 1% lauryl maltoside on ice for 30 min. Samples were centrifuged at 100,000 × g for 1 h at 4°C, and the supernatant was removed and quenched by the addition of 1 M Tris-HCl, pH 8.0, to a final concentration of 20 mM. Samples were immunoprecipitated and processed for Western blotting as described above.
Cell Lysis by Spheroplasting
Spheroplast formation and cell lysis were performed as described previously (Nunnari et al., 2002)
Sucrose Gradient Ultracentrifugation
Sucrose step gradients were prepared from bottom to top in a 2-ml TLS55 centrifuge tube, where each step contained the following volume of YEB and percentage of sucrose: 200 μl of 85.5%, 600 μl of 70%, 400 μl of 40%, and 400 μl of 30%. Extracts were overlayed and gradients were centrifuged in a TLS55 swinging bucket rotor in a tabletop ultracentrifuge at 100,000 × g for 16 h at 4°C. Fractions were collected from the top of the gradient in 200-μl aliquots and processed for Western blot analysis.
Northern Blots
Northern-blot analysis was performed as described previously (Powers and Walter, 1999). DNA probes were generated by PCR by using genomic DNA from strain S288c as template and specific primers (Research Genetics, Huntsville, AL) for individual genes (ACT1, RPL30, CIT2, DLD3, and GAP1). Blots were scanned using a STORM 860 imaging system (Amersham Biosciences, Sunnyvale, CA) and analyzed using software provided by the manufacturer.
Immunogold Electron Microscopy (IEM)
IEM was performed on ultrathin cryosections as described previously (Rieder et al., 1996)
RESULTS
Identification of Tor2p-associated Proteins
To identify proteins that associate with Tor2p, we used an approach described recently by Kellogg and coworkers for immunopurification of endogenous multiprotein complexes from yeast (Mortensen et al., 2002). Briefly, this method uses immobilized affinity-purified polyclonal antibodies raised against the HA epitope to isolate protein complexes where one of the proteins is tagged with HA. Using this approach, protein complexes can be isolated from relatively crude cell extracts and subsequently released by an excess of competitive HA dipeptide under mild elution conditions, allowing for the specific isolation of potentially labile complexes (Mortensen et al., 2002).
Accordingly, we constructed a yeast strain where the endogenous TOR2 gene was fused in frame after sequences coding for HA3, expressed under control of its own promoter and at its normal chromosomal position (Figure 1A). We chose to position the HA3 tag at the N terminus of Tor2p because a number of previous studies have shown that different N-terminal tags, including HA3, do not interfere with Tor2p function (e.g., Kunz et al., 2000). Moreover, this strategy ensured that only endogenous levels of Tor2p would be produced. The resulting strain, HA3-TOR2, grew as well as its parental wild-type strain on both agar plates as well as in liquid media (our unpublished data). To test the efficacy of the tag, whole cell extracts were prepared from this strain and Western blot analysis was performed, probing for the HA3 epitope (Figure 1B). The results showed that a single band occurred at the predicted molecular mass of Tor2p of ∼280 kDa, confirming that the tag was correctly fused to TOR2 and that full-length tagged Tor2p was produced.
Initial immunoprecipitation experiments demonstrated that very little HA3-Tor2p protein bound to our immobilized anti-HA antibody from whole cell extracts under native conditions, suggesting the epitope was in an environment not readily accessible to the antibody (our unpublished data). We therefore explored a variety of approaches in an effort to partially purify HA3-Tor2p such that interactions with other proteins might be maintained and yet the HA3 epitope would become accessible. The strategy that was ultimately successful, outlined in Figure 1C, was relatively simple and took advantage of two unique biochemical characteristics of HA3-Tor2p. First, we found that the majority of this protein in a clarified whole cell extract was effectively precipitated using 30% ammonium sulfate, whereas most other cellular proteins remained soluble (Figure 1D, compare lanes 5 and 6). Second, we found that the majority of enriched HA3-Tor2p was associated with a membranous fraction of very high buoyant density that could be pelleted after high-speed (100,000 × g) ultracentrifugation (see below). This observation is in agreement with the conclusion of Hall and coworkers that Tor2p is primarily a peripherally associated membrane protein (Kunz et al., 2000). Thus, through a combination of ammonium sulfate precipitation and centrifugation steps, we were able to isolate HA3-Tor2 in a high-speed pellet fraction, termed P100-AS (Figure 1E). Quantitative Western blot analysis indicated that the P100-AS fraction was ∼30-fold enriched for HA3-Tor2p, relative to the initial extract (our unpublished data).
Analysis of the P100-AS fraction indicated that the majority of HA3-Tor2p was indeed membrane associated. First, the protein could be converted into a soluble form after treatment with agents that disrupt membranes or membrane–protein interactions, including high concentrations of salt and/or a number of nonionic as well as ionic detergents (Figure 1G). Second, HA3-Tor2p localized to very dense fractions, corresponding to an equilibrium density of ∼70% sucrose, after ultracentrifugation of resuspended P100-AS on a sucrose step gradient (Figure 1F). We note that this measured equilibrium density of HA3-Tor2p in the P100-AS fraction is significantly greater than in whole cell extracts (Figure 2E). We attribute this difference to the likelihood that the purification scheme used has altered the protein and/or lipid content associated with HA3-Tor2p in the P100-AS fraction.
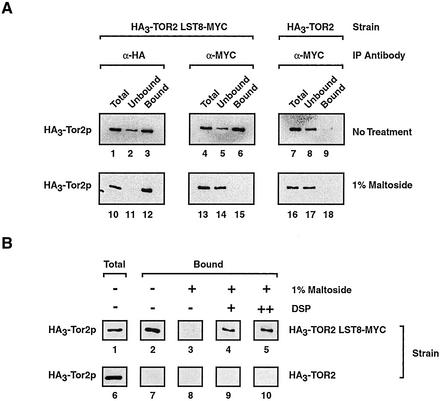
Figure 2.
Testing interactions between Lst8p and Tor2p. (A) Coimmunoprecipitation experiments demonstrating detergent sensitive interactions between Lst8p-Myc and HA3-Tor2p. P100-AS extracts were prepared from the indicated strains and immunoprecipitation reactions were performed using an antibody specific for one protein (IP Antibody). Western blot analysis was then performed to detect HA3-Tor2p. Where indicated, P100-AS extracts were treated with 1% maltoside before immunoprecipitation. (B) A cross-linking agent stabilizes interactions between Lst8p-Myc and HA3-Tor2p in the absence of membranes. Where indicated, DPS was added to a final concentration of 1 mM (+) or 1.8 mM (++) before detergent solubilization and immunoprecipitation by using anti-Myc antibody.
Approximately 20–50% of HA3-Tor2p from the P100-AS fraction now interacted with immobilized polyclonal anti-HA antibody (Figure 1H, compare lanes 5 and 6). Of this, >90% could be released upon incubation with competing HA dipeptide (Figure 1H, compare lanes 7 and 8). When the eluate was analyzed by SDS-PAGE, several distinct protein species were detected that coeluted with HA3-Tor2p (Figure 1I, lane 2). These proteins were specifically associated with HA3-Tor2p because they were not present when a control P100-AS extract, prepared from the parental untagged strain, was used instead for immunopurification (Figure 1I, lane 1). We noted that much of the eluted HA3-Tor2p seemed monomeric on a sucrose gradient, suggesting that the affinity purification step significantly disrupted interactions between HA3-Tor2 and another component(s) present in the P100-AS extract (our unpublished data).
To identify the proteins that copurified with HA3-Tor2p, the eluate was digested with trypsin and subjected to tandem mass spectrometric analysis (Carroll et al., 1998; Link et al., 1999). For comparison, the untagged control eluate was also analyzed. The results showed that, in addition to Tor2p, a predominant number of unique peptides corresponding to four different proteins were present in the eluate from the HA3-Tor2p sample (Table 1). Two of these (Avo1p and Avo3p) are encoded by essential genes and, along with Avo2p, were of previously unknown biological function. While this manuscript was in preparation, Hall and coworkers independently identified Avo1p-Avo3p as Tor2p-interacting proteins (Loewith et al., 2002). The fourth protein, Lst8p, is encoded by an essential gene and is the subject of further experiments described below.
Table 1.
Analysis of Tor2-associated proteins by mass spectrometry
| No. | Protein | ORF | MW | Unique Peptides | Total Peptides | Essential |
|---|---|---|---|---|---|---|
| 1 | Tor2 | YKL203C | 281,509 | 40 | 67 | Yes |
| 2 | Avo1 | YOL078W | 131,378 | 21 | 23 | Yes |
| 3 | Avo3 | YER093C | 164,368 | 11 | 12 | Yes |
| 4 | Avo2 | YMR068W | 47,140 | 8 | 9 | No |
| 5 | Lst8 | YNL006W | 34,034 | 7 | 7 | Yes |
ORF, open reading frame. Proteins listed correspond to abundant peptide species identified by mass spectrometry of trypsin-digested HA3-Tor2p copurifying material. Proteins are rank ordered according to the number of unique peptides obtained for each and were uniquely identified in the strain expressing HA3-Tor2p. Additional abundant peptides were obtained corresponding to a number of ribosomal proteins as well as TyB Gag-Pol proteins; however, many similar proteins were also identified in the control sample and their presence is thus considered spurious. Several additional proteins specifically coeluted with HA3-Tor2p; however, these proteins were associated with peptides present in lower amounts and have not been characterized further.
Detergent-sensitive Association between Lst8p and Tor2p
Lstp8p was identified originally in a genetic screen for components of the exocytic secretory pathway and has been implicated in the regulated intracellular sorting of the general amino acid permease Gap1p (Roberg et al., 1997). Butow and coworkers independently identified Lst8p as a negative regulator of RTG target gene expression (Liu et al., 2001). We as well as others have demonstrated that the Tor kinases also negatively regulate RTG target gene expression (Komeili et al., 2000; Shamji et al., 2000). Our identification of Lst8p as a protein that copurifies with Tor2p suggested that Lst8p might function as part of the Tor pathway. We therefore decided to explore this relationship in greater detail.
We first wanted to test whether Lst8p and Tor2p interact physically, as suggested by the results of mass spectrometry. We reasoned that we should be able to perform the reciprocal experiment and coimmunoprecipitate Tor2p by using an antibody specific for Lst8p. Accordingly, we introduced into the HA3-TOR2 strain a genomically integrated version of Lst8p that was carboxy terminally tagged with the Myc epitope (Lst8p-Myc). This strain grew as well as HA3-TOR2 and our untagged parental strain, indicating that the Myc tag did not interfere with the essential function of Lst8p (our unpublished data). We prepared a P100-AS extract and used an anti-Myc mAb to precipitate Lst8p-Myc, followed by Western blot analysis with an anti-HA mAb to detect HA3-Tor2p. The results demonstrated that HA3-Tor2p associated with Lst8p-Myc in the P100-AS extract (Figure 2A, lane 6). The amount of HA3-Tor2p that coprecipitated was similar to that observed when HA3-Tor2p was precipitated directly using an anti-HA antibody (Figure 2A, compare lanes 3 and 6). Moreover, the interaction was specific because no significant HA3-Tor2p was detected when an extract was used that was prepared from HA3-TOR2 (Figure 2A, lane 9). As an additional control, we performed a similar coimmunoprecipitation experiment by using an extract prepared from a strain that produced both HA3-Tor2p as well as a Myc-tagged version of Sec8p, a protein that was enriched in the P100-AS fraction but was not identified as an HA3-Tor2p copurifying protein (our unpublished data). Herein, no association was observed between HA3-Tor2p and Sec8p-Myc (our unpublished data).
Treating the P100-AS fraction with 1% lauryl maltoside, a nonionic detergent, before immunoprecipitation abolished interaction between HA3-Tor2p and Lst8p-Myc, demonstrating their association was stabilized by the presence of a membrane (Figure 2A, lane 15). As a control, we observed that under these conditions HA3-Tor2p was still efficiently precipitated with anti-HA antibody and Lst8p-Myc was efficiently precipitated with anti-Myc antibody, indicating that the detergent did not interfere with binding of the antibodies to their respective epitopes (Figure 2A, lane 12; our unpublished data). To test whether these proteins were in a complex within the membrane, we first cross-linked proteins in the P100-AS fraction by using the homobifunctional cross-linking agent DSP before detergent solubilization. Under these conditions, significant coprecipitation of HA3-Tor2p and Lst8p-Myc was observed (Figure 2B, compare lane3 with lanes 4 and 5). Because DSP is believed to primarily cross-link proteins that are in proximity (Deshaies et al., 1991), we conclude that Tor2p and Lst8p are likely to be in close physical contact within the context of a membrane.
Tor2p and Lst8p Colocalize In Situ
To further test whether Tor2p and Lst8p act together, we determined their localization within cells. To date, no information regarding the intracellular location of Lst8p has been reported. Hall and coworkers, on the other hand, have presented evidence, based on cell fractionation and indirect immunofluorescence studies, that Tor2p localizes both at the plasma membrane as well as within the cell interior (Kunz et al., 2000). One potential drawback to their study, however, was that Tor2p had to be overexpressed to be visualized by fluorescence microscopy. As an alternative approach, we explored the use of IEM to study the location of these proteins (Rieder et al., 1996). This approach has proven successful to localize epitope-tagged proteins expressed at endogenous levels (Rieder et al., 1996; Wang et al., 2001). Moreover, by examining protein localization at the ultrastructural level, additional insights into their physical environment in situ can often be obtained (Wang et al., 2001).
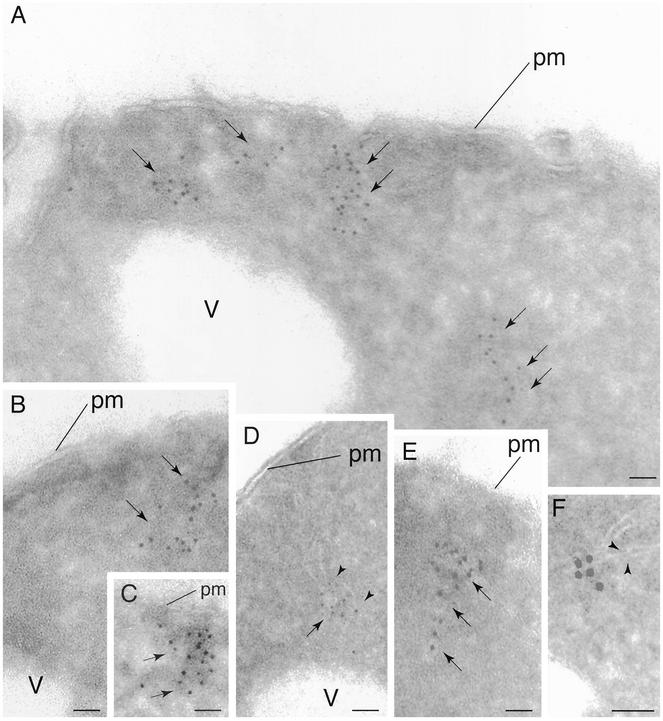
Ultrathin cryosections were prepared from fixed HA3-TOR2 cells and were incubated with anti-HA antibody. Samples were then incubated with 10-nm gold secondary antibody and visualized by electron microscopy. Clusters of gold particles were found primarily in electron dense regions within the cell (Figure 3, A–C, arrows). A significant fraction of these particles were adjacent to the cell periphery, yet at locations clearly distinct from the plasma membrane (40%, n = 225). The remaining particles were located primarily further within the cell periphery and, of these, a small number (∼12%, n = 65) were located in proximity to the vacuole. Very few particles were associated with the nucleus (3%, n = 16). In agreement with our biochemical analyses described above, many of the particles were in close association with membranes, which showed as characteristic tubule-like structures, or tracks, of lower electron density (Figure 3, D–F, arrowheads). These particles specifically labeled HA3-Tor2p because no such signal was detected when the untagged control strain was examined (our unpublished data). We conclude from this analysis that Tor2p associates with a membranous compartment(s) that is located both within the cell interior as well as adjacent to the plasma membrane.
Figure 3.
Localization of Tor2p by IEM. Cells expressing HA3-Tor2p were grown in rich media (YPD) and prepared for electron microscopy and probed with anti-HA antibody, followed by incubation with secondary antibody decorated with 5-nm gold particles. In A–F, arrows denote regions where gold particles are clustered. In D and F, arrowheads point out regions where membrane tracks are clearly evident. pm, plasma membrane; v, vacuole. Bar, 85 nm.
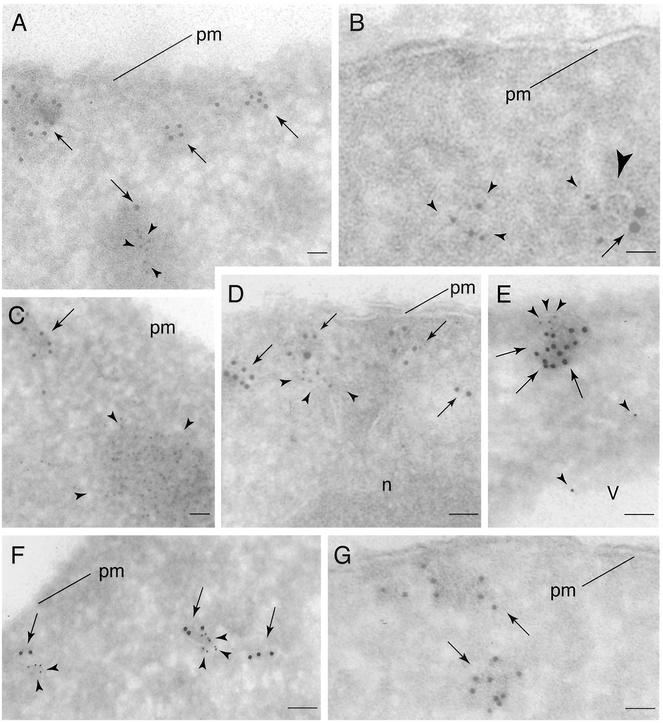
The presence of Tor2p in electron-dense regions close to the plasma membrane is reminiscent of previous descriptions of cortical actin patches (Mulholland et al., 1999). This was intriguing given the involvement of Tor2p in actin cytoskeletal dynamics (Schmidt et al., 1996, 1997; Schmelzle and Hall, 2000). Therefore, to determine the relationship between the Tor2p-labeled structures and actin patches, we performed a double labeling experiment where actin was labeled with 5-nm gold particles and Tor2p was labeled with larger 10-nm gold particles. This experiment took advantage of the fact that the majority of actin detected using this approach is located primarily within actin patches (Mulholland et al., 1999). Indeed, clusters of smaller gold particles were clearly observed in electron-dense stained structures near the cell periphery (Figure 4, A and C, arrowheads). These structures were clearly distinct from the HA3-Tor2p clusters, however, demonstrating that HA3-Tor2p does not localize to actin patches (Figure 4, A and C). Nevertheless, we noted several examples where actin that was not obviously associated with cortical patches colocalized with HA3-Tor2p, often in the vicinity of membrane tracks (Figure 4, B and D). Although the significance of this latter observation remains to be determined, it is consistent with the demonstrated functional relationship between actin organization and Tor2p signaling (Schmidt et al., 1996, 1997; Schmelzle and Hall, 2000).
Figure 4.
Visualizing Tor2p, Tor1p, Lst8p, and actin by IEM. (A–D) Double labeling of HA3-Tor2p (arrows) and actin (arrowheads) with 10- and 5-nm gold particles, respectively, in cryosections prepared from cells expressing HA3-Tor2p. In A and C, actin patches are clearly evident that are distinct from electron dense clusters of labeled HA3-Tor2p. In B and D, actin labeling is in proximity to HA3-Tor2p. The large arrowhead in B denotes a visible membrane track. (E and F) Double labeling of HA3-Tor2p (arrows) and Lst8p-Myc (arrowheads) with 10- and 5-nm gold particles, respectively, in cryosections prepared from cells expressing HA3-Tor2p and Lst8p-Myc. (G) Labeling of Tor1p in cells expressing HA3-Tor1p with anti-HA antibody, followed by incubation with secondary antibody decorated with 5-nm gold particles. In A–G, all cells were grown in YPD. n, nucleus. Other labels are as described in the legend to Figure 3.
We next prepared ultrathin cryosections from HA3-TOR2 LST8-MYC cells and performed a double-labeling experiment where HA3-Tor2p and Lst8p-Myc were labeled with 10- and 5-nm gold particles, respectively. We observed many cases where gold particles of both sizes were in proximity to each other, in electron dense stained regions near the plasma membrane as well as within the cell interior (Figure 4, E and F). Moreover, close inspection revealed the presence of membrane-like tracks at these sites of colocalization. We conclude from these results that Lst8p and Tor2p interact and colocalize to an intracellular membranous site(s) within the cell.
Lst8p Associates with Tor1p
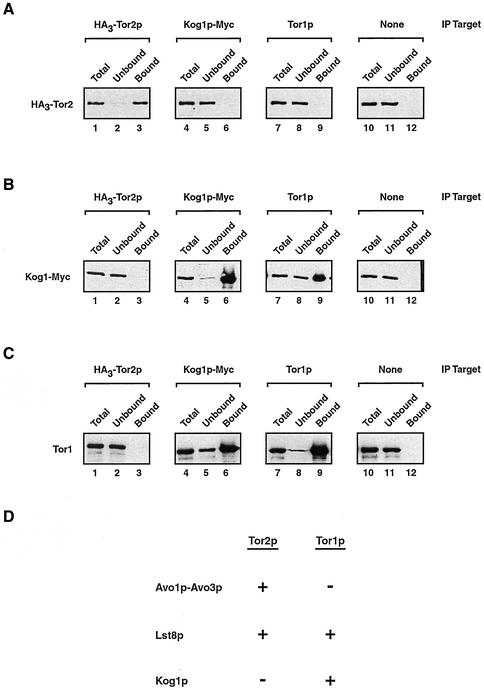
In addition to the observed colocalization of Tor2p and Lst8p in the IEM analysis described above, there were several instances where the signal from one protein was not within the vicinity of the other (Figure 4, E and F; our unpublished data). This result suggested that a portion of these proteins were not associated with one another. We reasoned that one possible explanation for this observation was that Lst8p might also interact with Tor1p. To test this idea directly, we performed a similar coprecipitation experiment to that described previously and immunoprecipitated Lst8p-Myc from a P100-AS extract and probed for the presence of Tor1p, by using a polyclonal antibody directed against this protein. As a control, we first confirmed that this anti-Tor1p antibody was specific for Tor1p in our strain background, because no signal corresponding to the predicted molecular weights of Tor1p or Tor2p could be detected upon deletion of the TOR1 gene (Figure 5A, compare lane 1 with lanes 2 and 3).
Figure 5.
Testing interactions between Tor1p and both Lst8p and Avo1p. (A) Demonstrating the specificity of an anti-Tor1p polyclonal antibody. Western blot analysis was performed on extracts prepared from the indicate strains. (B) Tor1p coprecipitates with Lst8p-Myc. Immunoprecipitation reactions were performed using anti-Myc antibodies and P100-AS fractions prepared from the indicated strains, followed by Western blot analysis to detect Tor1p. Where indicated, DSP was added at a final concentration of 1.8 mM before detergent solubilization. (C) Tor1p and Avo1p do not interact. Immunoprecipitation experiments were performed from extracts prepared from a strain that expressed Avo1p-Myc, by using either monoclonal anti-myc antibodies or polycolonal anti-Tor1p antibodies as indicated (IP target). Western blot analysis was then used to detect the presence of the indicated protein. Control beads indicates that Sepharose beads linked to protein G were used without a primary antibody. (D) Sedimentation profiles of Avo1p, Tor2p, Lst8p, and Tor1p. Clarified whole cell extracts were prepared from strain HA30TOR2 LST8-MYC or from strain AVO1-MYC and were subjected to ultracentrifugation of sucrose gradients. Fractions were then collected and analyzed by Western blotting to detect Avo1p-Myc, HA3-Tor2p, Lst8p-Myc, or Tor1p.
We observed that a portion of Tor1p indeed coprecipitated with Lst8p-Myc (Figure 5B, lane 2). The interaction was specific because no Tor1p coprecipitated from an extract prepared from a control strain producing untagged Lst8p (Figure 5B, lane 6). As with Tor2p and Lst8p, no association was evident if the P100-AS fraction was first solubilized with 1% maltoside, indicating that interaction between these proteins was detergent sensitive (Figure 5B, lane 3). Addition of the cross-linking agent DSP to the P100-AS fraction before detergent solubilization restored significant coprecipitation of Tor1p, indicating that Tor1p, like Tor2p, is likely to interact directly with Lst8p (Figure 5B, lane 4).
We also examined whether Tor1p interacted with another Tor2p-copurifying protein, Avo1p. For this, we constructed a genomically integrated version of Avo1p that was carboxy terminally tagged with the Myc epitope (Avo1p-Myc). Control experiments demonstrated that Avo1p-Myc was efficiently immunoprecipated from cell extracts by using monoclonal anti-Myc antibody (Figure 5C, lane 3). However, no coprecipitation of Tor1p was observed (Figure 5 C, lane 12). Similarly, no coprecipitation of Avo1p-Myc was observed when Tor1p was the target of immunoprecipitation (Figure 5C, lane 6). These results are in agreement with results of Hall and coworkers who have reported that Avo1, as well as Avo2 and Avo3, interact exclusively with Tor2 (Loewith et al., 2002).
In agreement with results from coprecipitation experiments, we observed that Lst8p-Myc displayed a sedimentation profile that was overlapping with respect to the profiles of Tor1p as well as HA3-Tor2p, when whole cell extracts were analyzed by equilibrium density ultracentrifugation (Figure 5D). In contrast, the sedimentation profile of Avo1-Myc was shifted to more dense fractions and seemed most similar to the profiles of HA3-Tor2p and, to a lesser extent, Lst8p-Myc (Figure 5D).
In Situ Localization of Tor1p
Given that their profiles on sucrose step gradients were only partially overlapping, we wished to determine whether Tor1p localized to a region of the cell that was fundamentally distinct from what we determined for Tor2p. Accordingly, we constructed strain HA3-TOR1 that produced a version of Tor1p under the control of its own promoter and tagged at its amino terminus with HA3, as described in MATERIALS AND METHODS. Ultrathin cryosections were prepared from fixed cells and HA3-Tor1p was visualized by IEM (Figure 4G). As was observed for HA3-Tor2p, HA3-Tor1p directed gold particles were observed both adjacent to the plasma membrane as well as within the cell interior (Figure 4, compare A and G). In many cases, these particles were clustered in electron dense regions and were often associated with characteristic membrane tracks (Figure 4G; our unpublished results). Thus, we conclude that at least a portion of Tor1p is in an environment very similar to that of Tor2p. Interestingly, however, we detected many unclustered Tor1p-directed gold particles dispersed throughout the cytoplasm (our unpublished data). Although the significance of this latter observation will require further investigation, it is conceivable that this more dispersed Tor1p signal corresponds to the population of Tor1p that occurred in fractions of lesser buoyant density on sucrose gradients (Figure 5D).
Interactions between Lst8p and Tor Proteins Withstand Rapamycin Treatment
To explore the functional relationship between Lst8p and Tor2p as well as Tor1p, we asked whether inhibition of Tor signaling by rapamycin affected their association. Accordingly, HA3-TOR2 LST8-MYC cells were treated with rapamycin or with drug vehicle alone (DMSO) for 30 min, followed by preparation of P100-AS fractions. As a control, Northern blot analysis demonstrated that the expression patterns of several Tor-regulated genes were affected in a characteristic manner by rapamycin, confirming the efficacy of the drug in this strain background (Figure 6A, compare lanes 1 and 2). Coprecipitation reactions were performed, where Lst8p-Myc was immunoprecipitated with anti-Myc antibody, followed by Western blot analysis to detect HA3-Tor2p or Tor1p. No significant difference in the amount of Tor1p or HA3-Tor2p coprecipitated with Lst8p-Myc was observed in rapamycin treated vs. untreated samples (Figure 6C, compare lanes 3 and 6 as well as lanes 15 and 18). In addition, we found that starvation for nitrogen as well as carbon sources also did not appreciably affect the association of these proteins (our unpublished data). Taken together, we conclude that Lst8p interacts stably with both Tor1p and Tor2p under at least these conditions known to influence Tor-dependent signaling.
Figure 6.
Testing the effects of rapamycin treatment on the association between Lst8p and Tor1p as well as Tor2p. (A) Control experiment demonstrating that previously characterized Tor-regulated target genes are influenced by rapamycin (Powers and Walter, 1999; Komeili et al., 2000; Shamji et al., 2000). Shown are results of Northern blot analysis, where RPL30 mRNA levels are decreased by rapamycin treatment, as described previously (Powers and Walter, 1999). In contrast, expression of CIT2 and DLD3, genes regulated by Rtg1p and Rtg3p, as well as GAP1, regulated by Gln3p, are all induced after drug treatment, as described previously (Komeili et al., 2000; Shamji et al., 2000). (B) Sedimentation profiles of Tor1p and Tor2p obtained from cells that were either treated or not treated with rapamycin. Clarified whole cell extracts were prepared from strain HA3-TOR2 after treatment with either rapamycin or drug vehicle alone and were subjected to ultracentrifugation on sucrose gradients. Fractions were then collected and analyzed by Western blotting to detect HA3-Tor2p or Tor1p. (C) Coimmunoprecipitation experiments demonstrating that the association of Lst8p with both Tor1p and Tor2p is stable to rapamycin treatment. The indicated strains were grown in YPD and treated with rapamycin, followed by preparation of P100-AS extracts. Lst8p-Myc was then immunoprecipitated and the presence of HA3-Tor2p and Tor1 was monitored by Western blot analysis.
Finally, we examined the sedimentation behavior of Tor1p and Tor2p on sucrose gradients in extracts prepared from either rapamycin treated or untreated cells (Figure 6B). No significant change in the sedimentation profile of either protein was evident, suggesting that their overall cellular distribution is unaffected by drug treatment. Similar results were observed when cells were starved for carbon as well as nitrogen sources (our unpublished data). These results suggest that changes in Tor signaling are unlikely to be accompanied by a dramatic relocalization of either Tor1p or Tor2p.
Kog1p Interacts Specifically with Tor1p
While our studies were in progress, two groups reported interactions between mTOR and a novel regulatory partner, termed Raptor (Hara et al., 2002; Kim et al., 2002). A homologue of this protein is identifiable in S. cerevisiae, encoded by an essential gene, YHR186C, recently named KOG1 (Loewith et al., 2002). We wanted to determine whether Kog1p was associated with Tor2p yet escaped detection by mass spectrometry. Accordingly, we constructed a strain that produced both HA3-Tor2p as well as a version of Kog1p that was tagged at its C terminus with multiple copies of the Myc-epitope (Kog1p-Myc). This strain displayed no growth defect, indicating that the Myc epitope did not interfere with the essential function of Kog1p (our unpublished observations). Extracts were prepared and HA3-Tor2p as well as Kog1p-Myc was immunprecipitated separately using anti-HA or anti-Myc monoclonal antibodies, respectively, followed by Western blot analysis. Each tagged protein was precipitated effectively using its corresponding antibody (Figures 7A, lane 3, and 6B, lane 6). In no case, however, was significant coprecipitation observed, indicating that Tor2p and Kog1p do not interact, at least under these experimental conditions (Figures 7A, lane 6, and 6B, lane 3).
Figure 7.
Kog1p associates with Tor1p. (A–C) Immunoprecipitation experiments were performed from extracts prepared from a strain that expressed both HA3-Tor2p as well as Kog1p-Myc, by using an antibody specific for a given protein (IP target). Western blot analysis was then used to detect the presence of the indicated protein. Control beads indicates that Sepharose beads linked to protein G were used without a primary antibody. (D) Summary of relationships established in this study, where a + indicates an association between a given protein with Tor1p and/or Tor2p was observed and a − indicates that no significant association is observed. Note that we have determined only that Avo1p does not interact with Tor1p and have not yet tested interactions between Avo2p, Avo3p, and Tor1p.
In contrast to these results, a significant amount of Tor1p coprecipitated with Kog1p-Myc (Figure 7C, lane 6). This interaction was specific because no Tor1p precipitated when control beads were used without antibody (Figure 7C, lane 12). Moreover, no Tor1p was precipitated by anti-Myc antibody in a strain that did not contain Myc-tagged Kog1p (our unpublished observations). To further examine their interaction, we performed the reciprocal experiment and determined that Kog1p-Myc was coprecipitated when antibodies directed against Tor1p were used for immunoprecipitation (Figure 7B, lane 9). During these experiments, we also monitored interactions between HA3-Tor2p and Tor1p; no significant coprecipitation was observed, indicating these proteins do not associate stably under these experimental conditions (Figures 7A, lane 9, and 6C, lane3).
DISCUSSION
Evidence for Distinct Tor Kinase Protein Complexes in S. cerevisiae
We have used a biochemical approach to identify a number of proteins that copurify with Tor2p, namely, Avo1p-Avo3p and Lst8p. Of these proteins, we have shown that Lst8p interacts directly with both Tor2p as well as Tor1p. Moreover, we have found that Avo1p associates with Tor2p but not with Tor1p. We also observed that Kog1p, the yeast homologue of the recently identified mTor regulatory protein Raptor, associates with Tor1p but not Tor2p. Finally, we find no evidence for a direct interaction between Tor1p and Tor2p. Taken together, these results provide evidence for the existence of Tor kinase signaling complexes that contain distinct yet overlapping components in S. cerevisiae (summarized in Figure 6D).
While this manuscript was in preparation, Hall and coworkers reported the identification of two distinct Tor-containing protein complexes (Loewith et al., 2002). Based on their observations, the Tor2p-associated proteins we have identified (Table 1; Figure 6D) most likely correspond to what they have termed Tor complex 2 (TORC2). Similarly, our described interactions between Tor1p, Lst8p, and Kog1p (Figure 6D) likely correspond to their described TORC1 (Loewith et al., 2002). In contrast to their findings, we did not detect a significant interaction between Kog1p and Tor2p. This discrepancy is likely due in part to the observation that, in mammalian cells, interactions between mTOR and Raptor are dynamic and potentially unstable (Hara et al., 2002; Kim et al., 2002).
We note that for three of the four Tor2p copurifying proteins we have identified, Avo1p, Avo3p, and Lst8p, their predicted molecular weights correspond closely to proteins identifiable by SDS-PAGE (Table 1 and Figure 1I). These proteins did not stain as intensely as Tor2p, however, nor could a specific band corresponding to the predicted molecular weight of Avo2p be detected. At present, there are several possible explanations for the apparent substoichiometric occurrence of these proteins relative to Tor2p, including the fact that our immunopurification of HA3-Tor2p partially disrupted interactions with other components. Future studies will be aimed at characterizing the precise stoichiometry and dynamics of these complexes. Moreover, at present it is unknown whether these Tor-associated proteins represent potential targets for Tor kinase activity or whether they instead modulate Tor activity.
Intracellular Membrane Association of Tor Kinases
A second major finding presented herein is that Tor1p, Tor2p, and Lst8p (and presumably Avo1p-Avo3p) all localize to membranous structures that are proximal to, yet distinct from, the plasma membrane and also within the cell interior. These findings are consistent with results pointing to the fact that in mammalian and yeast cells the Tor kinases are predominantly membrane associated (Sabatini et al., 1999; Kim and Chen, 2000; Zhang et al., 2002). However, they contrast with the plasma membrane localization reported by Hall and coworkers by using indirect immunofluorescence (Kunz et al., 2000). We attribute this difference to two possible factors. First, we have examined the localization of these proteins by IEM, which provides higher resolution information regarding subcellular localization, compared with immunofluorescence microscopy. Second, we monitored endogenous levels of the Tor proteins whereas Hall and coworkers monitored Tor proteins that were overexpressed by use of the strong GAL1/10 promoter, making it possible that these proteins were mislocalized to the plasma membrane (Kunz et al., 2000). Importantly, however, these disparate observations concerning Tor localization could point to the possibility that the Tor proteins cycle, perhaps through an endocytic pathway. This possibility would provide an explanation for the observation that a significant portion of Tor2p fractionates in a manner similar to plasma membrane proteins, which often partly localize to endomembrane structures (Shaw et al., 2001). In support of this idea, at the electron microscopy level these Tor-associated characteristic membranous tracks are morphologically most similar to membranes associated with the endocytic pathway (Rieder et al., 1996; Mulholland et al., 1999; Wang et al., 2001).
The possibility that the membranous structures associated with the Tor kinases are endosomal in origin is also appealing given the emerging connection between endocytosis and sphingolipid biosynthesis (Friant et al., 2001; Anderson and Jacobson, 2002; Dickson and Lester, 2002). Specifically, a number of recent studies indicate that sphingolipid-mediated signaling is linked to proper endocytosis as well as actin cytoskeletal organization (Friant et al., 2001; Anderson and Jacobson, 2002; Dickson and Lester, 2002). In this context, it is significant that Avo3p, one of the Tor2p copurifying proteins we identified, was discovered originally in a genetic screen for components involved in sphingolipid biosynthesis (Beeler et al., 1988) (note in this manuscript Avo3p/YER093C is named TSC11). Remarkably, TOR2 was also identified in this genetic screen, emphasizing a relationship between Tor signaling and processes connected, albeit indirectly, to endocytosis (Beeler et al., 1988). It is also becoming increasingly clear that the endocytic compartment is the site of a significant number of signaling events related to the control of cell growth and morphogenesis in yeast as well as in higher eukaryotes (Harsay and Schekman, 2002; Seto et al., 2002). Considering the multitude of processes affected by the Tor kinases, many of which are influenced by extracellular events, the endocytic compartment represents an attractive location to anchor Tor-dependent signaling.
Several additional observations are inconsistent, however, with an endosomal location for the Tor kinases or their associated proteins. First, in cell fractionation and equilibrium density ultracentrifugation studies, we failed to observe significant copurification of Tor2p, Tor1p, or Lst8p and markers of the endocytic pathway, such as Rsp5p and Pep12p, which are involved in early and late steps, respectively (Wang et al., 2001) (our unpublished data). Hall and coworkers also reported little similarity between the biochemical behavior of Tor2p and different endocytic marker proteins (Kunz et al., 2000). Moreover, attempts by these investigators to correlate Tor2p-dependent events and the activity of the endocytic pathway have yielded largely negative results (Beck et al., 1999; Kunz et al., 2000). These studies, however, do not exclude the possibility that the Tor-signaling complexes described here are nevertheless associated with a novel branch of the endocytic pathway that has escaped detection by experiments attempted thus far.
Another observation that potentially conflicts with an endosomal location for Tor is that Lst8p, which we have demonstrated interacts with both Tor1p as well as Tor2p, was originally identified in a genetic screen for components involved in the exocytic secretory pathway (Roberg et al., 1997). Analysis of an lst8-1 mutant suggested an involvement in regulated delivery of nutrient-regulated permeases, such as Gap1, to the plasma membrane (Roberg et al., 1997). As the initial screen was designed to identify components that interacted with Sec13p, a component of COPII transport vesicles, the proposed model was that Lst8p is involved in a post-Golgi step of the secretory pathway (Roberg et al., 1997). However, it was acknowledged that, because the effect of the lst8-1 allele on Gap1p trafficking is similar to growth on preferred nitrogen sources (e.g., glutamate), namely, increased trafficking of Gap1p to the vacuole, it thus remains equally possible that Lst8p is involved in a signaling pathway related to nitrogen metabolism. As detailed below, we believe that this latter possibility is likely to be the case. In any event, however, the precise identity of the membranous compartment(s) associated with the Tor complexes described herein must await future identification.
Tor Kinases and Lst8: Further Convergence of Tor and Retrograde Signaling
Additional mutations in the LST8 gene distinct from lst8-1, specifically the lst8-(2-5) alleles, were identified genetically by isolating cells that expressed the RTG target gene, CIT2, in the absence of Rtg2p (Liu et al., 2001). Further analysis showed that these mutations can alleviate the glutamate auxotrophy of an rtg2Δ strain and that Lst8p is a negative regulator of RTG target gene expression (Liu et al., 2001). This screen for Rtg2p bypass mutants also identified MKS1 as a negative regulator of the RTG target genes (Sekito et al., 2002). Both Lst8p and Mks1p have been placed within the context of mitochondrial retrograde signaling, possibly by linking intracellular glutamate levels to the activity of Rtg1p and Rtg3p (Liu et al., 2001; Sekito et al., 2002).
We as well as others have also identified Mks1p as a negative regulator of the RTG pathway (Dilova et al., 2002; Tate et al., 2002). Moreover, we have presented evidence that the phosphorylation state of Mks1p is influenced by Tor signaling (Dilova et al., 2002). Our finding herein that Lst8p interacts with both Tor1p as well as Tor2p suggests that Lst8 also is involved integrally in Tor signaling. Together these results substantiate the mechanistic similarities between retrograde control of RTG target gene expression and Tor-dependent control of these genes (Komeili et al., 2000; Sekito et al., 2000).
An exception to this overall agreement between the two responses concerns the phosphorylation state of Rtg3p, where retrograde induction of the pathway correlates with dephosphorylation of this protein (Sekito et al., 2000). In contrast, we observed that induction of RTG target gene expression by rapamycin treatment results in an apparent hyperphosphorylation of Rtg3p (Komeili et al., 2000). We have recently been able to attribute this discrepancy to differences in strain backgrounds as well as the precise composition of media used in these published studies (Dilova, unpublished data). We therefore believe that the fundamental mechanism(s) by which the RTG target genes are regulated is likely to be very similar between the Tor and retrograde pathways. We have found that interactions between Lst8 and the Tor kinases are not perturbed by rapamycin treatment or by nutrient starvation. It will be important to determine whether any of the identified Lst8p mutants affect interactions with Tor or, alternatively, with any of the Tor-associated proteins we have identified.
CONCLUSION
It has been suggested that the Tor kinases act as a “multichannel processor,” integrating diverse upstream nutrient-related signals to specific downstream responses (Shamji et al., 2000). For this to occur, the Tor proteins must be able to differentially control distinct downstream targets according to the precise nutritional state of the cell. The existence of distinct Tor1p and Tor2p containing multiprotein complexes in yeast suggests a mechanism for this differential regulation. We propose that unique Tor complex components function to specify the targets of Tor kinase activity, which to date remain largely unidentified. We also propose that their colocalization and shared components function to promote cross talk between these distinct Tor complexes. Finally, we are intrigued by the observation that the Tor kinases are associated with membranous structures located both proximal to the plasma membrane as well as within the cell interior. Given the emerging connection between intracellular signaling events and membrane-mediated protein trafficking, we believe this membrane association represents an important area for future investigation into the activity and regulation of the Tor kinases.
ACKNOWLEDGMENTS
We are indebted to D. Kellogg for advice and encouragement during identification of Tor2p-associated proteins. We also thank the Nunnari and Kaplan laboratories for advice and assistance during the course of this work. We thank M. Hall for support and for communication of results before publication. We are grateful to J. Nunnari for discussions and for critical comments on the manuscript. Finally, we thank Stephen Fairclough, Cory Iverson, Andrea Von Dollen, and Anthony Tam for technical assistance. This work was sponsored by a grant from the Cancer Research Coordinating committee of California, a Basil O'Connor Starter Research Award from the March of Dimes, and National Science Foundation grants MCB-1031221 (to T.P.) and DBI-0099706 (to J.M.M.), and by National Institutes of Health grant RR-11823 (to J.Y.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0609. Article and publication date are at http:// www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0609.
REFERENCES
- Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P. Heat repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of Ca2+-sensitive csg2Δ mutant. J Biol Chem. 1988;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan T-F, Zheng XFS. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cardenas M, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C, Altman R, Schieltz D, Yates J, Kellogg DR. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta. 2002;1583:13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- Dilova I, Chen C-Y, Powers T. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumor suppressor proteins antagonize amino-acid-TOR signaling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- Geitz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Harsay E, Schekman R. A subset of yeast vascuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan K-L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signaling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Alberts BM. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol Biol Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J. Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc Natl Acad Sci USA. 2000;97:14340–14345. doi: 10.1073/pnas.011511898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Komeili A, Wedaman KP, O'Shea EO, Powers T. Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liao X, Small WC, Srere PA, Butow RA. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotech. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Epstein CB, Butow RA. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 2001;20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie III A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mortensen EM, McDonald H, Yates J, III, Kellogg DR. Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Konopka J, Singer-Kruger B, Zerial M, Botstein D. Visualization of receptor-mediated endocytosis in yeast. Mol Biol Cell. 1999;10:799–817. doi: 10.1091/mbc.10.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Wong ED, Meeusen S, Wagner JA. Studying the behavior of mitochondria. Methods Enzymol. 2002;351:381–393. doi: 10.1016/s0076-6879(02)51859-0. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci USA. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7, and. LST8. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J, Heitman J, Cardenas ME. The TOR kinases link nutrient sensing to cell growth. J Biol Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Barrow RK, Blackshaw S, Burnett PE, Lai MM, Field ME, Bahr BA, Kirsch J, Betz H, Snyder SH. Interaction of RAFT1 with gephryin required for rapamycin-sensitive signaling. Science. 1999;284:1161–1164. doi: 10.1126/science.284.5417.1161. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Liu Z, Thornton J, Butow RA. RTG-dependent mitochondria-to-nucleus signaling is regulated by Mks1 and is linked to formation of yeast prion [URE3] Mol Biol Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Thorton J, Butow R. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto ES, Bellen HJ, Lloyd TE. When cell biology meets development: endocytic regulation of signaling pathways. Genes Dev. 2002;16:1314–1336. doi: 10.1101/gad.989602. [DOI] [PubMed] [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapmycin among the effectors of the Tor proteins. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- Shaw JD, Cummings KB, Huyer G, Michaelis S, Wendland B. Yeast as a model system for studying endocytosis. Exp Cell Res. 2001;271:1–9. doi: 10.1006/excr.2001.5373. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Tate JJ, Cox KH, Rai R, Cooper TG. Mks1p is required for negative regulation of retrograde gene expression in Saccharomyces cerevisiae but does not affect nitrogen catabolite repression-sensitive gene expression. J Biol Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, McCaffery JM, Wendland B, Dupre S, Haguenauer-Tsapis R, Huibregtse JM. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol Cell Biol. 2001;21:3564–3575. doi: 10.1128/MCB.21.10.3564-3575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Mikhailov A, Kallo-Hosein H, Hara K, Yonezawa K, Avruch J. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta. 2002;1542:41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shu L, Hosoi H, Murti KG, Houghton PJ. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem. 2002;31:28127–28134. doi: 10.1074/jbc.M202625200. [DOI] [PubMed] [Google Scholar]
- Zheng X-F, Fiorentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]