Abstract
We have shown previously that the transforming growth factor-β (TGFβ)-regulated Sma-Mad (Smad) protein 3 and Smad4 proteins transactivate the apolipoprotein C-III promoter in hepatic cells via a hormone response element that binds the nuclear receptor hepatocyte nuclear factor 4 (HNF-4). In the present study, we show that Smad3 and Smad4 but not Smad2 physically interact with HNF-4 via their Mad homology 1 domains both in vitro and in vivo. The synergistic transactivation of target promoters by Smads and HNF-4 was shown to depend on the specific promoter context and did not require an intact β-hairpin/DNA binding domain of the Smads. Using glutathione S-transferase interaction assays, we established that two regions of HNF-4, the N-terminal activation function 1 (AF-1) domain (aa 1–24) and the C-terminal F domain (aa 388–455) can mediate physical Smad3/HNF-4 interactions in vitro. In vivo, Smad3 and Smad4 proteins enhanced the transactivation function of various GAL4-HNF-4 fusion proteins via the AF-1 and the adjacent DNA binding domain, whereas a single tyrosine to alanine substitution in AF-1 abolished coactivation by Smads. The findings suggest that the transcriptional cross talk between the TGFβ-regulated Smads and HNF-4 is mediated by specific functional domains in the two types of transcription factors. Furthermore, the specificity of this interaction for certain target promoters may play an important role in various hepatocyte functions, which are regulated by TGFβ and the Smads.
INTRODUCTION
Transforming growth factor-β (TGFβ) is a pleiotropic cytokine that plays important roles in a plethora of biological processes, including cell growth, differentiation, apoptosis, and extracellular matrix production (Heldin et al., 1997; Massague, 1998; Massague and Wotton, 2000; ten Dijke et al. 2000; Moustakas et al. 2001). TGFβ binds to and activates two types of Ser/Thr kinase receptors (type I and type II TGFβ receptors) expressed in almost every tissue of the adult organism. After TGFβ stimulation, the type II receptor phosphorylates the type I receptor and the latter binds to and phosphorylates the pathway-restricted Smads 2 and 3. The phosphorylated Smads subsequently bind to the common partner Smad4 and translocate to the nucleus where they affect the transcription of target genes (Heldin et al., 1997; Massague, 1998; Massague and Wotton, 2000; ten Dijke et al. 2000; Moustakas et al. 2001). This is achieved through direct interactions of Smads with DNA sequence elements present on the promoters of these genes as well as through physical and functional interactions with other transcription factors or coactivators (Heldin et al., 1997; Massague, 1998; Massague and Wotton, 2000; ten Dijke et al. 2000; Moustakas et al. 2001).
Smads have two conserved domains, the N-terminal Mad homology 1 (MH1) and C-terminal Mad homology 2 (MH2) domains, which are separated by a middle, nonconserved linker region. The MH1 domain of Smads forms a β-hairpin structural element that mediates direct binding of Smads to DNA (Shi, 2001). This domain also includes a nuclear localization signal as well as surfaces for interaction with other transcription factors (Moustakas et al., 2001). The MH2 domain of Smads is a multifunctional domain that mediates Smad oligomerization, phosphorylation by the type I TGFβ receptor, and transactivation via coactivator recruitment (Moustakas et al. 2001).
Cross talking of the TGFβ/Smad signaling pathway with other signaling networks in the responsive cells may account for the pleiotropic activity of the TGFβ superfamily of ligands (ten Dijke et al. 2000). Positive and negative cross talking of Smads with members of the nuclear receptor superfamily have been reported. Nuclear receptors (NRs) are transcription factors that respond to a variety of hormonal and metabolic signals and affect such diverse aspects of life as embryogenesis, homeostasis, reproduction, and cell growth or death (Tsai and O'Malley, 1994; Mangelsdorf and Evans, 1995; Chambon, 1996; Moras and Gronemeyer, 1998; Shao and Lazar, 1999; Glass and Rosenfeld, 2000). This superfamlily includes ligand-dependent nuclear receptors as well as orphan receptors. Nuclear receptors are modular in nature and are composed of five domains. The N-terminal A/B domain harbors an autonomous transcriptional activation function (AF-1) which, when linked to a heterologous DNA binding domain, can activate transcription in a constitutive manner. This N-terminal domain has been found to be the subject of phosphorylation (Shao and Lazar, 1999). The highly conserved C domain encodes the DNA binding domain of nuclear receptors and confers sequence-specific DNA recognition. The D region of nuclear receptors is less conserved and can vary significantly in length. By linking the highly structured C and E domains, the hinge region may allow for flexibility in the conformation of the DNA binding and ligand binding domains. The ligand-dependent nuclear receptors also contain a ligand binding domain in the E region. The ligand binding domain harbors the ligand-dependent activation function 2 (AF-2) as well as a dimerization interface (Tsai and O'Malley, 1994; Mangelsdorf and Evans, 1995; Chambon, 1996; Moras and Gronemeyer, 1998; Shao and Lazar, 1999; Glass and Rosenfeld, 2000). Finally, some receptors possess a C-terminal F domain, which displays little evolutionary conservation and is present in some, but not all, receptors. The F domain might play a regulatory role in coactivator recruitment to the E domain (Peters and Khan, 1999; Sladek et al., 1999).
We have shown previously that TGFβ and its signaling effectors Smad3 and Smad4 proteins transactivate the liver-specific human apolipoprotein (apo)C-III gene promoter by synergizing with the orphan nuclear receptor hepatocyte nuclear factor 4 (HNF-4) (Kardassis et al., 2000). HNF-4 binds to hormone response elements (HREs) present in the promoters of a large network of genes expressed mainly or exclusively in the liver, including those of the apolipoproteins, a1-antitrypsin, transthyretin, hepatocyte nuclear factor-1, and others genes (Sladek, 1993). The critical role of HNF-4 for embryonic development and liver gene expression was established in studies where the HNF-4 gene was inactivated by homologous recombination in the mouse embryo (Li et al., 2000) or in the adult liver (Hayhurst et al., 2001). Other studies using transgenic mice carrying wild-type or mutant apoA-I/apoC-III gene cluster established the importance of the hormone response elements within the apoC-III enhancer and the proximal promoter that bind HNF-4 for the intestinal and hepatic expression of the genes of the apoA-I/apoC-III gene cluster (Zannis et al., 2001).
MATERIALS AND METHODS
Materials
The TNT quick-coupled transcription translation system and the luciferase assay kit were purchased from Promega (Madison, WI). Redivue l-[35S]methionine, [14C]chloramphenicol, protein A, protein G, and glutathione Sepharose 4B were purchased from Amersham Biosciences (Piscataway, NJ). The goat anti-HNF4α antibody (C-19) and the mouse anti-GAL4 antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The mouse anti-myc 9E10 and anti-FLAG M2 monoclonal antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Restriction and modification enzymes were from New England Biolabs (Beverly, MA) or Minotech (Heraklion, Greece). Other reagents were from common sources at the highest purity available.
Plasmid Constructions
The plasmids (−99/+24) apoCIII-CAT, (−268/+8) apoB CAT, (−268/+8) BM2 apoB CAT, (−911/+29) apoA-II-CAT, (−255/+5) apoA-I-CAT, pMT2-HNF4, and pMT2-EAR-3, have been described previously (Ladias et al., 1992; Kardassis et al., 2000). To generate the expression vector pCDNA1 amp/6myc-EAR-3, the full-length human EAR-3 cDNA was excised from the plasmid pMT2-EAR-3 by EcoRI and subcloned into the pcDNA1 amp/6-myc vector (Kardassis et al., 2000). The vector pEBG-Smad3 was constructed in two steps. First, the full-length human Smad3 cDNA was excised from the vector pGEX-2T-Smad3 by BamHI and EcoRI and was subcloned into the corresponding sites on pBluescript (Stratagene. La Jolla, CA). In the second step, the same fragment was excised from pBluescript by BamHI and KpnI and was subcloned into the vector pEBG, which is a modified version of vector pEF-BOS (Mizushima and Nagata, 1990). The pEBG-Smad3 plasmid expresses the Smad3 protein fused at the C terminus of glutathione S-transferase (GST) under the control of the human elongation factor 1α promoter and can be used for high levels of expression in mammalian cells.
For the construction of plasmids pEBG-Smad3-ΔMH1 and pEBG-Smad3-ΔMH2, the truncated Smad3 cDNAs were excised from pGEX-Smad3-ΔMH1 and pGEX-Smad3-ΔMH2 vectors by BamHI and NotI and subcloned into the corresponding sites of the pEBG vector described above. Plasmids pCDNA3/6myc-Smad3, pCDNA3/6myc-Smad4, p(CAGA)12 E1B-Luc and pCA-ALK5 have been described previously (Pardali et al., 2000). The vectors pGEX-Smad2, pGEX-Smad2-ΔMH1, pGEX-Smad2-ΔMH2, pGEX-Smad3, pGEX-Smad3-ΔMH1, pGEX-Smad3-ΔMH2, pGEX-Smad4, pGEX-Smad4-ΔMH1 and pGEX-Smad4-ΔMH2 have been described previously (Itoh et al., 2000). The pCDNA3/Flag-Smad3 (R74K, K81R) vector expressing Smad3 mutated in its DNA binding domain has been described previously (Moren et al., 2000). The plasmid pCDNA1-amp/6myc-Smad3 (1–374) was constructed by polymerase chain reaction (PCR) amplification of human Smad3 cDNA encoding for amino acids 1–374 and subsequent cloning at the EcoRI/NotI sites of vector pcDNA1-amp/6myc vector. The truncated forms of HNF-4 either in the vector pCDNA1-amp or in the vector pBX-G1 (as GAL4 fusions) have been described previously (Hadzopoulou-Cladaras et al., 1997; Kistanova et al., 2001). Plasmids pGEX-HNF-4 (130–455) and (−700/+10) apoA-IV-CAT were gifts of Dr. Iannis Talianidis (Institute of Molecular Biology and Biotechnology, Herakleion, Greece).
Cell Cultures, Transient Transfections, Chloramphenicol Acetyl Transferase (CAT), and Luciferase Assays
Human hepatoma HepG2 cells and monkey kidney COS-7 cells were cultured in DMEM supplemented with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Transient transfections were performed using the Ca3(PO4)2 coprecipitation method. Chloramphenicol acetyl transferase and β-galactosidase assays were performed as described previously (Kardassis et al., 2000). Luciferase assays were performed using the Luciferase assay kit from Promega.
Coimmunoprecipitation and Western Blotting
For coimmunoprecipitation of Smad3, Smad4, and HNF-4 proteins expressed endogenously in HepG2 cells or ectopically in COS-7 cells, cells were lysed in 500 μl of lysis buffer (20 mM Tris, pH 7.5, 0.15 M NaCl, 10% glycerol, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride) for 30 min on ice. Cell debris was removed by centrifugation at 13,000 rpm for 5 min. The supernatant (250 μl) was mixed with 1 μl of goat anti-HNF4 antibody in a rotator at 4°C for 2 h. Fifty microliters of protein G Sepharose (Amersham Biosciences) preequilibrated in lysis buffer was added to each sample and rotation continued for another 2 h at 4°C. Samples were boiled at 100°C for 10 min, and proteins were separated on 10% SDS-PAGE and transferred to two pieces of Hybond-C extra nitrocellulose (Amersham Biosciences) via a capillary diffusion in 1× transfer buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 2 mM EDTA, 0.5 mM β-mercaptoethanol) overnight. Proteins were detected by Western blotting with the anti-myc antibody followed by incubation with anti-mouse horseradish peroxidase-conjugated secondary antibody and home-made enhanced chemiluminescent assay on X-ray film (Fuji).
In Vitro Transcription and Translation
For in vitro transcription/translation, the TNT quick coupled transcription/translation systems (Promega) was used following the instructions of the manufacturer. For labeling of proteins, 20 μCi of Redivue L-[35S] methionine was included in each transcription/translation reaction.
Expression of GST Fusion Proteins in Escherichia coli or COS-7 Cells and GST Pull-Down Assays
The expression of proteins in E. coli cells was performed as described previously (Kardassis et al., 2000). For expression of proteins in COS-7 cells, 10 μg of pEBG-Smad3-ΔMH2 DNA or pEBG vector alone along with 2 μg of CA-ALK5 DNA were cotransfected into COS-7 cells in a 10-cm tissue culture dish. Forty hours posttransfection, COS-7 cells were harvested in phosphate-buffered saline, centrifuged, and lysed in 500 μl of lysis buffer as described above. Cell debris was removed by centrifugation and the supernatant was transferred to a new tube. One hundred microliters of glutathione Sepharose beads and 20 μl of in vitro synthesized HNF-4 or GAL4-HNF4 proteins were added to each vial and rotated at 4°C for 5 h. Beads were washed three times in lysis buffer and proteins bound to the beads were eluted by boiling at 100°C for 10 min. Proteins were separated on 12% SDS-PAGE, and the gel was dried using a vacuum pump and exposed to X-ray films for 3 d.
Adenoviral Infection
Adenoviruses expressing lac-Z, N-terminally flag-tagged Smad3 and Smad4 were donated by K. Miyazono (University of Tokyo, Tokyo, Japan) and were amplified and titrated as described previously (Fujii et al., 1999). Adenoviral transient infections of HepG2 cells with the specified multiplicity of infection (moi) were performed as described previously (Piek et al., 1999; Pardali et al., 2000). Essentially, 5 × 105 HepG2 cells growing in 3% fetal bovine serum-DMEM were infected with moi 200 of each Smad virus or with moi 400 of lac-z virus for 4 h at 37°C, and then the medium was changed and the next day 5 ng/ml TGF-β1 was added for 24 h. The end of the incubation corresponds to 40 h postinfection.
Reverse Transcription-PCR
Reverse transcription-PCR was performed as described previously (Valcourt et al., 2002). Briefly, total RNA was extracted from infected cells with the RNeasy kit (QIAGEN, Valencia, CA), and digested with DNase RQI (Promega) to remove any contaminating genomic DNA. For reverse transcription, a 40-μl reaction contained 1 μg of RNA, 12.5 ng/μl anchored oligo-dT17 primers (5′-AGCT17-3′), 500 μM each dNTP, 100 ng/μl bovine serum albumin, 10 mM dithiothreitol, 4 U of RNasin (Promega), and 200 U of SuperScript II RNase H− (Invitrogen). Reactions were carried out at 42°C for 50 min followed by inactivation of the enzyme at 70°C for 15 min. The cDNAs were then incubated with 4 U RNase H (Invitrogen) at 37°C for 30 min. Aliquots (2 μl) of the reverse-transcription reaction were used for PCR analyses. Routinely, each PCR amplification included 50 μM each dNTP, 0.2 μM each primer, 1.5 mM MgCl2, and 2.5 U of AmpliTaq Gold DNA polymerase. Amplification was performed in a T3 thermocycler (Biometra, Göttingen, Germany): an initial denaturation step at 95°C for 5 min, followed by 26–30 cycles of 30 s at 94°C, 30 s at optimal temperature and 30 s at 72°C, and a final elongation step at 72°C for 5 min. Specific primers were designed according to sequences available in the databanks or published by other authors. Primers for glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) were used to ascertain that an equivalent amount of cDNA was synthesized. The reverse transcription-PCR products were separated by electrophoresis on 2% agarose and stained with ethidium bromide. The sequence of the primers is as follows: GAPDH 5′ primer, 5′-ATCACTGCCACCCAGAAGAC-3′; GAPDH 3′ primer, 5′-ATGAGGTCCACCACCCTGTT-3′; ApoC-III 5′ primer, 5′-AGGAGTCCCAGGTGGCCCAGCAG-3′; and ApoC-III 3′ primer, 5′-CACGGCTGAAGTTGGTCTGACCTCA-3′.
The levels of adenoviral FLAG-tagged Smads were verified by anti-FLAG (M2; Sigma-Aldrich) Western blotting of whole cell extracts from duplicate six-well trays infected under identical conditions.
RESULTS
Physical and Functional Interactions between HNF-4, Smad3, and Smad4 Proteins Are Mediated by MH1 Domain of Smads
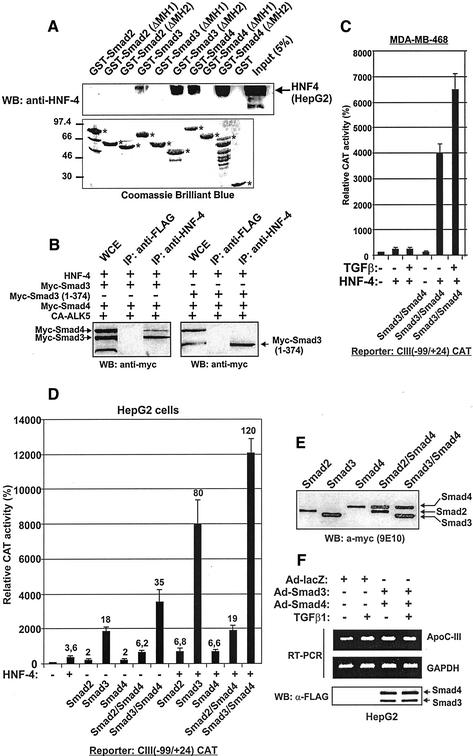
Direct physical interactions between Smad2, Smad3, and Smad4 proteins and HNF-4 were investigated in vitro by GST pull-down assays with a panel of wild-type (wt) or truncated Smad forms fused with GST. As shown in Figure 1A, HNF-4 expressed endogenously in HepG2 cells interacted with GST-Smad3 and GST-Smad4 proteins but failed to interact with GST-Smad2 or GST alone. Furthermore, deletion of the MH1 domain of Smad3 and Smad4 proteins abolished their physical interactions with HNF-4. In contrast, deletion of the MH2 domains of both proteins enhanced their physical interactions with HNF-4. These data indicate that direct physical interactions between Smad3 and Smad4 proteins and HNF-4 are mediated by the MH1 domain of Smads.
Figure 1.
(A–C) Smad3 and Smad4 proteins interact with HNF-4 and enhance apoC-III gene transcription in vitro and in vivo. (A) MH1 domain of Smad3 and Smad4 is an HNF-4 interacting domain. The indicated GST-Smad2, -Smad3, and -Smad4 fusion proteins or GST alone were isolated from E. coli, coupled to glutathione beads and allowed to interact with total cell extracts from HepG2 cells containing endogenous HNF-4. HNF-4 bound to GST-Smads was detected by immunoblotting with an anti-HNF-4 antibody as described in MATERIALS AND METHODS. The arrow shows the position of HNF-4. Input represents 5% of the initial cell extract used in the binding experiments. The expression of GST or the GST-Smad fusion proteins in bacteria was monitored by SDS-PAGE and Coomassie Brilliant Blue staining and is shown on the lower panel. The asterisks show the position of the expressed proteins. (B) Coimmunoprecipitation of Smads with HNF-4 expressed ectopically in COS-7 cells does not require the MH2 domain. Extracts from COS-7 cells transfected with the indicated combinations of expression vectors along with an expression vector for a constitutively active form of the type I TGFβ receptor (CA-ALK5) were immunoprecipitated with an anti-HNF-4 antibody or anti-FLAG antibody as control followed by SDS-PAGE and Western blotting with the anti-myc antibody that detects the transfected Smad3, Smad3 1–374, and Smad4 proteins. Arrows show the position of myc tagged Smad3, Smad3 1–374, and Smad4 proteins. Reprobing the membrane with an anti-HNF-4 antibody showed the presence of HNF-4 in the input and the anti-HNF-4 immunoprecipitated material, but not in the extracts that were immunoprecipitated with the anti-FLAG antibody (our unpublished data). WCE, whole cell extracts. (C) Smad proteins cannot transactivate the apoC-III promoter in the absence of HNF-4, whereas coexpression of HNF-4 restores transactivation. MDA-MB-468 Smad4 (−/−) cells were cotransfected with the (−99/+24) apoC-III CAT construct (1 μg) either alone or in combination with 1 μg each of the expression vectors for HNF-4, Smad3-FLAG, and Smad4-FLAG proteins as indicated at the bottom of the graph. Cells were treated with TGFβ (200 pM) or left untreated as indicated. The cytomegalovirus (CMV)-β galactosidase plasmid (1 μg) was included in each transfection for normalization of the transfection efficiency. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is shown in the form of a bar graph. (D) Potent synergistic transactivation of the apoC-III promoter by HNF-4 and Smad3/Smad4 heterodimers. HepG2 cells were cotransfected with the (−99/+24) apoC-III CAT construct (1 μg) either alone or in combination with 1 μg each of the expression vectors for HNF-4 and myc-tagged Smad3 and Smad4 proteins as indicated at the bottom of the bar-graph. The CMV-β galactosidase plasmid (1 μg) was included in each transfection for normalization of the transfection efficiency. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is shown in the form of a bar graph. The fold transactivation is shown on top of each bar. (E) Expression of transfected Smad proteins in HepG2 cells. The extracts of the transfected HepG2 cells of Figure 1D were analyzed for the levels of expression of Smad proteins by Western blotting by using the anti-myc mAb 9E10. The arrows show the position of the different Smads in the electrophoresis. (F) TGFβ and Smad3/Smad4 proteins enhance the transcription of the endogenous apoC-III gene in HepG2 cells. Reverse-transcription PCR was performed using total RNA extracted from HepG2 cells treated with TGFβ (5 ng/ml) for 24 h or left untreated or infected with recombinant adenoviruses expressing lacZ or Smad3 and Smad4 proteins as indicated on top of the photograph. The expression of FLAG-tagged Smad3 and Smad4 proteins in HepG2 cells was monitored by Western blotting by using a mouse monoclonal anti-FLAG antibody (M2).
The in vitro GST pull-down assays presented in Figure 1A were confirmed by coimmunoprecipitation assays in COS-7 cells. As shown in Figure 1B, myc-tagged Smad3 and Smad4 proteins expressed in COS-7 cells along with HNF-4 were immunoprecipitated by an antibody specific for HNF-4 (Figure 1B, left, anti-HNF-4). In contrast, a nonrelated antibody (anti-FLAG) failed to immunoprecipitate the Smad3 and Smad4 proteins. Furthermore, a Smad3 mutant lacking part of the MH2 domain, Smad3 (1–374), was efficiently coimmunoprecipitated with HNF-4 (Figure 1B, right, anti-HNF-4), confirming the findings of Figure 1A. Interestingly, Smad4 could not be coimmunoprecipitated with HNF-4 and the Smad3 (1–374) mutant. No immunoprecipitation of the Smad3 (1–374) mutant was observed using the nonrelated anti-FLAG antibody (Figure 1B, right, anti-FLAG). This finding, combined with the inability of the Smad3 (1–374) mutant to heterooligomerize with Smad4 (our unpublished data), suggested that, in vivo, Smad4 interacts physically with HNF-4 preferentially or exclusively in the form of Smad3/Smad4 heterooligomers.
Smad Proteins Cannot Transactivate the apoC-III Promoter in Cells That Do Not Express Endogenous HNF-4
To investigate further the importance of HNF-4 for the Smad-mediated transactivation of the apoC-III promoter, we performed cotransfection experiments in MDA-MB-468 breast cancer cells that do not express endogenous HNF-4 and have deletions of both alleles of the Smad4 gene (de Winter et al., 1997). In these cells, the TGFβ signaling pathway is inactive due to the absence of Smad4, whereas ectopic expression of Smad4 restores TGFβ signaling. As shown in Figure 1C, Smad3 and Smad4 proteins ectopically expressed in the MDA-MB-468 cells could not transactivate the apoC-III promoter in the absence of HNF-4. Importantly, ectopic expression of HNF-4 in these cells along with Smad3 and Smad4 strongly transactivated the apoC-III promoter by 40- and 65-fold in the absence or presence of added TGFβ, respectively (Figure 1C). The transactivation of the apoC-III promoter by Smad3 and Smad4 proteins in the absence of added TGFβ was possibly due to the overexpression of the proteins, a condition that confers Smads a constitutively active transactivation function as reported previously (Zhang et al., 1997; Moustakas and Kardassis, 1998). The above-mentioned findings confirmed the importance of HNF-4 in the transactivation of the apoC-III promoter by the TGFβ-regulated Smad proteins. Absence of transactivation of the apoC-III promoter by TGFβ and Smad3/Smad4 proteins was also observed in other cell lines that lack endogenous HNF-4 such as COS-7 kidney fibroblasts, human embryonic kidney-293 epithelial cells, or Drosophila SL2 cells. In all these cell lines, ectopic expression of HNF-4 restored the TGFβ and Smad3/Smad4-mediated transactivation of the apoC-III promoter (our unpublished data).
TGFβ and Smad3/Smad4 Proteins Enhance Transcription of Endogenous apoC-III Gene in HepG2 Cells
The data shown in Figure 1A indicated that Smad3 and Smad4 proteins have the ability to physically interact with HNF-4 in vitro, whereas Smad 2 does not. To investigate further the ability of different Smad family members to functionally synergize with HNF-4 in vivo and transactivate a target promoter, a series of transient transfection experiments were performed in HepG2 cells by using the −99/+24 apoC-III promoter as a reporter along with various combinations of Smad2, Smad3, and Smad4 proteins in the absence or presence of exogenous HNF-4. In parallel, we monitored the expression of the cotransfected Smad proteins by Western blotting analysis. HepG2 cells express endogenous HNF-4 at relatively high levels (Kardassis et al., 2000). As shown in Figure 1D, in the absence of exogenous HNF-4, Smad3 strongly transactivated the apoC-III promoter (18-fold, compared with the 3.6-fold transactivation achieved by HNF-4 or the twofold transactivation achieved by Smad2 and Smad4 proteins). Coexpression of Smad4 caused an additional twofold enhancement in the transactivation of the apoC-III promoter by Smad3 (35-fold). In contrast, the transactivation of the apoC-III promoter by Smad2 remained low even in the presence of Smad4 (6.2-fold). A similar pattern was observed in the presence of overexpressed HNF-4. As shown in Figure 1D, an increase in the intracellular concentration of HNF-4 along with Smad3 or Smad3/Smad4 caused a synergistic transactivation of the apoC-III promoter by 80- and 120-fold respectively, which could be attributed to the physical interactions between these proteins in vivo. This high level of transactivation by Smad3/Smad4 and HNF-4 (120-fold) exceeds the sum of the transactivations achieved by these proteins transfected independently (3.6- and 35-fold, respectively, sum = 38.6-fold). In these experiments, the ability of Smad3 to transactivate the apoC-III promoter was consistently 10–12 times stronger than the corresponding ability of Smad2 and Smad4, despite the fact that these proteins were expressed at equal levels in HepG2 cells (Figure 1E), suggesting a specificity in HNF-4/Smad cooperation for this specific Smad family member.
TGFβ caused an approximately twofold increase in the steady-state apoC-III mRNA levels in HepG2 cells (Figure 1F). Furthermore, using adenovirus-mediated gene transfer, we observed a similar induction of endogenous apoC-III gene expression in HepG2 cells by Smad3 and Smad4 proteins but not with a control adenovirus expressing β-galactosidase (Figure 1F). These data are consistent with our previous findings, which had shown that the addition of exogenous TGFβ to HepG2 cells caused a twofold transactivation of the −890/+24 apoC-III promoter, whereas a dominant negative form of Smad4 inhibited the basal apoC-III promoter activity by 50% in the same cells (Kardassis et al., 2000).
In conclusion, the findings of Figure 1, A–F, clearly indicate that Smad3/Smad4 heterodimers are the key effectors in the stimulation of apoC-III gene expression and apoC-III promoter activity by transforming growth factor-β. This stimulation requires HNF-4 and is mediated by physical interactions between Smads and HNF-4 involving the MH1 domain of Smads.
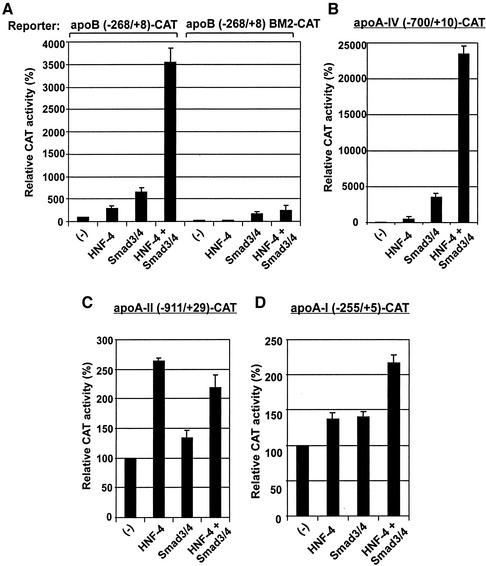
Smad3/Smad4 Proteins Transactivate Various Apolipoprotein Gene Promoters in a Promoter Context-dependent Manner
The physical and functional interactions between Smads and HNF-4 suggested that such interactions could modulate the transcription of other genes that contain HNF-4 binding sites in their promoters besides apoC-III. To test this hypothesis, additional apolipoprotein promoters shown previously to be regulated by HNF-4 were tested for their ability to be transactivated by Smads. As shown in Figure 2A, the proximal (−268/+8) apoB promoter, which also contains an HNF-4 binding site at position −79/−63 (Ladias et al., 1992) was transactivated by HNF-4 and Smad3/Smad4. Furthermore, coexpression of HNF-4 with Smads transactivated this promoter in a synergistic manner (∼35-fold). Importantly, the synergistic transactivation of the apoB promoter was abolished by point mutations that prevent binding of HNF-4 to its cognate site (mutation BM2; Kardassis et al., 1990) (Figure 2A). Similar synergistic transactivation by Smads and HNF-4 was observed using the promoter of the apolipoprotein A-IV gene, which also contains a binding site for HNF-4 (Ktistaki et al. 1994) (Figure 2B). In contrast, Smad3 and Smad4 proteins could not transactivate synergistically with HNF-4 the promoters of the apolipoprotein A-II and apolipoprotein A-I genes (Figure 2, C and D, respectively), which contain HNF-4 binding sites (Ladias et al., 1992; Tzameli and Zannis, 1996). The findings of Figure 2 indicate that the synergistic interactions between Smads and HNF-4 occur in a promoter context-dependent manner.
Figure 2.
Synergistic transactivation of various liver-specific promoters by Smads and HNF-4 depends on the proper promoter context. (A–D) HepG2 cells were cotransfected with the CAT reporter plasmids indicated on top of each graph (2 μg) either alone (−) or along with expression vectors for HNF-4 (1 μg), FLAG-tagged Smad3/Smad4 (1 μg each), or combination of them as indicated on the bottom of each graph. In each transfection, the CMV-β galactosidase vector was included for normalization of transfection variability. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is presented in the form of bar graphs.
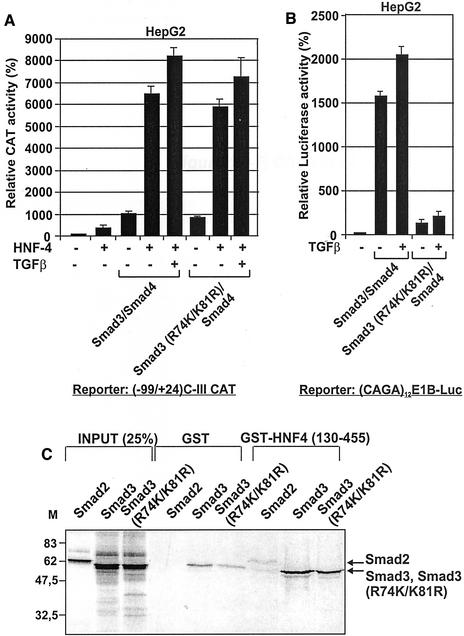
Mutations in DNA Binding Domain of Smad3 Do Not Affect Its Ability to Interact Physically and Functionally with HNF-4
The MH1 domain of Smad proteins, which was shown to mediate the Smad/HNF-4 interactions (Figure 1A), is also required for direct binding of Smads to the DNA (Heldin et al., 1997; Massague, 1998; Moustakas et al. 2001). The elucidation of the three-dimensional structure of Smad3 bound to DNA revealed the presence of a β-hairpin structure between amino acids 70–82, which makes DNA protein contacts (Shi, 2001). To assess the role of the β-hairpin structure of Smad3 in its physical and functional interactions with HNF-4, we performed GST-pull-down and transactivation experiments.
For this purpose, HepG2 cells, treated with TGFβ or left untreated, were cotransfected with the (−99/+24) apoC-III CAT reporter vector along with vectors expressing wild-type Smad3 or a Smad3 mutant containing a double amino acid substitution at residues 74 and 81 (Smad3 R74K/K81R) shown previously to abolish binding of Smad3 to DNA (Moren et al., 2000) along with Smad4 in the absence or presence of HNF-4. As shown in Figure 3A, both Smad3/Smad4 and Smad3 (R74K/K81R)/Smad4 proteins caused an equally strong transactivation of the (−99/+24) apoC-III promoter in the presence of HNF-4, which was further enhanced after TGFβ stimulation.
Figure 3.
(A–C) Mutations in the DNA binding domain of Smad3 proteins do not affect physical and functional interactions with HNF-4. (A and B) HepG2 cells were cotransfected with the (−99/+24) apoC-III CAT construct (1 μg) (A) or the (CAGA)12-E1B-luciferase construct (1 μg) (B) either alone or in combination with 1 μg each of the expression vectors for HNF-4, Smad3, Smad3 (R74K/K81R), and Smad4 proteins as indicated at the bottom of the graphs. Cells were treated with TGFβ (200 pM) or left untreated as indicated. The CMV-β galactosidase plasmid (1 μg) was included in each transfection for normalization of the transfection efficiency. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is shown in the form of a bar graph. (C) Indicated GST-HNF-4 (130–455) fusion protein or GST alone were isolated from E. coli, coupled to glutathione beads and allowed to interact with Smad2, Smad3, or Smad3 (R74K/K81R) proteins synthesized in vitro and labeled with [35S]methionine as described in MATERIALS AND METHODS. Bound proteins were analyzed by SDS-PAGE and detected by autoradiography. The arrow shows the position of Smad3 and Smad3 (R74K/K81R) proteins. Input represents 25% of the in vitro transcribed/translated proteins used in the binding experiments. M, protein molecular mass markers in kilodaltons.
In control experiments, Smad3/Smad4 proteins strongly transactivated an artificial promoter containing 12 tandem Smad binding elements fused with the E1B minimal promoter (CAGA12 E1B) (Dennler et al., 1998), whereas, as expected, the mutations in the DNA binding domain of Smad3 severely affected the Smad3/Smad4-mediated transactivation (Figure 3B).
For GST pull-down assays, glutathione Sepharose beads coupled with GST or a GST-HNF4 (130–455) fusion protein that binds efficiently Smad proteins (described below) were incubated with in vitro transcribed/translated and [35S]methionine-labeled wild-type Smad2, wild-type Smad3, or the Smad3 (R74K/K81R) mutant. Smad proteins eluted from the beads were analyzed by SDS-PAGE and autoradiography. As shown in Figure 3C, both Smad3 and Smad3 (R74K/K81R) specifically bound to HNF-4 (130–455) strongly but not to GST beads. Consistently with the findings of Figure 1A, no binding of Smad2 to HNF-4 (130–455) was observed.
The findings of Figure 3 suggested that mutations in the DNA binding domain of Smad3 do not affect the physical or the functional interactions between Smads and HNF-4 that account for the synergistic transactivation of the apoC-III promoter by these transcription factors.
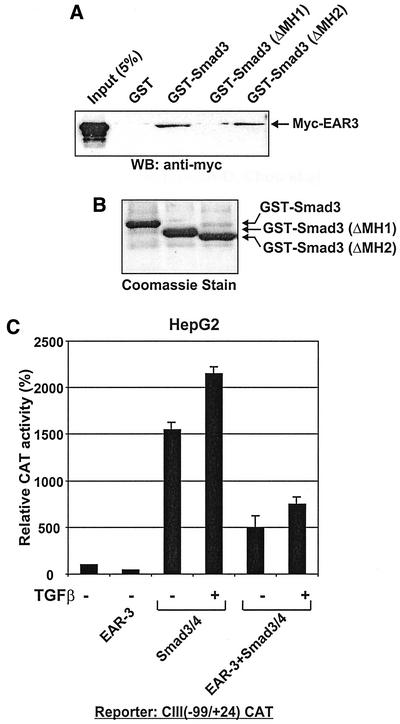
Transactivation of apoC-III Promoter by Smad Proteins Is Nuclear Receptor Specific
The proximal (−99/+24) apoC-III promoter contains an HRE between nucleotides −87/−63 that binds orphan (HNF-4, apolipoprotein AI regulatory protein 1, vErb-A related protein [EAR]-2, and EAR-3) as well as ligand-dependent (heterodimers of 9-cis retinoic acid receptor α with retinoic acid receptor α, thyroid hormone receptor β, and peroxisome proliferator activated receptor α) nuclear receptors (Ladias et al., 1992; Lavrentiadou et al., 1999). Previous studies have shown that HNF-4 transactivated eightfold, whereas apolipoprotein AI regulatory protein 1, EAR-2, and EAR-3 repressed by 90% the apoCIII promoter activity in HepG2 cells possibly due to the competition with HNF-4 for the same binding elements on the apoC-III promoter (Ladias et al., 1992). Physical and functional interactions between Smad3 and EAR-3 were investigated by GST pull-down and transactivation assays. Binding of EAR-3 to wild-type or truncated GST-Smad3 proteins was monitored by GST pull-down assays followed by immunoblotting by using an antibody that recognizes the myc epitope. In this analysis, equal amounts of glutathione Sepharose-bound GST-Smad3, GST-Smad3 ΔMH1, and GST-Smad3 ΔMH2 proteins were used (Figure 4B). This analysis showed that EAR-3 interacted with GST-Smad3 but not with GST, confirming the specificity of the Smad3/EAR-3 interaction (Figure 4A). Furthermore, deletion of the MH1 domain of Smad3 abolished the interaction with EAR-3, whereas deletion of the MH2 domain did not have any effect. The findings of Figure 3A indicate that, similar to HNF-4, EAR-3 protein interacts physically with Smad3 via the MH1 domain.
Figure 4.
(A–C) Smad proteins physically interact but do not functionally cooperate with the nuclear receptor EAR-3. (A) Indicated GST-Smad fusion proteins or GST alone were extracted from transfected COS-7 cells as described in MATERIALS AND METHODS, coupled to glutathione beads and allowed to interact with extracts from COS-7 cells transfected with an expression vector for myc-tagged EAR-3 protein. EAR-3 bound to GST-Smads was detected by immunoblotting with an anti-myc antibody. The arrow shows the position of the myc-EAR-3 protein. Input represents 5% of the initial cell extract used in the binding experiments. (B) Expression of GST or the GST-Smad fusion proteins used in the GST-pull-down assays shown in A. The indicated GST or GST-Smad proteins at the top of B were expressed in COS-7 cells, coupled to glutathione Sepharose beads, and an aliquot of the beads was analyzed for coupling efficiency by SDS-PAGE and Coomassie Brilliant Blue staining. (C) HepG2 cells were cotransfected with the (−99/+24) apoC-III CAT construct (1 μg) either alone or in combination with 1 μg each of the expression vectors for HNF-4, EAR-3, Smad3, and Smad4 proteins as indicated at the bottom of the graph. Cells were treated with TGFβ (200 pM) or left untreated as indicated. The CMV-β galactosidase plasmid (1 μg) was included in each transfection for normalization of the transfection efficiency. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is shown in the form of a bar graph.
Importantly, recruitment of Smad3/Smad4 proteins to the apoC-III promoter via EAR-3 does not result in the transactivation of this promoter in HepG2 cells. As shown in Figure 4C, ectopic expression of EAR-3 in HepG2 cells caused a 50% reduction in apoC-III promoter activity in accordance with previous reports (Ladias et al., 1992). Furthermore, no transcriptional synergism was observed by coexpressing EAR-3 with Smad3/Smad4 proteins.
The findings of Figure 4 suggested that the relative concentration and the identity of the nuclear receptors that bind to the hormone response element of the apoC-III promoter determines both the constitutive as well as the Smad-inducible activity of the apoC-III promoter in HepG2 cells. These findings also suggest that the mere recruitment of Smads to the apoC-III promoter by any nuclear receptor is not sufficient for transactivation. Rather, transactivation of the apoC-III promoter by Smads seems to be the result of functional cooperativity between Smads and specific nuclear receptors such as HNF-4, which bind to the apoC-III HRE.
Two Domains of HNF-4 Defined by Amino Acids 1–49 (Domains A/B) and 388–455 (Domain F) Mediate Physical Interactions with Smad3 In Vitro
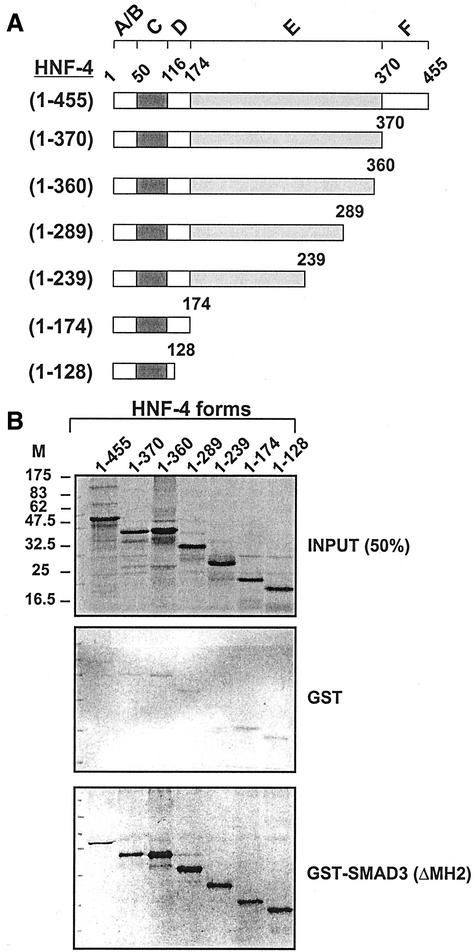
To identify the domains of HNF-4 that are required for physical interactions with Smad3, a series of truncated forms of HNF-4 were used in GST pull-down assays in vitro. The wild-type or truncated HNF-4 forms shown in Figure 5A were synthesized and labeled by in vitro transcription/translation in the presence of [35S]methionine (Figure 5B, top, designated INPUT) and tested for binding to GST-Smad3 (ΔMH2) protein. This analysis showed that the C-terminal truncations of HNF-4 extending to amino acids 370, 360, 289, 239, 174, and 128 did not affect the binding of HNF-4 to GST-Smad3 (ΔMH2) (Figure 5B, bottom, designated GST-Smad3 ΔMH2). None of the wild-type or truncated HNF-4 forms bound to GST (Figure 5B, middle, designated GST), confirming the specificity of the interaction. The analysis of Figure 5 identified the region of HNF-4 between amino acids 1–128 as a Smad3 binding domain without excluding the possibility that additional domain(s) in the 129–455 region of HNF-4 may participate in the Smad/HNF-4 interactions.
Figure 5.
(A and B) Identification of domains in HNF-4 that are required for binding to Smad3 by GST pull-down assays. (A) Schematic representation of the wt (1–455) or mutated HNF-4 proteins used in the GST pull-down experiments shown in B. Domains in HNF-4 are denoted by capital letters A–F. (B) Indicated wt (1–455) or mutant HNF-4 proteins were synthesized in vitro by coupled transcription/translation in the presence of [35S]methionine as described in MATERIALS AND METHODS and incubated with GST (GST) or GST-Smad3 (ΔMH2) Sepharose beads. Bound proteins were analyzed by SDS-PAGE and detected by autoradiography. The amount of proteins shown in the panel designated INPUT represent 50% of the amount used in the binding experiments of GST and GST-Smad3 (ΔMH2).
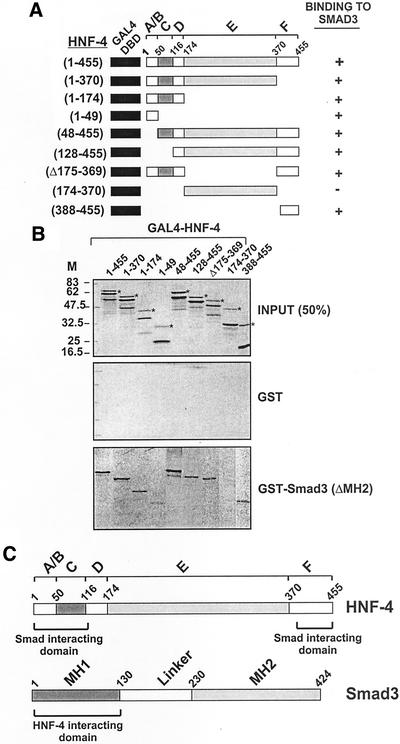
To confirm the findings of Figure 5 and to search for additional Smad binding domains in HNF-4, a second series of HNF-4 mutants, in this case fused with the DNA binding domain of GAL4, were used in GST pull-down (Figure 6) and transactivation (Figure 7) assays. First, the wild-type or the truncated GAL4-HNF-4 fusion proteins shown in Figure 6A were synthesized and labeled by in vitro transcription/translation in the presence of [35S]methionine (Figure 6B, top, designated INPUT) and tested for binding to GST or GST-Smad3 (ΔMH2) protein. As shown in Figure 6B, none of the GAL4-HNF-4 fusion proteins bound to the GST (Figure 5B, middle, designated GST). The wild-type GAL4-HNF-4 (1–455) as well as the C-terminal truncations of HNF-4 to amino acids 370 and 174 bound efficiently to Smad3 (ΔMH2) beads (Figure 6B, bottom, designated GST-SMAD3 ΔMH2) in agreement with the findings of Figure 5. Furthermore, an HNF-4 form containing amino acids 1–49, designated GAL4-HNF-4 (1–49), retained its capacity to interact physically with Smad3, thus mapping the Smad interaction domain within the first 49 amino acids of HNF-4 (domain A/B).
Figure 6.
(A–C) Two regions of HNF-4 defined by amino acids 1–49 (domain A/B) and 388–455 (domain F) mediate binding of HNF-4 to Smad3. (A) Schematic representation of the wt or mutated GAL4-HNF-4 fusion proteins used in the GST pull-down experiments shown in B. The domains in HNF-4 are denoted by capital letters A–F. Based on the data presented in B, the ability of the different GAL4-HNF-4 mutants to bind to Smad3 protein is indicated by a + sign on the right. Δ indicates a deletion. The GAL4 DNA binding domain (GAL4-DBD) is shown in black. (B) Indicated wt (1–455) or mutant GAL4-HNF-4 proteins were synthesized in vitro by coupled transcription/translation in the presence of [35S]methionine and incubated with GST or GST-Smad3 (ΔMH2) glutathione Sepharose beads. Bound proteins were analyzed by SDS-PAGE and detected by autoradiography. The amount of proteins shown in the top panel (designated INPUT) represent 50% of the amount used in the binding experiments of the middle panel (designated GST) and the lower panel [designated GST-SMAD3 (ΔMH2)]. (C) Schematic representation of the domains of HNF-4 and Smad3 involved in physical interactions among these two factors. The information is based on the data presented in Figures 1, 5, and 6 of this study.
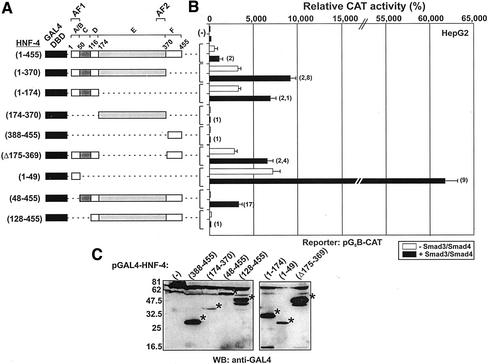
Figure 7.
(A–C) Coactivation of wt and mutant GAL4-HNF-4 fusion proteins by Smad3 and Smad4. (A) Schematic representation of the GAL4-HNF-4 fusion proteins used in the experiments of B and C. Domains in HNF-4 are labeled as in Figure 5A. (B) HepG2 cells were cotransfected with the reporter vector pG5B-CAT (1 μg) along with the indicated GAL4-HNF-4 expression vectors (100 ng) in the absence or presence of expression vectors for Smad3 and Smad4 (1 μg each). In each transfection, the CMV-β galactosidase vector was included for normalization of transfection variability. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is shown in the form of a bar graph. The transactivation of the pG5B promoter by GAL4-HNF-4 proteins in the absence or in the presence of Smad3 and Smad4 proteins is shown by gray and black bars, respectively. The fold enhancement of GAL4-HNF-4 transcriptional activity by Smads is shown in parentheses next to each bar. (C) Expression profile of the GAL4-HNF-4 proteins used in the transactivation experiments of B. COS-7 cells (100-mm dish) were transfected with expression vectors for the GAL4-HNF-4 forms indicated on top of the panel (17 μg each). Extracts from the transfected cells were analyzed by SDS-PAGE followed by Western blotting by using an antibody recognizing the GAL4 DNA binding domain (anti-GAL4). Asterisks show the position of the expressed proteins.
Interestingly, an HNF-4 mutant lacking the first 47 amino acids, GAL4-HNF-4 (48–455), or the first 127 amino acids, GAL4-HNF-4 (128–455), bound efficiently to GST-Smad3 (ΔMH2) (Figure 6B). This finding strongly suggested the presence of additional Smad3 binding domain(s) within the 128–455 region of HNF-4. This finding is in agreement with the data presented in Figure 2, which showed that the 130–455 region of HNF-4 interacted specifically with Smad3 but not with Smad2 in GST pull-down experiments. The existence of Smad binding domain(s) in the 128–455 region of HNF-4 was investigated further by using additional mutants of HNF-4. First, an HNF-4 mutant that lacks domain E, GAL4-HNF-4 (Δ 175–369), was used. This mutant bound efficiently to GST-Smad3 (ΔMH2), suggesting that domain E of HNF-4 (aa 174–370) is not required for interaction to Smad3. This was confirmed by using an HNF-4 mutant containing only domain E, GAL4-HNF-4 (174–370). This mutant was unable to interact with GST-Smad3 (ΔMH2) (Figure 6B). Finally, the GAL4-HNF-4 (388–455) mutant, which contains only domain F of HNF-4 (amino acids 388–455), interacted with GST-Smad3 (ΔMH2), indicating that a second Smad3 binding domain exists within the C-terminal domain F of HNF-4 (amino acids 388–455). Figure 6C is a schematic representation of the domains of HNF-4 and Smad3 that participate in physical interactions among these proteins. This information is based on Figures 1, 5, and 6 of this study.
Smad3/Smad4 Proteins Interact Functionally with the 1–128 Region of HNF-4 That Includes Activation Function 1 and DNA Binding Domain
The wild-type and truncated GAL4-HNF-4 proteins used in the GST pull-down assays (shown in Figures 6A and 7A) were used in transient cotransfections assays in HepG2 cells along with a GAL4-responsive promoter (pG5B-CAT) in the absence or presence of expression vectors for Smad3 and Smad4 proteins. In this analysis, we evaluated the ability of Smad3 and Smad4 proteins to function as coactivators of wild-type or mutant GAL4-HNF-4 fusion proteins by monitoring the increase in the activity of the pG5B-CAT reporter. As shown in Figure 7B, Smad3/Smad4 proteins enhanced twofold the activity of wild-type GAL4-HNF-4 (1–455). Control experiments showed that Smad3/Smad4 had no effect on the pG5B-CAT reporter in the presence of GAL4 alone (our unpublished data). Deletion of the C-terminal 371–455 or 175–455 regions of HNF-4 strongly enhanced basal HNF-4 activity in agreement with previous findings (Hadzopoulou-Cladaras et al., 1997) but did not affect the coactivation of these truncated HNF-4 forms by Smad3/Smad4 proteins (2.8- and 2.1-, onefold respectively). In contrast, the GAL4-HNF-4 forms 174–370 and 388–455 that contain the domains E and F, respectively, could not be coactivated by Smad3/Smad4 proteins. Furthermore, a GAL4-HNF-4 form lacking domain E (Δ175–369) was coactivated by Smads (2.4-fold). These observations confirmed that domain E of HNF-4, which includes the AF-2 core motif of the protein, is not required for coactivation by Smads.
To fine map the domains of HNF-4 that contribute to the Smad3/Smad4-mediated coactivation, additional GAL4-HNF-4 mutants were used. A GAL4-HNF-4 mutant containing the 1–49 region of HNF-4 had very high levels of basal activity due to the presence of a strong constitutively active AF-1 domain in the 1–24 region (Hadzopoulou-Cladaras et al., 1997) and was potently coactivated (ninefold) by Smad3/Smad4 proteins in HepG2 cells Figure 7B). Interestingly, an HNF-4 mutant lacking the 1–47 region, GAL4-HNF-4 (48–455), was also potently coactivated by Smad3/Smad4 (17-fold), whereas a GAL4-HNF-4 (128–455) mutant that lacks the 1–49 region and the adjacent 50–128 region was not coactivated by Smad3/Smad4 proteins. Figure 7C shows the expression levels of the GAL4-HNF-4 fusion proteins used in the transactivation experiments of Figure 7B.
In summary, the combined data of Figures 5–7 indicate that Smad3/Smad4 proteins coactivate the hepatocyte nuclear factor 4 by interacting physically and functionally with the 1–128 region that includes the activation function 1 and the DNA binding domain and with the C-terminal domain F (388–455).
Hydrophobic Residues in AF-1 Domain of HNF-4 Are Crucial for Functional Interactions between Smads and HNF-4
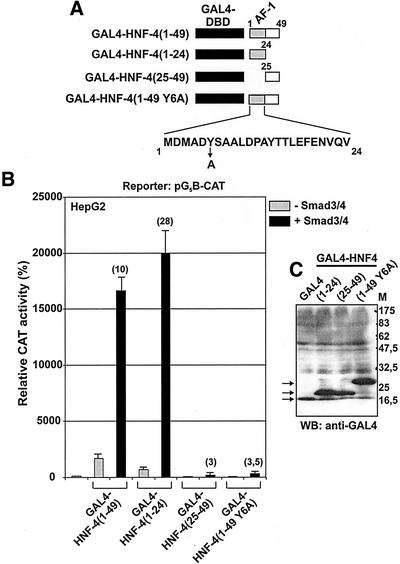
As shown in Figure 7, the 1–49 region of HNF-4 alone is sufficient to mediate coactivation by Smads. The first 24 amino acids of HNF-4 (aa 1–24) contain a strong transactivation domain designated AF-1 and is highly variable among the nuclear receptor superfamily. The AF-1 of HNF-4 functions as a constitutive autonomous activator of transcription (Hadzopoulou-Cladaras et al., 1997). AF-1 is rich in acidic and hydrophobic amino acids and is predicted to adopt an amphipathic α-helical secondary structure (Kistanova et al., 2001). Mutagenesis of acidic or hydrophobic residues within the AF-1 domain resulted in complete loss of transcriptional activity (Kistanova et al., 2001). As shown in Figure 8B, the GAL4-HNF-4 (1–24) form that contains only the AF-1 domain had very high levels of basal activity in HepG2 cells and this activity was enhanced greatly in the presence of Smad3/Smad4 proteins (28-fold). In contrast, a GAL4-HNF-4 mutant that lacks the AF-1 domain but contains the adjacent 25–49 region (GAL4-HNF-4 25–49) had almost undetectable basal activity in HepG2 cells and could not be coactivated by Smads. Furthermore, a GAL4-HNF-4 (1–49) mutant that contains a single amino acid substitution at residue 6 (a tyrosine was replaced by an alanine, Y6A) (Figure 8A) (Kistanova et al., 2001), had nearly background basal activity in HepG2 cells and could not be coactivated by Smad3/Smad4 proteins. This finding suggested that hydrophobic residues within the AF-1 domain of HNF-4 are crucial for the functional cooperativity between Smads and HNF-4 in vivo. All GAL4-HNF-4 proteins used in the transactivation experiments of Figure 8B were expressed at equal levels (Figure 8C).
Figure 8.
(A–C) Smad-mediated coactivation of HNF-4 was abolished by a specific amino acid substitution in the AF-1 domain of HNF-4. (A) Schematic representation of the GAL4-HNF-4 fusion proteins used in the transactivation experiments of B. The substitution of a tyrosine residue (Y) at amino acid position 6 of HNF-4 with an alanine (A) is shown with an arrow. (B) HepG2 cells were cotransfected with the reporter vector pG5B-CAT (1 μg) along with the indicated GAL4-HNF-4 expression vectors (100 ng) in the absence or presence of expression vectors for Smad3 and Smad4 (1 μg each). In each transfection, the CMV-β galactosidase vector was included for normalization of transfection variability. The normalized relative CAT activity (± SEM) of two independent experiments performed in duplicate is presented in the form of a bar graph. The transactivation of the pG5B promoter by GAL4-HNF-4 proteins in the absence or in the presence of Smad3 and Smad4 proteins is shown by gray and black bars, respectively. The fold enhancement of GAL4-HNF-4 transcriptional activity by Smads is shown in parentheses on top of each bar. Domains in HNF-4 are labeled as in Figure 5A. (C) Expression profile of the GAL4-HNF-4 proteins used in the transactivation experiments of B. COS-7 cells (60-mm dish) were transfected with expression vectors for the GAL4-HNF-4 forms indicated on top of the panel (6 μg each). Extracts from the transfected cells were analyzed by SDS-PAGE followed by Western blotting by using an antibody recognizing the GAL4 DNA binding domain (anti-GAL4). Arrows show the position of the expressed proteins.
In summary, this study established that the cross talk between the TGFβ-inducible Smad3 and Smad4 proteins and the orphan nuclear receptor HNF-4, which enhances the transcription of various apolipoprotein genes, requires the presence of an HNF-4 binding site and depends on the promoter context. The HNF-4/Smad interactions require the MH1 domain of Smads and both N-terminal and C-terminal domains of HNF-4.
DISCUSSION
Domains in Smad3 and Smad4 Proteins Required for Physical and Functional Interactions with Hepatocyte Nuclear Factor 4
We have shown previously that TGFβ and its signaling effectors Smad3 and Smad4 proteins transactivate the liver-specific apoC-III gene promoter by synergizing with an orphan nuclear receptor, the hepatocyte nuclear factor 4 (Kardassis et al., 2000). In the present study, we investigated in detail the domains of the two proteins involved in physical interactions as well as the mechanism of the functional cooperativity between Smads and HNF-4 and assessed its implication for hepatic gene expression.
It was found that Smad3 and Smad4 but not Smad2 interacted physically with HNF-4 expressed endogenously in HepG2 cells. The interactions between Smad3, Smad4, and HNF-4 were abolished by deletion of the MH1 domain but were not affected by deletion of the MH2 domain. These findings suggested that the MH1 domain of Smads is important for its physical and functional interaction with HNF-4 (Figure 1). The role of the MH1 domain of Smads in transcriptional activation and signaling cross talks has been studied previously (Moustakas et al., 2001, and references therein). It was found that the MH1 domain interacts with different transcription factors that cross talk with Smads such as the AP-1 family members ATF-2, c-Jun, JunB, and JunD; the ubiquitous transcription factor Sp1; the nuclear receptor for vitamin D; and nuclear factor YY1 among others (Moustakas et al., 2001). Furthermore, the MH1 domain interacts with the transcriptional corepressors HDAC and Hoxc-8 as well as the cytoplasmic adapters filamin and importin β1 (Moustakas et al., 2001). Thus, the MH1 domain of Smads seems to facilitate regulatory interactions between Smads and other proteins with diverse physiological roles.
In addition to the above-mentioned regulatory functions, the MH1 domain mediates direct binding of Smads to the DNA. The elucidation of the three-dimensional structure of the MH1 domain of Smad3 by x-ray crystallography in the presence of an eight-base pair Smad binding element (SBE) revealed the formation of a β-hairpin structure in the region defined by amino acids 70–82 (Shi, 2001). The structure also revealed that amino acid residues R74, Q76, and K81 of Smad3 contact directly DNA (Shi, 2001). Mutations in the above-mentioned residues abolished binding of Smads to Smad binding elements and the transactivation of Smad-responsive promoters in transient transfection assays (Moren et al., 2000).
In this study, mutations in the β-hairpin of Smad3 did not affect the physical interactions between Smads and HNF-4 and the Smad-mediated transactivation of the apoC-III promoter (Figure 3). The mutations in Smad3 that were used in this study (R74K/K81R) have been designed in such a way that they do not perturb the β-hairpin structure (Moren et al., 2000). This has been verified by computer modeling of the three-dimensional structure of the MH1 domain of Smad3 (Moustakas, personal communication). These amino acid substitutions only perturb the contact of Smads with SBEs. Furthermore, the swapping of lysine and arginine residues with the reciprocal amino acids has caused no change in the overall charge of the β-hairpin structure.
The data presented in this study favor a model according to which Smad-HNF-4 interactions on the promoters of target genes can occur even in the absence of direct binding of Smads to the DNA. This model should apply to all the promoters shown in the present study to be synergistically transactivated by Smad3/Smad4 and HNF-4 such as the apoC-III, apoB, and apoA-IV promoters. On the other hand, the possibility that Smads recognize with low-affinity DNA elements present in the vicinity of the hormone response elements in all the above-mentioned promoters cannot be excluded. However, this possibility is not supported by our previous observations that had shown that the transcriptional synergism between Smad3/Smad4 and HNF-4 is retained by transferring the hormone response elements of apoC-III, apoA-I, and apoC-II promoters to a heterologous promoter such as the minimal adenovirus major late promoter (Kardassis et al., 2000; our unpublished data) or by our inability to demonstrate direct binding of purified Smad3 protein to oligonucleotides covering the entire −90/−9 region of the apoC-III promoter in gel electrophoretic mobility shift assays (our unpublished data).
The importance of promoter context in the transactivation of HNF-4 responsive genes by the Smads and HNF-4 is also apparent by the results of Figure 2, which showed that the presence of an HNF-4 binding site on a promoter is not the only requirement for the responsiveness of this promoter to the TGFβ/Smad signaling pathway. Interestingly, the same HNF-4 binding site (element AID of the apoA-I promoter), which cannot be transactivated by Smad3/Smad4 and HNF-4 in the context of the natural −255/+5 apoA-I promoter (Figure 2D), can be strongly and synergistically transactivated by Smad3/Smad4 and HNF-4 when placed in a heterologous promoter (see Figure 4B of Kardassis et al. 2000). We are tempted to speculate that the specific multiprotein complexes formed on natural or artificial promoters (enhanceosomes), which strictly depend on the specific promoter context and on the identity of the transcription factors that bind to these promoters, may be favorable or unfavorable for Smad recruitment.
The present study also established that Smad2 does not interact with HNF-4 (Figures 1 and 3). Smad2, Smad3, and Smad4 proteins are highly homologous in their MH1 domains (>90% amino acid identity). The only exception is the presence of two stretches of unique amino acids/sequences in the MH1 domain of Smad2 that are absent from Smad3 and Smad4 proteins (Dennler et al., 1999; Yagi et al., 1999). In contrast to Smad3 and Smad4, Smad2 cannot contact DNA possibly due to the absence of the β-hairpin structure. Furthermore, an alternatively spliced variant of Smad2 with a deletion of exon 3 was recently identified in various cell types that binds DNA and transactivates TGFβ-responsive promoters as efficiently as wt Smad3 (Yagi et al., 1999). These data strengthen the hypothesis that the formation of the β-hairpin structure in the MH1 domain of Smad proteins may be crucial for the overall transactivation potential of Smad proteins and its interactions with other transcription factors.
Our coimmunoprecipitation and GST pull-down protein–protein interaction studies of Figure 1 showed that the direct physical interactions between Smads 3 and 4 and HNF-4 can occur even in the absence of exogenously added TGFβ or with nonphosphorylated Smads. However, this may be due to the in vitro conditions used. Hepatocyte nuclear factor 4 is a nuclear protein (Sladek, 1993; Soutoglou et al., 2000). Thus, to interact with HNF-4, Smads need to be phosphorylated by the receptor and be transported to the nucleus as heterodimers with Smad4. These Smad3/Smad4 heterodimers are the major Smad forms that interact with HNF-4 in vivo (see the coimmunoprecipitation experiment of Figure 1B). Thus, the contribution of the TGFβ receptor in vivo is to promote the formation of these heterodimers and their transport to the nucleus to interact with HNF-4.
Activation Function 1, the DNA Binding Domain and the C-Terminal Domain F of HNF-4 Contribute to Physical and Functional Interactions between Smad3 and HNF-4
The availability of a large panel of HNF-4 mutants allowed us to map the domains of HNF-4 involved in physical and functional interactions with Smad3. GST pull-down experiments revealed the existence of a Smad3 interaction domain at the N terminus of HNF-4 between amino acids 1 and 128, which includes domains A/B, C, and D (Figures 5 and 6), and a second interaction domain at the C terminus of HNF-4 between amino acids 388 and 455, which contains domain F (Figure 6). In contrast, the central 174–370 region of HNF-4, which contains domain E, did not bind to Smad3 in vitro (Figure 6).
The GAL4-HNF-4 mutants used in the in vitro protein–protein interaction experiments of Figure 6 were used in the transactivation experiments of Figure 7B. These analyses showed that Smad3 and Smad4 proteins could act as potent coactivators of HNF-4 and that the AF-1 domain (aa 1–24) was essential for the functional cooperativity with the Smads. The AF-1 domain of HNF-4 comprises a strong ligand-independent transactivation domain (Hadzopoulou-Cladaras et al., 1997). The AF-1 domain of HNF-4 binds factors of the basal transcriptional machinery such as TF-IIB as well as the ubiquitous coactivator protein cAMP response element-binding protein binding protein (CBP) (Dell and Hadzopoulou-Cladaras, 1999; Kistanova et al., 2001). Through these interactions, AF-1 seems to play an important role in the modulation of HNF-4 function in the nucleus. The mechanism of coactivation of HNF-4 by Smads via AF-1 is currently unknown. Interestingly, the CBP protein that was shown previously to coactivate HNF-4 via its AF-1 domain, also coactivates Smad3 and Smad4 proteins (Feng et al., 1998; Janknecht et al., 1998; Pouponnot et al., 1998). Whether the interaction of Smads with the AF-1 domain of HNF-4 results in more efficient recruitment of CBP or other coactivators to this region of HNF-4 remains to be determined.
Smad proteins failed to coactivate the GAL4-HNF-4 (174–370) form that contains domain E of HNF-4. Domain E of nuclear receptors contains the AF-2 core motif that is required for the recruitment of certain nuclear receptor coactivators (Moras and Gronemeyer, 1998). The above-mentioned finding suggested that Smads are not AF-2 coactivators. Smads lack a typical LXXLL motif (where L is leucine and X is any amino acid) that is characteristic of coactivators of nuclear receptors (Moras and Gronemeyer, 1998). Interestingly, Smad3 and Smad4 coactivated the GAL4-HNF-4 (48–455) mutant, which lacks the AF-1 domain whereas deletion of the adjacent DNA binding domain in this mutant abolished coactivation by Smads (Figure 7B). These findings suggested that, in addition to AF-1, the DNA binding domain of HNF-4 also seems to play a role in Smad/HNF-4 cooperation and functional synergism.
It was shown recently that the DNA binding domain of HNF-4 is acetylated by the CBP coactivator at specific lysine residues and that acetylation of this domain affected important HNF-4 properties such as DNA binding, nuclear localization, and interaction with coactivators (Soutoglou et al., 2000). Thus, binding of Smad proteins to the DBD of HNF-4 and/or to the adjacent AF-1 domain could enhance the recruitment of CBP to the DBD domain of HNF-4, thus modulating the acetylation state of this domain.
Role of C-Terminal Domain F of HNF-4 (Amino Acids 388–455) in Functional Interactions between HNF-4 and Smad3
The GST pull-down analyses presented in this study showed that domain F of HNF-4 (amino acids 388–455) also has the capacity to interact directly with Smads in vitro (Figure 6). Domain F is present in all nuclear receptors but is unusually large in HNF-4 and has been shown to act as a negative regulatory region that impedes access to coactivators such as GRIP-1 and SRC-1 (Sladek et al., 1999). Deletion of the F domain increased the HNF4-mediated transcriptional activation of the apoC-III promoter in HepG2 cells (Hadzopoulou-Cladaras et al., 1997). The role of this domain in HNF-4/Smad functional synergism as well as in other HNF-4 functions remains uncertain. It is possible that the F domain could serve as a tethering domain for other transcription factors such as Smads that modulate HNF-4 function via coactivator recruitment in response to extracellular or intracellular signals. Thus, this domain could inhibit HNF-4 transactivation function in the absence of the signal but could activate HNF-4 functions under certain stimulatory conditions. Thus, activation of HNF-4 could result from the interaction of Smad with coactivators that bind to the adjacent activation function-2 of HNF-4.
Interactions of Smads with Nuclear Receptors Could Lead to Transcriptional Activation or Repression
Four different nuclear receptors were found previously to interact with Smad proteins, the receptors for vitamin D (VitDR), glucocorticoids, androgens (AR), and estrogens (ER) (Song et al., 1999; Yanagi et al., 1999; Yanagisawa et al., 1999; Hayes et al., 2001; Matsuda et al., 2001; Chipuk et al., 2002). In the first case, Smad3 was found to act as a coactivator of the VitDR (Yanagisawa et al., 1999). The domains of the two proteins required for physical and functional interactions were mapped within the MH1 domain of Smad3 and part of the ligand binding domain of VitDR (Yanagisawa et al., 1999). In the same series of studies, Smad3 was shown to form a complex with the steroid receptor coactivator-1 protein (Src-1), a member of the p160 family of nuclear receptor coactivators (Yanagi et al., 1999; Yanagisawa et al., 1999). In another study, physical interactions between Smads and the glucocorticoid receptor were shown to inhibit the TGFβ responsiveness of the type-1 plasminogen activator inhibitor gene promoter (Song et al., 1999). In this case, the Smad MH2 domain and the ligand binding domain/activation function-2 of the glucocorticoid receptor were found to be essential for the protein–protein interactions.
Two studies investigated the mechanism by which TGFβ inhibits proliferation and induces apoptosis in prostate cancer cells (Hayes et al., 2001; Chipuk et al., 2002). It was found that TGFβ and Smad3 inhibited transcriptional activation mediated by the AR on two natural androgen-responsive promoters (Hayes et al., 2001). The transcription activation domain of AR and the MH2 domain of Smad3 were found to be responsible for the physical interactions between AR and Smad3 and for the repression of AR function in prostate cancer cells. Electrophoretic mobility shift assays indicated that the ligand-binding domain of the androgen receptor inhibits directly the association of Smad3 to the SBEs (Chipuk et al., 2002).
Finally, a two way cross talk between Smad3 and the ER was reported in human embryonic kidney cells (Matsuda et al., 2001). In this system, TGFβ signaling and Smad3-mediated transcriptional activation of a Smad-responsive promoter were repressed by ER only in the presence of estrogens whereas TGFβ and Smads induced ER signaling on an estrogen responsive promoter. This cross talk between Smads and ER was found to be mediated by the MH2 domain of Smad3. The domains of ER involved in these interactions were not investigated.
Smad/HNF-4 Cross Talk and Its Implications for Hepatic Gene Expression
The physical and functional interactions between HNF-4 and Smad3 in hepatocytes reported in this study could have important implications for hepatic gene expression. In transient transfection experiments in HepG2 cells, Smad3/Smad4 proteins transactivated a number of promoters that are regulated by HNF-4 such as the apolipoproteins C-III, B, and A-IV. In all these promoters, synergism between Smads and HNF-4 was observed. These findings suggested that the Smad/HNF-4 cross talk is not restricted to the apoC-III gene but may have wider implications for hepatic gene expression. Interestingly, no synergism was observed between Smads and HNF-4 in the transactivation of the apoA-I and apoA-II promoters, which also contain HNF-4 binding sites (Figure 2, C and D) (Ladias et al., 1992; Tzameli and Zannis, 1996). Furthermore, TGFβ-regulated Smads could not transactivate synergistically with HNF-4, a homopolymeric promoter consisting of three tandem HNF-4 binding sites derived from the apoA-II promoter (our unpublished data), whereas they transactivated artificial promoters consisting of HNF-4 binding sites derived from the apoC-III and the apoA-I promoters (Kardassis et al., 2000). The findings imply strongly that promoter context may play a very important role for the transcriptional cooperation between Smads and HNF-4. It seems that nuclear factors bound to elements adjacent to the HNF-4 binding sites may affect Smad/HNF-4 interactions in a positive or a negative manner. Alternatively, changes in the conformation of HNF-4 caused by its interaction with different hormone response elements could hinder or expose surfaces in HNF-4 that have the potential to interact with Smads such as the AF-1, the DNA binding domain or the F domain. The elucidation of the three-dimensional structure of HNF-4 will reveal very important and novel aspects of HNF-4 functions and regulation.
ACKNOWLEDGMENTS
We thank Dr. I. Talianidis for vectors used in this study and P. Papakosta for excellent technical assistance. This work was supported by grants from the Human Frontier Science Program (to D.K. and A.M.), National Institutes of Health grant HL-33952 (to V.I.Z.), and the Greek Ministry of Development (to D.K., M.H.-C., and V.I.Z).
Abbreviations used:
- apo
apolipoprotein
- CAT
chloramphenicol acetyl transferase
- CMV
cytomegalovirus
- CBP
cAMP response element-binding protein binding protein
- DBD
DNA binding domain
- EAR-3
vErb-A related protein 3
- GST
glutathione S-transferase
- HNF
hepatocyte nuclear factor
- HRE
hormone response element
- MH1
Mad homology domain 1
- Smad
Sma-Mad protein
- TGFβ
transforming growth factor β
- wt
wild-type
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0375. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0375.
REFERENCES
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- Chipuk JE, Cornelius SC, Pultz NJ, Jorgensen JS, Bonham MJ, Kim SJ, Danielpour D. The androgen receptor represses transforming growth factor-beta signaling through interaction with Smad3. J Biol Chem. 2002;277:1240–1248. doi: 10.1074/jbc.M108855200. [DOI] [PubMed] [Google Scholar]
- de Winter JP, Roelen BA, ten Dijke P, van der Burg B, van den Eijnden-van Raaij AJ. DPC4 (SMAD4) mediates transforming growth factor-beta1 (TGF-beta1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene. 1997;14:1891–1899. doi: 10.1038/sj.onc.1201017. [DOI] [PubMed] [Google Scholar]
- Dell H, Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- Dennler S, Huet S, Gauthier JM. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18:1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Takeda K, Imamura T, Aoki H, Sampath TK, Enomoto S, Kawabata M, Kato M, Ichijo H, Miyazono K. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell. 1999;10:3801–3813. doi: 10.1091/mbc.10.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M, Kistanova E, Evagelopoulou C, Zeng S, Cladaras C, Ladias JA. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J Biol Chem. 1997;272:539–550. doi: 10.1074/jbc.272.1.539. [DOI] [PubMed] [Google Scholar]
- Hayes SA, Zarnegar M, Sharma M, Yang F, Peehl DM, ten Dijke P, Sun Z. SMAD3 represses androgen receptor-mediated transcription. Cancer Res. 2001;61:2112–2118. [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Itoh S, Itoh F, Goumans MJ, ten Dijke P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardassis D, Hadzopoulou-Cladaras M, Ramji DP, Cortese R, Zannis VI, Cladaras C. Characterization of the promoter elements required for hepatic and intestinal transcription of the human apoB gene: definition of the DNA-binding site of a tissue-specific transcriptional factor. Mol Cell Biol. 1990;10:2653–2659. doi: 10.1128/mcb.10.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardassis D, Pardali K, Zannis VI. SMAD proteins transactivate the human ApoCIII promoter by interacting physically and functionally with hepatocyte nuclear factor 4. J Biol Chem. 2000;275:41405–41414. doi: 10.1074/jbc.M007896200. [DOI] [PubMed] [Google Scholar]
- Kistanova E, Dell H, Tsantili P, Falvey E, Cladaras C, Hadzopoulou-Cladaras M. The activation function-1 of hepatocyte nuclear factor-4 is an acidic activator that mediates interactions through bulky hydrophobic residues. Biochem J. 2001;356:635–642. doi: 10.1042/0264-6021:3560635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistaki E, Lacorte JM, Katrakili N, Zannis VI, Talianidis I. Transcriptional regulation of the apolipoprotein A-IV gene involves synergism between a proximal orphan receptor response element and a distant enhancer located in the upstream promoter region of the apolipoprotein C-III gene. Nucleic Acids Res. 1994;22:4689–4696. doi: 10.1093/nar/22.22.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladias JAA, Hadzopoulou-Cladaras M, Kardassis D, Cardot P, Cheng J, Zannis VI, Cladaras C. Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII, and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2, and EAR-3. J Biol Chem. 1992;267:15849–15860. [PubMed] [Google Scholar]
- Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrentiadou SN, Hadzopoulou-Cladaras M, Kardassis D, Zannis VI. Binding specificity and modulation of the human ApoCIII promoter activity by heterodimers of ligand-dependent nuclear receptors. Biochemistry. 1999;38:964–975. doi: 10.1021/bi981068i. [DOI] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Yamamoto T, Muraguchi A, Saatcioglu F. Cross-talk between transforming growth factor-beta and estrogen receptor signaling through Smad3. J Biol Chem. 2001;276:42908–42914. doi: 10.1074/jbc.M105316200. [DOI] [PubMed] [Google Scholar]
- Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Moren A, Itoh S, Moustakas A, Dijke P, Heldin CH. Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene. 2000;19:4396–4404. doi: 10.1038/sj.onc.1203798. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci USA. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali K, Kurisaki A, Morén A, ten Dijke P, Kardassis D, Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J Biol Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- Peters GA, Khan SA. Estrogen receptor domains E and F: role in dimerization and interaction with coactivator RIP-140. Mol Endocrinol. 1999;13:286–296. doi: 10.1210/mend.13.2.0244. [DOI] [PubMed] [Google Scholar]
- Piek E, Moustakas A, Kurisaki A, Heldin C-H, ten Dijke P. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci. 1999;112:4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- Pouponnot C, Jayaraman L, Massague J. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- Shao D, Lazar MA. Modulating nuclear receptor function: may the phos be with you. J Clin Invest. 1999;103:1617–1618. doi: 10.1172/JCI7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Structural insights on Smad function in TGFbeta signaling. Bioessays. 2001;23:223–232. doi: 10.1002/1521-1878(200103)23:3<223::AID-BIES1032>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Sladek FM. Orphan receptor HNF-4 and liver-specific gene expression. Receptor. 1993;3:223–232. [PubMed] [Google Scholar]
- Sladek FM, Ruse MD, Jr, Nepomuceno L, Huang SM, Stallcup MR. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Tian X, Gelehrter TD. Glucocorticoid receptor inhibits transforming growth factor-beta signaling by directly targeting the transcriptional activation function of Smad3. Proc Natl Acad Sci USA. 1999;96:11776–11781. doi: 10.1073/pnas.96.21.11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin C-H. Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Zannis VI. Binding specificity and modulation of the ApoA-I promoter activity by homo- and heterodimers of nuclear receptors. J Biol Chem. 1996;271:8402–8415. doi: 10.1074/jbc.271.14.8402. [DOI] [PubMed] [Google Scholar]
- Valcourt U, Gouttenoire J, Moustakas A, Herbage D, Mallein-Gerin F. Functions of transforming growth factor-beta family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J Biol Chem. 2002;277:33545–33558. doi: 10.1074/jbc.M202086200. [DOI] [PubMed] [Google Scholar]
- Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- Yanagi Y, Suzawa M, Kawabata M, Miyazono K, Yanagisawa J, Kato S. Positive and negative modulation of vitamin D receptor function by transforming growth factor-beta signaling through smad proteins. J Biol Chem. 1999;274:12971–12974. doi: 10.1074/jbc.274.19.12971. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono K, Kato S. Convergence of transforming growth factor-beta and vitamin D signaling pathways on SMAD transcriptional coactivators. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- Zannis VI, Kan HY, Kritis A, Zanni EE, Kardassis D. Transcriptional regulatory mechanisms of the human apolipoprotein genes in vitro and in vivo. Curr Opin Lipidol. 2001;12:181–207. doi: 10.1097/00041433-200104000-00012. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]