Abstract
The apoprotein corresponding to the mammalian photoreceptor rhodopsin has been expressed by using suspension cultures of HEK293S cells in defined media that contained 6-15N-lysine and 2-13C-glycine. Typical yields were 1.5–1.8 mg/liter. Incorporation of 6-15N-lysine was quantitative, whereas that of 2-13C-glycine was about 60%. The rhodopsin pigment formed by binding of 11-cis retinal was spectrally indistinguishable from native bovine rhodopsin. Magic angle spinning (MAS) NMR spectra of labeled rhodopsin were obtained after its incorporation into liposomes. The 15N resonance corresponding to the protonated retinylidene Schiff base nitrogen was observed at 156.8 ppm in the MAS spectrum of 6-15N-lysine-labeled rhodopsin. This chemical shift corresponds to an effective Schiff base-counterion distance of greater than 4 Å, consistent with structural water in the binding site hydrogen bonded with the Schiff base nitrogen and the Glu-113 counterion. The present study demonstrates that structural studies of rhodopsin and other G protein-coupled receptors by using MAS NMR are feasible.
Keywords: HEK293S cells, G protein-coupled receptor, signal transduction, 11-cis retinal, visual pigment
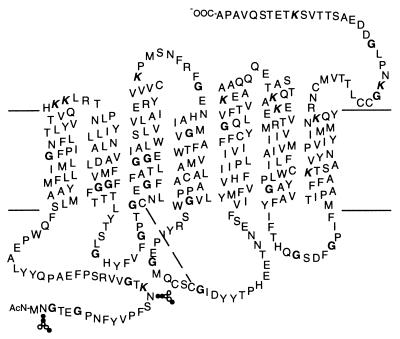
Considerable progress has been made in recent years in understanding structure-function relationships in rhodopsin, the mammalian photoreceptor, and other members of the G protein-coupled receptor family (1–3). These receptors share a common structural motif consisting of seven transmembrane helices (Fig. 1). Electron diffraction measurements of two-dimensional rhodopsin crystals have revealed the spatial arrangement of the seven transmembrane helices (4, 5), whereas a combination of systematic cysteine replacements in the cytoplasmic domain of the receptor followed by spin labeling and EPR measurements has begun to show the nature of the movements in the transmembrane helices that are involved in receptor activation (6–8). An essential feature of the activation mechanism that emerges from these studies is that structural changes in the transmembrane domain are tightly coupled to the conformations of the cytoplasmic and intradiscal (extracellular) loops of the protein.
Figure 1.
Secondary structure model of bovine rhodopsin showing the seven hydrophobic transmembrane helices. The retinal chromophore is attached to Lys-296 on helix 7 through a PSB bond. The amino acids labeled in this work are shown in bold (glycine) and bold italic (lysine).
In rhodopsin, receptor activation is controlled by the retinylidene chromophore. The 11-cis isomer of retinal is covalently attached as a protonated Schiff base (PSB) in the interior of the transmembrane domain. Photochemical isomerization of the retinal breaks helix–helix interactions, which, in the dark, lock rhodopsin in the inactive conformation. The idea that receptor-specific helix–helix interactions are involved in activation has arisen in studies on a number of G protein-coupled receptors (GPCRs) (2). Unfortunately, the lack of a high-resolution structure of rhodopsin, or of any other GPCR, has presented a formidable problem for establishing how the transmembrane helices pack and how structural changes in the transmembrane helices are coupled to ligand binding, retinal isomerization, or motion in the other domains of the receptor.
Magic angle spinning (MAS) NMR spectroscopy has been increasingly applied to investigate membrane proteins over the past 10 years. A number of MAS NMR approaches have been developed for measuring the magnitudes and orientations of chemical shift and dipolar interactions in membrane systems (9, 10). These measurements can be directly related to internuclear distances and torsion angles. The advantages of MAS NMR are considerable. Membrane proteins can be studied in their native membrane environment, reaction intermediates can be trapped at low temperature, and structural measurements are possible with ultra-high resolution. Internuclear distances can be measured with resolution on the order of 0.2 Å (11), while torsion angles can be determined to within ±10° (12). Such high-resolution measurements are well suited for establishing the key helix–retinal and helix–helix interactions that may be involved in receptor activation.
High-level expression of the rhodopsin gene in a suspension-adapted HEK293 stable cell line recently has allowed the preparation of milligram amounts of rhodopsin (13). By using defined media containing 6-15N-lysine and 2-13C-glycine, we now report that milligram quantities of isotopically labeled rhodopsin can be obtained in a similar fashion. This ability to incorporate isotope labels into rhodopsin on a large scale overcomes the major limitation for NMR structural measurements and opens the door for studies of this diverse and important G protein-coupled receptor family.
We present MAS NMR spectra of rhodopsin containing isotopically labeled amino acids. Incorporation of 6-15N-lysine into rhodopsin allows us to uniquely target the PSB bond that links the retinylidene chromophore to Lys-296 on transmembrane helix 7. The interaction between the PSB and the Glu-113 counterion is of considerable interest because of its critical role in spectral tuning (14, 15) and receptor activation (16). The 15N chemical shift at 156.8 ppm now reported is characteristic of a PSB nitrogen that has a weak counterion interaction. A long effective Schiff base–counterion distance suggested by the 15N chemical shift is consistent with a structural water molecule in the retinal binding site bridging the counterion and the Schiff base proton (17, 18).§
MATERIALS AND METHODS
Material.
The sources of all reagents for cell culture and rhodopsin purification have been described (13) except for the following. n-Octyl-β-d-glucoside was obtained from Anatrace (Maumee, OH). Custom-made DMEM lacking l-glutamine, glycine, l-arginine, l-histidine, l-lysine, sodium pyruvate, calcium, and sodium bicarbonate was obtained from Atlanta Biologicals (Norcross, GA). Cell culture grade calcium chloride, Pluronic F-68, and amino acids were from Sigma. Isotope-labeled amino acids were from MassTrace (Woburn, MA) and Cambridge Isotope Laboratories (Andover, MA). Both 2-13C-glycine and 6-15N-lysine were >99% labeled and used without further purification. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) was obtained from Avanti (Alabaster, AL).
Buffers.
The following buffers were used: buffer A, 137 mM NaCl/2.7 mM KCl/1.5 mM KH2PO4/8 mM Na2HPO4, pH 7.2; buffer B, buffer A with 4% (wt/vol) n-octyl-β-d-glucoside; buffer C, 10 mM 1,3-bis[tris(hydroxymethyl)-methylamino]-propane, pH 6.0 with 1.46% (wt/vol) n-octyl-β-d-glucoside; buffer D, buffer C + 100 μM nonapeptide; buffer E, buffer A with 1.46% (wt/vol) n-octyl-β-d-glucoside; buffer F, 10 mM Hepes/50 mM NaCl/1 mM EDTA/1 mM DTT, pH 7.1 with 4% (wt/vol) n-octyl-β-d-glucoside, and buffer G, 50 mM Tris/50 mM sodium acetate, pH 7.0.
Growth of Cells in Suspension.
The construction of the stable cell line (HEK293S) expressing the opsin gene has been described by Reeves et al. (13). Suspension cultures were set up by using 2-liter spinner flasks containing 500 ml of DMEM supplemented immediately before use with dialyzed (1-kDa cutoff) fetal bovine serum (FBS) (10%), Pluronic F-68 (0.1%), heparin (50 mg/liter), penicillin, streptomycin, and the missing amino acids (including l-glutamate) at the concentrations used in the original description of DMEM (19, 20). CaCl2 was added at a concentration of 340 μM based on growth optimization in this study. Cell lines first were grown to confluence in 15-cm culture dishes containing 30 ml of DMEM with the appropriate concentration of geneticin G418 (13). At about 70% confluence, cells were fed with the same medium containing 680 μM Ca2+ and grown to confluence. Spinner flasks were inoculated by using three such culture dishes (6–9 × 107 cells) and incubated at 37°C in a humidified incubator. After 6 days, the culture medium was further supplemented with 6 ml of 20% (wt/vol) glucose and 4 ml of 8% (wt/vol) NaHCO3 (21). Cells were harvested on day 7 and incubated with 11-cis retinal as described (13).
Immunoaffinity Purification of Rhodopsin.
Rhodopsin was purified by immunoaffinity chromatography with 1D4-Sepharose beads by using a modified procedure of Reeves et al. (13). The detergent dodecyl-β-d-maltoside was replaced by n-octyl-β-d-glucoside throughout. Buffers B, C, D, and E were used as solubilization buffer, first washing buffer, second washing buffer, and elution buffer, respectively, as described (13).
Reconstitution in DOPC.
The rhodopsin eluted in buffer D was concentrated by using concentration devices (Amicon Centricon, 10-kDa cutoff) to 1 mg/ml as determined by UV/Vis spectroscopy. A 300-fold molar excess of DOPC as a suspension in degassed buffer F was sonicated for 10–15 min. The purified rhodopsin and lipids then were combined and mixed end over end for 1 h before transferring into dialysis tubing (12- to 14-kDa cutoff). The rhodopsin in mixed micelles was dialyzed against 200 vol of buffer G at 4°C. The buffer was changed every 4 h over a 24-h period, and the resulting proteoliposomes were pelleted by centrifugation at 100,000 × g for 1 h at 4°C and loaded into the NMR rotor under dim red light.
Mass Spectrometric Analysis of 15N-Lysine Incorporation into Rhodopsin.
Rhodopsin (10 μg) reconstituted into phospholipid vesicles was resuspended in 1 ml of water to remove salts and buffers. After centrifugation at 12,800 × g for 15 min at room temperature, the pellet was resuspended in 10 μl of water to which endoproteinase Asp-N (0.08 μg/μl in 20 mM Tris, pH 8.0) was added such that the rhodopsin-to-enzyme ratio was 50:1 (wt/wt). After incubation at room temperature for 6 h, the suspension was centrifuged as above. One microliter of the supernatant was mixed with 0.1 μl of serial dilutions (1:1,000 to 1:10,000) of an internal control peptide (20 pmol/μl) with mass 1720.9 g/mol. This solution was mixed with 0.5 μl of 10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid and spotted onto the sample plate. The air-dried crystals were analyzed by matrix-assisted laser desorption ionization time of flight mass spectrometry (Voyager-DE STR Biospectrometry Workstation, PerSeptive Biosystems, Framingham, MA). Spectra were collected in the reflector mode.
Synthesis and NMR of the 11-cis Retinal Schiff Base Model Compounds.
11-cis-Retinylidene-butyl-15N-imide was synthesized by the method of Harbison et al. (22) but replacing all-trans retinal with 11-cis retinal. The resulting Schiff base was dissolved in CDCl3 and protonated with a 2-fold molar excess of either 37% HCl, 48% HBr, or 56% HI. The spectra for the model compounds were recorded at 25°C by using a Varian Inova 500 NMR spectrometer.
MAS NMR Spectroscopy of Rhodopsin.
MAS spectra were obtained on a Bruker Avance NMR spectrometer by using double-resonance probes from Doty Scientific (Columbia, SC). The 13C, 15N, and 1H frequencies were 90.4, 36.4, and 359.4 MHz, respectively. The protein spectra were obtained by using a ramped-amplitude cross polarization sequence (23) at −60°C. The 13C spectrum was obtained by using a 4-μs 1H 90° pulse length, 3-ms contact pulse, 20-μs dwell time, 41-ms acquisition time, and 5.8-kHz MAS speed. The 15N spectrum was obtained by using a 4-μs 1H 90° pulse length, 3-ms contact pulse, 50-μs dwell time, 51-ms acquisition time, and 4-kHz MAS speed.
RESULTS
Large-Scale Expression of Isotope-Labeled Rhodopsin.
Reeves et al. (13) demonstrated that milligram quantities of wild-type rhodopsin could be obtained from suspension cultures by using a HEK293S cell line stably expressing the opsin gene. The medium formulation was optimized to allow efficient incorporation of labeled amino acids into the expressed opsin. The medium designed for isotope labeling of opsin was deficient in five amino acids (Arg, Glu, Gly, His, and Lys) and calcium. The absence of five amino acids allows any combination of them to be added in a labeled form. Limiting the media to only five deficient amino acids decreases the number of parameters that must be optimized for maximum opsin production. All five amino acids, labeled or unlabeled, were added back to the medium to allow cell growth while calcium was added back in a controlled fashion. HEK293S cells were grown in various concentrations of calcium (0–6.8 mM) and assessed for growth on plates after 5 days of incubation (data not shown). CaCl2 concentrations below 34 μM and above 3,400 μM were found to be toxic. Suspension growth was supported with calcium levels between this range, but was sensitive to the exact concentration. CaCl2 concentrations of 100–340 μM supported cell growth and promoted cell detachment from the culture dish. Animal serum was used as a source of essential growth factors, lipids, and other micronutrients that are required for the growth of animal cells. However, serum also contains amino acids that would interfere with specific labeling. These amino acids therefore were removed by dialysis by using tubing with a 1-kDa cutoff. The efficiency of rhodopsin production using media supplemented with either dialyzed or nondialyzed FBS at a final concentration of 10% was tested as a function of CaCl2 concentration (Table 1). Optimal rhodopsin production was achieved by using 340 μM CaCl2 and dialyzed FBS. These conditions subsequently were used for the preparative scale labeling experiments.
Table 1.
Rhodopsin yields from HEK293S suspension growth (0.5 liter) under different medium conditions
| Medium DMEM base | cCaCl2/μM | Wet cell mass/g⋅L−1 | Yield rhodopsin/mg⋅L−1 |
|---|---|---|---|
| Standard | 1,360 + FBS* | 1.7 | 1.54 |
| Custom made | 100 + FBS* | 3.1 | 1.77 |
| Custom made | 170 + dialyzed FBS | 3.2 | 1.04 |
| Custom made | 340 + dialyzed FBS | 4 | 1.78 |
Includes roughly 140 μM CaCl2 contribution from FBS.
The preparative biosynthetic incorporation of 6-15N-lysine and 2-13C-glycine into opsin was carried out by using 12 0.5-liter cultures as described in Materials and Methods. Upon treatment of cells with 11-cis retinal the estimated total yield of rhodopsin, as determined by UV/Vis difference spectroscopy, was 10 mg.
Purification and Reconstitution of Rhodopsin into DOPC.
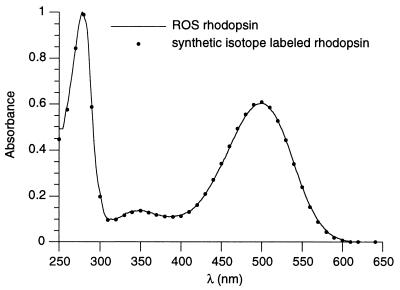
After generation of the rhodopsin pigment by the addition of 11-cis retinal, the whole cells were pelleted and solubilized with octyl-β-glucoside. Cleared lysates from two 500-ml cultures were combined and passed over a 2-ml Sepharose-1D4 column. The column then was washed, and rhodopsin was eluted as described (13) but by using the buffers described in Materials and Methods. The total yield of pure rhodopsin obtained from the 12 500-ml cultures was 9 mg. The labeled rhodopsin prepared in this manner was indistinguishable from rhodopsin purified from rod outer segments (ROS) by all criteria applied. In Fig. 2 the UV/Vis absorption spectrum of the purified HEK293S rhodopsin (A280/A500 ratio of 1.65) is compared with rhodopsin purified from the ROS.
Figure 2.
Absorption spectra of native (line) and synthetic isotope-labeled rhodopsin (dots). Native rhodopsin was obtained from bovine retinas and purified by using the 1D4 immunoaffinity column in a manner parallel to the purification of isotope-labeled rhodopsin expressed in HEK293S cells. The spectra were normalized to the absorption at 280 nm.
For MAS NMR spectroscopy experiments, the labeled rhodopsin was concentrated and reconstituted into dioleolyl phosphatidylcholine vesicles by octyl-β-d-glucoside dialysis in the presence of the phospholipid at a 300:1 molar ratio of lipid to protein (24–26). The extent of reconstitution was determined by UV/Vis spectroscopy on samples resolubilized with detergent. The final amount of rhodopsin reconstituted into liposomes and subsequently used for the MAS NMR experiments was 6.5 mg.
13C MAS NMR Spectroscopy of 2-13C-Glycine, 6-15N-Lysine Rhodopsin.
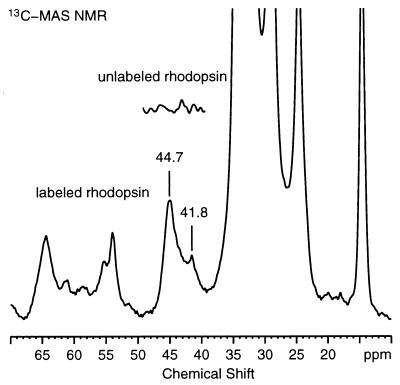
Fig. 3 presents the 13C MAS spectrum of 2-13C-glycine, 6-15N-lysine rhodopsin in DOPC. The 23 glycine resonances in the protein are largely not resolved. An intense 2-13C glycine resonance is observed at 44.7 ppm, and a smaller resonance is observed at 41.8 ppm. The inset shows the spectrum of unlabeled rhodopsin and clearly shows that 2-13C glycine has been incorporated into the opsin apoprotein. Other resonances in the spectrum result from natural abundance 13C of the protein and lipid. The intense peaks between 20 and 40 ppm are caused by signals from methylene groups, while the resonances between 13 and 16 ppm originate from methyl groups.
Figure 3.
13C MAS spectrum of 6.5 mg of 2-13C-glycine, 6-15N-lysine rhodopsin in DOPC membranes. The 13C-resonances from 2-13C glycine are observed between 40 and 47 ppm. (Inset) The spectrum of unlabeled rhodopsin. The spectrum shown corresponds to 36,864 acquisitions. The 13C chemical shifts are reported relative to aqueous tetramethylsilane (TMS). The MAS spectrum was referenced to the carbonyl resonance of external glycine (176.02 ppm relative to TMS). The spectrum was processed with 20 Hz of exponential line broadening.
The efficiency of incorporation of isotope labels into the opsin protein was determined by using MAS NMR. The natural abundance resonance from the backbone and side-chain carbonyls at 175 ppm provides an internal standard, and its integrated intensity was compared with the integrated intensity of the labeled resonance(s). This comparison is possible because the glycine CH2 resonance is separated from the other backbone α-carbons. Quantitation in the MAS experiment is improved by using ramped-amplitude cross polarization (23). The ratio of the integrated intensities of the labeled glycine resonance at 40–47 ppm and natural abundance carbonyl resonances at 175 ppm is approximately 3, corresponding to an incorporation of roughly 60%.
15N MAS NMR Spectroscopy of 2-13C-Glycine, 6-15N-Lysine Rhodopsin.
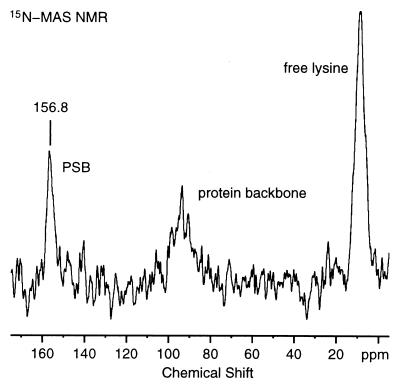
Fig. 4 presents the 15N MAS spectrum of rhodopsin labeled with 2-13C-glycine and 6-15N-lysine. The resonance at 8.7 ppm corresponds to the 10 free lysines in the protein. The natural abundance 15N resonance from the peptide backbone is observed at 93.6 ppm. Finally, the intense resonance at 156.8 ppm is assigned to the 15N resonance of Lys-296, which forms a PSB linkage with the retinylidene chromophore. The spectrum was obtained at 4.5 kHz and is dominated by the isotropic 15N resonances. The ratio of the natural abundance 15N resonance (93.6 ppm) to the 15N labeled PSB resonance (156.8 ppm) is 1.3 ± 0.2. Complete labeling would result in a ratio of 1.32. This ratio indicates that the PSB nitrogen is quantitatively labeled with 15N. This result was confirmed by mass spectrometric analysis of a peptide derived from the rhodopsin C terminus by enzymatic cleavage (residues 330–348, see Fig. 1) containing a single lysine residue (K339). The mass spectrum of the peptide derived from unlabeled rhodopsin showed peaks at m/z 1938, m/z 1939, m/z 1940, and m/z 1941 corresponding to the monoisotopic peaks expected from the calculated mass of the peptide [M + H]+. In the spectrum of labeled rhodopsin, all peaks were shifted by +1, and negligible intensity was found at m/z 1938. These results confirmed that 15N-lysine was quantitatively incorporated into rhodopsin. In the NMR spectrum, however, the ratio of the free lysine signal at 8.7 ppm to the natural abundance 15N signal is lower than expected. The most likely explanation for the lower ratio of the free lysine peak relative to the natural abundance 15N signal is that motion of free lysine residues reduces the efficiency of cross polarization. Even though we have used ramped-amplitude cross polarization to compensate for cross polarization mismatches, it was not possible to optimize the cross polarization conditions on the actual sample because of the extremely low sensitivity.
Figure 4.
15N MAS spectrum of 6.5 mg of 2-13C-glycine, 6-15N-lysine rhodopsin in DOPC membranes. The 15N PSB resonance is observed at 156.8 ppm. The 15N signals corresponding to the free lysines and natural backbone of the protein are observed at 8.7 and 93.6 ppm, respectively. The spectrum shown corresponds to 122,880 acquisitions. The 15N chemical shifts are reported relative to external 5.6 M aqueous 15NH4Cl. The spectrum was processed with 20 Hz of exponential line broadening.
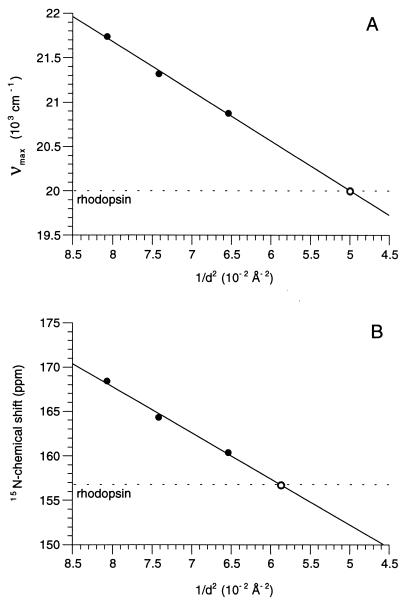
The Schiff base–counterion distances in the protonated 11-cis retinylidene-butyl-15N-imides (Cl−, Br−, I−) were used to estimate the counterion distance in rhodopsin. Both the absorption and the 15N NMR spectra of these model compounds were recorded in chloroform. The frequency of the absorption maximum in wave numbers and the 15N chemical shifts were plotted versus the inverse of the center-to-center distance squared (1/d2) of the crystallographic radii of N3− and the halide counterion (27) as described for bacteriorhodopsin (28, 29) (Fig. 5). The solid line shows the linear regression of the experimental points, while the dotted line shows the corresponding values for rhodopsin. The PSB–counterion distance is estimated to be 4.4 Å based on the correlation of absorption maxima (Fig. 5A) and is estimated to be 4.1 Å based on the correlation of 15N chemical shifts (Fig. 5B).
Figure 5.
Correlation of the frequency of the absorption maximum in wave numbers (A) and the 15N chemical shifts (B) of the protonated 11-cis retinylidene-butyl-15N-imides (Cl−, Br−, I−) versus the inverse of the center-to-center distance squared (1/d2) of the crystallographic radii of N3- and the halide counterion. The solid lines show the linear regression of the experimental points, and the dotted lines show the corresponding values for rhodopsin.
DISCUSSION
Biosynthetic Incorporation of Isotope Labels into Rhodopsin.
The suspension-adapted HEK293 mammalian stable cell lines previously were found from comprehensive investigations of expression systems to yield the highest expression levels of rhodopsin and its mutants (13). A major advantage of this cell line is its ability to efficiently perform the specific posttranslational modifications that are needed for correct folding of the protein and insertion into membranes in a fully functional form. After binding of 11-cis retinal, expressed opsin was indistinguishable from rhodopsin isolated from bovine retinas based on several criteria, including absorption spectra obtained in the dark and after photobleaching, the initial rate of transducin activation, and the rate of phosphorylation by rhodopsin kinase (13). We now show that HEK293S cells can be grown in defined media that contain isotopically labeled amino acids. HEK293S cells grown in suspension allow the production of labeled opsin in the range of 10 mg by using culture sizes of 5–7 liters. This system has provided the amounts of the receptor that are required for biophysical studies, such as those involving MAS NMR. Although the amounts obtained in this study are adequate, clearly more effort is possible in scaling up rhodopsin production by further optimizing cell culture conditions, receptor purification, and incorporation into membranes.
Effective Counterion-Retinal PSB Interaction.
The ability to incorporate 15N-lysine into rhodopsin provides a direct probe of the Schiff base environment by using MAS NMR. The interaction between the Schiff base and its counterion is involved in spectral tuning (14, 15) and receptor activation (16). A salt bridge between the positively charged PSB and its negatively charged Glu-113 counterion has been proposed as part of the mechanism for locking rhodopsin in the inactive conformation in the dark (30). Receptor activation in wild-type rhodopsin is triggered by transfer of the Schiff base proton to Glu-113 in the formation of metarhodopsin II (31), whereas constitutive activation occurs in the E113Q mutant where the negatively charged counterion is neutralized (16, 30). In a low dielectric environment, the neutral species would be favored over the charged ion pair (32). Moreover, the Schiff base pKa of >16 is unusual and reflects an extremely stable complex that is not accessible to the bulk water (18, 33). As a result, the “salt bridge” in rhodopsin is clearly not a simple ion pair. Chemical shift measurements of the protein-bound retinal chromophore previously were used to localize the Glu-113 interaction near C12 of the retinylidene chain (17, 34). Yet, this position of the counterion appears to be inconsistent with the high C = N stretching frequency of the retinal Schiff base linkage that is similar to 11-cis retinal PSB model compounds in methanol and has long been interpreted in terms of a strong Schiff base hydrogen bond (35).
The 15N chemical shift of the imine nitrogen involved in the PSB linkage is unique because it is part of the conjugated retinylidene polyene chain. It is also sensitive to the electrostatic environment of the retinal binding site. DeGroot et al. (36) have shown that the range of 15N chemical shifts in a series of halide PSB model compounds (Cl−, Br−, I−) stretches over 20 ppm because of differences in the counterion strength. Herzfeld and coworkers (28) have correlated the 15N chemical shift with charge delocalization along the retinal chain and the effective Schiff base–counterion distance. These studies have been used for correlating 15N chemical shifts with absorption maxima and Schiff base–counterion distances in bacteriorhodopsin and its photointermediates (29).
For rhodopsin, we report here a 15N chemical shift for the Schiff base of 156.8 ppm. This shift clearly shows that the retinal Schiff base is protonated. Of significance is that the correlation between 15N chemical shifts and absorption maxima observed for all-trans PSB model compounds and bacteriorhodopsin also holds true for rhodopsin (Fig. 5). This correlation supports the finding that the 15N chemical shift generally tracks with charge delocalization along the retinal chain and Schiff base–counterion separation. Based on the correlation between the absorption maxima and counterion separation, the effective counterion distance in rhodopsin is roughly 4.4 Å (Fig. 5A). A slightly shorter separation, 4.1 Å, is predicted from the observed isotropic 15N chemical shift (Fig. 5B). These distances are close to the 4.3-Å distance obtained previously by modeling the 13C chemical shifts along the conjugated retinal chain and retinal absorption spectrum (17, 34). The distance predicted by the 15N chemical shift is also close to that previously predicted for the effective separation between the “complex” counterion in bacteriorhodopsin and the PSB nitrogen of its retinal chromophore (29). The red-shifted absorption maximum observed in bR568, relative to rhodopsin, consequently can be attributed largely to the 6-s-trans conformation of the retinylidene chromophore when bound to the bacterial protein.
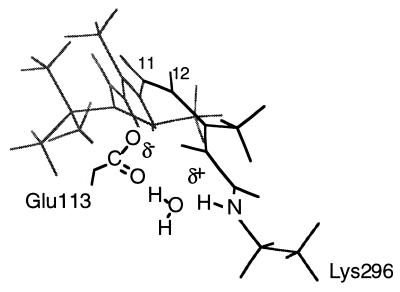
The long effective Schiff base–counterion distance suggested by the 15N chemical shift can be reconciled with the high C = N frequency by the inclusion of structural water in the retinal binding site. A structural water molecule is thought to bridge and be tightly associated with the counterion and the Schiff base proton (17, 18, 33, 37). This Schiff base–counterion geometry explains the high pKa observed for the rhodopsin PSB (18) and reproduces the 500-nm rhodopsin absorption maximum (17). A model of this geometry with a bridging water between the PSB and its counterion is shown in Fig. 6.
Figure 6.
Proposed interaction between the retinal PSB and Glu-113 counterion based on the 156.8 ppm 15N chemical shift. Structural water is thought to mediate the interaction between the positively charged Schiff base proton and the negatively charged Glu-113 carboxyl group. This geometry is consistent with an effective Schiff base–counterion distance of >4 Å based on 13C NMR chemical shift measurements showing a direct interaction between Glu-113 and the C12 position of the retinal (17) and the high C = N stretching frequency characteristic of a strong hydrogen bonding interaction (35).
CONCLUSIONS
The ability to incorporate isotope labels into the opsin protein opens up an experimental approach for establishing the proximity of protein groups within rhodopsin and for probing the structural changes that occur upon receptor activation. Measurements of specific retinal-protein distances are possible by using isotopically labeled opsin regenerated with labeled retinal. Also, because the chemical shifts of Glu, Asp, and His residues are sensitive to protonation state, protonation changes, which occur in the formation of metarhodopsin II, can be measured. Finally, both solid-state and high-resolution solution NMR spectroscopy studies are possible by incorporating selectively labeled amino acids into the cytoplasmic domain of rhodopsin. Such studies should address how structural changes in the transmembrane helices are coupled to motion in the cytoplasmic loops and the binding of the G protein, transducin.
Acknowledgments
We thank A. Maeda and D. Raleigh for their critical comments on the manuscript and Judith Klein Seetharaman for the mass spectroscopy analysis of the labeled rhodopsin samples and numerous discussions. We also thank Francis Picart for the solution NMR measurements on the 11-cis model compounds. This work was supported by grants from the National Institutes of Health to S.O.S. (GM 41412) and H.G.K. (GM 28289).
ABBREVIATIONS
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- FBS
fetal bovine serum
- MAS
magic angle spinning
- PSB
protonated Schiff base
Footnotes
This is paper no. 28 in the series “Structure and Function in Rhodopsin.” Paper no. 27 is ref. 8.
References
- 1.Ji T H, Grossmann M, Ji I. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 2.Gether U, Kobilka B K. J Biol Chem. 1998;273:17979–17982. doi: 10.1074/jbc.273.29.17979. [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 4.Schertler G F, Villa C, Henderson R. Nature (London) 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 5.Unger V M, Hargrave P A, Baldwin J M, Schertler G F X. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 6.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 7.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J M, Altenbach C, Thurmond R L, Khorana H G, Hubbell W L. Proc Natl Acad Sci USA. 1997;94:14273–14278. doi: 10.1073/pnas.94.26.14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith S O, Aschheim K, Groesbeek M. Q Rev Biophys. 1996;29:395–449. doi: 10.1017/s0033583500005898. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths J M, Griffin R G. Anal Chim Acta. 1993;283:1081–1101. [Google Scholar]
- 11.Peersen O B, Groesbeek M, Aimoto S, Smith S O. J Am Chem Soc. 1995;117:7228–7237. [Google Scholar]
- 12.Feng X, Verdegem P J E, Lee Y K, Sandstrom D, Eden M, Boveegeurts P, Degrip W J, Lugtenburg J, Degroot H J M, Levitt M H. J Am Chem Soc. 1997;119:6853–6857. [Google Scholar]
- 13.Reeves P J, Thurmond R L, Khorana H G. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1991;88:3079–3083. doi: 10.1073/pnas.88.8.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson P R, Cohen G B, Zhukovsky E A, Oprian D D. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 17.Han M, Smith S O. Biochemistry. 1995;34:1425–1432. doi: 10.1021/bi00004a037. [DOI] [PubMed] [Google Scholar]
- 18.Steinberg G, Ottolenghi M, Sheves M. Biophys J. 1993;64:1499–1502. doi: 10.1016/S0006-3495(93)81518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dulbecco R, Freeman G. Virology. 1959;8:396–397. doi: 10.1016/0042-6822(59)90043-1. [DOI] [PubMed] [Google Scholar]
- 20.Smith J D, Freeman G, Vogt M, Dulbecco R. Virology. 1960;12:185–180. [Google Scholar]
- 21.Garnier A, Cote J, Nadeau I, Kamen A, Massie B. Cytotechnology. 1994;15:145–155. doi: 10.1007/BF00762389. [DOI] [PubMed] [Google Scholar]
- 22.Harbison G S, Herzfeld J, Griffin R G. Biochemistry. 1983;22:1–4. doi: 10.1021/bi00270a600. [DOI] [PubMed] [Google Scholar]
- 23.Metz G, Wu X, Smith S O. J Magn Reson. 1994;110:219–227. [Google Scholar]
- 24.Jackson M L, Litman B J. Biochemistry. 1982;21:5601–5608. doi: 10.1021/bi00265a033. [DOI] [PubMed] [Google Scholar]
- 25.Wiedmann T S, Pates R D, Beach J M, Salmon A, Brown M F. Biochemistry. 1988;27:6469–6474. doi: 10.1021/bi00417a041. [DOI] [PubMed] [Google Scholar]
- 26.Brown M F. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 27.Weast R C, editor. Handbook of Chemistry and Physics. Cleveland, OH: Chemical Rubber Co.; 1970. [Google Scholar]
- 28.Hu J, Griffin R G, Herzfeld J. Proc Natl Acad Sci USA. 1994;91:8880–8884. doi: 10.1073/pnas.91.19.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J G, Griffin R G, Herzfeld J. J Am Chem Soc. 1997;119:9495–9498. [Google Scholar]
- 30.Cohen G B, Oprian D D, Robinson P R. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 31.Jäger F, Fahmy K, Sakmar T P, Siebert F. Biochemistry. 1994;33:10878–10882. doi: 10.1021/bi00202a005. [DOI] [PubMed] [Google Scholar]
- 32.Beppu Y, Kakitani T, Tokunaga F. Photochem Photobiol. 1992;56:1113–1117. [Google Scholar]
- 33.Deng H, Huang L, Callender R, Ebrey T. Biophys J. 1994;66:1129–1136. doi: 10.1016/S0006-3495(94)80893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han M, DeDecker B S, Smith S O. Biophys J. 1993;65:899–906. doi: 10.1016/S0006-3495(93)81117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livnah N, Sheves M. J Am Chem Soc. 1993;115:351–353. [Google Scholar]
- 36.De Groot H J M, Harbinson G S, Herzfeld J, Griffin R G. Biochemistry. 1989;28:3346–3353. doi: 10.1021/bi00434a033. [DOI] [PubMed] [Google Scholar]
- 37.Nagata T, Terakita A, Kandori H, Kojima D, Shichida Y, Maeda A. Biochemistry. 1997;36:6164–6170. doi: 10.1021/bi962920t. [DOI] [PubMed] [Google Scholar]