Abstract
Mutations in NPHS2, encoding podocin, have been identified in childhood onset focal and segmental glomerulosclerosis (FSGS). The role of NPHS2 in adult disease is less well defined. We studied 30 families with FSGS and apparent autosomal recessive inheritance and 91 individuals with primary FSGS. We screened family members for NPHS2 mutations. NPHS2 mutations appeared to be responsible for disease in nine of these families. In six families, the affected individuals were compound heterozygotes for a nonconservative R229Q amino acid substitution. This R229Q variant has an allele frequency of 3.6% in a control population. In these families, R229Q was the only mutation identified on one of the two disease-associated NPHS2 alleles. We used in vitro–translated podocin and purified nephrin to investigate the effect of R229Q on their interaction and found decreased nephrin binding to the R229Q podocin. These data suggest that this common polymorphism contributes to the development of FSGS. Chromosomes bearing the R229Q mutation share a common haplotype defining an approximately 0.2-Mb region. R229Q appears to enhance susceptibility to FSGS in association with a second mutant NPHS2 allele. Identification of R229Q mutations may be of clinical importance, as NPHS2-associated disease appears to define a subgroup of FSGS patients unresponsive to corticosteroids.
Introduction
Focal and segmental glomerulosclerosis (FSGS) presents both as a primary (idiopathic) condition and as a secondary reaction to a number of common underlying disorders, including obesity, HIV infection, reflux nephropathy, diabetes, renal mass ablation, and hypertension (1, 2). FSGS is a common pattern of glomerular injury, defined by segmental sclerotic lesions involving only a subpopulation of glomeruli. Affected patients frequently progress to end-stage renal failure. While generally regarded as an idiopathic disease, inherited forms of nephrotic syndrome and FSGS have been increasingly recognized. Recently, substantial progress has been made in defining the molecular basis of inherited forms of nephrotic syndrome and FSGS (3). As kidney disease caused by different underlying genetic defects may respond differently to various interventions (e.g., immunosuppression, angiotesin convertin enzyme inhibition, and renal transplantation), defining the molecular basis of this set of diseases is clinically important.
Defects in both NPHS1 alleles, encoding nephrin, cause congenital nephrotic syndrome of the Finnish type (CNF) (4). Nephrin, a putative transmembrane protein, is located at the slit diaphragm of the glomerular epithelial cell (or podocyte). Given the massive proteinuria in CNF infants and the localization of nephrin, this protein clearly plays an essential and nonredundant role in the normal glomerular filtration barrier (5, 6). A much milder and slowly progressive form of familial proteinuric renal dysfunction is characterized by dominant inheritance, FSGS, slowly progressive renal disease, and mutations in α-actinin-4 (7). A second dominant FSGS locus maps to chromosome 11q21-22. The responsible gene has not yet been identified (8).
Boute et al. (9) recently demonstrated that mutations in NPHS2, which encodes podocin, cause a recessive form of steroid-resistant nephrotic syndrome. Affected patients were characterized by early-onset disease (age 6 years or less) and, in most cases, FSGS on renal biopsy (10). Other disease-associated mutations have now been reported in children with familial as well as sporadic disease (11–15). Recent genetic and biochemical studies have suggested an interaction between nephrin and podocin (16–18). Previously, we studied six families with recessive FSGS and adult or adolescent onset (average age of disease onset 24 years). Five of these families showed evidence of linkage to the NPHS2 locus on chromosome 1q25-31 (19). To investigate the possibility that NPHS2 mutations might be responsible for milder, adult-onset disease, we performed mutational analysis of NPHS2 in affected members of these and other families with FSGS in which disease segregation was consistent with recessive inheritance. We followed these family-based studies with analyses in patients with sporadic FSGS.
Methods
Patients and families.
We studied a total of 30 multiplex families, including several families reported previously (FS-W, FS-I, FG-BU, FG-XB, FS-G, and FG-BL) (19). From all families enrolled in our study of familial FSGS, we selected, by review of available clinical information, families with disease segregation pattern consistent with autosomal recessive transmission. Most families were ascertained after referral by a nephrologist caring for one or more of the family members. Autosomal recessive transmission was supported by (a) absence of disease in parents of affected subjects, (b) no disease transmission through multiple generations, and (c) the occurrence of disease in two or more siblings in each family. The diagnosis of FSGS in the index cases was demonstrated histologically by renal biopsy. Families with radiologic, clinical, or histopathologic findings suggesting secondary forms of FSGS were not included. Mean age of disease onset in affected members of these families was 21.8 years. Family members were evaluated by measurement of serum creatinine and urinary microalbumin excretion. Subsequently, we studied 91 adults with (sporadic) primary FSGS. Informed consent was obtained in accordance with protocols approved by the Human Research Committees of the responsible institutions.
Sequence analysis and analysis of segregation.
Genomic DNA was extracted from peripheral blood cells using the QIAamp DNA blood kit (QIAGEN Inc., Valencia, California, USA). Oligonucleotide primers of eight exons encoding NPHS2 were designed based on reported human NPHS2 cDNA sequence (GenBank accession no. NM_014625) and available genomic sequence (Homo sapiens chromosome 1 BAC clone RP11-545A16, available genomic sequence (GenBank AL160286). Genomic DNA (25–50 ng) was subjected to 30–35 cycles of PCR amplification in a volume of 25 μl including 10 mM Tris HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 mM dNTP, and 0.01 U Taq polymerase. Products were gel-purified, and 25–50 ng of DNA was sequenced directly by use of the ABI PRISM dye terminator (Applied Biosystems, Foster City, California, USA). An ABI DNA Sequencer (model 377) was used to perform sequence analysis. Sequence was determined from both strands.
Mutational analysis using denaturing HPLC (DHPLC) was performed using primers optimized for this technology: exon 1, GCAGCGACTCCACAGGGACT and TCCACCTTATCTGACGCCCC; exon 2, AGAATTGGACCAACAGATGC and AAGTGAGAATGGGCATGGTG; exon 3, GCCCCCGCCGTCTTATGCCAAGGCCTTTTGAAGAC and GGGTTGAAGAAATTGGCAAGTCAGG; exon 4, AAGGTGAAACCCAAACAGC and CGGTAGGTAGACCATGGAAA; exon 5, AGGATTTACCACAGGATTAAGTTGTGCA and TAGCTATGAGCTCCCAAAGGGATGG; exon 6, TATTATAAATAAGGCACTGTGAAGTTAAATACAAC AND CCCCCGCCCGCCAGAATATTTTCCTTTATCATACAG; exon 7, GAGGCTTGCAAGTCTGTGTGAAAGC and AGGAAGCAAAGGGGAAATGTTCTCC; exon 8, CGTCCCGCGTGAAGCCTTCAGGGAATGAAGAAC AND GCGTCCCGTTCTATGGCAGGCCCCTTTACAGTC. PCR-amplified DNA was analyzed for sequence alterations by DHPLC (Transgenomic Inc.; Omaha, Nebraska, USA), following the manufacturer’s recommendations, after determination of melting profile.
Segregation of the mutations with disease was assessed by either sequencing or restriction-enzyme digestion of PCR-amplified exons from all available family members. ClaI digestion of exon 5 PCR products (545 bp) normally produces two fragments of 364 and 181 bp. An R229Q mutation (G→A transition at nucleotide 755) was detected by a loss of the ClaI digestion site. An R291W mutation (C→T transition at nucleotide 941) created a new PflMI digestion site and was detected by PflMI digestion of exon 7 PCR products (283 bp) into two fragments (155 and 128 bp).
Mutational analysis of transcript.
RNA was extracted from peripheral lymphocytes using Trizol reagent and reverse-transcribed with random hexamer priming using a SuperScript kit (Invitrogen Corp., Carlsbad, California, USA). Primers ATAGATGCCATTTGCTACTACCG and ACAGCCAGTGAGTGCTGAAG, derived from NPHS2 exons 5 and 7, were designed to amplify a 274-bp amplicon from the NPHS2 transcript containing the sequence encoding R229. RT-PCR–amplified product from lymphocytes from two control individuals (without the R229Q DNA variant) and two individuals heterozygous for R229Q was digested with ClaI and analyzed by agarose gel electrophoresis.
Genotype analysis.
We genotyped family members using known chromosome 1q markers (D1S2883, D1S2640, D1S215, and D1S2751) as well as markers neighboring the NPHS2 gene locus recently reported by Boute et al. (9) (D1S3760 and D1S3758). We found additional CA repeat markers by searching the genomic sequence near NPHS2, published by the Sanger Institute (http://www.sanger.ac.uk/HGP/Chr1). CA126 was found in BAC sequence (GenBank AL160286) and amplified using primers 5′-TTTCATAACTACTTAATGCTGGCAGGC-3′ (sense) and 5′-GGTACACTGGATACAACCCAATTGATA-3′ (antisense). CA1419 was found in BAC RP11-177A2 (GenBank AL139132) and was PCR-amplified using oligonucleotides 5′-CGGATTATATTTTTTAGGTCCAGGATTTC-3′ (sense) and 5′-CCTCTCAGCCAAGGAATTCAAAATAGC-3′ (antisense). Genotypes were determined by PCR amplification, with one of the primers labeled with either [32P]-dATP or a fluorescent dye, under standard conditions, followed by gel electrophoresis of the product. The marker order and the approximate distances were obtained from databases of the Whitehead Institute/MIT Center for Genome Research, Cambridge, Massachusetts; http://www.genome.wi.mit.edu) and from the Genome Database (http://gdbwww.gbd.org). Haplotypes were determined manually by inspection of genotypes determined in parents and/or siblings.
We screened 91 adult individuals with sporadic FSGS for R229Q polymorphisms. We also screened DNA from several control populations: (a) 124 individuals without known kidney disease (primarily spouses of FSGS patients), (b) 16 African and 16 African-American individuals (because of the high incidence of FSGS in these populations), and (c) 49 individuals from Brazil (as a control for the largest family, FS-W).
Nephrin-binding experiment.
We subcloned a full-length nephrin cDNA clone into the pcDNA3.1/V5/His-TOPO vector (Invitrogen Corp.). We transfected human embryonic kidney 293 cells with the cDNA construct. Transfected HEK293 cells were lysed with buffer A (containing 1% Triton X-100, 10 mM imidazole, and protease inhibitors). Purification of the His-tagged protein was performed using Ni-NTA (QIAGEN Inc.) agarose following the manufacturer’s protocol. The concentrated protein was recognized by a polyclonal anti-nephrin antibody (described in ref. 20).
We produced 35S-labeled wild-type, R229Q mutant, and R291W mutant podocin by in vitro transcription/translation as described in ref. 7. The mutant podocin clones were derived from a full-length human NPHS2 cDNA clone by site-directed mutagenesis using a QuickChange kit (Stratagene, La Jolla, California, USA) and verified by sequencing. In vitro transcription/translation products were normalized to equal counts of translation product per volume. We used equal volume (20 μl) of labeled in vitro–translated wild-type, R229Q, and, in two experiments, R291W podocin as well. To each 20-μl reaction we added 800 μl of precipitation immunomix, consisting of 0.2% (wt/vol) SDS, 10% (wt/vol) BSA, 1 mM EDTA, 1 mM PMSF, and 5 mM indole acetic acid in wash immunomix (1% Triton X-100, 0.5% sodium deoxycholate in PBS). After preabsorption by incubation with 10 μl of preimmune rabbit serum for 2 hours at 4°C, we added 50 μl of protein A-Sepharose CL-4B beads (Amersham Biosciences, Buckinghamshire, UK) to the reaction to eliminate nonspecific binding. After gentle rocking at 4°C for 2 hours, the beads were removed by brief centrifugation. This incubation with beads and washing was repeated. The supernatants were then incubated overnight with anti-nephrin antibody (20) and 1 μg of the purified nephrin. We added 50 μl of protein A-Sepharose CL-4B beads, and the samples were incubated 2 hours at 4°C with gentle rocking. The beads were collected by centrifugation at 16,000 g at 4°C for 20 minutes. The pellet was washed with a series of buffers to eliminate nonspecific binding. The series of washings was as follows: once with wash immunomix, then with 2 M KCl in wash immunomix, then with Neufeld buffer (0.05% [wt/vol] NP-40, 0.1% [wt/vol] SDS, 0.6 M NaCl, 10 mM Tris-HCl [pH 8.5]), and then twice with 1:10 diluted PBS. The pellet was collected by centrifugation. The immune complexes were extracted from the pellet by heating at 95°C for 5 minutes with 60 μl of Laemmli buffer followed by centrifugation at 13,000 rpm for 30 minutes. The clear supernatant was subjected to 10% PAGE in the presence of SDS under reducing conditions. The gel was transferred onto nitrocellulose and exposed to film for autoradiography. The same blot was used for the immunodetection of the amount of nephrin protein precipitated in each reaction. As a negative control, this experiment was also performed in the absence of added nephrin. Densitometry was performed using NIH Image software (21).
Results
Sequence analysis of NPHS2 gene.
We analyzed PCR amplicons of each of the eight exons of NPHS2 by direct sequencing and/or DHPLC, using genomic DNA as template. We found that all affected individuals in a large family (FS-W) are compound heterozygotes for two independent missense substitutions: a G→A transition at nucleotide 755, predicting an Arg→Gln substitution at codon 229 (exon 5, R229Q), and a C→T transition at nucleotide 941, predicting an Arg→Trp substitution at codon 291 (exon 7, R291W; Figure 1). The former is a missense nucleotide substitution; the latter has been previously described in early-onset FSGS (9). We then examined these mutations in members of this family by restriction endonuclease digestion of PCR products. As shown in Figure 1b, all affected members of this family are compound heterozygotes for these two mutations. No unaffected individual in the family carries the two mutations, consistent with cosegregation of the sequence variants with disease under a recessive model.
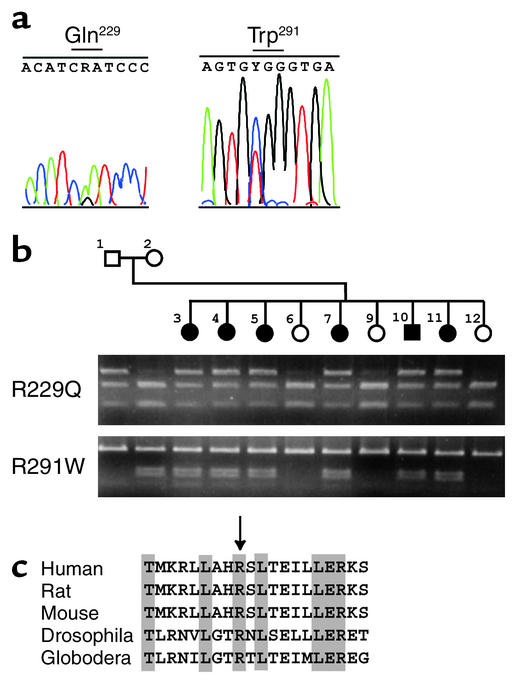
Figure 1.
NPHS2 missense mutations in family FS-W. (a) DNA sequence chromatograms. Left: DNA sequence analysis of exon 5 amplified from an affected individual in family FS-W. A G→A transition predicts an Arg→Gln substitution. Right: DNA sequence analysis of exon 7 shows a C→T transition resulting in an Arg→Trp substitution. (b) Cosegregation of mutations with disease. Affected individuals are showed by filled circles (female) and squares (male). Agarose gel electrophoresis demonstrates the presence or absence of mutations within family FS-W. Top: R229Q mutations detected by ClaI digestion. The point mutation G755A results in a loss of ClaI site, and mutant R229Q alleles are shown by the top bands (545 bp), representing PCR products missing a ClaI site. Bottom: R291W mutations identified by PflMI digestion. A C941T mutation creates a PflMI restriction site and is visualized as two bands of 155 bp and 128 bp. Affected individuals are compound heterozygotes for R229Q and R291W. (c) Alignment of species orthologs of podocin. Amino acid sequences encoded by human NPHS2 (positions 221–240, GenBank NP_055440), rat NPHS2 (GenBank AF309631), mouse EST (GenBank AI552534), Drosophila melanogaster EST (GenBank AA141235), and Globodera rostochiensis EST (GenBank AW505792) were aligned by the PILEUP program (Wisconsin package version 10.0 [Unix]; Genetic Computer Group, Accelrys, San Diego, California, USA). Identical amino acid residues are indicated by shading. The arginine residue at position 229 is highlighted by an arrow.
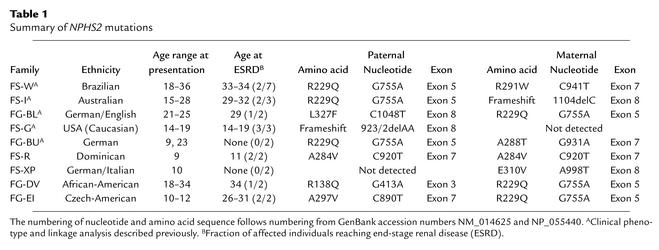
We analyzed affected members of additional FSGS families and found missense mutations (R138Q, A284V, A288T, A297V, E310V, and L327F) and two novel minor deletions (923/2delAA and 1104delC) in segregation patterns consistent with recessive disease inheritance (Table 1). Two of these mutations have been described earlier (9, 22). R229Q has been identified as a polymorphism (22). The 2-bp deletion at nucleotide 923 (923/2delAA) causes a frameshift and alters the amino acid sequence of exon 7 after codon 285. A single-nucleotide deletion, 1104delC, in exon 8 results in a frameshift and premature termination at codon 347, thereby truncating the C-terminal 36 amino acids. We found mutations cosegregating with disease in a total of nine families. In six families (FS-W, FS-I, FG-BL, FG-BU, FG-DV, FG-EI), the affected individuals are compound heterozygotes, and in one (FS-R), the affected individuals are homozygous. In two families (FS-G and FS-XP), only a single NPHS2 mutation was found; mutations were not detected in the second NPHS2 allele. In these patients, we sequenced the entire coding regions as well as the intron-exon boundaries. We suspect either that coding sequence mutations escaped our detection (this is unlikely), or that mutations in noncoding regulatory regions are present in the second allele. In 21 other FSGS families tested, we found no likely disease-causing NPHS2 mutations.
Table 1.
Summary of NPHS2 mutations
Cosegregation of mutations with disease was established either by direct sequencing or by restriction-enzyme digestion of PCR products (data not shown). Of note, the mutations observed appear to cluster in the carboxyl terminal portion of podocin encoded by exons 5–8 (with one exception). Four missense mutations (A284V, A288T, A297V, and E310V) are located in an alanine- and glutamate-rich region that is highly conserved within the stomatin protein family. This distribution of the mutations appears to contrast with the widely distributed location of mutations in families with early-onset disease (9, 14, 15, 22).
R229Q mutations in familial FSGS.
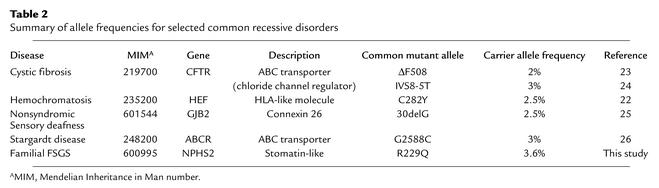
The R229Q mutation was detected in six of the nine FSGS families with NPHS2 alterations. We screened R229Q mutation in several groups of control individuals by means of ClaI digestion of PCR-amplified exon 5. We found the R229Q mutation present in the populations screened with the following allele frequencies: 0.016 (1 of 64 alleles from individuals of African descent), 0.031 (3 of 98 alleles from Brazilian individuals), 0.036 (9 of 248 alleles from an ethnically diverse but largely North American and Western European panel of DNA samples), and 0.058 (6 of 104 alleles from North American individuals). Analysis of available sequence from species orthologs of NPHS2 genes shows that the arginine residue at position 229 is evolutionarily conserved from invertebrates (Drosophila melanogaster and Globodera rostochiensis) to vertebrates (human, rat, and mouse), suggesting an important role of Arg 229 for structure and function (Figure 1). Based on the high frequency of the R229Q allele in FSGS families and the evolutionary conservation of this residue, we believe the R229Q allele to be a relatively common disease-causing recessive allele, rather than a benign polymorphism. All of the other mutations identified (Table 2) were absent in 144 control chromosomes examined.
Table 2.
Summary of allele frequencies for selected common recessive disorders
To examine the possibility that the R229Q variant altered transcript expression or stability, we performed RT-PCR using peripheral lymphocytes RNA from R229Q subjects to amplify a 274-bp fragment including the R229Q-encoding nucleotide change, followed by ClaI digestion in order to discriminate between the two alleles. Approximately 50% of the RT-PCR product was cleaved by ClaI (compared with complete digestion of the RT-PCR product from control lymphocytes), suggesting that both the R229Q and the non-R229Q alleles are expressed.
Haplotype analysis of R229Q allele.
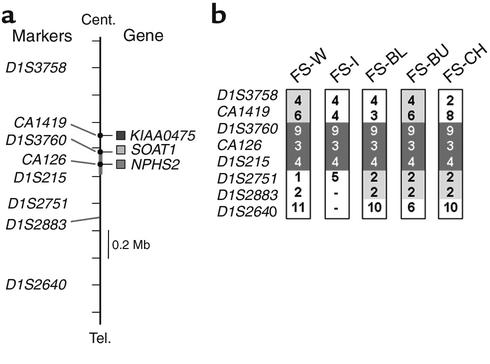
To investigate whether the R229Q allele is derived from a common founder, we genotyped markers placed within 1 centimorgan of NPHS2 and constructed haplotypes by inspection of segregation within families. CA126 and NPHS2 are located on a common BAC clone (GenBank AL160286) and therefore lie within approximately 100 kb of NPHS2. D1S3760 and D1S215 are also located in close proximity to the NPHS2 gene, 100 and 200 kb centromeric and telomeric, respectively. We found a common disease haplotype with allele numbers arbitrarily designated 9-3-4 (based on (CA)n repeat number of 192 bp, 156 bp, and 206 bp) at markers D1S3760, CA126, and D1S215 in all five families tested that carried the R229Q mutation (Figure 2). We extended these studies to include nine R229Q-heterozygous individuals identified from the controls studied and found that these individuals also share the 9-3-4 disease-associated haplotype. The frequencies of these R229Q-associated alleles as assayed in a control population were found to be 0.181 (for allele 9 of marker D1S3760; 94 chromosomes tested), 0.179 (for allele 3 of marker D1S215; 140 chromosomes tested), and 0.28 (for allele 4 of marker CA126; 134 chromosomes tested). The marker distances define common inheritance of a shared region of chromosome 1 approximately 0.2 Mb in size. This suggests that the R229Q variant arose in a common ancestor.
Figure 2.
Haplotype analysis for 1q25 markers surrounding the NPHS2 gene. Haplotypes of chromosomes bearing the R229Q-encoding mutations are shown. (a) The genomic context of the markers used. (b) Portions of the haplotypes that are totally conserved (dark gray) and partially shared (light gray) are indicated. The orders and intervals of the frame markers are determined based on Genemap ’99 (www.ncbi.nlm.nih.gov/genemap), the Sanger Center chromosome database. The markers CA126, CA1419, and D1S3760 are derived from the BACs harboring NPHS2 and two neighboring genes SOAT1 (sterol-O-acetyltransferase) and KIAA0475. Cent., centromere; Tel., telomere.
R229Q in sporadic FSGS.
We next genotyped DNA from 91 adults with primary FSGS for the R229Q podocin variant. The majority of these individuals were ascertained through presentation to nephrologists in the northeastern US. We identified one R229Q allele in 11 of these individuals (allele frequency 6.0%). We observed no homozygous individuals. We also genotyped DNA from three control groups. We found the R229Q mutation present in the populations screened with allele frequencies ranging from 1.6% (1 of 64 alleles from individuals of African descent) to 3.1% (3 of 98 alleles from Brazilian individuals) to 3.6% (9 of 248 alleles from a largely Western panel of DNA samples). This slight increase in the frequency of R229Q in FSGS patients does not reach statistical significance.
Because this slight increase in R229Q frequency in FSGS patients could be explained by a subset of these patients having recessive disease (i.e., an NPHS2 mutation on the second allele as well), we used DHPLC and direct sequencing to look for other NPHS2 mutations in these 11 sporadic FSGS patients with the R229Q polymorphism. In two of these patients, we identified a second NPHS2 mutation, in both cases the previously described A284V substitution (9). In nine patients, only the single R229Q polymorphism was observed.
Nephrin binding.
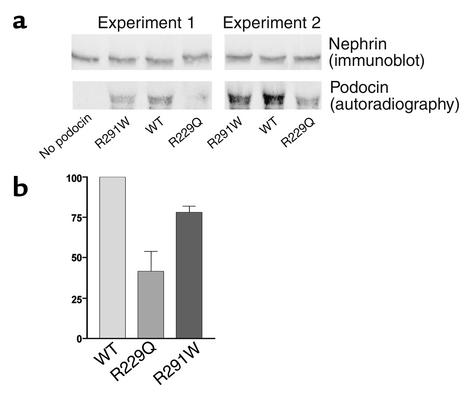
Podocin has been shown to localize to the glomerular slit diaphragm and to interact directly with nephrin, a major slit diaphragm protein (4, 5, 17, 18). In order to demonstrate a biologic difference between the R229Q and wild-type peptides, we performed nephrin-binding assays using purified nephrin, labeled in vitro–translated R229Q, wild-type podocin, and, in two sets of experiments, podocin bearing the previously identified R291W mutation (9). In each of seven pairs of experiments, we observed more wild-type podocin than R229Q mutant podocin in the nephrin pulldown assays (Figure 3a). On average, the normalized intensity of R229Q mutant podocin detected was 41.5% of wild-type (n = 7 paired comparisons, SEM 12.3%, P = 0.003 in a paired two-tailed t test). In both assays in which R291W mutant podocin was also included as an additional control, signal intensity was less for R291W than for wild-type podocin. R291W binding was not as diminished as with R229Q (78% of wild-type, SD 4.0; Figure 3), and this difference was not statistically significant (P = 0.11). In the absence of nephrin, podocin did not immunoprecipitate. Western blot analysis showed that equivalent amounts of nephrin were immunoprecipitated in each reaction (Figure 3b).
Figure 3.
Nephrin binding assay. (a) The bottom autoradiograph shows in vitro–translated and labeled mutant and wild-type (WT) podocin after incubation with purified nephrin, immunoprecipitation with an anti-nephrin antibody, transfer to nitrocellulose, and exposure to radiographic film (as described in detail in Methods). Shown above is the result of Western analysis of the same blots using the anti-nephrin antibody for immunodetection. (b) Densitometry of the R229Q and R291W bands from repeated paired experiments. Intensity (with error bars indicating SEM) is given as a percentage of wild-type intensity, which is set by definition at 100%.
Discussion
In 30 families with adolescent or adult-onset recessive FSGS, we found seven NPHS2 mutations, of which two are novel. These mutations appear to be responsible for disease in nine of these families. Our data suggest that NPHS2 alleles have considerable heterogeneity in their biologic effect and cause FSGS with a quite broad spectrum of severity and age of onset. In this group of families, the R229Q podocin variant was quite common. Several lines of evidence lead us to believe that the R229Q allele is disease-causing, rather than a benign polymorphism: (a) there is evolutionary conservation of the Arg 229 residue in podocin homologs; (b) arginine to glutamine is a nonconservative amino acid change; (c) R229Q segregates with disease; and (d) the biologic properties of the encoded peptide are altered. In contrast with other disease-causing NPHS2 variants, which are rare, R229Q alleles are common in the general populations, with an observed frequency similar to those of other common recessive diseases, including cystic fibrosis and hemochromatosis (23) (Table 2). The R229Q mutation alone is insufficient to cause FSGS but appears to enhance susceptibility to renal injury in the compound heterozygote state in association with a second mutant NPHS2 allele. Heterozygosity for R229Q was found at a slightly greater frequency in FSGS patients than in control groups. In two of 11 of these patients, the presence of a second A284V NPHS2 mutation on the other allele suggests that compound heterozygosity for these sequence changes is the likely cause of disease. The recent observation that, in one neonate, a single R229Q allele may have modified a congenital nephrotic phenotype (due to NPHS1 defects) to produce a neonatal focal segmental glomerulosclerosis phenotype further supports the notion that the R229Q variant has a biologic effect (16). This observation also raises the related possibility that variants in other genes (including NPHS1) may facilitate the phenotypic effect of the common R229Q podocin variant.
It is likely that the phenotype resulting from homozygosity for the R229Q variant is different from the effect of compound heterozygosity for R229Q and another disease-associated allele. More noteworthy than the very small increase in the frequency of R229Q in the sporadic FSGS group is the fact that it is not seen with much greater frequency than in controls: Because of the high frequency of R229Q in the population, we would naively expect that R229Q/R229Q homozygosity would be among the most common causes of FSGS, if this genotype were associated with FSGS. There are several plausible explanations for the absence of this finding: (a) R229Q is benign; (b) R229Q/R229Q causes either no phenotype or an exceedingly mild phenotype; (c) R229Q/R229Q causes a severe, perhaps embryonic-lethal, phenotype. We reject the first explanation on the basis of the data presented in this paper. We believe that the third explanation (early lethality) is much less likely than the second (mild or no phenotype), but we cannot reject it on the basis of the data we have obtained to date. Assuming that the carrier allele frequency is 0.036 and that the observed control population is under Hardy-Weinberg equilibrium, we would expect to see R229Q homozygotes at a frequency of approximately 1:1,000. Since the incidence of FSGS is generally thought to be on the order of 1:105, it is reasonable to speculate that homozygotes for R229Q do not develop clinical findings sufficient to bring them to medical attention, at least in the absence of an additional renal insult. Certainly, it is possible that R229Q homozygotes do have a mild phenotype that generally escapes medical attention (e.g., asymptomatic proteinuria without renal insufficiency). We cannot formally exclude the possibility that the R229Q variant is in linkage disequilibrium with additional NPHS2 locus variants that cause or contribute to disease. However, data presented above support the role of the R229Q substitution itself in disease pathogenesis.
The high frequency of R229Q suggests an old mutation. The relatively short length of the chromosomal region in which linkage disequilibrium is conserved strongly supports the idea of an ancient origin for R229Q mutations. The contribution of common alleles to human disease has been extensively studied in other autosomal recessive disorders. Similar carrier frequencies have been reported for well-known common diseases (Table 2), including ΔF508, and IVS8-5T in the cystic fibrosis gene CFTR (24, 25); C282Y in HFE in hemochromatosis (23); 30delG in GJB2 in nonsyndromic sensory deafness (26); and G2588C in ABCR in Stargardt disease (27). The high frequency of particular mutations among carriers may reflect low selective pressure due to their modest pathogenic effects. As has been described in hemoglobin S and malaria, high prevalence may be caused by a selective advantage of heterozygote carriers. The selective advantage of harboring R229Q is not obvious.
The biochemical function of podocin and its role in FSGS pathogenesis remain to be investigated. Podocin has 46% amino acid identity to human stomatin and 40% identity to Caenorhabditis elegans MEC-2 (28) and Unc1 (29). Amino acid sequence suggests that NPHS2 has a structure similar to stomatin and has a single transmembrane domain. Stomatin is a 31-kDa integral membrane protein and is absent in red blood cells in the rare human hemolytic anemia hereditary stomatocytosis (30). Red cells lacking this protein show a high permeability for monovalent cations, suggesting the possibility that the protein has a role in the regulation of ion transport. In immunocytochemical studies with the human amniotic cell line UAC, stomatin concentrates preferentially to plasma membrane folds and protrusions and appears to be colocalized to the cortical actin cytoskeleton (31), suggesting a role in cell morphogenesis. The data suggest that in patients with NPHS2 mutations, FSGS may be due to disrupted structural integrity of podocyte cytoskeleton.
In contrast to the NPHS2 mutations reported in the severe childhood form of steroid-resistant nephrotic syndrome (9), the NPHS2 mutations we report display some distinct characteristics that appear to be associated with later onset of disease. The NPHS2 mutations we report are either missense mutations or nucleotide deletions. The absence of obvious null mutations in our report suggests that the encoded peptide may retain some function and thus result in a milder phenotype. Second, we found that these mutations cluster in the C-terminal part of podocin. By contrast, mutations associated with severe childhood-onset are dispersed throughout the encoded protein but, in general, are more N-terminal in location than the mutations reported here. This raises the possibility that such N-terminal mutations may represent particular alleles causing protein misfolding or altered protein processing, thereby causing a more severe clinical phenotype. The presence of common variants such as R229Q will improve carrier detection and genetic counseling, as well as identification of appropriate therapy and elimination of corticosteroids as a therapeutic option (10). The G→A substitution abolishes a ClaI restriction site, which allows rapid screening for this mutation. Further investigation is required to determine whether a mild phenotype may be associated with heterozygosity for NPHS2 variants, as well as whether homozygotes for the common R229Q variant have any kidney dysfunction. The severity of clinical disease associated with NPHS2 alterations is highly variable. This suggests that other factors (genetic and environmental) are involved in the development of FSGS. Identification of such factors will improve diagnosis and treatment of nephrotic syndrome. Common NPHS2 polymorphisms may be among those factors that alter the susceptibility to various forms of secondary renal injury.
Acknowledgments
This work was supported by grants from the Uehara Medical Foundation (to H. Tsukaguchi), the NIH (DK-54931 to M.R. Pollak; DK-51711 and DK-55001 to R. Kalluri), and the National Kidney Foundation (to M.R. Pollak); a Kidney and Urology Foundation of America Clinical Fellowship (to J.A. Schwimmer); and grants from the Glomerular Institute at Columbia University and Zo’s Fund for Life (to G.B. Appel). We thank the study subjects for their participation.
Footnotes
Hiroyasu Tsukaguchi and Akulapalli Sudhakar contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: focal and segmental glomerulosclerosis (FSGS); denaturing HPLC (DHPLC).
References
- 1.D’Agati V. The many masks of focal segmental glomerulosclerosis. Kidney Int. 1994;46:1223–1241. doi: 10.1038/ki.1994.388. [DOI] [PubMed] [Google Scholar]
- 2.Ichikawa I, Fogo A. Focal segmental glomerulsoclerosis. Pediatr Nephrol. 1996;10:374–391. doi: 10.1007/BF00866790. [DOI] [PubMed] [Google Scholar]
- 3.Somlo S, Mundel P. Getting a foothold in nephrotic syndrome. Nat Genet. 2000;24:333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 4.Kestila M, et al. Positionally cloned gene for a novel glomerular protein — nephrin — is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 5.Holzman LB, et al. Nephrin localizes to the slit pore of the glomerular epithelial cell. Kidney Int. 1999;56:1481–1491. doi: 10.1046/j.1523-1755.1999.00719.x. [DOI] [PubMed] [Google Scholar]
- 6.Ruotsalainen V, et al. Nephrin is specifically located at the slit diaphragm of glomerular podocytes. Proc Natl Acad Sci USA. 1999;96:7962–7967. doi: 10.1073/pnas.96.14.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan JM, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 8.Winn MP, et al. Linkage of a gene causing familial focal segmental glomerulosclerosis to chromosome 11 and further evidence of genetic heterogeneity. Genomics. 1999;58:113–120. doi: 10.1006/geno.1999.5828. [DOI] [PubMed] [Google Scholar]
- 9.Boute N, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 10.Fuchshuber A, et al. Mapping a gene (SRN1) to chromosome 1q25-q31 in idiopathic nephrotic syndrome confirms a distinct entity of autosomal recessive nephrosis. Hum Mol Genet. 1995;4:2155–2158. doi: 10.1093/hmg/4.11.2155. [DOI] [PubMed] [Google Scholar]
- 11.Wu MC, Wu JY, Lee CC, Tsai CH, Tsai FJ. Two novel polymorphisms (c954T>C and c1038A>G) in exon8 of NPHS2 gene identified in Taiwan Chinese. Hum Mutat. 2001;17:237. doi: 10.1002/humu.13. [DOI] [PubMed] [Google Scholar]
- 12.Wu MC, Wu JY, Lee CC, Tsai CH, Tsai FJ. A novel polymorphism (c288C>T) of the NPHS2 gene identified in a Taiwan Chinese family. Hum Mutat. 2001;17:81–82. doi: 10.1002/1098-1004(2001)17:1<81::AID-HUMU31>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Fuchshuber A, et al. Clinical and genetic evaluation of familial steroid-responsive nephrotic syndrome in childhood. J Am Soc Nephrol. 2001;12:374–378. doi: 10.1681/ASN.V122374. [DOI] [PubMed] [Google Scholar]
- 14.Caridi G, et al. Prevalence, genetics, and clinical features of patients carrying podocin mutations in steroid-resistant nonfamilial focal segmental glomerulosclerosis. J Am Soc Nephrol. 2001;12:2742–2746. doi: 10.1681/ASN.V12122742. [DOI] [PubMed] [Google Scholar]
- 15.Frishberg Y, et al. Mutations in NPHS2 encoding podocin are a prevalent cause of steroid-resistant nephrotic syndrome among Israeli-Arab children. J Am Soc Nephrol. 2002;13:400–405. doi: 10.1681/ASN.V132400. [DOI] [PubMed] [Google Scholar]
- 16.Koziell A, et al. Genotype/phenotype correlations of NPHS1 and NPHS2 mutations in nephrotic syndrome advocate a functional inter-relationship in glomerular filtration. Hum Mol Genet. 2002;11:379–388. doi: 10.1093/hmg/11.4.379. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz K, et al. Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest. 2001;108:1621–1629. doi: 10.1172/JCI12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber TB, Kottgen M, Schilling B, Walz G, Benzing T. Interaction with podocin facilitates nephrin signaling. J Biol Chem. 2001;276:41543–41546. doi: 10.1074/jbc.C100452200. [DOI] [PubMed] [Google Scholar]
- 19.Tsukaguchi H, et al. A locus for adolescent and adult onset familial focal segmental glomerulosclerosis on chromosome 1q25-31. J Am Soc Nephrol. 2000;11:1674–1680. doi: 10.1681/ASN.V1191674. [DOI] [PubMed] [Google Scholar]
- 20.Hamano Y, et al. Determinants of vascular permeability in the kidney glomerulus. J Biol Chem. 2002;277:31154–31162. doi: 10.1074/jbc.M204806200. [DOI] [PubMed] [Google Scholar]
- 21.Maeshima Y, et al. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science. 2002;295:140–143. doi: 10.1126/science.1065298. [DOI] [PubMed] [Google Scholar]
- 22.Karle SM, et al. Novel mutations in NPHS2 detected in both familial and sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2002;13:388–393. doi: 10.1681/ASN.V132388. [DOI] [PubMed] [Google Scholar]
- 23.Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 24.Chillon M, et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N Engl J Med. 1995;332:1475–1480. doi: 10.1056/NEJM199506013322204. [DOI] [PubMed] [Google Scholar]
- 25.Chu CS, Trapnell BC, Curristin S, Cutting GR, Crystal RG. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 26.Morell RJ, et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339:1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- 27.Maugeri A, et al. The 2588G—>C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet. 1999;64:1024–1035. doi: 10.1086/302323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- 29.Rajaram S, Sedensky MM, Morgan PG. Unc-1: a stomatin homologue controls sensitivity to volatile anesthetics in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:8761–8766. doi: 10.1073/pnas.95.15.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart GW. The membrane defect in hereditary stomatocytosis. Baillieres Clin Haematol. 1993;6:371–399. doi: 10.1016/s0950-3536(05)80151-9. [DOI] [PubMed] [Google Scholar]
- 31.Snyers L, Thines-Sempoux D, Prohaska R. Colocalization of stomatin (band 7.2b) and actin microfilaments in UAC epithelial cells. Eur J Cell Biol. 1997;73:281–285. [PubMed] [Google Scholar]