Abstract
Familial hypercholesterolemia is an autosomal dominant disorder with a gene-dosage effect that is usually caused by mutations in the LDL receptor gene that disrupt normal clearance of LDL. In the homozygous form, it results in a distinctive clinical phenotype, characterized by inherited hypercholesterolemia, cholesterol deposition in tendons, and severe premature coronary disease. We described previously two families with autosomal recessive hypercholesterolemia that is not due to mutations in the LDL receptor gene but is characterized by defective LDL receptor–dependent internalization and degradation of LDL by transformed lymphocytes from the patients. We mapped the defective gene to chromosome 1p36 and now show that the disorder in these and a third English family is due to novel mutations in ARH1, a newly identified gene encoding an adaptor-like protein. Cultured skin fibroblasts from affected individuals exhibit normal LDL receptor activity, but their monocyte-derived macrophages are similar to transformed lymphocytes, being unable to internalize and degrade LDL. Retroviral expression of normal human ARH1 restores LDL receptor internalization in transformed lymphocytes from an affected individual, as demonstrated by uptake and degradation of 125I-labeled LDL and confocal microscopy of cells labeled with anti–LDL-receptor Ab.
Introduction
Familial hypercholesterolemia (FH) is characterized by increased levels of plasma LDL cholesterol that leads to the formation of tendon xanthomas, accelerated atherosclerosis, and premature coronary heart disease. In most cases, FH is an autosomal dominant disorder caused by mutations in the LDL receptor gene that lead to defective clearance of plasma LDL. There is a strong gene-dosage effect, and homozygous FH patients exhibit a severe and highly characteristic clinical phenotype (1).
We described previously two kindreds with a clinical diagnosis of homozygous FH whose Epstein-Barr virus–transformed lymphocytes (EBV-lymphocytes) in culture showed defective LDL receptor–dependent internalization of LDL, despite normal expression of LDL receptor mRNA and protein (2). The disorder was clearly inherited, but as an autosomal recessive, rather than dominant, trait, suggesting that the probands were homozygous for a defective gene whose product is involved in internalization or trafficking of the LDL. We mapped the defect in the two families to chromosome 1p36 (3), a region that has since been shown to harbor a novel gene (ARH1), mutations in which cosegregate with autosomal recessive hypercholesterolemia (ARH) in several families of different ethnic origin (4). The gene encodes a putative adaptor protein that was reported to bind to the cytoplasmic tail of the LDL receptor (4). However, although earlier studies in vivo showed that some of the patients with mutations in this gene exhibit defective clearance of LDL from the circulation, no defect in LDL receptor function has been observed in their cultured skin fibroblasts (5).
We now show that our original patients, as well as patients in a third kindred of English origin, are homozygous for mutations in ARH1 that are all predicted to result in synthesis of truncated forms of the protein. We have confirmed, by repairing the cellular defect by retroviral expression of normal ARH1 cDNA, that defective LDL receptor–dependent internalization and degradation of LDL by EBV-lymphocytes from the patients are caused by defects in ARH1.
Methods
Subjects.
The patients in families 1 and 2 have been described in detail previously (2, 3). Two further patients, brothers of English origin, first presented at Great Ormond Street Hospital for Sick Children at the ages of 9 and 7 years with a clinical diagnosis of “pseudo” homozygous FH. DNA samples and cultured skin fibroblasts were sent to us by J.V. Leonard (Great Ormond Street Hospital for Sick Children, London, United Kingdom) for investigation of possible mutations in the LDL receptor gene. All individuals gave informed consent.
Nucleotide sequencing of ARH1.
Total RNA was isolated from cultured EBV-lymphocytes (6). Overlapping fragments of ARH1 cDNA were amplified by RT-PCR, and their sequences (ABI3700) were compared with ARH1 cDNA (GenBank accession no. AL117654). Genomic fragments comprising exons or pairs of exons were amplified from genomic DNA isolated from whole blood or cells (7), with primers located in the introns at least 50 bp from the intron/exon junction. Primers for exons 5–9 were designed from the sequence of ARH1 in BAC clone AL606491; others were as published (8, 9) (for all primer sequences and PCR conditions see supplementary data at http://www.jci.org/cgi/content/full/110/11/1695/DC1).
Genotyping.
Polymorphic markers flanking ARH1 were selected from the Ensembl database and genotyped as described previously (3).
Fluorescent in situ hybridization.
Metaphase spreads (10) from EBV-lymphocytes of affected individual FH3.1 were hybridized with DIG-labeled probes to ARH (DIG-Nick Translation Mix; Roche Diagnostics Ltd., Lewes, United Kingdom) and a biotin-labeled probe to chromosome 1 α-satellite (Qbiogene-Alexis Ltd. Nottingham, United Kingdom) (10). ARH probe 1 was an 11-kb Eag1-Pac1 fragment of BAC clone AL031280 (Sanger Centre, Hinxton, Cambridge, United Kingdom), and ARH probe 2 comprised three PCR products amplified from BAC clone 121-03 (ResGen Invitrogen Corp., Paisley, United Kingdom), resulting in a 9.5-kb probe encompassing exons 2–7 of ARH1 (see supplementary data). ARH probes were detected with FITC anti-DIG and α-satellite with Cy3 anti-biotin Ab (Sigma-Aldrich, Poole, Dorset, United Kingdom) and viewed under an Olympus BX40 microscope with the CytoVision system (Applied Imaging International Ltd., Newcastle Upon Tyne, United Kingdom).
Cell culture.
Skin fibroblasts and EBV-lymphocytes were maintained as described (6, 11). Mononuclear cells were isolated from 20–30 ml of blood, seeded at 2.5 × 106 cells per 4.5-cm-diameter well in 12-place multiwell dishes (Linbro; ICN Pharmaceuticals Ltd., Basingstoke, United Kingdom), incubated for 1.5 hours, and washed to remove nonadherent lymphocytes (11). Adherent monocytes were incubated for 7 days in RPMI-1640 medium (GIBCO BRL; Life Technologies, Paisley, United Kingdom) containing autologous serum (20% vol/vol) or in serum-free medium (Macrophage-SFM; GIBCO BRL; Life Technologies) containing human recombinant GM-CSF (0.1 μg/ml; Sigma-Aldrich). PA317 amphotropic retroviral packaging cells (ECACC/89032007) were grown in DMEM supplemented with GlutaMAX (GIBCO BRL; Life Technologies), 4.5 g/l D-glucose, and 10% FCS.
For measurement of uptake or degradation of labeled LDL, cells were preincubated for 16 hours in medium containing 10% (vol/vol) lipoprotein-deficient serum (LPDS). Degradation of 125I-labeled LDL was determined as described (6, 11). Western blotting of cell extracts to detect c-myc-ARH1 was as described previously for the LDL receptor, with the exception that the primary Ab was mouse monoclonal anti–c-myc (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) diluted 1:3,000 (2).
Measurement of ARH1 mRNA by real-time PCR.
Cells were preincubated in medium containing LPDS (10% vol/vol) and compactin (0.1 μg/ml; Sigma-Aldrich) for 16 hours before isolation of total RNA with RNA-Bee (Biogenesis Ltd., Poole, Dorset, United Kingdom). ARH1 mRNA was assayed by real-time PCR using an ABI PRISM Sequence Detection System (Applied Biosystems, Warrington, United Kingdom) (for probes and primers see supplementary data). Primers and probes for GAPDH mRNA were included in each assay as an internal standard. All assays were carried out in triplicate, and all values were related to a standard curve generated from control mRNA, combined from two normal cell lines.
Retroviral expression of c-myc-ARH in EBV-transformed B cells.
ARH1 cDNA was amplified from plasmid DKFZp586D0624 (Deutsches Ressourcenzentrum für Genomforschung GmbH, Berlin, Germany) with primers that introduced a c-myc tag at the amino-terminus, and cloned into the HindIII/ClaI sites of the LNCX retroviral vector (12). PA317 cells (5 × 104 cells/ml) were transfected with LNCX/c-myc-ARH1 using LipofectAMINE PLUS reagent (GIBCO BRL; Life Technologies). Virus-containing medium was harvested 48 hours later, filtered (0.45 μm), and incubated with 5 μg/ml DEAE dextran (Sigma-Aldrich) for 20 minutes and then with an equal volume of EBV-lymphocytes (1 × 106 cells/ml) for 48 hours before selection for neomycin resistance (0.6 mg/ml G418; GIBCO BRL; Life Technologies). The cells were maintained in 40% conditioned medium containing 15% FBS and 0.6 mg/ml G418. After 1 month, G418-resistant cells started to divide, while uninfected cells failed to survive.
Microscopy.
EBV-lymphocytes were preincubated for 16 hours in medium containing 10% (vol/vol) LPDS and compactin (0.1 μg/ml), and allowed to adhere to poly-L-lysine–coated (Sigma-Aldrich) coverslips (Scientific Laboratory Supplies Ltd., Nottingham, United Kingdom). Cells were blocked with 1% (wt/vol) BSA in PBS (wash buffer) and incubated at 4°C for 1 hour with rabbit anti–LDL receptor Ab (diluted 1:200 in PBS; Progen Biotechnik GmbH, Heidelberg, Germany). To examine internalization of the LDL receptor, the washed cells were incubated at 37°C for 10 minutes. Cells were fixed in 4% (wt/vol) paraformaldehyde, permeabilized in PBS containing 0.1% Triton X-100 and 10 mM glycine, incubated sequentially for 1 hour at ambient temperature with mouse monoclonal anti–α-adaptin Ab (Santa Cruz Biotechnology Inc.; diluted 1/100), Alexa 568–conjugated goat anti-rabbit IgG (highly cross-absorbed; Molecular Probes Europe BV, Leiden, The Netherlands; diluted 1/100), and Alexa 488–conjugated goat anti-mouse IgG (highly cross-absorbed; Molecular Probes, diluted 1/100), and then mounted on slides with VECTASHIELD plus 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) (Vector Laboratories, Peterborough, United Kingdom). Cells were viewed on a Leica confocal microscope using the ×100 oil objective, and the images were analyzed using Leica confocal software (Leica Microsystems Ltd., Milton Keynes, United Kingdom).
Preparation of fluorescently-labeled LDL.
Human LDL was labeled with the fluorescent probe 1,1-dioctylacyl 3,3,3′,3′-tetramethylindocyanine perchlorate (DiI) as described (13). Monocyte-derived macrophages on coverslips were preincubated for 16 hours with medium containing 10% LPDS and then for 3 hours at 37°C with DiI-LDL (7.5 μg/ml) in the presence or absence of an excess of unlabeled LDL (1.0 mg/ml). Cells were fixed and mounted as described above.
Results and Discussion
Clinical characteristics of the patients.
Two of the families, one of Turkish and the other of Asian Indian origin, have been described in detail previously (2, 3); their pedigrees are shown in Figure 1. Two further patients, brothers of English origin, presented at the ages of 9 and 7 years with a clinical phenotype of “pseudo” homozygous FH. There was no evidence of consanguinity (family 3, Figure 2). The elder brother (3.1) presented with xanthomata on his elbows and wrists. These were confirmed by biopsy to contain cholesterol, and at that time his total serum cholesterol was 14.2 mmol/l. The younger brother (3.2) was found to have total cholesterol of 13.4 mmol/l but did not have any xanthomata or other cutaneous lesions. The past medical history of patient 3.1 is unremarkable. In 1999 he had an exercise electrocardiogram, which was normal. The echocardiography performed at that time showed that, apart from having a tricuspid aortic valve and a mild central jet of aortic regurgitation, he had no evidence of aortic stenosis. Both brothers are completely asymptomatic from the cardiovascular point of view. They respond well to treatment with statins. We had failed previously to detect any mutations in the LDL receptor or apoB genes of these two brothers (unpublished data), but this family was not included in our earlier gene-mapping study (3), because it was unlikely that they were homozygous for any mutant allele.
Figure 1.
Pedigrees of family 1 (a) and family 2 (b). The plasma cholesterol concentration (mmol/l) is shown below each symbol. Filled symbols, homozygous for the Q136X (a, family 1) or the delGG86,87 (b, family 2) mutations in ARH1; half-filled symbols, heterozygous carriers of the mutation, confirmed by sequencing amplified fragments of genomic DNA. Individuals whose cells were analyzed for LDL receptor activity or ARH1 mRNA levels are identified by a number in italics.
Figure 2.
Pedigree and haplotype analysis of family 3. The plasma cholesterol (chol.) concentration (mmol/l) is shown below each symbol. Filled symbols, homozygous for the insC mutation in ARH1; half-filled symbols, heterozygous carriers of the mutation, confirmed by sequencing amplified fragments of genomic DNA. Haplotypes for the ARH1 locus are shown for polymorphic markers flanking the gene, known polymorphisms (8), and the two novel base substitutions within the coding region of ARH1; the position of each marker on Ensembl is shown (Mb). A recombination in the paternal uncle of the proband is indicated (X). The maternal allele carrying the insC620 mutation is boxed; the paternal allele carrying the putative deletion is boxed and shaded. The haplotype in the father, shown in italics, was deduced. Individuals whose cells were analyzed for LDL receptor activity or ARH1 mRNA levels are identified by a number in italics.
Characterization of point mutations in ARH1.
The sequence of ARH1 cDNA, amplified from EBV-lymphocyte mRNA, was determined. The proband of Turkish origin (individual 1.1, Figure 1a), her affected sibling (1.2), and her affected cousin (1.3) were found to be homozygous for a single base substitution of C427 to T (nucleotide numbering based on GenBank accession no. AL117645). This mutation has previously been described in a Lebanese proband and is predicted to introduce a premature termination codon (Q136X) in exon 4 (4). Further investigation revealed that antecedents in the Turkish family were Lebanese, and thus it is likely that this allele was inherited from a common ancestor. Sequencing of genomic DNA confirmed that the three affected individuals were homozygous for the mutation and revealed that both parents and several other apparently unaffected siblings were heterozygous carriers (Figure 1a), as expected from their previously determined haplotypes (3).
The two affected siblings of Asian Indian origin (2.1 and 2.2, Figure 1b) were found to be homozygous for a 2-bp deletion of GG in a run of seven consecutive G nucleotides (bp 86–92) in exon 1. The resultant frameshift is predicted to introduce a premature termination codon eight residues after Gly23. This mutation has not been described previously, but two others have been identified within this short section (4). Both parents were heterozygous for the mutation, as were two apparently unaffected siblings in the family (Figure 1b), in agreement with previous genotyping (3).
The two affected English siblings (3.1 and 3.2, Figure 2) were both found to be homozygous in ARH1 mRNA for two single base substitutions. The first was an insertion of C in a run of eight consecutive C residues (bp 620–627) in exon 6 that results in a frameshift and is predicted to give rise to a premature termination codon 17 residues after Pro202. The second was substitution of C733 with T in exon 7 and is predicted to change residue Arg238 to Trp. When genomic DNA was sequenced, both affected siblings were homozygous for both variants. This was somewhat surprising, because there was no suggestion of consanguinity in the family. Their unaffected sister carried neither mutation and their mother was heterozygous for both, confirming that both mutations lay on the same allele. The father of the affected boys was deceased, but neither of the paternal grandparents carried the mutant allele (Figure 2) and thus the origin of the second mutant allele in the probands was not apparent.
One possible explanation was that the second allele carried a deletion of the ARH1 gene encompassing at least exons 6 and 7. However, Southern blotting of genomic DNA digested with several enzymes and hybridized with cDNA probes representing the entire transcript did not reveal any additional bands in the probands (data not shown). This suggested that any deletion must encompass the majority of the ARH1 gene, a view supported by haplotype analysis (Figure 2). The pattern of inheritance of polymorphic markers flanking and within ARH1 implied that the probands inherited an allele with a deletion at this locus between D1S2674 and D1S2639 from their paternal grandmother (Figure 2), and this was demonstrated by fluorescent in situ hybridization (FISH). When metaphase chromosomes from affected individual 3.1 were hybridized with a probe extending from exon 5 to 7 kb beyond the 3′ end of ARH1 (Figure 3a, probe 1), both copies of chromosome 1, as visualized with a chromosome 1 α-satellite probe, were labeled (Figure 3b, probe 1). However, when hybridized with a probe comprising exons 2–7 of ARH1 (Figure 3a, probe 2), only one copy of chromosome 1 was labeled (Figure 3b, probe 2). Thus, as implied by the sequence data (Figure 2), the deletion includes at least exons 2–7 but does not extend far downstream beyond exon 7.
Figure 3.
Identification of a partial deletion of ARH1 in family 3. (a) Diagram showing the approximate size of the ARH1 DNA probes used for fluorescent in situ hybridization (FISH). The vertical black bars represent the nine exons of ARH1. (b) Metaphase chromosomes from individual FH3.1 were hybridized with either DIG-labeled ARH1 DNA probe 1 or probe 2 as indicated (detected with FITC-labeled anti-DIG Ab; small filled arrows) and biotin-labeled chromosome 1–specific α-satellite probe DNA (detected with Cy3-labeled anti-biotin Ab; large open arrows).
Expression of ARH1.
The amount of ARH1 mRNA in EBV-lymphocytes was measured by real-time RT-PCR (Table 1). In EBV-lymphocytes from individual 2.1 (ARH1 delGG86,87), ARH1 mRNA was not significantly reduced compared with normal cells, but in cells from individual 1.1 (ARH1 Q136X), there was an approximately 50% reduction. In cells from individuals 3.1 and 3.2 (ARH1 insC620), there was a 90% reduction in ARH1 mRNA. Even when the absence of mRNA due to the deletion of one allele is taken into account, the insC620 mutation has the most marked effect on cellular ARH1 mRNA, reducing it to one-fifth of the level expected from that allele. The different effects of the three mutations in ARH1 on mRNA levels is a little surprising; the mutations might be expected to induce nonsense-mediated decay to a similar extent, since all three introduce a premature termination codon in the mRNA 5′ to the 50 nucleotides that precede the final exon/intron boundary (14). Despite near-normal levels of mRNA in cells from the probands in family 2, there is no doubt that ARH1 protein is nonfunctional in all three families, because the mutations are predicted to result in the production of a severely truncated protein. ARH1 mRNA levels, expressed relative to GAPDH mRNA, were identical in EBV-lymphocytes and skin fibroblasts from subject 3.1 (data not shown).
Table 1.
Determination of ARH1 mRNA levels by quantitative PCR
LDL receptor function in ARH1-negative cells.
We have previously shown that EBV-lymphocytes from the probands in families 1 and 2 are unable to take up and degrade LDL (2). We have recently obtained an explant from the proband in family 1 (1.1) and now show that cultured skin fibroblasts from this subject exhibit the same saturable, high-affinity uptake and degradation of LDL as do fibroblasts from a normolipemic individual (Figure 4a). Others have also reported the absence of any defect in LDL receptor function in skin fibroblasts from patients homozygous for mutations in ARH1 (4, 8). EBV-lymphocytes from a heterozygous sibling of the proband (individual 1.4 in Figure 1) and from another sibling who does not carry the mutant allele (1.5) degrade LDL normally (Figure 4b). This finding is supported by the observation that the heterozygous carriers of a mutant allele of ARH1 in the three families have no obvious clinical phenotype. The same pattern was observed with cells from individual 3.1 in family 3, in that cultured skin fibroblasts were able to degrade LDL normally (Figure 5b), while no degradation occurred in EBV-lymphocytes from this proband or his affected brother (Figure 5a).
Figure 4.
Degradation of 125I-labeled LDL by cultured skin fibroblasts and EBV- lymphocytes from individuals in family 1. Cells were preincubated for 16 hours in medium containing LPDS and then for 4 hours with 125I-labeled LDL. Saturable degradation of LDL was determined as the difference in the amount of trichloroacetic acid–soluble (TCA-soluble), non-iodide radioactivity in the medium of cells incubated in the presence and absence of an excess of unlabeled LDL (1 mg/ml); values are the mean of duplicate dishes. Nonsaturable degradation of LDL by normal cells was always less than 5% of the total. Data shown are representative of at least two separate experiments. (a) Cultured skin fibroblasts from the proband in family 1 (ARH–/–), and from a normolipemic control (ARH+/+). (b) EBV-lymphocytes from the proband in family 1 (ARH–/–), her heterozygous sibling (ARH+/–), and a sibling who does not carry the mutant allele (ARH+/+).
Figure 5.
Degradation of 125I-labeled LDL by skin fibroblasts, EBV-lymphocytes, and monocyte-derived macrophages from individuals in family 3. Cells were preincubated for 16 hours in medium containing LPDS and then for 4 hours with 125I-labeled LDL. Saturable degradation of LDL was determined as the difference in the amount of TCA-soluble, non-iodide radioactivity in the medium of cells incubated in the presence and absence of an excess of unlabeled LDL (1 mg/ml); values are the mean of duplicate dishes. Nonsaturable degradation of LDL by normal cells was always less than 5% of the total. Data shown are representative of at least two separate experiments. (a) EBV-lymphocytes, (b) skin fibroblasts, and (c) monocyte-derived macrophages, from probands 3.1 (filled triangles) and 3.2 (open triangles) in family 3 and from unrelated normolipemic controls (filled circles).
To determine whether the cellular phenotype in EBV-lymphocytes was a consequence of their transformation with EBV, we also determined the ability of monocyte-derived macrophages from ARH1-defective individuals to degrade LDL. Blood monocytes were isolated and maintained in culture for 7 days, during which time they developed into macrophages (15). Even after upregulation of LDL receptor expression, monocyte-derived macrophages from the proband in family 3 (3.1) were totally unable to degrade 125I-labeled LDL (Figure 5c), confirming that internalization of the LDL receptor was defective in these cells. Somewhat surprisingly, monocyte-derived macrophages from his affected brother, 3.2, who was homozygous for the same mutation(s) in ARH1, showed saturable, high-affinity degradation of LDL that was approximately 20% of that exhibited by normal cells. Identical results were obtained with two different preparations of cells from both individuals.
In preliminary investigations, microscopy of monocyte-macrophages incubated with fluorescently-labeled LDL suggested that the low level of degradation of 125I-labeled LDL observed in individual 3.2 was due to normal uptake by a few of the cells in the culture, and not to a low level of uptake by all cells (data not shown). There were no obvious morphological differences between cells that were able to take up LDL and those that were not, but further studies to investigate different cell types in the culture were hampered by strictly limited availability of the cells. We conclude that a proportion of the cells in the monocyte-macrophage cultures from individual 3.2 shared a phenotype with fibroblasts, in that LDL receptor function was independent of ARH1 function. The underlying reason for this is currently unknown.
Expression of exogenous ARH1 in mutant EBV-lymphocytes.
A retroviral vector containing a cDNA for human ARH1 tagged with c-myc at its amino-terminus was transfected into PA317 cells. The transfected viral-packaging cells produced a myc-tagged protein of approximately 37 kDa (Figure 6, lane 2), the expected size for c-myc-ARH1 (lane 1), that was absent in nontransfected cells (lane 3). EBV-lymphocytes from proband 1.1 (ARH1 hmz Q136X) were infected with medium from the PA317 producer cells, and cells expressing viral genes were selected for neomycin resistance. These cells also expressed a myc-tagged protein of the expected size (lane 4) that was absent from nontransfected cells (lane 5), although the level of expression declined with prolonged time in culture (lane 7). Maximum expression was restored by incubation of the infected cells for 16 hours with 0.3 μM trichostatin A, a histone deacetylase inhibitor (16) (lane 8), showing that the viral construct had not been lost, but unfortunately this treatment reduced LDL receptor protein levels (lanes 9 and 10) and was not useful for future experiments.
Figure 6.
Effect of expression of c-myc-ARH1 on LDL receptor activity in mutant EBV-lymphocytes. (a and b) Cells were preincubated for 16 hours with LPDS before preparation of cell extracts. Proteins were fractionated on nonreduced SDS-polyacrylamide gels (13%), transferred to nylon membranes, and immunoblotted with anti–c-myc (lanes 1–8) or anti–LDL receptor Ab (lanes 9 and 10). Bound Ab was detected with peroxidase-conjugated anti-mouse IgG and chemiluminescence. (a) Whole-cell extracts (approximately 50 μg of protein per lane) of Chinese hamster ovary (CHO) cells transiently transfected with pcDNA3-c-myc-ARH1PA317 (lane 1), PA317 cells transfected with ARH1 retroviral construct (lane 2), PA317 cells (lane 3), EBV-lymphocytes from affected individual 1.1 (ARH1.1 cells) 1 month after infection with c-myc-ARH1 retrovirus (lane 4), and uninfected EBV-lymphocytes from affected individual 1.1 (lane 5). (b) Whole-cell extracts (approximately 50 μg of protein per lane) of EBV-lymphocytes from affected individual 1.1 (lane 6), the same cells 3 months after stable infection with c-myc-ARH1 retrovirus (lanes 7 and 9), and the same infected cells after preincubation for 16 hours with 0.3 μM trichostatin A (lane 8 and 10). (c) Virus-infected (ARH–/c-myc-ARH) and uninfected (ARH–) EBV-lymphocytes from individual 1.1 were preincubated for 16 hours in medium containing LPDS and then for 4 hours with 125I-labeled LDL. Saturable degradation of LDL was determined as the difference in the amount of TCA-soluble, non-iodide radioactivity in the medium of cells incubated in the presence and absence of an excess of unlabeled LDL (1 mg/ml); values are the mean of duplicate dishes. Data shown are representative of two separate experiments.
Unlike the mutant cells, infected cells were able to take up and degrade 125I-labeled LDL (Figure 6c). This showed that LDL receptor activity in the internalization-defective cells could be restored in the mutant cells by expression of c-myc-ARH1 and demonstrated that a defect in this gene is responsible for the phenotype in the patients. The apparent affinity of the LDL receptor for labeled LDL was lower in the mutant cells expressing c-myc-ARH1 than that in control cells, which may be due to a rate-limiting level of expression of exogenous ARH1 or to the c-myc tag interfering with normal function to some extent. We are unable to distinguish between these possibilities at present.
The subcellular localization of the LDL receptor in EBV-lymphocytes in which LDL receptor expression was upregulated was examined by confocal microscopy. No LDL receptor staining was observed with cells that had not been preincubated with LPDS or with cells incubated with second Ab alone (data not shown). In normal and mutant EBV-lymphocytes incubated with anti–LDL receptor Ab at 4°C, LDL receptor protein (red) was only visible on the cell surface (Figure 7, a and b). In LDL receptor Ab–labeled cells incubated at 37°C, LDL receptor protein was visible as a punctate pattern inside control cells (Figure 7d) but remained on the surface of ARH1-negative cells, presumably because LDL receptor internalization is defective (Figure 7e). In ARH1-negative cells expressing c-myc-ARH1, LDL receptor protein was visible on the surface of cells incubated at 4°C (Figure 7c), and as a punctate pattern similar to that in normal cells after incubation at 37°C (Figure 7f), showing that the LDL receptor had been internalized.
Figure 7.
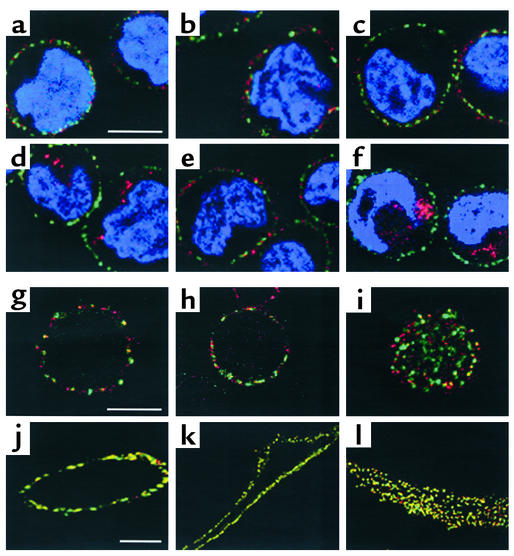
Confocal microscopy of cells labeled with anti–LDL receptor Ab. (a–f) EBV-lymphocytes were incubated with rabbit anti–LDL receptor (red) at 4°C, washed at 4°C, and either directly permeabilized (a–c) or incubated for 10 minutes at 37°C before permeabilization to allow internalization of LDL receptor/Ab complexes (d–f). Permeabilized cells were then incubated with mouse anti–α-adaptin (AP2), washed, and incubated with Alexa 568–conjugated goat anti-rabbit IgG (LDL receptors, red) and Alexa 488–conjugated goat anti-mouse IgG (AP2, green). Nuclei were stained with DAPI. The plates shown are an overlay of red and green images. The bar represents 5.0 μm. (a and d) Control cells; (b and e) cells from proband 1.1; (c and f) cells from proband 1.1 expressing viral c-myc-ARH. (g–l) EBV-lymphocytes (g–i) or cultured skin fibroblasts (j–l) from three different control subjects were incubated with anti–LDL receptor Ab at 4°C, permeabilized, and then incubated with anti–α-adaptin Ab (AP2) as described for a–f above. The bars represent 5.0 μm (in g for g–i, and in j for j–l).
LDL receptor–mediated endocytosis is thought to occur via clathrin-coated pits, of which the adaptor complex AP2 is a component (17). However, in normal or ARH1-negative EBV-lymphocytes, α-adaptin–stained AP2 was visible as punctate staining on the inner cell membrane (green, Figure 7, a–f) but did not colocalize with the LDL receptor (no yellow visible). Similar results were obtained with cells from three different individuals (Figure 7, g–i); very little of the LDL receptor colocalized with AP2 at the cell membrane when viewed either in cross section (Figure 7, g and h) or at the surface (Figure 7i). In marked contrast, in three normal skin fibroblast cell lines, the majority of the LDL receptor colocalized with AP2 (Figure 7, j–l). The surprisingly small degree of colocalization between the LDL receptor and AP2 observed in lymphocytes is consistent with previous observations in hepatocytes from transgenic mice expressing the human LDL receptor (18), suggesting that EBV-lymphocytes might present a better model for hepatic LDL receptor–mediated endocytosis than do skin fibroblasts.
ARH resembles a putative adaptor protein, since it contains a phosphotyrosine-binding (PTB) domain similar to that found, for example, in the Drosophila numb protein (4). In many adaptor proteins, the PTB domain binds to a phosphorylated NPXpY motif (where X is any amino acid and pY is phosphotyrosine), but in some, such as Disabled-1 and Disabled-2, it binds with high affinity to a nonphosphorylated NPXY sequence (19). The PTB domain of ARH1 has recently been shown to interact in vitro with the NPVY internalization sequence in the cytoplasmic tail of the LDL receptor (20). It was not possible to examine interaction of ARH1 with the LDL receptor in EBV-lymphocytes expressing c-myc-ARH1, because retroviral expression of c-myc ARH1 declined to almost undetectable levels in the cells. Although preincubation of the cells with trichostatin A increased expression of c-myc-ARH1, this resulted in very low levels of LDL receptor protein. As a result, the two proteins could not both be detected in any one cell by confocal microscopy (data not shown).
In this study we have shown that the defect in LDL receptor internalization that we have observed previously in EBV-lymphocytes from patients with autosomal recessive hypercholesterolemia is due to defects in ARH1 and can be corrected in these cells by retroviral expression of normal ARH1. We have further shown that ARH1 is required for LDL receptor function in normal macrophages, but not in skin fibroblasts from the same individuals. Our observation that the LDL receptor colocalizes with AP2 at 4°C in fibroblasts, but not in EBV-lymphocytes, suggests a possible mechanism for the difference in LDL receptor phenotype between EBV-lymphocytes and fibroblasts lacking ARH1 and clearly warrants further investigation.
Supplementary Material
Acknowledgments
We are very grateful to the members of the families for their willing cooperation in these studies and to J.V. Leonard for providing samples from family 3. We are indebted to Nina Krausewicz (Medical Research Council, Clinical Sciences Centre) for suggesting the use of trichostatin A. Bruce Pottinger provided excellent help with tissue culture. Simon Gregory (Sanger Centre, Hinxton, Cambridge) kindly provided BAC clone AL031280. This work was supported in part by a project grant from the British Heart Foundation (PG 98062).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: familial hypercholesterolemia (FH); Epstein-Barr virus (EBV); autosomal recessive hypercholesterolemia (ARH); lipoprotein-deficient serum (LPDS); 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI); 1,1-dioctylacyl 3,3,3′,3′-tetramethylindocyanine perchlorate (DiI).
References
- 1.Goldstein, J.L., Hobbs, H., and Brown, M.S. 1995. Familial hypercholesterolemia. In The metabolic and molecular bases of inherited disease. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill. New York, New York, USA. 1981–2030.
- 2.Norman D, et al. Characterization of a novel cellular defect in patients with phenotypic homozygous familial hypercholesterolemia. J Clin Invest. 1999;104:619–628. doi: 10.1172/JCI6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eden ER, Naoumova RP, Burden JJ, McCarthy MI, Soutar AK. Use of homozygosity mapping to identify a region on chromosome 1 bearing a defective gene that causes autosomal recessive homozygous hypercholesterolemia in two unrelated families. Am J Hum Genet. 2001;68:653–660. doi: 10.1086/318795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 5.Zuliani G, et al. Characterization of a new form of inherited hypercholesterolemia: familial recessive hypercholesterolemia. Arterioscler Thromb Vasc Biol. 1999;19:802–809. doi: 10.1161/01.atv.19.3.802. [DOI] [PubMed] [Google Scholar]
- 6.Sun XM, Patel DD, Knight BL, Soutar AK. Comparison of the genetic defect with LDL-receptor activity in cultured cells from patients with a clinical diagnosis of heterozygous familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 1997;17:3092–3101. doi: 10.1161/01.atv.17.11.3092. [DOI] [PubMed] [Google Scholar]
- 7.Maniatis, T. 1989. Analysis of genomic DNA by Southern hybridization. In Molecular cloning: a laboratory manual. 2nd edition. J. Sambrook, E.F. Fritsch, and T. Maniatis, editors. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 9.31–9.57.
- 8.Al-Kateb H, et al. Mutation in the ARH gene and a chromosome 13q locus influence cholesterol levels in a new form of digenic-recessive familial hypercholesterolemia. Circ Res. 2002;90:951–958. doi: 10.1161/01.res.0000018002.43041.08. [DOI] [PubMed] [Google Scholar]
- 9.Arca M, et al. Autosomal recessive hypercholesterolaemia in Sardinia, Italy, and mutations in ARH: a clinical and molecular genetic analysis. Lancet. 2002;359:841–847. doi: 10.1016/S0140-6736(02)07955-2. [DOI] [PubMed] [Google Scholar]
- 10.2001. Human cytogenetics. I. Constitutional analysis. D.E. Rooney, editor. Oxford University Press. Oxford, United Kingdom. 39–188.
- 11.Knight BL, Soutar AK. Changes in the metabolism of modified and unmodified low-density lipoproteins during the maturation of cultured blood monocyte-macrophages from normal and homozygous familial hypercholesterolaemic subjects. Eur J Biochem. 1982;125:407–413. doi: 10.1111/j.1432-1033.1982.tb06698.x. [DOI] [PubMed] [Google Scholar]
- 12.Miller A, Rosman G. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 13.Pitas RE, Innerarity TL, Mahley RW. Foam cells in explants of atherosclerotic rabbit aortas have receptors for beta-very low density lipoproteins and modified low density lipoproteins. Arteriosclerosis. 1983;3:2–12. doi: 10.1161/01.atv.3.1.2. [DOI] [PubMed] [Google Scholar]
- 14.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 15.Knight BL, Soutar AK. Degradation by cultured fibroblasts and macrophages of unmodified and 1,2-cyclohexanedione-modified low-density lipoprotein from normal and homozygous familial hypercholesterolaemic subjects. Biochem J. 1982;202:145–152. doi: 10.1042/bj2020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaetano C, et al. Transcriptionally active drugs improve adenovirus vector performance in vitro and in vivo. Gene Ther. 2000;19:1624–1630. doi: 10.1038/sj.gt.3301296. [DOI] [PubMed] [Google Scholar]
- 17.Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu Rev Cell Dev Biol. 1999;15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- 18.Pathak RK, et al. Tissue-specific sorting of the human LDL receptor in polarized epithelia of transgenic mice. J Cell Biol. 1990;111:347–359. doi: 10.1083/jcb.111.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra SK, et al. Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J. 2002;21:4915–4926. doi: 10.1093/emboj/cdf487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He G, et al. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin and AP-2. J Biol Chem. 2002;277:44044–44049. doi:10.1074 /jbc.M208539200. doi: 10.1074/jbc.M208539200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.