Abstract
Using microarray analysis, we have detected downregulation of several components of the cGMP signaling pathway during replicative senescence of primary human diploid fibroblasts (HDFs). Therefore, the effect of pharmacological inhibition of cGMP synthesis was analyzed in HDFs. Treatment with 6-anilino-5,8-quinolinequinone (LY83583, referred to as LY hereafter), a previously described inhibitor of guanylate cyclase, induced cellular senescence. Microarray analysis revealed that LY treatment induced the Cdk inhibitor p21WAF1/SDI/CIP1. In colorectal cancer cells, transcription of p21 was induced by LY in a p53-independent manner. Furthermore, p21, but not p53, was required for inhibition of proliferation by LY. The lack of p53 involvement suggests that LY does not induce DNA damage. Growth inhibition was also observed in malignant melanoma and breast cancer cell lines. Functional inactivation of the retinoblastoma tumor-suppressor protein, an effector of p21-mediated cell-cycle inhibition, converted LY-induced growth arrest to apoptosis. These results suggest that LY, or derivatives, may be useful therapeutic agents for the treatment of tumors.
Introduction
Several cellular mechanisms have evolved to prevent proliferation of cells that have acquired mutations with carcinogenic potential. One mechanism is represented by apoptosis: cells with deregulated expression of oncogenes are removed through this route (1). Another cellular response that has gained more attention recently is senescence, a state of terminal arrest in which cells are metabolically active for extended periods but are unable to re-enter the cell cycle after mitogenic stimulation (2, 3). Several tumor-suppressor genes (e.g., p53 and p16INK4A) are involved in the induction and maintenance of senescence, which strongly suggests that senescence is a tumor-suppressive mechanism (4, 5). Supporting this notion on the organismal level, mice that have been engineered to display elevated p53 activity show a decreased rate of tumor formation and premature aging (6).
Replicative senescence results from progressive shortening of chromosomes, which occurs during every cell division. After 50–70 cell divisions, the loss of telomeric DNA repeats results in unprotected chromosome ends, which generate a DNA-damage signal resulting in activation of the tumor-suppressor gene product p53. Therefore, several downstream targets of the p53 transcription factor are induced during replicative senescence — for example, p21WAF1/SDI/CIP1 and 14-3-3sigma (7, 8). Consistent with an essential function of these p53 target genes during senescence, deletion of p21 through homologous recombination is sufficient to bypass replicative senescence in human diploid fibroblasts (HDFs) (9). Similarly, inactivation of 14-3-3sigma leads to immortalization of primary human keratinocytes (8).
In contrast to telomere-dependent replicative senescence, cellular senescence — also referred to as premature senescence — is a response to diverse stimuli (e.g., oncogenic signaling, suboptimal tissue-culture conditions) and generally involves induction of the tumor suppressor gene p16INK4A. Cellular senescence accompanied by induction of p16 is also observed after serial cultivation of epithelial cells under standard-tissue culture conditions. Interestingly, the p16 gene is not induced in epithelial cells when these are cultivated on feeder layers: under these conditions, epithelial cells have a greatly elevated replicative potential and terminally arrest because of shortened telomeres —that is, they undergo replicative senescence (10). In contrast to epithelial cells, HDFs undergo replicative senescence under standard tissue-culture conditions (11). These examples point to the existence of cell-type–dependent signaling pathways that activate the senescence program.
DNA-damaging agents commonly used for cancer therapy induce a senescence-like state in normal and malignant cells (12, 13). However, since these substances also induce DNA damage in normal proliferating cells, numerous unwanted side effects are observed during chemotherapy (14). Furthermore, these agents generate mutations in precancerous cells, which may result in secondary cancer. In this study, we aimed to identify substances that activate cellular senescence without inducing DNA damage. We hypothesized that pharmacological inhibition of signaling pathways that are specifically downregulated during replicative senescence may result in the reactivation of the senescence program in tumor cells. The ideal drug target for this strategy would be an enzyme encoded by a gene that is repressed during senescence. Inhibition of such an enzyme by a small, membrane-permeable drug molecule in early-passage or tumor cells should theoretically be sufficient to induce cellular senescence. In order to detect genes and pathways repressed during replicative senescence, the gene-expression pattern of senescent HDFs was compared with the expression signature of confluent early-passage cells. We thereby identified a pharmacological substance that induces cellular senescence.
Methods
Cell culture and drug treatments.
Neonatal skin HDFs were obtained from Clonetics (San Diego, California, USA) and cultivated in DMEM (Invitrogen Corp., Carlsbad, California, USA) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, Missouri, USA). To obtain senescent HDFs, the cells were diluted every 3 days in a ratio of 1:10 (equal to 1 passage) until they ceased to proliferate.
HCT116 cells were cultured in McCoy’s medium (Invitrogen Corp.) supplemented with 10% FBS. A-375, HeLa, HEK293, p16–/– mouse embryo fibroblasts (MEFs) and NIH3T3-L1 derivatives were kept in DMEM containing 10% FBS. 6-Anilino-5,8-quinolinedione (LY83583, referred to as LY hereafter; Calbiochem, San Diego, California, USA) was dissolved in DMSO (Sigma-Aldrich) at a concentration of 300 μM (∼300× solution). As a control, cells were treated with equal volumes of DMSO (<1%). The LY concentration was restored at intervals of 24 hours by media exchange.
Microarray analysis of gene expression.
RNA was isolated using RNAgents reagents (Promega Corp., Madison, Wisconsin, USA). After mRNA isolation, integrity and enrichment was ensured using Northern blot analysis. Six hundred nanograms of poly-A mRNA was converted to cDNA with incorporation of Cy3- or Cy5-labeled deoxynucleotide-triphosphates (dNTPs). Hybridization to arrays coated on glass, quality control, and normalization were performed by IncyteGenomics (Palo Alto, California, USA). The “Human Unigene 1” array contained cDNA probes representing 8,392 annotated genes/expressed sequence tag (EST) clusters and 74 nonannotated genes/ESTs.
Northern blot analysis.
RNA was isolated using the RNAgents kit. A Northern probe directed against the 3′-untranslated region of elongation factor 1α (ELF1α) mRNA was generated by PCR, using an EST as the template, and subsequent gel purification. For p21, a probe corresponding to the open reading frame of p21 was used. A probe corresponding to the 5′ region of soluble guanylate cyclase β3 (sGC-β3) was amplified from HDF cDNA, confirmed by sequencing, and subcloned. Hybridizations were performed in QuickHyb according to the manufacturer’s instructions (Stratagene, La Jolla, California, USA).
Quantitative real-time PCR.
Quantitative real-time PCR (qPCR) was performed using the LightCycler and the FastStart DNA Master SYBR Green 1 kit (Roche Diagnostics GmbH, Mannheim, Germany) as previously described (15). For qPCR of cDNA, primer pairs were designed to generate intron-spanning products of 100–200 bp. Primer sequences were as follows: GUCY1A3, sense 5′-TTCAGAGGAGGCAGCAGG-3′ and antisense 5′-GCAACATTCAGCCGTTCAA-3′ (annealing temperature, 62°C); and GUCY1B3, sense 5′-AGGAATCACGCATCAGCC-3′ and antisense 5′-TATGAGGACGAACCAGCGA-3′ (annealing temperature, 62°C).
cDNA was generated using the RevertAid First Strand cDNA synthesis kit (MBI Fermentas, St. Leon-Rot, Germany). The generation of specific PCR products was confirmed by melting-curve analysis and gel electrophoresis. Each primer pair was tested with a logarithmic dilution of a cDNA mix to generate a linear standard curve (crossing point [CP] plotted vs. log of template concentration), which was used to calculate the primer-pair efficiency (E= 10(–1/slope)). ELF1α mRNA was used as an external standard, since its expression was not altered significantly in senescent versus early passage confluent HDF (data not shown). For data analysis, the second-derivative maximum method was applied, and induction of a cDNA species (geneX) was calculated according to Pfaffl (16) as follows:
 |
1 |
Measurement of DNA content and apoptosis by flow cytometry.
Cells were trypsinized. Both adherent and floating cells were washed once with PBS and fixed on ice in 70% ethanol for over 2 hours, washed once with PBS, and incubated for 30 minutes at room temperature in staining solution containing 50 μg/ml of propidium iodide (PI), 0.2 mg/ml of RNase A, and 0.1 % (v/v) Triton X-100 in PBS. Quantification of apoptotic cells was performed using the Annexin V-FITC apoptosis detection kit (BD Pharmingen, San Diego, California, USA). Samples were analyzed with a FACScan unit (Becton Dickinson, Mountain View, California, USA). 1 × 104 cells were analyzed for each assay.
Proliferation assay.
Cells were seeded in equal numbers in six-well plates 24 hours before the addition of LY. Cells were treated in triplicates (3 replicas of the same experiment) with daily exchange of medium containing drug or drug-free vehicle. For each time point, cells were trypsinized and cell proliferation was assessed using a Z1 Coulter Counter (Coulter Electronics, Beds, United Kingdom).
cGMP assay.
Cells of early and late passages were seeded in equal numbers in six-well plates. Twenty-four hours later, sodium nitroprusside (SNP, Sigma-Aldrich) dissolved in complete medium was added at a final concentration of 100 μM. Controls received the same volume of drug-free medium. After 2 hours of incubation, medium was removed and cells were lysed by addition of 400 μl of 0.1 M HCl per well for 10 minutes. cGMP concentration in lysates was determined using a competitive immunoassay according to the manufacturer’s instructions (Sigma-Aldrich). A plate reader, LAMBDA E (MWG-Biotech AG, Ebersberg, Germany), was used to detect the signal at 405 nm. Experiments were performed in triplicates, with all samples measured twice.
Senescence-associated β-galactosidase staining.
Cells were fixed by incubation in 0.5% glutaraldehyde in PBS for 5 minutes at room temperature and stained for senescence-associated β-galactosidase (SA-β-gal) at pH 6.0 as described (17).
Detection of DNA synthesis.
Cells were plated on CELLocate glass grids (Eppendorf) and labeled for 6 hours using the 5′-bromo-2′-deoxyuridine Labelling and Detection Kit I (Roche Diagnostics GmbH). After staining with a FITC-labeled anti- bromodeoxyuridine (anti-BrdU) antibody, positive cells were detected by fluorescence microscopy.
Western blot analysis.
Cells were lysed in 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 100 mM NaF, 10 mM Na4P2O7, 1 μM PMSF, and 1 μM Na3VO4 (all chemicals from Sigma-Aldrich). Protein concentration was determined using a Bradford assay (Bio-Rad Laboratories, Hercules, California, USA). Twenty micrograms of protein were diluted in Laemmli buffer, separated on SDS-polyacrylamide gels, and transferred to Immobilon-P membranes (Millipore, Bedford, Massachusetts, USA). Antibodies against p53 (Pab 1801), p21 (c-19), p16 (c-20), and α-tubulin (TU-02) were from Santa Cruz Biotechnology Inc. (Santa Cruz, California, USA). Anti-pRB antibodies (G3-245) were from BD Pharmingen. Enhanced chemiluminescence generated by horseradish peroxidase–coupled secondary antibodies was detected with a Image Station 440CF (Eastman Kodak Co., New Haven, Connecticut, USA).
Luciferase assay.
HCT116 cells were cotransfected with pWWW-Luc (18) and pCMV–β-galactosidase (BD Biosciences Clontech, Palo Alto, California, USA) in triplicates using Lipofectamine reagent (Invitrogen Corp.). After 24 hours, cells were exposed to 1.5 μM LY or an equal volume of DMSO for 12 hours. Luciferase activity was measured according to the manufacturer’s instructions (Luciferase Assay System kit, Promega Corp.). β-galactosidase activity was measured in the same lysates using Galacto-Light reagents (Tropix, Bedford, Massachusetts, USA). Luciferase and β-galactosidase determination were performed on a MicroLumatPlus LB96V luminometer (EG&G Berthold, Bad Wildbad, Germany).
Microscopy.
For phase contrast and β-galactosidase staining, an Axiovert 25 microscope (Carl Zeiss Co., Oberkochen, Germany) equipped with a HyperHAD CCD camera (Sony, Tokyo, Japan) and ImageBase software (Kappa Optoelectronics GmbH, Gleichen, Germany) was used. In vivo expression of histone-H2B–enhanced green fluorescent protein (histone-H2B-eGFP) was documented on an inverted Axiovert 200M microscope (Carl Zeiss Co.) equipped with a CCD camera (CoolSNAP-HQ, Photometrics, Tucson, Arizona, USA) and Metamorph software (Universal Imaging Corp., Downingtown, Pennsylvania, USA).
Results
Microarray analysis of senescence-specific gene expression.
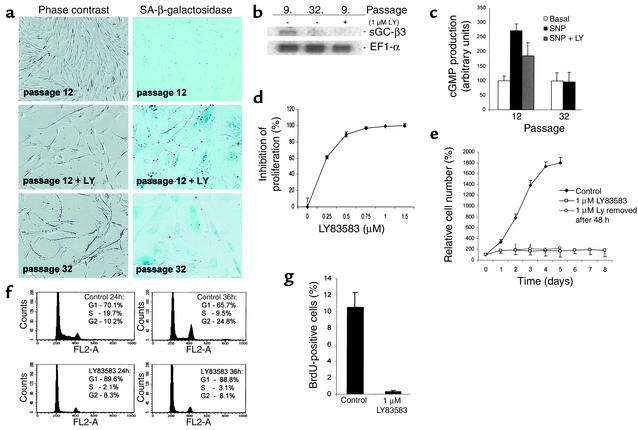
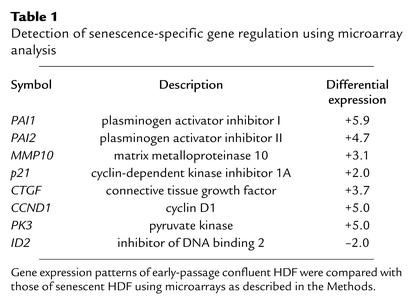
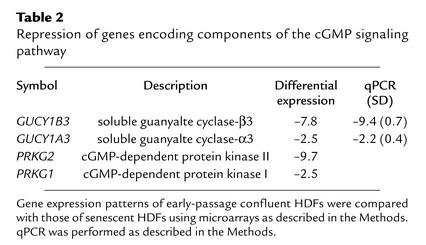
We used microarray analysis to identify genes and pathways involved in the induction and maintenance of replicative senescence. Primary HDFs were cultivated until proliferation of all cells ceased because of replicative senescence (n ∼ 75 population doublings). At this point, the HDFs showed characteristic signs of senescence: cellular enlargement, SA-β-gal staining at pH 6 (Figure 1a), and terminal arrest in the presence of high serum concentration (mainly in the G1 phase as determined by flow cytometry) (data not shown). mRNA was isolated from senescent HDFs and subjected to analysis with a microarray, which allows detection of 8,392 annotated cDNAs (for details, see Methods). Presenescent HDF, which had been arrested in the G1 phase by confluence at population doubling 12, were used as a reference. mRNA was isolated 48 hours after HDF had reached confluence and more than 95% of the cells were in the G1 phase, as determined by flow cytometry (data not shown). This state was selected to avoid a comparison of nonproliferating senescent HDF with proliferating early-passage HDF, which would presumably result in the detection of a large number of differences in gene expression secondary to growth arrest. Several genes previously identified as consistently up- or downregulated during senescence of HDFs (19–21) were detected as regulated in a similar fashion in the system used here (Table 1): for example, PAI-1 was previously detected as upregulated in senescent HDFs and was induced 5.9-fold in senescent HDFs in this study (Table 1). Among the genes repressed during senescence were four genes encoding enzymatic components of the cGMP pathway (Table 2): soluble guanylate cyclase α3 (sGC-α3), sGC-β3, and cGMP-dependent protein kinase I and II. Differential regulation of sGC-α3 and sGC-β3 during senescence was confirmed by qPCR (Table 2). Downregulation of sGC-β3 during senescence was also detected using Northern blot analysis (Figure 1b). sGC-α3 and sGC-β3 represent the large and small subunit of soluble guanylate cyclase, which converts GTP to 3′5′-cyclic GMP and pyrophosphate (22). It is interesting to note that both genes are located in near proximity on chromosome 4q31.3-q33 (23), which could provide the basis for the coregulation observed here.
Figure 1.
Modulation of the cGMP pathway in HDFs and effects of LY. (a) Shown is a comparison of HDFs after reaching replicative senescence with HDFs after LY treatment. The left panel shows morphology (magnification, ×100) and the right panel shows detection of SA-β-gal at pH 6 (magnification, ×200) using phase-contrast microscopy. (b) Northern blot analysis of GUCY1B3 expression in HDFs. Total RNA was isolated from early- (“9.”) and late-passage (“32.”) HDFs and HDFs (“9. passage”) treated with 1 μM LY for 4 days; 2.5 μg of total RNA was loaded per lane. GUCY1B3 and, as a control, ELF1α mRNA were detected with 32P-labeled probes. (c) Response of early-passage and senescent HDFs to activation of cGMP synthesis. Cells were treated with 100 μM SNP for 2 hours in the absence or presence of LY. cGMP levels were determined as described in the Methods. Treatments were performed in triplicates and measured twice. (d) Inhibition of proliferation by LY. Subconfluent HDFs were treated with the indicated concentrations of LY for 3 days. Cell numbers were determined in triplicates. (e) Irreversible effect of LY on proliferation of HDFs. Cells were treated with 1 μM LY for the indicated periods. After trypsinization, cell numbers were determined in triplicates. (f) FACS analysis of HDFs treated with LY. HDFs were synchronized by confluence and released by trypsinization. After 24 and 36 hours of incubation in media containing 1 μM LY or vehicle, cells were collected for FACS analysis. For details, see the Methods. (g) Rate of DNA synthesis after LY treatment. BrdU incorporation was determined after incubation of HDFs in 10 μM BrdU for 6 hours. For details, see the Methods.
Table 1.
Detection of senescence-specific gene regulation using microarray analysis
Table 2.
Repression of genes encoding components of the cGMP signaling pathway
Induction of senescence by LY.
The coordinated repression of two genes encoding the subunits of soluble guanylate cyclase, which generates cGMP in response to signals — for example, extracellular nitric oxide (NO) (reviewed in 24) — suggested that senescent cells may have an attenuated response to NO donors such as SNP. Indeed, exposure to SNP did not result in elevated cGMP levels in senescent HDFs, whereas early-passage HDFs showed a threefold increase in cGMP levels after the addition of SNP (Figure 1c). To specifically inhibit the cGMP signaling pathway, we used a competitive inhibitor of soluble guanylate cyclase, LY, with an IC50 of 2 μM (25). Addition of 1 μM LY to HDFs significantly inhibited the SNP-induced synthesis of cGMP by 50% (Figure 1c), showing that LY had the expected inhibitory effect on guanylate cyclase. We next asked whether addition of LY would affect the proliferation of early-passage HDFs, as we would expect according to our working hypothesis. The effect of LY was tested at concentrations ranging from 0.25–1.5 μM (Figure 1d). Indeed, treatment of early-passage fibroblasts with a concentration of 1 μM of LY was sufficient to completely inhibit proliferation, and the presence of 250 nM LY led to a 50% reduction in proliferation of HDFs after 3 days (Figure 1d). Strikingly, the inhibition of proliferation of early-passage HDFs by LY was accompanied by morphological changes characteristic of senescence and by SA-β-gal staining at pH 6 (Figure 1a). Consistent with the induction of cellular senescence, inhibition of proliferation by LY was irreversible after a treatment period of more than 2 days (Figure 1e). Extended treatment of HDFs with LY for 8 days did not cause significant reduction of cell number due to cell death but resulted in stable inhibition of proliferation (Figure 1e). In order to characterize the cell-cycle arrest induced by LY, HDFs were synchronized in the G1 phase by confluence (91.2% G1 phase and 2% S phase) and then released by trypsinization (Figure 1f); untreated cells entered the S phase at a high percentage. Addition of LY almost completely blocked the entry of HDFs into the S phase, suggesting that LY acts during the G1 phase of the cell cycle. Furthermore, addition of 1 μM LY to synchronized HDFs caused complete inhibition of DNA synthesis as determined by BrdU incorporation (Figure 1g).
Microarray analysis of LY-regulated genes.
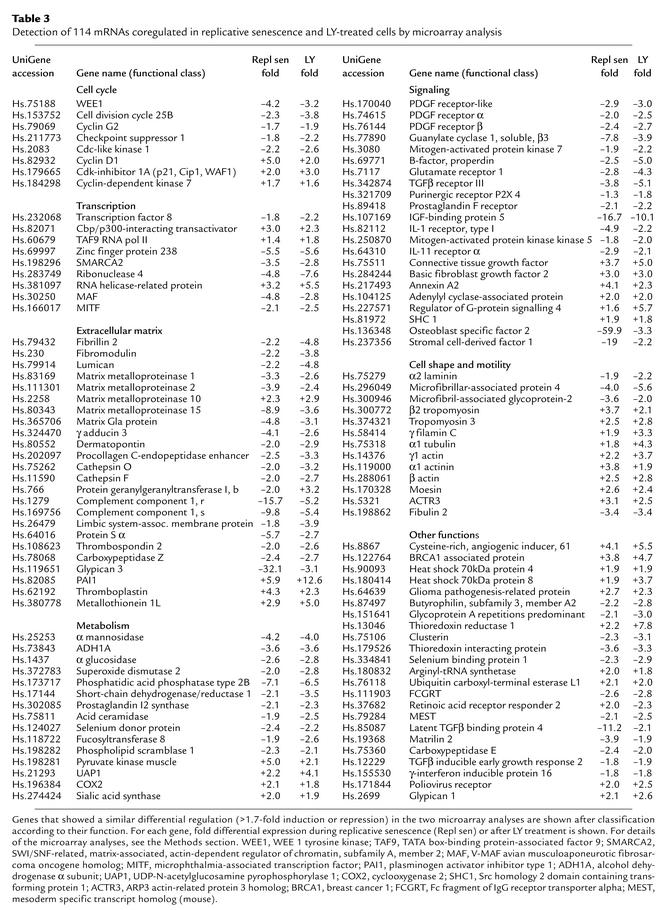
In order to identify the downstream mediators of the LY-induced cell-cycle arrest and senescence, a microarray analysis of LY-treated HDFs was performed with the same arrays used for the analysis of replicative senescence in HDFs described above (for details, see Methods). RNA was isolated 36 hours after exposure of early-passage HDFs to LY. The control RNA was isolated from confluent early-passage HDFs. In HDFs that had reached replicative senescence, 216 transcripts were induced significantly, whereas 272 mRNAs were induced after LY treatment. Repression was observed for 266 mRNAs in HDFs undergoing replicative senescence, whereas 294 mRNAs were repressed after LY treatment. There was a substantial overlap in the differentially expressed genes observed during replicative senescence and during LY-induced senescence, with 114 transcripts differentially regulated in a similar manner (Table 3). Among these were genes that had been previously identified as differentially regulated during replicative senescence: PAI-1, matrix metalloproteinase 10, fibrillin, cdc25b, cyclin D1, fibromodulin, and osteoblast specific factor 2 (11, 20, 21). However, most of the genes identified here represent new additions to the growing number of genes identified as components of the senescence program. Supporting this notion, many of the coregulated genes have functions that may contribute to the phenotype of terminally arrested cells. For example, downregulation of the PDGF receptor α and β chains may contribute to the refractory state of senescent cells, which do not respond to mitogenic stimulation with growth factors (Table 2). The changes in expression observed in genes encoding components of the cytoskeleton (e.g., α1-tubulin, β2-tropomyosin, γ-filamin C, α-actinin) may contribute to the flattened and enlarged shape characteristic of senescent HDFs (Figure 1a). Interestingly, the sGCβ3 mRNA was down-regulated in LY-treated HDFs (Table 3 and Figure 1b).
Table 3.
Detection of 114 mRNAs coregulated in replicative senescence and LY-treated cells by microarray analysis
Rapid induction of p21WAF1/CIP1/SDI by LY.
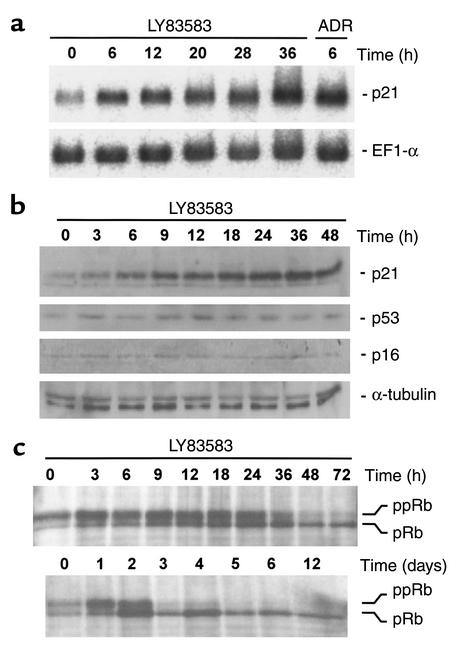
Among the genes induced by both replicative senescence and LY was p21WAF1/CIP1/SDI (Table 3), which encodes an inhibitor of cyclin-dependent kinases (Cdk’s). Cdk/cyclin complexes drive cell-cycle progression and proliferation by phosphorylation of key substrates (reviewed in 26). Since induction of p21 could potentially explain the antiproliferative effect of LY, we analyzed the connection between LY and p21 in more detail. Six hours after treatment with LY, the levels of p21 mRNA in HDFs were similar to the levels observed 6 hours after addition of adriamycin, as shown by Northern blot analysis (Figure 2a). However, after treatment with LY, the level of p53 protein, which consistently increases after generation of DNA damage and mediates transactivation of p21, did not increase (Figure 2b), indicating that LY does not induce DNA damage. The induction of p21 mRNA by adriamycin demonstrates that the signaling pathways necessary for p53 activation were still intact in the HDFs used (Figure 2a). Induction of p21 protein was detectable between 3 and 6 hours after addition of LY, whereas the Cdk inhibitor p16INK4A was not induced even after 48 hours of treatment with LY (Figure 2b). This result was unexpected, since p16INK4A is commonly induced in scenarios that lead to premature senescence — for example, ectopic expression of an activated ras gene (27) or suboptimal cell-culture conditions (10).
Figure 2.
Effects of LY on the expression and phosphorylation status of cell-cycle regulatory proteins. (a) Northern blot analysis of LY-treated HDFs. Total RNA was isolated from HDFs treated with 2 μM LY or 0.2 μg/ml adriamycin for the indicated periods; 7.5 μg of total RNA was loaded per lane. p21WAF1/CIP1/SDI and, as a control, ELF1α mRNA were detected with 32P-labeled probes. (b) Western blot analysis of HDFs treated with LY. Lysates were obtained from HDFs treated with LY for the indicated periods; 20 μg of protein was loaded per lane. After separation on 4–20% gradient gels and transfer to PVDF membranes, proteins were detected with antibodies against human p21, p53, p16, and — as a loading control — α-tubulin. (c) pRb phosphorylation status after treatment with LY. After addition of LY for the indicated periods, cell lysates were prepared and proteins were separated on a 7–12% gradient denaturing PAGE gel. For details, see the Methods; ppRb, hyperphosphorylated pRb; pRb, hypophosphorylated pRb.
The main cell-cycle–relevant targets of the Cdk inhibitor p21 are cyclin-dependent kinases 2, 4, and 6 (28, 29), which keep the pocket-proteins pRb, p107, and p130 in an inactive hyperphosphorylated state during cell-cycle progression (reviewed in 30).
Therefore, p21-mediated inhibition of G1/S phase–associated Cdk/cyclin complexes leads to hypophosphorylation of pocket proteins, which allows their subsequent binding and inhibition of E2F transcription factors. The association between pocket proteins and E2Fs results in inhibition of G1/S progression and proliferation (reviewed in 31). Consistent with the induction of p21 by LY, the phosphorylation status of pRb changed rapidly after addition of LY; after only 6 hours the ratio of phosphorylated to hypophosphorylated, active pRb was clearly shifted toward the hypophosphorylated form, and by 19 hours pRb was almost completely hypophosphorylated (Figure 2c). Even after extension of the LY treatment to 12 days, the pRb hypophosphorylation was not reversed (Figure 2c). These results show that LY inhibits proliferation by inducing p21 to levels that are sufficient to inhibit Cdk activity and activate the cell-cycle inhibitory function of pRb.
Effect of LY on cancer cell proliferation.
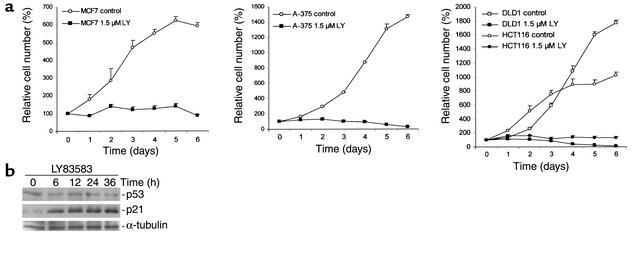
In order to determine whether LY may be useful to terminate cancer cell proliferation, colorectal cancer cell lines (HCT116, DLD1), a breast cancer–derived cell line (MCF7), and a melanoma cell line (A-375) were analyzed after addition of LY; all cell lines showed complete cessation of cell proliferation (Figure 3a). Consistent with the findings in HDFs, HCT116 cells showed induction of p21 at the protein level after addition of LY (Figure 3b). Furthermore, p21 mRNA was induced by LY in MCF7 cells (data not shown).
Figure 3.
Effects of LY on epithelial cancer cells. (a) Growth curves of MCF7, A-375, HCT116, and DLD1 cells. Cells were trypsinized at the indicated time points and cell numbers were determined. (b) Induction of p21 protein after treatment with LY. HCT116 colorectal cancer cells were treated with 1.5 μM LY for the indicated periods. Cell lysates were prepared and subjected to Western blot analysis. For details, see the Methods.
Requirement of p21, but not p53, for inhibition of proliferation by LY.
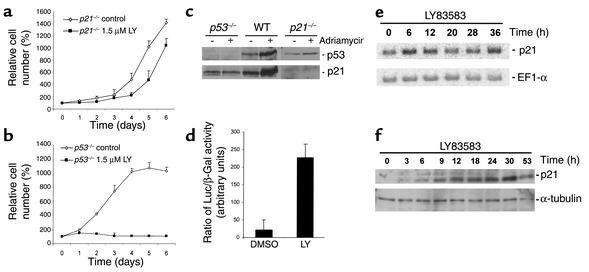
To determine whether induction of p21 is essential for inhibition of proliferation by LY, p21-deficient HCT116 cells, generated through homologous recombination by Waldman et al. (32), were analyzed (Figure 4a); p21-deficient HCT116 cells were largely resistant to LY, whereas wild-type HCT116 cells showed complete cessation of proliferation (Figure 3a). These results show that p21 is required for the inhibitory effects of LY on cellular proliferation.
Figure 4.
Requirement of p21, but not p53, for inhibition of proliferation by LY. (a) Response of HCT116 colorectal cancer cells deficient for p21 to LY. Cells were treated with LY or, as a control, with vehicle (DMSO), and cell numbers were assessed. Measurements were performed in triplicates. (b) Response of p53+/+ and p53–/– HCT116 cells to LY. Cells were treated with LY or, as a control, with vehicle (DMSO), and cell numbers were assessed. Measurements were performed in triplicates. (c) Analysis of HCT116-derived knockout cell lines. Shown are the results of Western blot analysis of p53 and p21 protein levels after addition of adriamycin at 0.2 μg/ml for 8 hours. (d) p21 promoter reporter activity in p53-deficient HCT116 cells after treatment with 1.5 μM LY for 12 hours. The experiment was performed in triplicates. For details, see the Methods. (e) p53-independent induction of p21 mRNA by LY. Shown are the results of Northern blot analysis with RNA isolated at the indicated time points after addition of 1.5 μM LY to p53-deficient HCT116 cells. (f) p53-independent induction of p21 protein levels by LY. Shown are the results of Western blot analysis with lysates from p53–/– HCT116 cells treated with 1.5 μM LY. Membranes were probed with antibodies specific for p21 and, as a loading control, α-tubulin. For details, see the Methods.
Consistent with the absence of p53 accumulation in LY-treated HDFs, p53-deficient HCT116 cells engineered by Bunz et al. (33) showed a complete block of proliferation after addition of LY (Figure 4b). The identities of the three different HCT116 cell lines used in this study were confirmed by treatment with adriamycin, which induces DNA double-strand breaks (Figure 4c). As expected, only wild-type HCT116 cells showed induction of p21 protein after exposure to adriamycin (Figure 4c).HCT116 cells deficient in p53 and transiently transfected with a reporter construct of human p21 promoter fused to the luciferase gene showed a more than 10-fold increase in luciferase activity after 12 hours of LY treatment (Figure 4d). The levels of p21 mRNA and protein of p53–/– HCT116 cells after treatment with LY were induced with kinetics similar to those observed in HDFs (Figure 4, e and f). These results exclude p53 and thereby induction of DNA damage as mediators of cell-cycle arrest by LY and induction of p21 expression. Furthermore, LY treatment did not lead to induction of the Cdk inhibitors p27 and p15 (data not shown). In addition, proliferation of p16INK4A-deficient MEFs was inhibited by LY, showing that p16INK4A is not required for the effects of LY on the cell cycle (data not shown). Supporting this conclusion, the HCT116 colorectal cancer cell lines used in this study do not express functional p16INK4A; one p16INK4A allele carries an inactivating point mutation, and the other p16INK4A allele is completely silenced because of cytosine-phosphate-guanosine dinucleotide methylation (34). Taken together, these results show that the inhibitory effects of LY on proliferation are mediated through induction of p21.
Conversion of LY-induced arrest to apoptosis in pRb-negative cells.
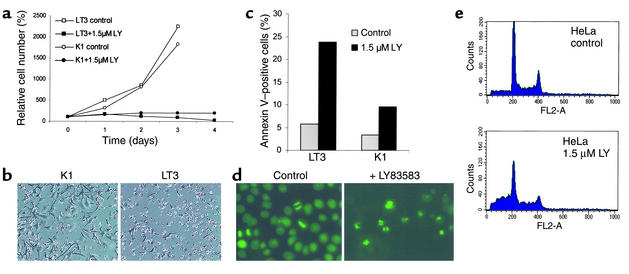
Since the antiproliferative effect of p21 is mediated through pocket proteins (pRb, p107, p130) (reviewed in 30), we analyzed the response of cells with nonfunctional pocket proteins to treatment with LY. Isogenic NIH3T3-L1 cell lines (described in 35) stably expressing either SV40 large T antigen or an SV40 large T-antigen point mutant (K1) (36) deficient in binding and inactivating pocket proteins were treated with LY. NIH3T3-L1 cells expressing the K1-mutant showed inhibition of proliferation, whereas NIH3T3-L1 cells expressing wild-type SV40 large T antigen underwent apoptosis (Figure 5, a and b). Induction of apoptosis by LY was further quantified using annexin V staining (Figure 5c).
Figure 5.
Effect of LY on cells with inactivated pRb pathways. (a) Response of cell lines with lesions in the pRb pathway to LY. NIH3T3-L1 cells expressing SV40 large T antigen (LT3) and NIH3T3-L1 cells expressing a point mutant of SV40 large T antigen unable to bind to pRb-like proteins (K1) were treated with 1.5 μM LY. Cell numbers were determined at the indicated time points in triplicates. (b) Morphology of NIH3T3-L1 cells after LY treatment. Phase-contrast microscopy was performed after incubation of NIH3T3-L1-K1 (K1) and NIH3T3-L1-LT3 (LT3) cells in 1.5 μM LY for 3 days (magnification, ×100). (c) Quantification of early stages of apoptosis. K1 and LT3 cells (control cells and cells treated with LY for 48 hours) were subjected to double staining with anti-annexin V and PI to discriminate living, early apoptotic, and late apoptotic/necrotic populations and were analyzed by flow cytometry. The percentage of PI-negative, annexin V–positive cells is shown. (d) Induction of apoptosis by LY. HeLa cells expressing a histone H2B-eGFP fusion protein (kindly provided by G. Wahl) were treated with 2 μM LY for 48 hours, and the chromatin morphology in living cells was analyzed at a wavelength of 495 nm. (e) Quantification of apoptosis by detection of cells with sub-G1 DNA content. HeLa cells were treated for 60 hours with LY. DNA content was determined after PI staining as described in the Methods.
In order to extend these observations to human cancer cells, the cell lines HeLa and HEK293 were analyzed. The cervical cancer cell line HeLa expresses the human papilloma virus (HPV) 18 encoded E7 protein, which binds to and inactivates pocket proteins. In HEK293 cells, the adenoviral E1A protein performs analogous functions. HeLa and HEK293 cells did not arrest after treatment with LY but instead underwent cell death, which was accompanied by cell shrinkage suggesting apoptosis (data not shown). HeLa cells expressing a histone H2B-eGFP fusion protein allowed us to confirm the induction of apoptosis by LY (Figure 5d); treatment with LY led to chromatin condensation, which is characteristic of apoptosis. Furthermore, flow cytometry analysis revealed a substantial increase of cells with a sub-G1 DNA content after addition of LY (Figure 5e).
Discussion
The results described here demonstrate that LY induces cellular senescence in HDFs and cell-cycle arrest in several human and murine cell types. One requirement for these effects of LY is the transcriptional induction of the Cdk inhibitor p21, since p21-deficient cells are resistant to LY-induced arrest. Furthermore, in the absence of functional pocket proteins (pRb, p107, p130), which are downstream effectors of p21 function, cell-cycle arrest is converted to induction of apoptosis in a p53-independent manner. We assume that in this case apoptosis is a cellular response to conflicting signals: negative signals due to LY-induced p21 and resulting cdk inhibition versus cell-cycle–driving signals due to the lack of active pocket proteins, which allow high levels of active E2F transcription factors.
The induction of p21 is not mediated through the DNA-damage/p53 pathway, since p53-deficient cells also show upregulation of p21 by LY and subsequent inhibition of cell proliferation. Many tumor cells lose pRb function, either through mutation of pRb (37) or through expression of viral proteins. For example, in cervical cancer, expression of E7 by HPV16/18 leads to inactivation of pRb (reviewed in 38). In HPV16/18-infected cells, treatment with LY would presumably induce apoptosis. Since LY induces p21, cell-cycle arrest, and apoptosis in a p53-independent manner, it may prove useful for the treatment of cancers harboring p53 mutations, which represent more than half of all human cancers (reviewed in 39).
In order to induce senescence in early-passage cells, we decided to inhibit the cGMP pathway, since components of this pathway were transcriptionally suppressed during senescence. For this purpose, LY was selected, since it specifically inhibits soluble guanylate cyclase (40). LY was first described as an inhibitor of leukotriene release (25). Later, it was found to lower cGMP levels by inhibiting soluble guanylate cyclase (41). In the experiments presented here, LY was able to mimic cellular senescence in primary HDFs and inhibited proliferation in several cancer cell lines. However, it is unclear whether inhibition of cGMP generation is the only pathway by which LY achieves its effects. It is possible that LY has additional, unknown properties that participate in the induction of cellular senescence and cell-cycle arrest. It has been proposed that the quinone structure of LY may lead to the generation of superoxide anions (42). However, a recent report that used the colorectal cancer cell line HCT116 and the isogenic p21- or p53-deficient cells also analyzed in this study showed that reactive oxygen species (ROS) generated by hyperoxic conditions induce expression of p21 in a p53-dependent manner (43). Furthermore, proliferation of HCT116 cells is inhibited by hyperoxic conditions in a p21- and p53-independent manner (43). In contrast to induction of p21 by ROS, induction of p21 by LY was not mediated by p53. Furthermore, inhibition of proliferation by LY was dependent on p21. These results suggest that LY does not act by generating ROS, which in turn mediate cell-cycle arrest or senescence.
In the future, it will be important to analyze whether LY or derived substances can be used to prevent tumor growth in vivo using animal models. The results provided in this study may serve as a basis for further characterization and optimization of LY-derived drugs as therapeutic agents for the treatment of tumors. An obvious advantage of LY is its ability to interfere with cellular proliferation and to induce apoptosis in pRb-negative cells without inducing DNA damage. These effects of LY are not mediated by the Cdk inhibitor p16INK4A, which is an advantage given the frequent inactivation of p16INK4A in human cancer. On the other hand, the p21 gene, which presumably mediates the effects of LY, has not been reported to be inactivated in human cancer. Commonly used chemotherapeutic agents, like adriamycin, generate DNA damage and thereby selectively induce apoptosis in cancer cells, which lack cell-cycle check points (reviewed in 44). However, DNA-damaging substances have major side effects on proliferating tissues (45, 46) and may induce further mutations in precancerous and healthy cells, which lead to secondary cancer (47, 48).
The strategy used in this study — that is, identification of pathways that are repressed during senescence and their inhibition in tumor cells by synthetic drugs — may also prove useful for the identification of other inhibitors of tumor cell proliferation.
Note added in proof.
Global gene expression data recently obtained for other cell types undergoing senescence, e.g., for prostate epithelial cells (49), may prove helpful for similar approaches.
Acknowledgments
We thank Geoffrey Wahl for providing H2B-eGFP–expressing HeLa cells, Bert Vogelstein for HCT116 derivatives, Manuel Serrano for p16-deficient MEFs, Jens Oliver Funk and Axel Ullrich for cell lines, and Ansgar Resch for plasmids. We also thank members of the Molecular Oncology lab for comments and discussions. This work was supported by the Max Planck Society.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.Nonstandard abbreviations used: human diploid fibroblast (HDF); 6-anilino-5,8-quinolinequinone (LY83583); LY83583 (LY); mouse embryo fibroblasts (MEFs); elongation factor 1α (ELF1α); soluble guanylate cyclase β3 (sGC-β3); quantitative real-time PCR (qPCR); crossing point (CP); propidium iodide (PI); sodium nitroprusside (SNP); senescence-associated β-galactosidase (SA-β-gal); bromodeoxyuridine (BrdU); soluble guanylate cyclase α3 (sGC-α3); nitric oxide (NO); cyclin-dependent kinase (Cdk); human papilloma virus (HPV); reactive oxygen species (ROS).
References
- 1.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol. 2000;1:72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- 4.Serrano M, Blasco MA. Putting the stress on senescence. Curr Opin Cell Biol. 2001;13:748–753. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:27–31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 6.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 7.Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 8.Dellambra E, et al. Downregulation of 14-3-3sigma prevents clonal evolution and leads to immortalization of primary human keratinocytes. J Cell Biol. 2000;149:1117–1130. doi: 10.1083/jcb.149.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez RD, et al. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristofalo VJ, Volker C, Francis MK, Tresini M. Age-dependent modifications of gene expression in human fibroblasts. Crit.Rev.Eukaryot. Gene Expr. 1998;8:43–80. doi: 10.1615/critreveukargeneexpr.v8.i1.30. [DOI] [PubMed] [Google Scholar]
- 12.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 13.Chang BD, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;26:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 14.Bast, R.C., et al., editors. 2000. Cancer medicine. 5th edition. BC Decker Inc. Hamilton, Canada. 2546 pp.
- 15.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome using SAGE: Identification and characterization of c-MYC-target genes. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2003–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad SciUSA. 1995;92:9363–9637. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 19.West MD, Shay JW, Wright WE, Linskens MH. Altered expression of plasminogen activator and plasminogen activator inhibitor during cellular senescence. Exp. Gerontology. 1996;31:175–193. doi: 10.1016/0531-5565(95)02013-6. [DOI] [PubMed] [Google Scholar]
- 20.Ly DH, Lockhart DJ, Lerner RA, Schultz PG. Mitotic misregulation and human aging. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- 21.Shelton DN, Chang E, Whittier PS, Choi D, Funk WD. Microarray analysis of replicative senescence. Curr Biol. 1999;9:939–945. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 22.Giuili G, Scholl U, Bulle F, Guellaen G. Molecular cloning of the cDNAs coding for the two subunits of soluble guanylyl cyclase from human brain. FEBS Lett. 1992;304:83–88. doi: 10.1016/0014-5793(92)80594-7. [DOI] [PubMed] [Google Scholar]
- 23.Giuili G, Roechel N, Scholl U, Mattei M-G, Guellaen G. Colocalization of the genes coding for the alpha-3 and beta-3 subunits of soluble guanylyl cyclase to human chromosome 4 at q31.3-q33. Hum Genet. 1993;91:257–260. doi: 10.1007/BF00218267. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann F, Ammendola A, Schlossmann J. Rising behind NO: cGMP-dependent protein kinases. J Cell Sci. 2000;113:1671–1676. doi: 10.1242/jcs.113.10.1671. [DOI] [PubMed] [Google Scholar]
- 25.Fleisch JH, et al. Pharmacologic analysis of two novel inhibitors of leukotriene (slow reacting substance) release. J Pharmacol Exp Ther. 1984;229:681–689. [PubMed] [Google Scholar]
- 26.Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000;100:71–78. doi: 10.1016/s0092-8674(00)81684-0. [DOI] [PubMed] [Google Scholar]
- 27.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 28.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 29.Harper JW, et al. Inhibition of cyclin-dependent kinases by p21. Mol.Biol. Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–830. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 32.Waldman T, Lengauer C, Kinzler KW, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 33.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 34.Myohanen SK, Baylin SB, Herman JG. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- 35.Hermeking H, et al. Role of c-myc in simian virus 40 large tumor antigen-induced DNA synthesis in quiescent 3T3-L1 mouse fibroblasts. Proc Natl Acad Sci USA. 1994;91:10412–10416. doi: 10.1073/pnas.91.22.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeCaprio JA, et al. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1998;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 37.Horowitz JM, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad SciUSA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.zur Hausen H. Oncogenic DNA viruses. Oncogene. 2001;20:7820–7823. doi: 10.1038/sj.onc.1204958. [DOI] [PubMed] [Google Scholar]
- 39.Hollstein M, et al. New approaches to understanding p53 gene tumor mutation spectra. Mutat Res. 1999;431:7–12. doi: 10.1016/s0027-5107(99)00162-1. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt MJ, Sawyer BD, Truex LL, Marshall WS, Fleisch JH. LY83583: an agent that lowers intracellular levels of cyclic guanosine 3′,5′-monophosphate. J Pharmacol Exp Ther. 1985;232:764–769. [PubMed] [Google Scholar]
- 41.Mulsch A, Busse R, Liebau S, Forstermann U. LY 83583 interferes with the release of endothelium-derived relaxing factor and inhibits soluble guanylate cyclase. J Pharmacol Exp Ther. 1988;247:283–288. [PubMed] [Google Scholar]
- 42.Cherry PD, Omar HA, Farrel KA, Stuart JS, Wolin MS. Superoxide anion inhibits cGMP-associated bovine pulmonary arterial relaxation. Am J Physiology. 1990;259:1056–1062. doi: 10.1152/ajpheart.1990.259.4.H1056. [DOI] [PubMed] [Google Scholar]
- 43.Helt CE, Rancourt RC, Staversky RJ, O’Reilly MA. p53-dependent induction of p21(Cip1/WAF1/Sdi1) protects against oxygen-induced toxicity. Toxicol Sci. 2001;63:214–222. doi: 10.1093/toxsci/63.2.214. [DOI] [PubMed] [Google Scholar]
- 44.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 45.Batchelor D. Hair and cancer chemotherapy: consequences and nursing care — a literature study. Eur J Cancer Care. 2001;10:147–163. doi: 10.1046/j.1365-2354.2001.00272.x. [DOI] [PubMed] [Google Scholar]
- 46.Patridge AH, Burstein HJ, Winer EP. Side effects of chemotherapy and combined chemohormonal therapy in women with early-stage breast cancer. J Natl Cancer Inst Monogr. 2001;30:135–142. doi: 10.1093/oxfordjournals.jncimonographs.a003451. [DOI] [PubMed] [Google Scholar]
- 47.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 48.Travis LB. Therapy-associated solid tumors. Acta Oncol. 2002;41:323–333. doi: 10.1080/028418602760169361. [DOI] [PubMed] [Google Scholar]
- 49.Untergasser G, Koch HB, Menssen A, Hermeking H. Characterization of epithelial senescence by serial analysis of gene expression: identification of genes potentially involved in prostate cancer. Cancer Res. 2002;62:6255–6262. [PubMed] [Google Scholar]