Abstract
Two related integral membrane proteins, claudin-1 and -2, recently were identified as novel components of tight junction (TJ) strands. Here, we report six more claudin-like proteins, indicating the existence of a claudin gene family. Three of these were reported previously as RVP1, Clostridium perfringens enterotoxin receptor, and TMVCF, but their physiological functions were not determined. Through similarity searches followed by PCR, we isolated full length cDNAs of mouse RVP1, Clostridium perfringens enterotoxin receptor, and TMVCF as well as three mouse claudin-like proteins and designated them as claudin-3 to -8, respectively. All of these claudin family members showed similar patterns on hydrophilicity plots, which predicted four transmembrane domains in each molecule. Northern blotting showed that the tissue distribution pattern varied significantly, depending on claudin species. Similarly to claudin-1 and -2, when these claudins were HA-tagged and introduced into cultured Madin–Darby canine kidney cells, all showed a tendency to concentrate at TJs. Immunofluorescence and immunoelectron microscopy with polyclonal antibodies specific for claudin-3, -4, or -8 revealed that these molecules were exclusively concentrated at TJs in the liver and/or kidney. These findings indicated that multiple claudin family members are involved in the formation of TJ strands in various tissues.
The tight junction (TJ) is a specialized membrane domain at the most apical region of polarized epithelial and endothelial cells that not only creates a primary barrier to prevent paracellular transport of solutes (barrier function) but also restricts the lateral diffusion of membrane lipids and proteins to maintain the cellular polarity (fence function) (for reviews, see refs. 1–4). Freeze fracture techniques demonstrate that TJs represent a continuous network of interconnected rows of intramembranous particles that appear as strands with complementary grooves, which are thought to be directly involved both in barrier and fence functions of TJs (5, 6). To date, several unique peripheral membrane proteins have been identified as concentrated at the cytoplasmic surface of TJs: ZO-1 (7–9), ZO-2 (10, 11), ZO-3 (12, 13), cingulin (14), 7H6 antigen (15), and symplekin (16). Among these, ZO-1, ZO-2, and ZO-3 are related to each other and belong to the membrane-associated guanylate kinase homologues gene family, which contains PDZ, SH3, and GUK (guanylate kinase-like) domains (17–19).
By contrast, the TJ-specific integral membrane protein, i.e., the components of TJ strands, only recently were identified. As the first component of TJ strand, occludin, an ≈65-kDa integral membrane protein bearing four transmembrane domains, was identified (20, 21). There is now accumulating evidence that occludin is a component of the TJ strand itself (22, 23) and that occludin is directly involved in the barrier as well as fence functions of TJs (24–27). Recently, gene knockout of occludin was performed successfully, but, unexpectedly, occludin-deficient epithelial cells still demonstrated a well developed network of TJ strands (28).
Most recently, as the second and third components of TJ strand, claudin-1 and -2, ≈22-kDa integral membrane proteins that are structurally related (38% identical at the amino acid sequence level), were identified (29). These two proteins also bear four transmembrane domains but do not show any sequence similarity to occludin. Furthermore, it was shown that claudin-1 and -2 have an ability to induce the formation of networks of strands/grooves at cell–cell contact sites when introduced into fibroblasts lacking TJs (30). Because occludin induced only a small number of short strands at cell–cell contact sites in fibroblasts (30), these findings suggest that claudin-1 and -2 function as major structural components of TJ strands and that occludin is an accessory protein in terms of TJ strand formation.
So far, intensive efforts have failed to identify isotypes of occludin in any species. In contrast, identification of claudin-1 and -2 indicated the existence of a gene family that could be called the “claudin family.” Questions, then, naturally have arisen as to how many members constitute this gene family and whether all of the members are directly involved in TJ formation. To date, three full length cDNAs, RVP1, Clostridium perfringens enterotoxin receptor (CPE-R), and TMVCF, with sequence similarity to claudin-1 and -2 cDNAs, have been reported, although physiological functions have not been determined for any of their products. Rat RVP1 was isolated as a cDNA expression that was elevated in regressing ventral prostate by subtraction cDNA cloning (31), and its human homologue also was identified recently (32, 33). Monkey CPE-R was shown to encode a receptor for the Clostridium perfringens enterotoxin (CPE) (32), and, recently, human/mouse CPE-R as well as human RVP1 products were shown to specifically bind to CPE (32). The human TMVCF (transmembrane protein deleted in Velo-cardio-facial syndrome) gene was localized to chromosome 22q11, which frequently is deleted in Velo-cardio-facial/DiGeorge syndrome patients (35). Furthermore, several sequences similar to claudin-1 and -2 were found in expressed sequence tag (EST) databases. In this study, we isolated mouse RVP1, CPE-R, and TMVCF and three full length cDNAs that encoded members of the claudin family. Similarly to claudin-1 and -2, when HA-tagged mouse RVP1, CPE-R, TMVCF, and three other claudin members were introduced into Madin–Darby canine kidney (MDCK) cells, all of them showed a tendency to concentrate at preexisting TJs. Furthermore, we obtained polyclonal antibodies (pAbs) specific for three members of the claudin family, which specifically labeled TJs in the liver and/or kidney. These results indicate that claudin family members play a central role in TJ formation in various tissues.
MATERIALS AND METHODS
Antibodies and Cells.
Rat anti-mouse occludin mAb (MOC37) was raised and characterized as described (36). Rat anti-mouse E-cadherin mAb (ECCD2) kindly was provided by M. Takeichi (Kyoto University). Mouse anti-HA-tag mAb (12CA5) was purchased from Boehringer Mannheim. MDCK cells were grown in DMEM supplemented with 10% fetal calf serum.
cDNA Cloning and Sequencing.
Mouse liver or kidney total RNA was isolated according to the method described by Chomczynski and Sacchi (37). Poly(A)+ RNA was prepared from the total RNA by using oligo(dT)-cellulose (New England Biolabs). First strand cDNA was prepared from this poly(A)+ RNA with Superscript II reverse transcriptase (GIBCO/BRL) and was used for PCR.
As described in detail in Results, 15 mouse EST clones with sequence homology to claudin-1 were found, which enabled us to amplify cDNAs encoding ORFs of claudin-3, -5, -6, -7, and -8 from mouse liver or kidney first strand DNA by PCR. Claudin-4 cDNA also was amplified from mouse kidney first strand DNA by PCR using the previously reported sequence of mouse CPE-R (32). Furthermore, human claudin-5 (human TMVCF) was obtained from human genomic DNA isolated from peripheral blood cells by PCR. The amplified cDNAs were subcloned into the pGEM-T Easy Vector (Promega). To ensure that no sequence errors occurred during PCR, the nucleic acid sequence was determined from three independently amplified fragments. DNA sequence analysis was performed by using a Dye Terminator Cycle Sequence Kit (Applied Biosystems).
Northern Blotting.
The expression of claudin-3 to -8 in various mouse tissues was examined by Northern blotting by using Mouse Multiple Tissue Northern Blot (CLONTECH). DNA fragments of mouse claudins-3 (3′-terminal region of ORF + 3′ untranslated region), -4 (3′-terminal region of ORF + 3′ untranslated region), -5 (entire ORF), -6 (3′-half of ORF), -7 (entire ORF), and -8 (entire ORF) were radiolabeled with [32P]dCTP and were used as probes for Northern blotting. Hybridization was performed in ExpressHyb Hybridization Solution (CLONTECH) at 68°C for 12 h. The membranes were washed with 2× standard saline citrate (SSC) containing 0.1% SDS at room temperature for 30 min and then with 0.1× SSC containing 0.1% SDS at 50°C for 30 min. The membranes were exposed to imaging plates for 12 h, and the signals were visualized by using a Bio-Imaging Analyzer BAS2000 (Fuji).
Mammalian Expression Vectors and Transfection.
Mouse claudin-3, -4, -6, -7, and -8 and human claudin-5 were tagged with HA-peptide at their COOH-termini. To construct HA-tagged claudin-expression vectors, EcoRI sites were introduced at 3′ ends of claudin cDNAs by PCR, and amplified fragments were subcloned into pBluescript SK(-)-HA-tag. The inserts were excised by SaII-XbaI digestion followed by blunting with T4 polymerase and then were introduced into pCAGGS neodel EcoRI (38) provided by J. Miyazaki (Osaka University). MDCK cells were used for transfection as described (29), and the clones expressing tagged proteins were screened by fluorescence microscopy with anti-HA mAb.
Polyclonal Antibody Production.
The polypeptides CPRSTGPGTGTGTAYDRKDYV, CDKPYSAKYSAARSVPASNYV, and CQRSFHAEKRSPSIYSKSQYV, corresponding to the COOH-terminal cytoplasmic domains of mouse claudin-3, -4, and -8, respectively, were synthesized and coupled via cysteine to keyhole limpet hemocyanin. These conjugated peptides were used as antigens to generate pAbs in rabbits. The antiserum specific for claudin-3, -4, or -8 was affinity-purified with glutathione S-transferase (GST) fusion protein with the COOH-terminal cytoplasmic domain of claudin-3, -4, or -8, respectively. To express the GST/claudin fusion proteins in Escherichia coli, DNA fragments were amplified by PCR from cDNAs encoding COOH-terminal cytoplasmic domains of mouse claudins, and the amplified fragments were subcloned into pGEX4T-1 (Pharmacia) in frame.
Immunofluorescence Microscopy and Immunoelectron Microscopy.
Stable MDCK transfectants plated on glass coverslips and frozen sections of mouse liver and kidney were stained immunofluorescently as described (20, 30). Immunoelectron microscopy to examine freeze-fracture replicas was performed as described (22, 29, 30).
RESULTS
Cloning and Characterization of cDNAs Encoding Claudin Family Members.
Claudin-1 and -2 were identified (29). As mentioned above, similarity searches identified three previously reported full length cDNAs encoding integral membrane proteins that showed homology to claudin-1 and –2: RVP1 (31, 33), CPE-R (32, 34), and TMVCF (35), which are designated here as claudin-3, -4, and -5, respectively. Among these, only CPE-R cDNA was isolated from mouse (ORF = 210 amino acids; calculated molecular mass = 22.3 kDa) whereas RVP1 and TMVCF cDNAs were obtained from rat/human and human, respectively. We then searched for mouse RVP1 (mRVP1) sequences in databases and identified four overlapping mouse EST clones (AA537925, AB000715, W11468, and W30232), which were almost identical in deduced amino acid sequences to rat/human RVP1. By using these EST sequences, a full length cDNA encoding claudin-3/mRVP1 was amplified by PCR from mouse liver first strand DNA. Its ORF encoded a protein of 219 amino acids with a calculated molecular mass of 23.3 kDa (Fig. 1). As a mouse homologue of TMVCF (mTMVCF), two overlapping mouse EST clones were obtained (AA542175, AA572018), which allowed us to amplify claudin-5/mTMVCF cDNA by PCR from mouse kidney first strand DNA. Its ORF encoded a protein of 218 amino acids, with a calculated molecular mass of 23.1 kDa (Fig. 1).
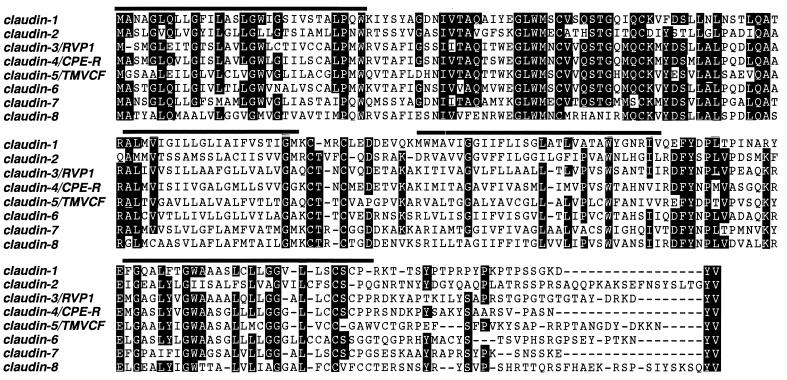
Figure 1.
Multiple sequence alignments for the members of the claudin family. Putative membrane-spanning domains are represented by bars. Residues conserved in at least six of the eight sequences are highlighted with black backgrounds. The amino acid sequence was fairly conserved in the first and fourth transmembrane domains as well as in the first and second extracellular loops but was diversified in the second and third transmembrane domains. Note that all of the claudin family members end in -Y-V at their COOH-termini. The nucleotide sequence data of mouse claudin-1 to -8 are available from European Molecular Biology Laboratory/GenBank database/DNA Data Base in Japan under accession numbers AF072127, AF072128, AF087821, AF087822, AF087823, AF087824, AF087825, and AF087826, respectively.
In addition to these 6 mouse EST clones corresponding to claudin-3 and -5, similarity searches of databases identified nine more mouse EST clones, which showed sequence similarity to claudin-1 and -2 but were distinct from claudin-1, -2, -3, -4, or -5. Four (W35896, AA450511, AA271859, AA537008) and three (AA240230, AA624235, W36669) of them overlapped, leading to the isolation of cDNAs encoding two claudins, claudin-6 and -7, by PCR from mouse kidney and liver first strand DNA, respectively. They encoded proteins of 219 and 211 amino acids with calculated molecular masses of 23.4 and 22.6 kDa, respectively (Fig. 1). Although the remaining two EST clones (AA510770, AA167924) did not overlap, PCR using both EST sequences as primers amplified a cDNA encoding a claudin, claudin-8, from mouse kidney first strand DNA. This cDNA encoded a protein of 225 amino acids with a calculated molecular mass of 24.9 kDa (Fig. 1).
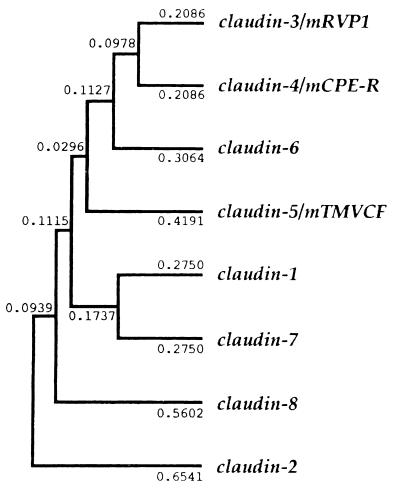
Hydrophilicity analyses predicted that, like claudin-1 and -2, all of the identified claudins (claudin-3 to -8) contained four transmembrane domains with both NH2- and COOH-termini located in the cytoplasm (data not shown). The putative first extracellular loop of each claudin was longer and more hydrophobic than the putative second extracellular loop. Multiple sequence alignments carried out for all claudin family members revealed that the amino acid sequence was fairly conserved in the first and fourth transmembrane domains as well as in the first and second extracellular loops but diversified in the second and third transmembrane domains (Fig. 1). Although the amino acid sequences of COOH-terminal cytoplasmic domains were also fairly diversified, COOH-termini of all of the claudins ended in -Y-V. Finally, to deduce the possible evolutionary relationships among the claudin family members, the amino acid sequences were compared quantitatively, and a phylogenetic tree was constructed by using the unweighted pair–group method (39–41) (Fig. 2). This tree suggested that claudin-2 and the primordial form of the other members were generated first and that claudin-3 and -4 had diversified most recently.
Figure 2.
A potential phylogenetic tree of the claudin gene family. The tree was constructed by the unweighed pair group method (39–41) using the calculated genetic distances (presented numerically) between pairs of members.
Tissue Distribution of Claudin Family Members.
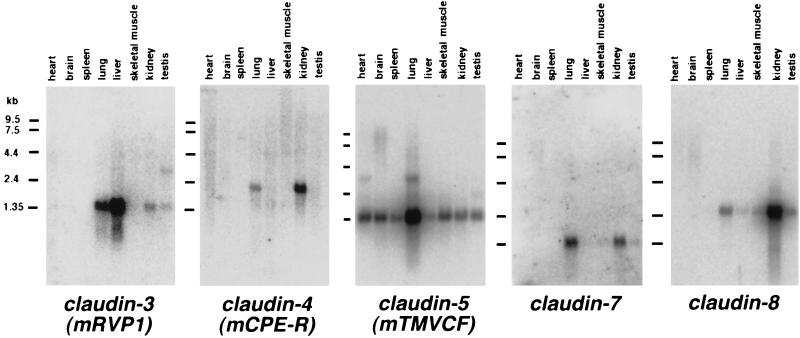
From cDNAs of claudin-3, -4, -5, -6, -7, and -8, we designed specific probes for Northern blotting and examined the expression of each member of claudin family in various tissues (Fig. 3). Tissue distribution patterns of each claudin were distinct. Claudin-3 mRNA was detected in large amounts in lung and liver and in small amounts in kidney and testis. Claudin-4, -7, and -8 were expressed primarily in lung and kidney. In contrast, claudin-5 was expressed in all of the tissues examined, at especially high levels in the lung. Of interest, claudin-6 expression was not detected in the adult tissues examined (data not shown), although claudin-6 cDNA was amplified by reverse transcription–PCR from mouse kidney. Because all of the EST sequences corresponding to claudin-6 were identified from embryos, it is possible that the expression of claudin-6 is regulated developmentally.
Figure 3.
Northern blots of mouse claudin-3, -4, -5, -7, and -8 expression. Mouse Multiple Tissue Northern Blot (CLONTECH) was probed with DNA fragments of claudins (see Materials and Methods). Tissue distribution patterns were distinct between claudin species. The expression of claudin-6 was not detected by Northern blotting, although it was detected by reverse transcription–PCR at least in mouse kidney.
Introduction of HA-Tagged Claudin Family Members into Cultured MDCK Cells.
Previously, it was found that when FLAG-tagged claudin-1 and -2 were introduced into cultured MDCK cells, they were selectively targeted to and incorporated in the pre-existing TJ strands (29). We then transfected cDNAs encoding HA-tagged claudin-3, -4, -5, -6, -7, and -8 into MDCK cells and obtained stable transfectants. The subcellular distributions of these tagged proteins were examined by confocal microscopy with anti-HA mAb in comparison with those of occludin. As shown in Fig. 4, all of the HA-tagged members of the claudin family examined were coconcentrated with occludin at the level of TJs, although the efficiency of their targeting to TJs varied between claudin species. Computer-generated cross-sectional images revealed that all of the HA-tagged members of the claudin family examined were colocalized precisely with occludin at the most apical part of the lateral membranes of confluent MDCK transfectants.
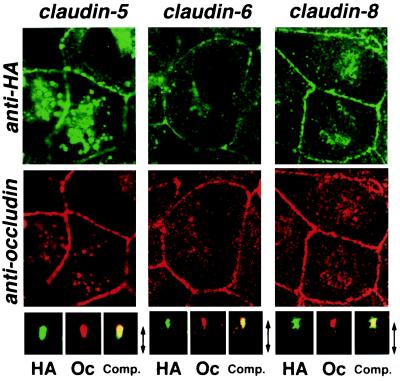
Figure 4.
Concentration of HA-tagged claudins at the occludin-positive tight junctions in MDCK transfectants. Confluent cultures of MDCK transfectants expressing HA-tagged claudin-5, -6, or -8 (claudin-5, claudin-6, claudin-8) were doubly stained with anti-HA mAb (anti-HA) and anti-occludin mAb (anti-occludin). Images were obtained at the focal plane of the most apical region of lateral membranes by confocal microscopy. HA-tagged claudins were coconcentrated with occludin at the level of TJs. As shown in the bottom panels, in computer-generated cross-sectional views, HA-claudins (HA) and occludin (Oc) were highly concentrated at the most apical portion of lateral membranes of MDCK cells, and overlaid images showed that HA-claudins were precisely colocalized with occludin (Comp.). The thickness of each cellular sheet is indicated by arrows. The same results were obtained from MDCK transfectants expressing HA-tagged claudin-3, -4, and -7 (data not shown).
Immunolocalization of Claudin-3/mRVP1, Claudin-4/mCPE-R, and Claudin-8.
As discussed previously, it was difficult to obtain antibodies specific to claudin-1 or -2, probably because of the small size and/or low antigenicity of extramembrane portions of these molecules (29). However, we succeeded in raising pAbs specific to claudin-3/mRVP1, claudin-4/mCPE-R or claudin-8 in rabbits by using synthesized polypeptides corresponding to their COOH-terminal cytoplasmic domains as antigens. These domains are not conserved among claudin family members. By immunoblotting, each affinity-purified pAb specifically recognized the GST fusion protein with COOH-terminal cytoplasmic domain of the corresponding member of the claudin family that was produced in E. coli (Fig. 5A).
Figure 5.
Subcellular distribution of endogenous claudin-3/mRVP-1, claudin-4/mCPE-R, and claudin-8. (A) Specificity of anticlaudin-3, -4, and -8 pAbs. Immunoblots of total lysates of E. coli expressing GST fusion proteins with cytoplasmic domains of all claudin family members (arrow) confirmed their specificities. CBB, Coomassie brilliant blue staining. (B) Frozen sections of mouse liver (a and b) and kidney (c–f) were stained with affinity-purified anti-claudin-3 pAb (a), anti-claudin-4 pAb (c), or anti-claudin-8 pAb (e). All sections were counter-stained with anti-occludin mAb (b, d, and f). Arrows, distal tubules; asterisks, proximal tubules. (Bar = 10 μm.)
Because claudin-3/mRVP1 was expressed in liver and because claudin-4/mCPE-R and claudin-8 were expressed in kidney (see Fig. 3), frozen sections from liver and kidney were doubly stained with anti-claudin-3 pAb/anti-occludin mAb, anti-claudin-4 pAb/anti-occludin mAb, or anti-claudin-8 pAb/anti-occludin mAb (Fig. 5B). In liver, claudin-3 and occludin were colocalized precisely along bile canaliculi. In distal tubules of the kidney, claudin-4/mCPE-R and claudin-8 were colocalized exclusively and precisely with occludin at their junctional complex region. In proximal tubules, occludin as well as claudins signals were detected weakly in the junctional regions. Finally, the liver was analyzed by the immunofreeze fracture replica method by using affinity-purified anti-claudin-3/RVP1 pAb. As shown in Fig. 6, in liver, TJ strands themselves were labeled heavily and specifically with anti-claudin-3/RVP1 pAb.
Figure 6.
Immunoelectron microscopic images of freeze-fracture replicas of mouse liver with affinity-purified anti-claudin-3 pAb. Tight junction strands themselves (T) were specifically labeled with anticlaudin-3 pAb. (Bar = 0.2 μm.)
DISCUSSION
Previously, it was found that, in liver, at least three integral membrane proteins with four transmembrane domains, occludin, claudin-1, and claudin-2, constitute TJ strands (29). Furthermore, when these claudins were introduced into cultured fibroblasts, they reconstituted TJ strands (30). In this study, six full length cDNAs encoding integral membrane proteins showing similarity to claudin-1 and -2 were isolated, and these were designated as claudin-3 to -8. All of these identified claudins appeared to bear four transmembrane domains and were targeted to TJs when tagged with HA and introduced into cultured MDCK cells. Furthermore, the pAbs specific for claudin-3/mRVP1, claudin-4/mCPE-R, or claudin-8 specifically stained TJs in liver and/or kidney by immunofluorescence microscopy, and anti-claudin-3/mRVP1 pAb exclusively labeled TJ strands themselves in liver by immunoreplica electron microscopy. Considering that phylogenetically claudin-3 and -4 were generated most recently and that claudin-8 (and claudin-2) was related most distantly to claudin-3 and -4 (see Fig. 2), these findings indicated that there is a new multigene family, the claudin family, and that at least eight distinct members of this family are involved directly in the formation of TJ strands in various tissues.
The present results indicating the involvement of a multigene family in the formation of TJ strands were reminiscent of another multigene family, the connexin family, which constitutes gap junctions (for review see refs. 42 and 43). The connexin family of proteins is encoded by at least 13 genes in rodents. Each connexin bears four transmembrane domains like claudins, and a single gap junction channel (connexon) is composed of six connexin molecules. Most cells express more than one connexin species, and it is now believed that connexins can be not only homohexameric, containing only a single connexin species, but also heterohexameric, containing different connexins. Complexity is generated further when two connexons in apposed membranes are associated to form “homotypic” or “heterotypic” communicating channels between adjacent cells. Although the manner of aggregation of four-transmembrane domain proteins in junctions may be different between gap junctions and TJs, the existence of the claudin multigene family as well as the characteristic tissue distribution pattern of each claudin species (see Fig. 3) suggested that similar complexity can also be expected in TJs. Furthermore, occludin, another four-transmembrane domain protein with no sequence similarity to claudins, also is incorporated into TJ strands (20–22), although occludin knockout analysis as well as claudin-1/-2 transfection experiments in fibroblasts revealed that the TJ strand itself can be formed without occludin (28, 30). The existence of occludin would make the molecular architecture of TJ strands more complex than that of gap junctions. It is now necessary to clarify how multiple four-transmembrane domain proteins constitute TJ strands and how the complexity in the molecular architecture of TJ strands contributes to the generation of functional diversity of TJs in vivo.
The interactions between the cytoplasmic domains of claudins and TJ-specific peripheral membrane proteins also remain elusive. ZO-1 is associated directly with the COOH-terminal ≈150 amino acids of occludin (44), but, in epithelial cells differentiated from occludin-knockout embryonic stem cells, ZO-1 still was localized exclusively at TJs (28). Because ZO-1 is a multidomain protein consisting of one GUK (guanylate kinase-like), SH3, and three PDZ domains (8, 9, 45), it is possible that ZO-1 is tethered directly or indirectly not only to the cytoplasmic domain of occludin but also to those of claudins through its distinct domains. It should be noted here that all of the claudin family members end in -Y-V at their COOH-termini (see Fig. 1). It is well known that the COOH-terminal -E-S/T-D-V motif in the cytoplasmic tails of ion channels such as Shaker K+ channel (-ETDV) and NMDA R2 subunit (-ESDV) is specifically recognized by the PDZ domains of the Dlg/PSD-95 family of proteins (46–48). Recently, however, some PDZ domains were shown to have binding specificities for COOH-terminal sequences that are distinct from -E-S/T-D-V. Of interest, the PDZ domain of LIN-2/CASK binds to the COOH-terminal tails of neurexins, which ends in -E-Y-Y-V (49). Therefore, it is tempting to speculate that the COOH-termini of claudins binds to PDZ domains of ZO-1 directly or indirectly through association with PDZ domains of ZO-2, ZO-3, or other PDZ-containing proteins. Yeast two-hybrid analyses using the COOH-terminal sequences of claudins as bait or in vitro binding analyses using GST fusion proteins with the cytoplasmic domains of claudins should be useful for evaluation of this speculation.
Another issue that we should discuss here is that claudin-4 had been identified as a receptor for CPE (32, 34). Katahira et al. (34) cloned a cDNA encoding the receptor for CPE (CPE-R) from an expression library of enterotoxin-sensitive monkey Vero cells. Of interest, they noticed that the previously reported RVP1 showed marked similarity to CPE-R and found that RVP1 (claudin-3) also functioned as a receptor for CPE (32). It is thus possible that other members of the claudin family, if not all, also bind to CPE. Further analyses on the interaction of claudins with CPE should provide a way to utilize CPE as a tool to modulate TJ functions in general.
In this study, we identified a multigene family, the claudin family, consisting of at least eight related integral membrane proteins with four transmembrane domains and demonstrated that all of the members are components of TJ strands in various tissues. Considering that claudin-1 and -2 can singly reconstitute TJ strands in fibroblasts (30), it is easily expected that the other 6 claudins also have an ability to form TJ strands within the plasma membrane. We believe that this study raises many intriguing questions regarding claudin itself as well as the structure and functions of TJs, especially regarding the physiological relevance of the existence of many related claudins as well as how multiple four-transmembrane proteins such as claudins and occludin are integrated into TJ strands. Studies along these lines currently are being conducted in our laboratory.
Acknowledgments
We thank Drs. J. Katahira and Y. Horiguchi (Osaka University) for discussion regarding the Clostridium perfringens enterotoxin receptor. Our thanks are also due to all of the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University) for helpful discussions. This study was supported in part by a Grant-in-Aid for Cancer Research and a Grand-in-Aid for Scientific Research (A) from the Ministry of Education, Science and Culture of Japan.
ABBREVIATIONS
- TJ
tight junction
- MDCK cells
Madin–Darby canine kidney cells
- CPE
Clostridium perfringens enterotoxin
- CPE-R
CPE receptor
- pAb
polyclonal antibody
- EST
expressed sequence tag
- GST
glutathione S-transferase
Footnotes
References
- 1.Schneeberger E E, Lynch R D. Am J Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- 2.Gumbiner B. Am J Physiol. 1987;253:C749–C758. doi: 10.1152/ajpcell.1987.253.6.C749. [DOI] [PubMed] [Google Scholar]
- 3.Gumbiner B. J Cell Biol. 1993;123:1631–1633. doi: 10.1083/jcb.123.6.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J M, van Itallie C M. Am J Physiol. 1995;269:G467–G475. doi: 10.1152/ajpgi.1995.269.4.G467. [DOI] [PubMed] [Google Scholar]
- 5.Staehelin L A. J Cell Sci. 1973;13:763–786. doi: 10.1242/jcs.13.3.763. [DOI] [PubMed] [Google Scholar]
- 6.Staehelin L A. Int Rev Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- 7.Stevenson B R, Siliciano J D, Mooseker M S, Goodenough D A. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita S, Tsukita S. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willott E, Balda M S, Fanning A S, Jameson B, van Itallie C, Anderson J M. Proc Natl Acad Sci USA. 1993;90:7834–7831. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumbiner B, Lowenkopf T, Apatira D. Proc Natl Acad Sci USA. 1991;88:3460–3464. doi: 10.1073/pnas.88.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jesaitis L A, Goodenough D A. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balda M S, González-Mariscal L, Matter K, Cereijido M, Anderson J M. J Cell Biol. 1993;123:293–302. doi: 10.1083/jcb.123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haskins J, Gu L, Wittchen E S, Hibbard J, Stevenson B R. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citi S, Sabanay H, Jakes R, Geiger B, Kendrick-Jones J. Nature (London) 1988;333:272–276. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Y, Saitoh T, Minase T, Sawada N, Enomoto K, Mori M. J Cell Biol. 1993;120:477–483. doi: 10.1083/jcb.120.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keon B H, Schäfer S, Kuhn C, Grund C, Franke W W. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods D A, Bryant P J. Mech Dev. 1993;44:85–89. doi: 10.1016/0925-4773(93)90059-7. [DOI] [PubMed] [Google Scholar]
- 18.Kim S K. Curr Opin Cell Biol. 1995;7:641–649. doi: 10.1016/0955-0674(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 19.Anderson J M, Fanning A S, Lapierre L, van Itallie C M. Biochem Soc Trans. 1995;23:470–475. doi: 10.1042/bst0230470. [DOI] [PubMed] [Google Scholar]
- 20.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Itoh M, Yonemura S, Furuse M, Tsukita S. J Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimoto K. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 23.Furuse M, Fujimoto K, Sato N, Hirase T, Tsukita S, Tsukita S. J Cell Sci. 1996;109:429–435. doi: 10.1242/jcs.109.2.429. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy K M, Skare I B, Stankewich M C, Furuse M, Tsukita S, Rogers R A, Lynch R D, Schneeberger E E. J Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- 25.Balda M S, Whitney J A, Flores C, González S, Cereijido M, Matter K. J Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y-H, Merzdorf C, Paul D L, Goodenough D A. J Cell Biol. 1997;138:891–899. doi: 10.1083/jcb.138.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong V, Gumbiner B M. J Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, Takano H, Noda T, Tsukita S. J Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuse M, Fujita T, Hiiragi T, Fujimoto K, Tsukita S. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuse M, Sasaki H, Fujimoto K, Tsukita S. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briehl M M, Miesfeld R L. Mol Endocrinol. 1991;5:1381–1388. doi: 10.1210/mend-5-10-1381. [DOI] [PubMed] [Google Scholar]
- 32.Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. J Biol Chem. 1997;272:26652–26658. doi: 10.1074/jbc.272.42.26652. [DOI] [PubMed] [Google Scholar]
- 33.Peacock R E, Keen T J, Inglehearn C F. Genomics. 1997;46:443–449. doi: 10.1006/geno.1997.5033. [DOI] [PubMed] [Google Scholar]
- 34.Katahira J, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. J Cell Biol. 1997;136:1239–1247. doi: 10.1083/jcb.136.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirotkin H, Morrow B, Saint-Jore B, Puech A, Das Gupta R, Patanjali S R, Skoultchi A, Weissman S M, Kucherlapati R. Genomics. 1997;42:245–251. doi: 10.1006/geno.1997.4734. [DOI] [PubMed] [Google Scholar]
- 36.Saitou M, Ando-Akatsuka Y, Itoh M, Furuse M, Inazawa J, Fujimoto K, Tsukita S. Eur J Cell Biol. 1997;73:222–231. [PubMed] [Google Scholar]
- 37.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 38.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 39.Fitch W M, Margoliash E. Science. 1967;155:361–376. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 40.Doolittle R F. In: The Plasma Proteins. Putnam F W, editor. IV. Orlando, FL: Academic; 1984. pp. 317–360. [Google Scholar]
- 41.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. pp. 293–298. [Google Scholar]
- 42.Kumar N M, Gilula N B. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 43.Jiang J X, Goodenough D A. Proc Natl Acad Sci USA. 1996;93:1287–1291. doi: 10.1073/pnas.93.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukita S, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. J Cell Biol. 1993;123:1049–1053. doi: 10.1083/jcb.123.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 47.Komau H-C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 48.Niethammer M, Kim E, Sheng M. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hata Y, Butz S, Sudhof T C. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]