Abstract
Background
Eukaryotic initiation factor 4E (eIF4E) is essential for cap-dependent initiation of translation. Cell proliferation is associated with increased activity of eIF4E and elevated expression of eIF4E leads to tumorigenic transformation. Many tumors express very high levels of eIF4E and this may be a critical factor in progression of the disease. In contrast, overexpression of 4EBP, an inhibitor of eIF4E, leads to cell cycle arrest and phenotypic reversion of some transformed cells.
Results
A constitutively active form of 4EBP-1 was inducibly expressed in the human breast cancer cell line MCF7. Induction of constitutively active 4EBP-1 led to cell cycle arrest. This was not associated with a general inhibition of protein synthesis but rather with changes in specific cell cycle regulatory proteins. Cyclin D1 was downregulated while levels of the CDK inhibitor p27Kip1 were increased. The levels of cyclin E and CDK2 were unaffected but the activity of CDK2 was significantly reduced due to increased association with p27Kip1. The increase in p27Kip1 did not reflect changes in p27Kip1 mRNA or degradation rates. Rather, it was associated with enhanced synthesis of the protein, even though 4EBP-1 is expected to inhibit translation. This could be explained, at least in part, by the ability of the p27Kip1 5'-UTR to mediate cap-independent translation, which was also enhanced by expression of constitutively active 4EBP-1.
Conclusions
Expression of active 4EBP-1 in MCF7 leads to cell cycle arrest which is associated with downregulation of cyclin D1 and upregulation of p27Kip1. Upregulation of p27Kip1reflects increased synthesis which corresponds to enhanced cap-independent translation through the 5'-UTR of the p27Kip1 mRNA.
Background
Most eukaryotic mRNAs are translated through a cap-dependent mechanism of initiation. In this process, eIF4E, which functions by binding the 7-methylguanosine cap, is the rate-limiting factor due to its low abundance [1,2]. In normal cells, elevated eIF4E activity is associated with proliferation and is regulated by multiple mechanisms. eIF4E gene transcription is enhanced in response to mitogenic stimulation and may be mediated by c-myc [3]. Phosphorylation of eIF4E by the protein kinase Mnk1 increases its affinity for the cap, and thus increases cap-dependent initiation of translation [4-6]. An inhibitor of eIF4E, 4E binding protein (4EBP), is also regulated by phosphorylation. Phosphorylation of 4EBP disrupts its ability to bind eIF4E, freeing eIF4E to bind eIF4G, which functions to recruit additional components of the initiation complex [7,8]. The net result is that phosphorylation of 4EBP enhances cap-dependent translation initiation. 4EBP phosphorylation is thought to be mediated by mammalian target of rapamycin (mTOR) in mitogen stimulated cells [9-11].
Numerous studies have implicated eIF4E in formation and progression of tumors. Overexpression of eIF4E leads to deregulated growth and malignant transformation of a variety of cultured cell lines [12-14]. Moreover, elevated levels of eIF4E are commonly found in solid tumors, especially in breast, colon, and head and neck tumors [15]. Clinical studies indicate that eIF4E gene amplification and protein overexpression is associated with malignant progression in these tumors [16]. High eIF4E levels are also associated with a higher rate of cancer recurrence and cancer-related death [17]. In contrast to eIF4E, overexpression of 4EBP inhibits cell proliferation [18] and 4EBP-1 expression levels are inversely correlated with the progression of certain types of tumors [19].
The findings summarized above indicate that elevated eIF4E activity plays a fundamental role in cancer formation and progression and suggest the possibility that eIF4E could be exploited as a therapeutic target. This is supported by several recent investigations. Inhibiting the expression of eIF4E by expressing antisense RNA reverses ras-mediated transformation of cultured cells as indicated by decreased efficiency of growth in soft agar and tumor formation in nude mice [20]. Similarly, antisense-mediated reduction of eIF4E in breast cancer cells inhibits both their tumorigenic and angiogenic properties [21]. On the other hand, overexpression of 4EBP reverses transformation mediated by v-src [18].
Rapamycin and its analogs are potent inhibitors of mTOR and lead to cell cycle arrest. Thus there is interest in using this class of inhibitors for chemotherapeutic treatment of cancer and several phase I trials have been carried out [22]. Treatment of mammalian cells with rapamycin leads to dephosphorylation and activation of 4EBP, which in turn binds to and inhibits eIF4E. However, several other targets downstream of mTOR are also affected by rapamycin. These include effects on other translational control proteins including p70 S6 kinase (p70S6K), eIF4G, eIF4B, and eEF2 (reviewed in [23]). At present neither the targets that are essential for inhibition of cell proliferation by rapamycin nor the mechanism by which these targets mediate cell cycle arrest are completely understood.
In the present study an MCF7 breast cancer cell line was developed which can be induced to express a mutant form of 4EBP-1 in which five amino acids that are targets for phosphorylation have been changed to alanines [24]. Since the protein cannot be inactivated by phosphorylation it constitutively binds to and inhibits eIF4E. Thus expression of the mutant mimics the effects of rapamycin on 4EBP but does not affect the other targets of mTOR. Induction of the constitutively active 4EBP-1 leads to cell cycle arrest which correlates with loss of cyclin D1 expression and increased levels of the cyclin dependent kinase inhibitor p27Kip1. The increase in p27Kip1 is mediated, at least in part, by enhanced synthesis of the protein and corresponds to activation of the p27Kip1 mRNA 5'-untranslated region (UTR) and its ability to mediate cap-independent initiation of translation.
Results
Expression of constitutively active 4EBP-1 inhibits the proliferation of MCF7 cells
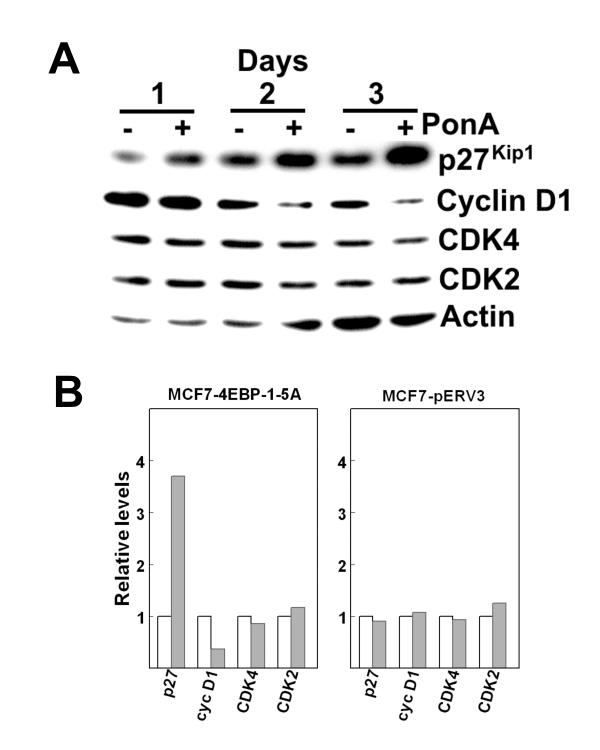
To test the effect of 4EBP-1 on the proliferation of human breast cancer cells, an expression vector encoding the constitutively active 4EBP-1 mutant (4EBP-1-5A) was stably transfected into MCF7 cells. The 4EBP-1-5A cDNA was placed under the control of a promoter that is responsive to the insect hormone ponasterone A (pon A), an ecdysone analog. In the absence of pon A an ecdysone receptor/VP16 fusion protein actively represses the promoter leading to very low basal levels of expression. In the presence of pon A the ecdysone receptor recruits coactivators that lead to elevated expression. The stably transfected MCF7 line (MCF7-4EBP-1-5A) was treated with pon A and at various times cells were harvested for analysis of 4EBP-1 expression by Western blotting (Fig. 1A). Several forms of 4EBP-1 are present in the stably transfected MCF7 cells. The slower migrating bands represent endogenous 4EBP-1 that is phosphorylated at various sites. The fastest migrating band corresponds to hypophosphorylated 4EBP-1. Treatment with pon A leads to a rapid increase in the intensity of the fastest migrating band, which corresponds to the 4EBP-1-5A mutated form of the protein which cannot be phosphorylated. In cells treated with pon A, the fastest migrating form of 4EBP-1 is the predominant band at all time points. Expression of this form of 4EBP-1 continues to increase for at least five days after treatment with pon A. In the control cells, which were not treated with pon A, phosphorylated, inactive 4EBP-1 is the predominant form of the protein (Fig. 1A). Treatment of untransfected MCF7 cells with pon A had no effect on expression or phosphorylation status of 4EBP-1 (data not shown). The expressed 4EBP-1-5A protein is active as determined by a large increase in the amount of 4EBP-1 bound to eIF4E in pon A treated MCF7-4EBP-1-5A cells (Fig. 1B).
Figure 1.
Expression of 4EBP-1-5A inhibits proliferation of MCF-7 cells. (A) MCF7-4EBP-1-5A cells were treated with or without pon A for up to 5 days as indicated. Cell extracts representing 2 × 105 cells were used for Western blotting with anti-4EBP-1. The lower band (arrow) represents endogenous, hypophosphorylated 4EBP-1 and 4EBP-1-5A induced by pon A. (B) MCF7-4EBP-1-5A cells were cultured in the presence or absence of pon A for 2 days. Cell extracts were prepared and equal amounts of protein were incubated with 7-methyl-GTP-Sepharose as described in Experimental Procedures. Bound eIF4E and associated 4EBP-1 were detected by Western blotting. (C) MCF7-4EBP-1-5A (left) or MCF-7 (right) cells were grown in the presence (open circles) or absence (closed circles) of pon A. Cell numbers were counted on days 1, 3 and 5 as indicated. Each point represents the mean (± standard error) of either 4 (left panel) or 3 (right panel) independent determinations of cell number.
Expression of constitutively active 4EBP-1, through inhibition of eIF4E activity, would be expected to slow the rate of proliferation of MCF7 cells. To test this, the growth of MCF7-4EBP-1-5A breast cancer cells was determined by counting cell number in cultures treated with or without pon A (Fig. 1C). In the absence of pon A, the cells continued to proliferate for at least five days. However, induction of 4EBP-1-5A by the addition of pon A almost completely blocked cell proliferation. There was no indication of cell death in the pon A treated cultures and pon A had no effect on the untransfected parental MCF7 cell line (Fig. 1C). These results indicate that expression of constitutively active 4EBP-1 leads to cell cycle arrest of MCF7 cells.
Expression of 4EBP-1-5A results in enhanced p27Kip1 expression and downregulation of cyclin D1
Elevated levels of eIF4E are associated with tumorigenic transformation and enhanced expression of specific cell cycle regulatory proteins such as cyclin D1. Expression of constitutively active 4EBP-1 should lead to a global decrease in cap-dependent translation which could lead to cell cycle arrest. However, it is also possible that constitutively active 4EBP-1 inhibits proliferation through specific effects on cell cycle regulatory proteins. Therefore, a number of G1 cell cycle regulatory molecules were examined in MCF7-4EBP-1-5A cells that had been treated with or without pon A. Addition of pon A altered the levels of two critical G1 regulatory proteins. There was a significant increase in the level of p27Kip1 observed at every time point up to three days after addition of pon A (Fig. 2A and 2B). At the same time there was a substantial decrease in the level of cyclin D1. No difference was observed in the protein levels of CDK4 or CDK2 between pon A treated and untreated cells. Treatment of MCF7-pERV3 cells, which express the ecdysone receptor/VP16 fusion protein but not the inducible constitutively active 4EBP-1 (see Methods), with pon A had no effect on the expression of any of these proteins (Fig. 2B).
Figure 2.
Expression of 4EBP-1-5A enhances expression of p27Kip1 and inhibits expression of cyclin D1. (A) MCF7-4EBP-1-5A cells were treated with or without pon A and harvested 1, 2 or 3 days after treatment for analysis of cellular proteins by Western blotting. Each lane represents proteins from 2 × 105 cells. The same membrane was used to detect p27Kip1, cyclin D1, CDK2, CDK4 and β-actin. (B) MCF7-4EBP-1-5A (left) or MCF7-pERV3 (right) cells treated with ponA (gray bars) or left untreated (open bars) for 3 days were analyzed as described in A. After Western blotting, protein levels were estimated by densitometry and normalized using β-actin as a standard.
Constitutively active 4EBP-1 does not alter p27Kip1 and cyclin D1 turnover rates
The results above suggest that cell cycle arrest of MCF7 cells in response to 4EBP-1-5A expression involves specific effects on expression of cyclin D1 and p27Kip1. The most plausible explanation is that the observed changes are mediated by altered rates of translation. However, others have shown that eIF4E overexpression enhances cyclin D1 mRNA transport from the nucleus to the cytosol [25] and it is possible that 4EBP-1 affects this process. Also, p27Kip1 expression is actually enhanced while 4EBP-1 would be expected to inhibit translation rates. Therefore it was of interest to examine the mechanism by which 4EBP-1-5A expression modulates p27Kip1 and cyclin D1 levels.
First, to determine if 4EBP-1 led to a change in degradation of either p27Kip1 or cyclin D1, turnover rates were estimated in MCF7-4EBP-1-5A cells that had been treated with or without pon A for 24 hours. Cycloheximide was added for various times and p27Kip1 and cyclin D1 levels were estimated by western blotting (Fig. 3). Neither the rate of p27Kip1 turnover nor that of cyclin D1 was significantly altered by pon A-induced expression of 4EBP-1-5A. Thus, modulation of p27Kip1 and cyclin D1 levels in response to expression of constitutively active 4EBP-1 does not appear to be mediated by changes in protein degradation.
Figure 3.
Effect of constitutively active 4EBP-1 on turnover of p27Kip1 and cyclin D1. MCF7-4EBP-1-5A cells were cultured in the presence (open circles) or absence (closed circles) of pon A for 24 hours at which time cycloheximide (100 μg/ml) was added. At the indicated time cells were harvested for analysis of p27Kip1 (left) or cyclin D1 (right) protein levels by Western blotting. Protein levels were then estimated by densitometry.
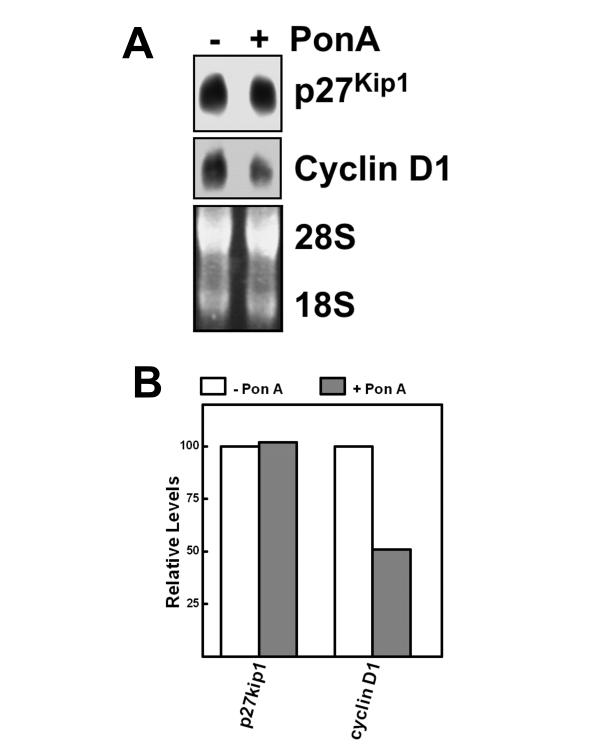
Expression of constitutively active 4EBP-1 does not alter p27Kip1 mRNA levels but decreases cyclin D1 mRNA levels
To examine whether 4EBP-1 could regulate p27Kip1 or cyclin D1 at the transcriptional level or through changes in message stability, the levels of p27Kip1 and cyclin D1 mRNA levels were examined by Northern blotting (Fig. 4A and 4B) There was no difference in mRNA levels encoding p27Kip1 after treatment with pon A. However, pon A treatment led to an approximately 50% decrease in the levels of cyclin D1 mRNA (Fig. 4B). This may contribute to the downregulation of cyclin D1 protein and is consistent with a previous report showing an increase in cyclin D1 mRNA in cells overexpressing eIF4E [26].
Figure 4.
Effect of constitutively active 4EBP-1 on p27Kip1 and cyclin D1 mRNA levels. (A) MCF7-4EBP-1-5A cells were cultured with or without pon A for 2 days and then the levels of mRNA encoding p27Kip1 (top panel) or cyclin D1 (middle panel) were determined by Northern blotting. Total RNA was detected by staining the gel with Sybr Gold prior to transfer (bottom panel). (B) Densitometric quantitation of the Northern blot data shown in A. White bars denote RNA levels in the absence of pon A and black bars denote synthesis in the presence of pon A.
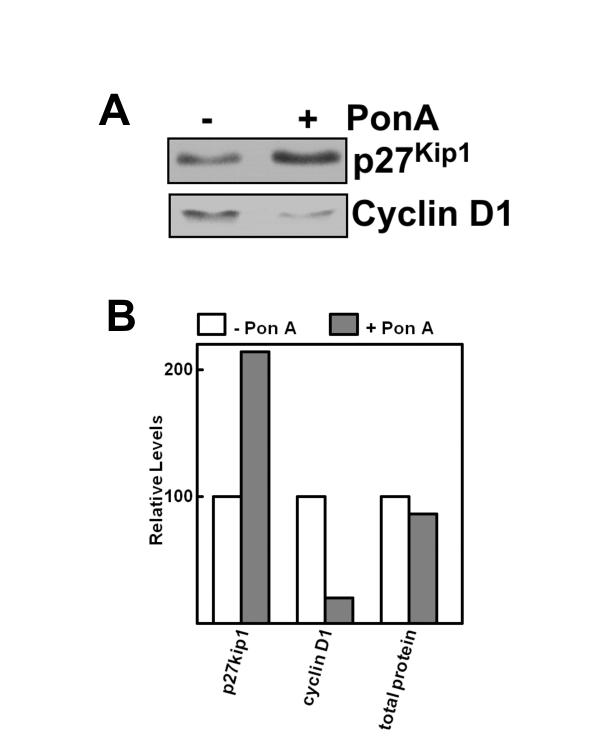
Expression of 4EBP-1-5A enhances de novo p27Kip1 protein synthesis
The observation that expression of constitutively active 4EBP-1 leads to increased p27Kip1, but does not significantly affect protein turnover or mRNA levels suggests that synthesis of p27Kip1 is affected. Therefore, the effect of 4EBP-1-5A on the de novo synthesis of p27Kip1 was tested by metabolic labeling with [35S]-labeled amino acids. After treatment of MCF7-4EBP-1-5A cells with or without pon A for 48 hours the cells were pulse labeled for 1.5 hours. Half of the cellular extract was used to immunoprecipitate cyclin D1 and the other half to immunoprecipitate p27Kip1 (Fig. 5A and 5B). The results demonstrate an increase in p27Kip1 protein synthesis of approximately two-fold in cells expressing 4EBP-1-5A. Since no changes in p27Kip1 mRNA levels or protein turnover were observed, the increased synthesis of p27Kip most likely reflects increased rates of translation. In the same experiment a substantial decrease in de novo synthesis of cyclin D1 was observed, which could be due, in part, to lower cyclin D1 mRNA levels (see Fig. 4). Interestingly, total protein synthesis, as determined by trichloroacetic acid (TCA) precipitation of total cellular proteins following pulse labeling, was decreased only about 15% by expression of constitutively active 4EBP-1 (Fig. 5B). This is consistent with reports that changes in the levels of active eIF4E have a much greater effect on the translation of specific classes of mRNA than on general translation rates [15,27].
Figure 5.
Effect of constitutively active 4EBP-1 on synthesis p27Kip1 and cyclin D1. (A) MCF7-4EBP-1-5A cells were cultured with or without pon A for 2 days. The cells were then pulse-labeled for 1.5 hours with 35S-amino acids in the presence of the proteasome inhibitor MG-132 to limit degradation of newly synthesized protein. Labeled cyclin D1 and p27Kip1 were immunoprecipitated, separated by SDS-PAGE, and detected by autoradiography. (B) The levels of newly synthesized p27Kip1 and cyclin D1, as detected in A, were estimated by densitometry. The level of total protein synthesis was determined by TCA precipitation of a portion of the same extracts prior to immunoprecipitation.
Expression of 4EBP-1-5A leads to inhibition of CDK2 activity
A critical factor in p27Kip1-mediated cell cycle arrest is inhibition of CDK2. 4EBP-1-5A expression did not significantly change CDK2 protein levels (see Fig. 2A). However, there was a substantial loss of CDK2 activity as determined by an immunoprecipitation kinase assay (Fig. 6). This loss of CDK2 activity correlated with an increase in the amount of p27Kip1 that co-immunoprecipitated with CDK2 (Fig. 6). At the same time, the association of p27Kip1 with CDK4 was dramatically decreased (Fig. 6). These results suggest that 4EBP-dependent cell cycle arrest involves p27Kip1-mediated inhibition of CDK2 kinase activity.
Figure 6.

Effect of constitutively active 4EBP-1 on p27Kip1 association with CDK2 and CDK4. MCF7-4EBP-1-5A cells were cultured with or without pon A for 2 days. Cell extracts were prepared as described in Experimental Procedures. A portion of each extract, containing equal amounts of protein, was subjected to immunoprecipitation with anti-CDK2 (left) or anti-CDK4 (right) antibodies. After immunoprecipitation a portion of the sample was used to detect p27Kip1 and CDK2 by Western blotting. For the CDK2 immunoprecipitate the remainder of the precipitate was used to assay CDK2 kinase activity with histone H1 as a substrate.
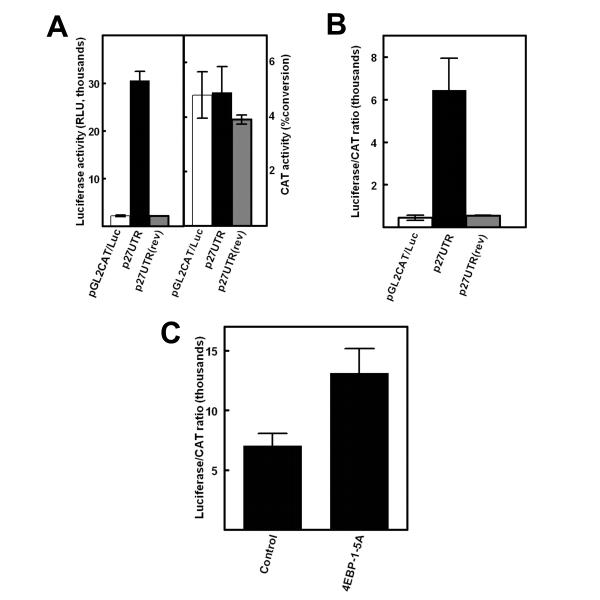
Expression of constitutively active 4EBP-1-5A enhances cap-independent translation through the p27Kip1 5'-UTR
A novel finding of the experiments above is that synthesis of endogenous p27Kip1 is enhanced under conditions that are expected to reduce cap-dependent translation. One possibility is that the p27Kip1 mRNA is translated through a cap-independent mechanism that does not require eIF4E. It was recently demonstrated that elements within a 217 nucleotide sequence from the 5'-UTR of the mouse p27Kip1 mRNA is able to mediate cap-independent translation of a reporter gene in NIH3T3 and D6P2T cells [28]. There is a high level of sequence homology between the mouse and human 5'-UTRs and the major transcriptional start site is also conserved [29]. Therefore it is likely the 5'-UTR of the human p27Kip1 message is also able to mediate cap-independent translation initiation. To test this, a 472 nucleotide fragment, representing the full-length human p27Kip1 5'-UTR derived from transcription initiation at the major start site, was inserted into the bicistronic expression vector pGL2CAT/Luc [30]. In this vector, the first cistron, encoding chloramphenicol acetyltransferase (CAT), is proximal to the mRNA's cap structure and therefore expected to be translated by the conventional cap-dependent translation mode. The second cistron, encoding luciferase, is downstream of the CAT stop codon and therefore must be translated by a cap-independent mechanism mediated by sequence elements inserted between the two cistrons. Bicistronic constructs without an insert or with the human p27Kip1 5'-UTR inserted in either the correct or reverse orientation were transiently expressed in MCF7 cells. The cells were harvested after one day and both CAT and luciferase activities were analyzed (Fig. 7A and 7B). Expression of CAT was nearly identical with all three constructs and was therefore independent of an insert between the two coding regions. In contrast, only the construct carrying the p27Kip1 5'-UTR insert in the correct orientation expressed a significant level of luciferase. The ratio of luciferase/CAT was enhanced approximately 12-fold by the human 5'-UTR (Fig. 7B). Thus the full-length human p27Kip1 5'-UTR is able to mediate cap-independent translation in a manner similar to that previously shown for the 217 nucleotide sequence of the mouse 5'-UTR [28].
Figure 7.
Constitutively active 4EBP-1 stimulates p27 Kip1 mRNA 5'-UTR-mediated cap-independent translation. (A) MCF7 cells were transfected with the bicistronic expression vector pGL2CAT/Luc, pGL2CAT/Luc-p27UTR in which the full-length human p27Kip1 5'-UTR is inserted between the two cistrons in the forward direction, or pGL2CAT/Luc-p27UTR(rev) in which the 5'-UTR is inserted in the reverse orientation. After one day, the levels of both luciferase (left) and CAT (right) activities were determined. (B) The relative levels of cap-independent translation from the transfections in A were estimated from the luciferase/CAT ratio. (C) The construct pGL2CAT/Luc-p27UTR was cotransfected with an expression vector encoding 4EBP-1-5A or an empty vector as a control. One day after transfection luciferase and CAT activities were assayed and the luciferase/CAT ratio was calculated. All data represent the mean of a minimum of three replicates plus or minus the standard error.
The effect of constitutively active 4EBP-1 on the activity of the p27Kip1 5'-UTR was determined by cotransfecting the bicistronic construct together with an expression vector encoding 4EBP-1-5A into MCF7 cells. Expression of 4EBP-1-5A led to a two-fold increase in the luciferase/CAT ratio (Fig. 7C). Interestingly, this is approximately the same level of increase that is observed in synthesis of endogenous p27Kip1 protein (see Fig. 5B).
Discussion
In normal mitogen-activated cells eIF4E activity is enhanced due to elevated transcription of the eIF4E gene, a phosphorylation-dependent increase in affinity for the 7-methylguanosine cap, and phosphorylation-dependent inactivation of the inhibitor protein 4EBP. In many tumor cells eIF4E levels are elevated and overexpression of the protein in normal cultured cells causes transformation. Elevated eIF4E levels enhance translation of specific mRNAs. Many of these mRNAs are involved in cell cycle progression. They have long GC-rich 5'-UTRs that can form extensive secondary structure and have been classified as "weak" mRNAs because they are translated inefficiently when eIF4E levels are limiting (reviewed in [15]). It has been proposed that when eIF4E levels are elevated these mRNAs can more effectively recruit eIF4E and its associated factors including eIF4G, a scaffold protein essential in recruiting the small ribosomal subunit, and eIF4A, a helicase essential for scanning through secondary structures in the 5'-UTR [15,27].
In the experiments described here we have inducibly expressed a constitutively active form of 4EBP-1 in the human breast cancer cell line MCF7. This leads to nearly complete inhibition of cell proliferation which is associated with an increase in binding of 4EBP-1 to eIF4E. This does not lead to a major drop in total protein synthesis as indicated by metabolic labeling with 35S-amino acids. However, there are major changes in expression of two cell cycle regulatory proteins, cyclin D1 and p27Kip1. Cyclin D1 levels decline to about 30% of that observed in uninduced cells and this appears to be due to a reduction in cyclin D1 mRNA levels and protein synthesis, since there was no evidence for changes in cyclin D1 turnover. The observed decline in cyclin D1 is expected since constitutively active 4EBP should reverse cyclin D1 expression mediated by eIF4E overexpression. Interestingly, others have demonstrated that eIF4E has an additional role in the nucleus. A portion of the cellular eIF4E is imported into the nucleus through a mechanism that requires the protein 4E-T [31]. In the nucleus eIF4E is localized to specific nuclear bodies and associates with promyelocytic leukemia protein (PML, [32]). Nuclear eIF4E appears to be involved in export of cyclin D1 mRNA to the cytosol [25]. Both 4E-T and PML interact with the same domain of eIF4E that binds 4EBP [32]. Thus expression of constitutively active 4EBP may block nuclear functions of eIF4E, one consequence being a reduction in cyclin D1 mRNA that can be translated in the cytoplasm.
In contrast to cyclin D1, p27Kip1 levels increase when 4EBP-1-5A is expressed. An interesting note is that the p27Kip1 mRNA has a long 5'-UTR that is 65% GC and is predicted to form stable secondary structures [29]. Thus, even though it shares these characteristics with the putative "weak" mRNAs that require elevated eIF4E activity for efficient translation, p27Kip1 responds in an opposite manner. Part of this can be explained by the fact that the p27Kip1 5'-UTR is able to promote cap-independent translation. When placed between the CAT and luciferase coding regions in a bicistronic mRNA, the 5'-UTR stimulates high level expression of the downstream coding region (luciferase). This property is shared by the highly homologous mouse p27Kip1 5'-UTR [28]. Coexpression of 4EBP-1-5A with the bicistronic construct containing the p27Kip1 5'-UTR leads to an approximately two-fold increase in cap-independent expression. This increase is similar to that observed for endogenous p27Kip1 in cells induced to express 4EBP-1-5A. The mechanism by which 4EBP-1-5A could mediate enhanced translation through the p27Kip1 5'-UTR is not presently known. One possibility is that binding of 4EBP-1 to eIF4E releases other initiation factors that are in limited supply but also required for cap-independent initiation.
Inhibition of cell proliferation by 4EBP-1-5A correlates with a large decrease in CDK2 activity. This is not due to a decline in CDK2 or cyclin E protein levels. Rather it corresponds to enhanced binding of p27Kip1 to the cyclin E/CDK2 complex. This is probably mediated by two separate mechanisms. First, downregulation of cyclin D1 leads to release of p27Kip1 sequestered by the cyclin D1/CDK4 complex. This is indicated by a large decrease in co-immunoprecipitation of p27Kip1 with CDK4 upon induction of 4EBP-1-5A. Second, de novo synthesis contributes to the pool of cellular p27Kip1 that is available to bind the cyclin E/CDK2 complex.
Given its role in control of cell proliferation and in tumorigenesis, there is interest in targeting the pathways that control eIF4E activity for cancer chemotherapy. Clinical trials using the rapamycin analog CCI-779 show promise for this strategy [22]. Rapamycin and its synthetic analogs block mTOR kinase activity leading to dephosphorylation of 4EBP and other mTOR substrates. MCF7 cells are sensitive to rapamycin analogs in the low nM range [33]. A potential problem with the use of rapamycin and its analogs is that some tumors are intrinsically resistant to its effects [33,34]. Resistance in mammalian cells has been associated with decreased expression of 4EBP-1 [35], mutations in the ATM protein [36], and low level expression of p27Kip1 due to enhanced turnover of the protein [37]. In addition, it has been demonstrated that fibroblasts from p27Kip1 knockout mice are more resistant to the effects of rapamycin than normal cells [37]. Thus p27Kip1 is an important mediator of cell growth inhibition in response to rapamycin. Many tumor cells express very low levels of p27Kip1 and this is correlated with poor prognosis. If rapamycin and its analogs are to be developed as useful antitumor agents, it will be important to determine how p27Kip1 levels affect responsiveness to these compounds.
Rapamycin and its analogs affect all of the downstream targets of mTOR, including p70 S6 kinase and other components of the translation machinery. The data presented here indicate that direct inhibition of eIF4E may be more effective and a more specific means of targeting tumor cells than treatment with rapamycin. It will be important to determine if direct inhibition of eIF4E is able to arrest the cell cycle in rapamycin resistant tumors. If so, it may be possible to develop reagents that target eIF4E activity, thereby bypassing the upstream signaling events in the pathway.
Methods
Cell culture
MCF7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, and 100 μg/ml streptomycin. The inducible MCF7 cell line expressing 4EBP-1-5A was established using the Complete Control™ Inducible Mammalian Expression System (Stratagene). First, MCF7 cells were transfected with the pERV3 vector encoding both the RXR protein and an ecdysone/glucocorticoid receptor fusion protein using GenePorter (Gene Therapy Systems). Stably transfected colonies were selected using G418 (400 μg/ml). These colonies were further screened for the ability to inducibly express luciferase from the pEGSH-luc construct in response to pon A in transient transfection assays. A responsive cell line derived in this manner (designated MCF7-pERV3) was further transfected with pEGSH-4EBP-1-5A, a construct in which a constitutively active form of 4EBP-1 is inserted into the pon A-inducible expression cassette of pEGSH. Cell clones were selected using hygromycin B (100 μg/ml). The resulting colonies were isolated, expanded, and tested for pon A-inducible expression of 4EBP-1-5A. A single stably transfected cell line designated MCF7-4EBP-1-5A was used in the experiments described.
Transfections and reporter assays
Cells growing in 35 mm plates were transfected using GenePorter. One day after transfection cells were lysed using reporter lysis buffer (Promega) and scraped from the dishes. Luciferase activity was determined using the Steady-Glo substrate (Promega) according to the manufacturer's protocol, using 50 μl of cellular extract. CAT enzyme activity was determined using 14C-labeled chloramphenicol as described previously [38].
Plasmid constructs
The human p27Kip1 5'UTR was amplified by PCR using primers CCCAAGCTTTCTCCCGGGTCTGCACGACCGCCTCT and CCCAAGCTTCTTCGTCAGCCTCCCTTCCAC. The PCR product was digested with Hind III and ligated into the Hind III site of pGL2CAT/Luc (a gift of R.E. Rhoads). To subclone 4EBP-1-5A into pEGSH the plasmid pcDNA3.1-4EBP-1-5A was digested with Hind III, the ends were filled using Klenow fragment, and then the insert was released by digesting with Xba I. The resulting fragment was ligated into pEGSH that had been digested with Eco RV and Xba I.
Cell proliferation assay
MCF7 and MCF7-4EBP-1-5A cells were treated with pon A (10 μM) or the same volume of ethanol for 5 days. The medium was changed every day. After 1, 3, or 5 days incubation, cells were harvested using trypsin and the cell number was determined using a hemacytometer.
Western blotting
Cells were harvested by trypsinization and the cell number was determined using a hemacytometer. The cells were pelleted and washed with phosphate-buffered saline (PBS). The cell pellets were resuspended in sodium dodecylsulfate (SDS)-sample buffer at 1 × 104 cells per μl and then sonicated to shear nucleic acids. Samples representing 2 × 105 cells were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilion P membranes. Membranes were probed with the appropriate antibodies. The p27Kip1 and eIF4E monoclonal antibodies were purchased from Transduction Laboratories. Anti-PHAS1 (4EBP-1) polyclonal antibody was purchased from Zymed. Anti-β-actin monoclonal antibody was purchased from Sigma. Antibodies against cyclin D1, CDK2, and CDK4 were purchased from Santa Cruz Biotechnology. Secondary horseradish peroxidase-linked anti-mouse or anti-rabbit IgG antibodies were purchased from Santa Cruz Biotechnology. Bands were visualized using the SuperSignal West Pico chemiluminescence detection system (Pierce).
Protein turnover analysis
MCF7-4EBP-1-5A cells were cultured in the presence or absence of pon A (10 μM) for 24 hours. The medium was then changed with readdition of pon A or an equal volume of ethanol and the protein synthesis inhibitor cycloheximide (100 μg/ml, Sigma) was added. Cells were incubated for the indicated times and then harvested in SDS-sample buffer as described above for Western blotting. Cyclin D1 and p27Kip1 proteins were detected by Western blotting and the levels of each protein were estimated by densitometry using a ChemiImager system (Alpha Innotech).
Cap-binding assay
MCF7-4EBP-1-5A cells were cultured in the presence or absence of pon A (10 mM) for 48 hours. Cell monolayers were washed with PBS three times and then lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 10 μg/ml leupeptin, 2 μg/ml Aprotinin, and 0.1 mM PMSF) by subjecting the cells to 3 cycles of freezing in liquid nitrogen and thawing in a 37°C water bath. The lysate was then centrifuged at full speed in a microcentrifuge at 4°C for 15 min. The supernatants were incubated with 50 μl of 7-methyl-GTP-Sepharose (Amersham Pharmacia Biotech, Inc.) at 4°C for 2 hours on a rotator. The resin was washed three times with lysis buffer and resuspended in 50 μl of SDS-sample buffer. Proteins were resolved by SDS-PAGE and transferred to Immobilon P membranes. eIF4E and 4EBP-1 were detected by Western blotting.
Immunoprecipitation, pulse-labeling and CDK2 kinase assays
Cell extracts were prepared using a lysis buffer consisting of 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% Tween-20, 1 mM DTT, 1 mM NaF, 0.1 mM Na3VO4, 10 μg/ml leupeptin, 2 μg/ml Aprotinin, and 0.1 mM PMSF. Extracts were briefly sonicated and then centrifuged at full speed in a microcentrifuge at 4°C for 15 min. Protein concentration was determined using the Bio-Rad protein assay solution.
For pulse-labeling, the cells were washed three times with DMEM lacking Met and Cys and then incubated in the same medium containing 80 μCi/ml of 35S-labeled Met and Cys (Tran35S-label, ICN) and the proteasome inhibitor MG-132 (10 μM, Calbiochem) for 1.5 hours. The cells were washed three times with ice-cold PBS and lysed as above. A portion of the supernatant was precipitated with TCA to estimate the level of total protein synthesis. The remainder of the cell extract was used for immunoprecipitation. Equal amounts of total protein from each extract (0.5 to 1 mg) were used for analysis. Cell lysates were incubated with 2 μg of rabbit polyclonal antibody overnight at 4°C on a rotator. Protein A-conjugated agarose beads (30 μl) were added, and the incubation continued for 1 hour on a rotator. The beads were pelleted and washed five times with lysis buffer. Next, 30 μl of 2X SDS-sample buffer was added to elute immunoprecipitated proteins. Labeled proteins were resolved by SDS-PAGE and detected by autoradiography.
For CDK2 kinase assays, lysates (100 μg protein) prepared as described above were used to immunoprecipitate CDK2 with a polyclonal antibody. The beads with the bound proteins were washed and then resuspended in 30 μl of kinase buffer [50 mM HEPES, pH 7.4, 2.5 mM EGTA, 10 mM β-glycerophosphate, 0.1 mM vanadate, 1 mM NaF, 1 mM DTT, 10 mM MgCl2, 20 μM ATP, 50 μCi γ-32P-ATP (7000 Ci/mmol, ICN), and 1 μg histone H1]. Reaction mixtures were incubated for 15 min at 30°C and stopped by adding the same volume of 2X SDS-sample buffer. The phosphorylated histone H1 was resolved by SDS-PAGE and detected by autoradiography using Kodak XAR-5 film.
For co-immunoprecipitation of p27Kip1 with CDK4 or CDK2, immunoprecipitations were performed as described above with either anti-CDK4 or anti-CDK2. The precipitated proteins were dissolved in SDS-sample buffer, separated by SDS-PAGE and then p27Kip1 and CDK2 were detected by Western blotting.
Authors' Contributions
HJ participated in manuscript preparation and design and performance of the experiments. JC constructed and characterized the bicistronic reporter constructs and aided with manuscript preparation. RM and WKM conceived of the project, directed the performance of the experiments, and prepared the final manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by NIH grant CA84325.
Contributor Information
Hong Jiang, Email: hjiang@usd.edu.
Jennifer Coleman, Email: jcoleman@usd.edu.
Robin Miskimins, Email: rmiskim@usd.edu.
W Keith Miskimins, Email: kmiskimi@usd.edu.
References
- Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. 1985;260:7843–7849. [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Flynn A, Waskiewicz AJ, Webb BL, Vries RG, Baines IA, Cooper JA, Proud CG. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J Biol Chem. 1998;273:9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- von Manteuffel SR, Dennis PB, Pullen N, Gingras AC, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Sonenberg N. The mRNA 5' cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol Cell Biol. 1992;12:1234–1238. doi: 10.1128/mcb.12.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A, Joshi B, Graff JR, Zimmer SG. CHO cells transformed by the translation factor eIF4E display increased c-myc expression but require overexpression of Max for tumorigenicity. Mol Cell Differ. 1994;2:247–371. [Google Scholar]
- De Benedetti A, Harris AL. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31:59–72. doi: 10.1016/S1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- Sorrells DL, Meschonat C, Black D, Li BD. Pattern of amplification and overexpression of the eukaryotic initiation factor 4E gene in solid tumor. J Surg Res. 1999;85:37–42. doi: 10.1006/jsre.1999.5653. [DOI] [PubMed] [Google Scholar]
- Li BD, McDonald JC, Nassar R, De Benedetti A. Clinical outcome in stage I to III breast carcinoma and eIF4E overexpression. Ann Surg. 1998;227:756–756l. doi: 10.1097/00000658-199805000-00016. discussion 761–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau D, Gingras AC, Pause A, Sonenberg N. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene. 1996;13:2415–2420. [PubMed] [Google Scholar]
- Martin ME, Perez MI, Redondo C, Alvarez MI, Salinas M, Fando JL. 4E binding protein 1 expression is inversely correlated to the progression of gastrointestinal cancers. Int J Biochem Cell Biol. 2000;32:633–642. doi: 10.1016/S1357-2725(00)00007-8. [DOI] [PubMed] [Google Scholar]
- Rinker-Schaeffer CW, Graff JR, De Benedetti A, Zimmer SG, Rhoads RE. Decreasing the level of translation initiation factor 4E with antisense RNA causes reversal of ras-mediated transformation and tumorigenesis of cloned rat embryo fibroblasts. Int J Cancer. 1993;55:841–847. doi: 10.1002/ijc.2910550525. [DOI] [PubMed] [Google Scholar]
- Nathan CA, Carter P, Liu L, Li BD, Abreo F, Tudor A, Zimmer SG, De Benedetti A. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene. 1997;15:1087–1094. doi: 10.1038/sj.onc.1201272. [DOI] [PubMed] [Google Scholar]
- Hidalgo M, Rowinsky EK. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene. 2000;19:6680–6686. doi: 10.1038/sj.onc.1204091. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci U S A. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC., Jr Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol. 2000;20:3558–3567. doi: 10.1128/MCB.20.10.3558-3567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E, Lazaris-Karatzas A, Karatzas C, Zhao X. Overexpressing eukaryotic translation initiation factor 4E stimulates bovine mammary epithelial cell proliferation. Int J Biochem Cell Biol. 2001;33:133–141. doi: 10.1016/S1357-2725(00)00089-3. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. mRNA 5' cap-binding protein eIF4E and control of cell growth. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Miskimins WK, Wang G, Hawkinson M, Miskimins R. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol Cell Biol. 2001;21:4960–4967. doi: 10.1128/MCB.21.15.4960-4967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J, Hawkinson M, Miskimins R, Miskimins WK. The major transcription initiation site of the p27Kip1 gene is conserved in human and mouse and produces a long 5'-UTR. BMC Mol Biol. 2001;2:12. doi: 10.1186/1471-2199-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Celle ML, Rhoads RE. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5' cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 2001;20:4547–4559. doi: 10.1093/emboj/20.16.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Toral-Barza L, Discafani C, Zhang WG, Skotnicki J, Frost P, Gibbons JJ. mTOR, a novel target in breast cancer: the effect of CCI-779, an mTOR inhibitor, in preclinical models of breast cancer. Endocr Relat Cancer. 2001;8:249–258. doi: 10.1089/10665270152530845. [DOI] [PubMed] [Google Scholar]
- Hosoi H, Dilling MB, Liu LN, Danks MK, Shikata T, Sekulic A, Abraham RT, Lawrence JC, Jr, Houghton PJ. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol Pharmacol. 1998;54:815–824. doi: 10.1124/mol.54.5.815. [DOI] [PubMed] [Google Scholar]
- Dilling MB, Germain GS, Dudkin L, Jayaraman AL, Zhang X, Harwood FC, Houghton PJ. 4E-binding proteins, the suppressors of eukaryotic initiation factor 4E, are down-regulated in cells with acquired or intrinsic resistance to rapamycin. J Biol Chem. 2002;277:13907–13917. doi: 10.1074/jbc.M110782200. [DOI] [PubMed] [Google Scholar]
- Beamish H, Williams R, Chen P, Khanna KK, Hobson K, Watters D, Shiloh Y, Lavin M. Rapamycin resistance in ataxia-telangiectasia. Oncogene. 1996;13:963–970. [PubMed] [Google Scholar]
- Luo Y, Marx SO, Kiyokawa H, Koff A, Massague J, Marks AR. Rapamycin resistance tied to defective regulation of p27Kip1. Mol Cell Biol. 1996;16:6744–6751. doi: 10.1128/mcb.16.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q, Bommakanti M, Miskimins WK. A mitogen-responsive promoter region that is synergistically activated through multiple signalling pathways. Mol Cell Biol. 1993;13:1796–1804. doi: 10.1128/mcb.13.3.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]