Abstract
β-Catenin is a multifunctional molecule that is activated by signaling through WNT receptors. β-Catenin can also enhance the transcriptional activity of some steroid hormone receptors such as the androgen receptor and retinoic acid receptor α. Androgens can affect nuclear translocation of β-catenin and influence its subcellular distribution. Using mammalian two-hybrid binding assays, analysis of reporter gene transcription, and coimmunoprecipitation, we now show that β-catenin binds to the androgen receptor ligand-binding domain (LBD) and modulates the transcriptional effects of TIF2 and the androgen receptor N-terminal domain (NTD). In functional assays, β-catenin bound to androgen receptor only in the presence of ligand agonists, not antagonists. β-Catenin binding to the androgen receptor LBD was independent of and cooperative with the androgen receptor NTD and the p160 coactivator TIF2, both of which bind to the activation function 2 (AF-2) region of the androgen receptor. Different mutations of androgen receptor helix 3 amino acids disrupted binding of androgen receptor NTD and β-catenin. β-Catenin, androgen receptor NTD, and TIF2 binding to the androgen receptor LBD were affected similarly by a subset of helix 12 mutations, but disruption of two sites on helix 12 affected only binding of β-catenin and not of TIF2 or the androgen receptor NTD. Mutational disruption of each of five LXXLL peptide motifs in the β-catenin armadillo repeats did not disrupt either binding to androgen receptor or transcriptional coactivation. ICAT, an inhibitor of T-cell factor 4 (TCF-4), and E-cadherin binding to β-catenin also blocked binding of the androgen receptor LBD. We also demonstrated cross talk between the WNT and androgen receptor signaling pathways because excess androgen receptor could interfere with WNT signaling and excess TCF-4 inhibited the interaction of β-catenin and androgen receptor. Taken together, the data show that β-catenin can bind to the androgen receptor LBD and modulate the effects of the androgen receptor NTD and TIF2 on transcription.
Prostate cancer is an androgen-dependent malignancy that usually undergoes clinical remission in response to hormone deprivation but eventually relapses after cancer cells develop the ability to grow in the presence of very low levels of androgen or in response to adrenal steroids (25, 26). Prostate cancer cells can develop resistance to hormone deprivation by augmenting signaling pathways activated by the androgen receptor (AR). For example, a substantial fraction of prostate cancers amplify the AR gene after treatment with hormone ablation (27, 33, 53). Hormone deprivation and exposure to antiandrogens can also lead to selection for AR gene mutations that alter response to antiandrogens and broaden the spectrum of ligand agonists (4, 8, 10, 30, 36, 45, 47, 48, 50, 58, 59).
Androgen receptor activity can also undergo pathological modulation in androgen insensitivity syndromes by disruption of androgen receptor interaction with coactivator proteins that mediate amplification of the androgen receptor transcription signal or by mutations in the coactivator proteins themselves (2, 39). In prostate cancer, mutations in the COOH-terminal region of the androgen receptor can affect the binding of p160 coactivator molecules (18). Hormone-resistant prostate cancer cells may also overexpress coactivators important for androgen receptor signaling (17). Activating mutations in β-catenin, a multifunctional molecule responsible for transcriptional activation in WNT signaling, are found in 5% of prostate cancers (3, 9, 34, 56). The potential significance of this finding was underscored by the finding that β-catenin interacts with the androgen receptor to enhance androgen receptor transcriptional activation and liberalize ligand specificity (52).
β-Catenin armadillo repeats, particularly repeat six, were shown to bind androgen receptor in a yeast two-hybrid interaction (62). Moreover, androgen caused nuclear localization of β-catenin and enhanced β-catenin-mediated transcription (35, 38). In this report, we show that β-catenin binds to the AF-2 region of the androgen receptor. Furthermore, binding is complementary, not competitive, with both the androgen receptor N-terminal domain (NTD) and the p160 coactivator TIF2 and results in amplified transcriptional signals.
MATERIALS AND METHODS
Plasmids and plasmid construction.
pGALD-H encodes the DNA binding domain of the Saccharomyces cerevisiae GAL4 protein (amino acid residues 1 to 147) and the androgen receptor (amino acid residues 624 to 919) (we renamed it GAL4/AR ligand-binding domain [LBD]), and pNLVP-hAR encodes the transcriptional activation domain of the herpes simplex virus VP16 protein (residues 411 to 456) and the full-length androgen receptor (we renamed it VP16/AR) (28). VP16/AR1-660 and its deletion mutants (Δ9-28, Δ179-199, Δ394-405, Δ429-439, and Δ339-499) code for the herpes simplex virus VP16 protein transactivation domain expressed as a fusion protein with the wild-type or deletion mutated androgen receptor NH2-terminal and DNA binding domain (DBD; residues 1 to 660) (20). GAL4/AR LBD-K720A and GAL4/AR LBD-V716R have been described (19). All other individual point mutations in GAL4/AR LBD were made with the Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Mutations were confirmed by nucleotide sequencing.
pCMVhAR, pCMVhARΔ14-150, pCMVhARΔ142-337, and pCMVhARΔ339-499 are expression vectors for wild-type full-length androgen receptor or androgen receptor deletion mutants. pCMVhAR-(507-919) and pCMVhAR-(507-919)-V716R encode the human androgen receptor DNA and ligand binding domains without and with a single mutation in helix 3 of the androgen receptor LBD, respectively. pCMVhAR-(1-503) encodes the N terminus of the androgen receptor (43). Individual mutations in pCMVhAR-(507-919) were made with the Quick Change site-directed mutagenesis kit as described above. The primers were the same as listed above.
pcDNA3HA-AR-LBD has been described before (19) and was used for in vitro translation of wild-type androgen receptor LBD peptides. β-Catenin and its individual LXXLL→AXXAA mutants were cloned in the pcDNA3.1 vector.
The mammalian two-hybrid system was used to assay in vivo protein-protein interactions. Because preliminary experiments indicated that VP16 and GAL4 fusions with the C terminus of β-catenin interfered with binding activity, we made constructs in which the fusion partner was added to the N terminus of β-catenin. To construct VP16/β-catenin, full-length β-catenin was amplified by PCR from pcDNA-β-catenin-Flag and cloned into the pCMX-VP16 vector with 5′-GAGCTCGGATCCAGCCCGATCGGTACCT-3′ and 5′-ACTAGTGGATCCTTAGGCGTAGTCGGGGA-3′ as the forward and reverse primers, respectively. The primers have BamHI sites at both ends, and β-catenin was cloned into pCMX-VP16 at the BamHI site. The orientation and reading frame of the resultant vector were confirmed by sequencing. To generate GAL4/β-catenin and Gal4/ARM-β-catenin expression vectors, full-length protein and the armadillo repeat region of β-catenin were amplified by PCR with 5′-GAGCTCGGATCCACGCCCGATCGGTACCT −3′ and 5′-ACTAGTGGATCCTTAGGCGTAGTCGGGGA-3′ for full-length β-catenin and 5′-ATCCCGGGAAACTATCAAGATGATGCAGAA-3′ and 5′-ATGGATCCTTGTGGCTTGTCCTCAGACATT-3′ for the armadillo repeat region. Both full-length β-catenin and the ARM region of β-catenin were cloned downstream of the GAL4 DNA binding sequences of the pCMX-GAL4 vector with the BamHI sites for full-length β-catenin and SmaI and BamHI sites for the armadillo repeat region. pCMX-VP16 and -GAL4 vectors were kindly provided by Ronald Evans.
K435A mutations in pcDNA-β-catenin and VP16/β-catenin were made with the Quick Change site-directed mutagenesis kit as described above. pCS2+/βS33A-VP16, which was renamed VP16/β-catenin(S33A), pCS2+/βΔN-VP16, which was renamed VP16/β-catenin(Δ15-552), and pCS2+/βS33AΔC-VP16, which was renamedVP16/β-catenin(Δ576-781) (S33A), were kindly provided by Andreas Hecht (21). In these constructs, β-catenin S33A carries four alanine substitutions in the glycogen synthase kinase 3β recognition site. pGEX-KT-β-catenin, pGEX-KT-β-catenin(1-257), and pGEX-KT-β-catenin(418-781) encode glutathione S-transferase (GST) fusion proteins of full-length, N-terminal, and C-terminal β-catenin. These vectors were kindly provided by David L Rimm, Yale University, New Haven, Conn. (40).
TIF2, TIF2.5, and TIF2m1,2,3 have been described (54). pFR-LUC, pM, pVP16, pSG5 and pBSK+ vectors were from Stratagene (La Jolla, Calif.). The Renilla null luciferase reporter was from Promega (Madison, Wis.). MMTV-luc has been described before (52). pcDNA3.1(+) was from Invitrogen (Carlsbad, Calif.). TCF4 plasmid and OT/OF reporter vectors were provided by Bert Vogelstein, Johns Hopkins Oncology Center, Baltimore, Md. pSVLGR encoding rat glucocorticoid receptor (GR) was kindly provided by Stoney Simons (National Institutes of Health, Bethesda, Md.). ICAT was kindly provided by T. Nakamura, Institute of Molecular and Cellular Biosciences, University of Tokyo, Tokyo, Japan.
Cell culture and transfection.
CV-1 cells were routinely maintained in modified Iscove's minimal essential medium supplemented with 5% fetal bovine serum. Twenty-four hours before transfection, cells were plated in 24-well microtiter plates (Falcon) at a cell density of 4 × 104 cells per well. Transfection was performed with Lipofectamine Plus reagent (Life Technologies, Inc.) with 10 ng of receptor-containing plasmid, 100 ng of reporter, 10 ng of Renilla null luciferase, and the indicated amounts of coregulators (TIF2, β-catenin, or androgen receptor N-terminal domain [NTD]). Unless otherwise stated in the figure legends, transfection was done with the total transfected DNA brought up to 300 ng/well with pBSK+ DNA.
In experiments with various amounts of coregulator cDNA plasmids, equimolar amounts of the same plasmid vector were cotransfected to control for artifacts of the vector DNA. The presence or absence of plasmid vector DNA added to equalize the total DNA added to the culture had no effect on binding or transcription assay. After incubation for 16 h, the cells were washed, and phenol red-free medium supplemented with 5% dextran charcoal-stripped fetal calf serum (CSS) containing hormones as indicated in the figure legends, or vehicle was added. The final concentration of vehicle ethanol was 0.1%. Hydroxyflutamide was a gift from Rudolph Neri, Schering Corporation. RU-486 was obtained from Sigma Chemical Co. After a further 24 h, cells were lysed in 1× passive lysis buffer (100 μl/well; Promega), and 30 μl of the cell lysates was used to assay for luciferase activity with the dual-luciferase assay system from Promega (Madison, Wis.) according to the supplier. The data were then normalized for the cotransfected Renilla activity. All samples were in triplicate, and each experiment described was repeated at least two times.
To determine the effects of β-catenin on TCF-dependent transactivation, CV-1 cells were plated in 24-well plates and cotransfected as described above with 10 ng of a TCF4 expression plasmid, 100 ng of the reporter plasmid OT (containing three TCF binding sites upstream of the firefly luciferase cDNA), and the indicated amount of β-catenin or VP16/β-catenin or their respective mutants, with or without increasing amounts of wild-type GAL4/AR LBD or GAL4/AR LBD(V889M) and 10 ng of Renilla null luciferase. Transfections were completed to 300 ng/well with pBSK+ DNA and performed in triplicate. Firefly and Renilla luciferase activities were measured, and TCF reporter activity was corrected for transfection efficiency by determining the ratio of firefly and Renilla luciferase activities.
GST pulldown assays.
In vitro interactions between the androgen receptor LBD and β-catenin were examined by the GST pulldown assay. GST, GST-β-catenin, GST-β-catenin(amino acids 1 to 257), and GST-β-catenin(418 to 781) fusion proteins were expressed in the CAG456 strain of Escherichia coli. CAG456 is a derivative of a protease-deficient line and was used to minimize degradation of the fusion protein products. GST fusion proteins were purified on glutathione-Sepharose beads (Amersham Pharmacia Biotech, Arlington Heights, Ill.). Androgen receptor LBD was translated in vitro with the TNT coupled transcription-translation system (Promega Corp., Madison, Wis.) in the presence of [35S]methionine. In the GST pulldown reactions, the translated proteins were incubated for 30 min at room temperature in the presence or absence of R1881 (10−6 M) before the addition of fusion proteins. The reactions were incubated for 2 h at room temperature in NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 0.5% Nonidet P-40), and the same amounts of ligand as above with occasional mixing. The beads were then washed five times with NETN buffer, and the bound 35S-labeled receptor was analyzed by sodium dodecyl sulfate (SDS) gel electrophoresis, followed by autoradiography.
Immunoprecipitation and Western blotting.
Antibodies were obtained as follows. TIF2 antibody was from Transduction Laboratories. Antibody to GRIP-1 (M-343) was a rabbit polyclonal antibody to amino acids 787 to 1129 of murine GRIP-1 from Santa Cruz Biotechnology. Antibody M-343 reacts with GRIP-1 of mouse, rat, and human origin by Western blotting. Anti-androgen receptor antibody (PG-21) was from Upstate Biotechnology. Androgen receptor (441) antibody was from Santa Cruz Biotechnology. Anti-β-catenin antibody was from Transduction Laboratories. GAL4 DBD (RK5C1) antibody was from Santa Cruz. Green fluorescent protein (GFP) antibody was from Clontech. β-Actin antibody was from Sigma.
For detection of the expression of GAL4/AR LBD mutants, COS-7 cells (1.5 × 106) were plated overnight in 100-mm2 dishes. Cells were transiently cotransfected with 3 μg each of wild-type GAL4/AR LBD or different mutants, together with 3 μg of GFP vector with a Lipofectamine method according to the manufacturer's protocol. For detection of the expression of β-catenin and LXXLL mutants, SKBR3 cells (2 × 106) were plated overnight in 100-mm2 dishes. Cells were transiently transfected with 3 μg each of β-catenin and LXXLL mutants together with GFP vector. Forty-eight hours after transfection, cells were collected, washed with ice-cold phosphate-buffered saline (PBS), and lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 120 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM Na3VO4, supplemented with protease inhibitor mixture [Roche Molecular Biochemicals]). The lysates were cleared by centrifugation (14,000 × g, 10 min at 4°C).
The total protein concentration of the extracts was measured with the DC protein assay (Bio-Rad, Hercules, Calif.); 30 μg of protein was loaded into a precast 4 to 20% Tris-glycine gel (Invitrogen), followed by electrophoresis, and subsequently transferred onto a nitrocellulose membrane (Bio-Rad). The membranes were blocked overnight at 4°C with 5% milk in PBST (PBS containing 0.1% Tween 20). The membranes were washed three times for 10 min each with PBST, followed by incubation at room temperature with a primary antibody in PBST containing 1% milk. Antibodies for Western blot detection were the Gal4 DBD antibody (1:500), β-catenin antibody (1:2,000), anti-GFP antibody (1:5,000), β-actin antibody (1:5,000), or antibody to α-tubulin (1:1,000). After an hour of incubation, the membranes were washed three times for 10 min each with PBST, followed by a secondary anti-mouse (GAL4, β-catenin, β-actin, and α-tubulin) or anti-rabbit (GFP) immunoglobulin-horseradish peroxidase antibody (1:5,000) incubation for another hour at room temperature. Signal detection was performed with Supersignal West Pico chemiluminescent substrate (Pierce).
Coimmunoprecipitation was done with transfected COS-7 cells (1.5 × 106) plated overnight in 100-mm2 dishes. Cells were transiently cotransfected with 5 μg each of androgen receptor, β-catenin, and TIF2 vectors with a Lipofectamine method. In some cases, COS-7 cells were transfected with β-catenin together with or without androgen receptor and/or TIF2. Sixteen hours after transfection, cells were treated with 10 nM R1881 or ethanol. Twenty-four hours later, cells were chilled on ice, washed with PBS, and lysed in lysis buffer. For the coimmunoprecipitation assay, 30 μl/sample of 50% slurry protein A/G agarose beads (Oncogene) was rinsed with PBS and incubated with 4 μg of either anti-androgen receptor antibody (PG-21) or anti-GRIP-1 antibody in 0.5 ml of lysis buffer at 4°C for 1 h. The beads were washed three times with ice-cold lysis buffer and then incubated with 500 μg of cell lysates at 4°C overnight. The beads were washed five times with 1 ml of ice-cold lysis buffer. The immune complexes were collected by adding 30 μl of 2× SDS sample buffer. Samples were separated by electrophoresis with 4 to 20% Tris-glycine gels and transferred to nitrocellulose. Western blot detection was done as described above. Antibodies for Western blot detection were anti-androgen receptor antibody 411 (1:500), β-catenin antibody (1:2,000), and TIF2 antibody (1:250).
Radioligand binding assay.
Androgen receptor binding was determined in COS-1 cells with a whole-cell binding assay (63). COS-1 cells were seeded into 24-well plates at a cell density of 6 × 104 cells/well in phenol red-free medium containing 5% CSS. Forty-eight hours later, cells were transfected with 100 ng/well of wild-type GAL4/AR LBD or various point mutants together with 400 ng/well of pBSK+ with the Lipofectamine method. Sixteen hours after transfection, cells were cultured in fresh phenol red-free medium containing 5% CSS and maintained for 24 h. Then cells were cultured in phenol red-free, serum-free medium containing increasing concentrations (0.15625 to 5 nM) of [3H]R1881 (methyltrienolone, 83.5 Ci/mmol; Perkin Elmer Life Sciences) in the absence or presence of a 200-fold molar excess of unlabeled R1881 (each with 0.14% ethanol) for 90 min at 37°C. The binding was terminated by the addition of 1 ml of ice-cold PBS. Cells were washed three times with ice-cold PBS, each for 3 min, and cells were collected in 0.1 N NaOH and 1% Triton X-100 (0.1 ml/well) for scintillation counting. The specific binding was calculated by subtracting the nonspecific binding (200-fold R1881) from the total [3H]R1881 binding. The binding capacity and affinity were determined by Scatchard plot analysis.
RESULTS
Androgen receptor LBD is necessary and sufficient for androgen receptor/β-catenin interaction.
To clarify the role of β-catenin in mediating ligand-dependent transcription, we utilized constructs containing the androgen receptor DBD and LBD and a fusion protein of herpes simplex virus VP16 and β-catenin. The VP16/β-catenin fusion protein can mediate R1881-dependent transcription initiated by binding the androgen receptor LBD to the androgen-responsive murine mammary tumor virus (MMTV) long terminal repeat (LTR) promoter (Fig. 1A). β-Catenin alone increased androgen-dependent transcriptional activity three- to fivefold, as we had previously reported (Fig. 1B) (52). Because of the magnitude of the effect, VP16/β-catenin was used in subsequent transfection experiments to study the androgen receptor/β-catenin interaction.
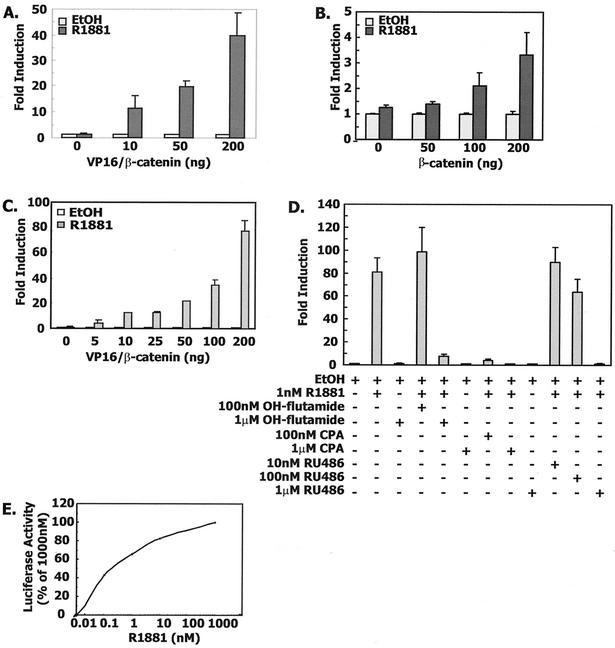
FIG. 1.
Agonist-dependent interaction of β-catenin with androgen receptor LBD. (A) VP16/β-catenin enhances androgen receptor LBD-mediated transactivation in CV-1 cells. Transient transfection of CV-1 cells was done with 10 ng of AR(507-919), 100 ng of MMTV-LUC, 10 ng of Renilla null luciferase reporter, and increasing amounts of VP16/β-catenin (0 to 200 ng/well) adjusted with VP16 expression vector to produce equimolar amounts of VP16 vectors. The total amount of DNA in each well was brought to 320 ng with pBSK+. (B) Similar experiment with β-catenin instead of VP16/β-catenin. EtOH, ethanol. (C) The androgen receptor LBD interacts with β-catenin in a dose-dependent manner. Mammalian two-hybrid assay in CV-1 cells with 100 ng of the GAL4-responsive reporter plasmid pFR-LUC, 10 ng of GAL4/AR LBD, and increasing amounts of VP16/β-catenin. Empty VP16 expression vector was used to maintain equimolar concentrations across all cultures. (D) The interaction of the androgen receptor LBD and β-catenin is agonist dependent, and antagonists can block the interaction. CV-1 cells were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, 10 ng of Renilla null luciferase reporters, and 50 ng of VP16/β-catenin as described above. Vehicle, R1881, and antihormones were added as described in Materials and Methods. CPA, cyproterone acetate. (E) Androgen receptor LBD interaction with β-catenin is proportional to the concentration of R1881. CV-1 cells were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, 10 ng of Renillla luciferase expression plasmid, and 50 ng of VP16/β-catenin and treated with increasing amounts of R1881.
To address more directly the interaction of the androgen receptor LBD with β-catenin, the VP16/β-catenin fusion construct was then used to mediate a mammalian two-hybrid interaction with a GAL4/AR LBD fusion protein. As shown earlier, expression of GAL4/AR LBD shows no induction of transcriptional activity, indicating the absence of AF-2 activity. However, when cells were cotransfected with VP16/β-catenin, the agonist R1881 induced GAL4-dependent transcription. Both binding and transcriptional activity were ligand dependent and proportional to the amount of VP16/β-catenin plasmid added to the culture (Fig. 1C). That the exogenous expression of VP16/β-catenin can rescue the agonist-dependent AF-2 activity in the androgen receptor LBD complements our previous findings that β-catenin can enhance androgen receptor-dependent transcription (52). This suggests a model in which binding of a ligand agonist to the androgen receptor LBD changes its conformation to form a surface corresponding to the AF-2 domain, which in turn accommodates coactivator binding (31, 42). Only about three- to fourfold induction of luciferase activity was observed with the reciprocal chimeras GAL4/β-catenin and VP16/AR-LBD, suggesting a preferential orientation for fusion protein interaction (data not shown).
Binding of β-catenin was dependent on agonist-mediated folding of the androgen receptor LBD around a ligand, since antiandrogens such as hydroxyflutamide and cyproterone acetate were unable to mediate binding, as was true for the antiprogestin RU-486, which can also act as an antiandrogen (Fig. 1D) (51). In addition, excess antagonists were able to compete with agonist occupancy of the receptor and disrupt the binding of VP16/β-catenin to the GAL4/AR LBD. Androgen receptor binding of β-catenin was mediated by physiologic concentrations of androgen and increased at a slower rate for R1881concentrations above 1 nM (Fig. 1E).
To ask if the N terminus of the androgen receptor is also involved in the interaction with β-catenin, we carried out the mammalian two-hybrid assay with GAL4/β-catenin and VP16/AR(1-660) or deletion mutants (Δ9-28, Δ179-199, Δ394-405, Δ429-439, and Δ339-499). The results showed that there was no detectable induction of the reporter with any of the AR(1-660) constructs compared with the VP16 control, suggesting that the N terminus of the androgen receptor, including the DNA binding domain of the androgen receptor, is not needed for the interaction with β-catenin (data not shown).
Interaction of androgen receptor LBD with β-catenin and androgen receptor NTD.
The androgen receptor NTD binds to the androgen receptor AF-2 region in the LBD via hydrophobic domains (19, 20). In the presence of ligand, full-length androgen receptor was able to mediate the two-hybrid interaction with GAL4/AR LBD and initiate GAL4-dependent transcription (Fig. 2A). Full-length androgen receptor was more active than VP16/β-catenin in this assay and was able to synergize with VP16/β-catenin to activate a very high level of transcription. Androgen receptor mutants ARΔ14-150 and ARΔ339-499 had reduced interaction with GAL4/AR LBD, to levels 9% and 31% of that of the wild-type androgen receptor, respectively. These two mutants had lost either the 23FQNLF27 or the 433WHTLF437 sequences that are required for interaction of the androgen receptor LBD and NTD (20).
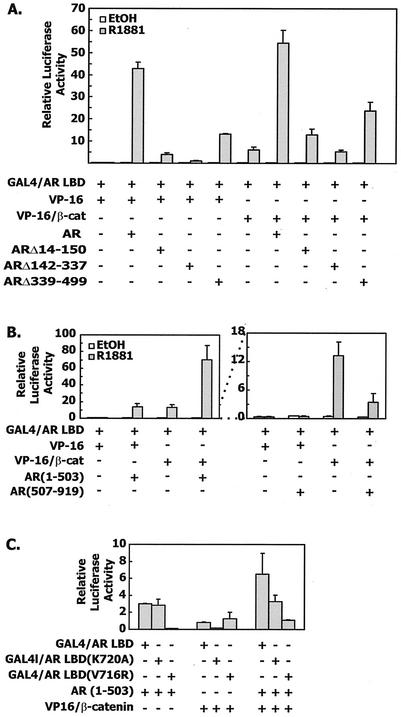
FIG. 2.
Synergistic effect of VP16/β-catenin and androgen receptor NTD on androgen receptor AF-2. (A) CV-1 cells were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, 50 ng of pCMVhAR or the indicated deletion mutants together with 50 ng of VP16/β-catenin or 30 ng of VP16 expression vector. (B) CV-1 cells were cotransfected as described for panel A except that pCMVhAR1-503 or pCMVhAR507-919 was used. (C) VP16/β-catenin and the androgen receptor NTD require different amino acids for binding to the androgen receptor LBD. CV-1 cells were transfected as described for panel B except that both wild-type and mutant GAL4/AR LBD (K720A and V716R) were used.
In the presence of VP16/β-catenin, GAL4-dependent transcription was still lower in the presence of these two deletion mutants than in that of the wild-type androgen receptor. However, there was a mild interaction of the androgen receptor deletion mutants and VP16/β-catenin that was proportional to the activities seen in the presence of VP16 alone, suggesting that the domains responsible for N/C interaction of the androgen receptor are required for the efficient interaction of androgen receptor and VP16/β-catenin. Deletion of the AF-1 function in ARΔ142-337 caused almost total loss of GAL4-dependent transcription in response to R1881. Even though ARΔ142-337 could still interact efficiently with the androgen receptor LBD (19), it could not activate the AF-2 function of the GAL4/AR LBD chimera. Indeed, cotransfection of ARΔ142-337 with VP16/β-catenin produced the same level of GAL4-dependent transcription as with VP16/β-catenin alone.
We next asked if the androgen receptor NTD was necessary and sufficient for the synergistic effects with VP16/β-catenin. AR(1-503) was cotransfected with the GAL4/AR LBD together with or without VP16/β-catenin. As expected, AR(1-503) mediated GAL4-dependent transcription (Fig. 2B). Moreover, cotransfection of AR(1-503) and VP16/β-catenin induced a synergistic effect on reporter expression. Since there was no direct interaction between AR(1-503) and VP16/β-catenin in the mammalian two-hybrid assay, we conclude that AR(1-503) and VP16/β-catenin might be able to bind to the androgen receptor LBD independently, resulting in a synergistic enhancement of transcription. As expected, the androgen receptor LBD alone had a dominant negative effect on the binding of GAL4/AR LBD and VP16/β-catenin, consistent with the interpretation that the androgen receptor LBD contained the β-catenin binding site (Fig. 2B, right panel).
We examined the relationship of the androgen receptor NTD and β-catenin binding to the androgen receptor LBD by assaying their interaction with two mutants of the androgen receptor LBD that contained single amino acid alterations in helix 3. GAL4/AR LBD (V716R) is able to bind ligand but does not interact with the androgen receptor NTD or the p160 coactivator TIF2 (19). We confirmed that this mutant had no interaction with the androgen receptor NTD but was still able to interact with VP16/β-catenin (Fig. 2C). In contrast, GAL4/AR LBD (K720A) reduced binding to the androgen receptor NTD by 50%, but abrogated binding to VP16/β-catenin. Androgen receptor LBDs with either V716R or K720A mutations are capable of agonistic binding with dihydrotestosterone that may have been enhanced by our use of R1881 (19). VP16/β-catenin and the androgen receptor NTD acted independently in the presence of the V716R and K720A mutant constructs, suggesting that the synergistic effects of β-catenin and the androgen receptor NTD were mediated by independent binding of each to the androgen receptor LBD (Fig. 2C, right side).
Interaction of androgen receptor LBD with β-catenin and TIF2.
Binding studies with the helix 3 point mutants demonstrated that there were differences between β-catenin and androgen receptor NTD binding to the androgen receptor LBD, consistent with our finding that the two interacted synergistically with the androgen receptor LBD. Since the androgen receptor NTD and the p160 coactivator TIF2 bind to similar regions of the androgen receptor LBD, we examined the interaction of β-catenin and the androgen receptor LBD in the presence of TIF2. We found that TIF2 was a potent activator of GAL4-dependent transcription, and therefore the mammalian two-hybrid experiments were done with TIF2 instead of a VP16/TIF2 hybrid (Fig. 3A). The mutant TIF2.5 has deletions of the entire N- and C-terminal regions of TIF2 to leave only the nuclear receptor-interacting domain (amino acids 624 to 869) and is unable to mediate transcription. TIF2.5 was inactive in the mammalian two-hybrid assay, as was TIF2m123, which has all three NR boxes mutated (LXXLL→LXXAA) to eliminate binding to nuclear receptors (Fig. 3) (54).
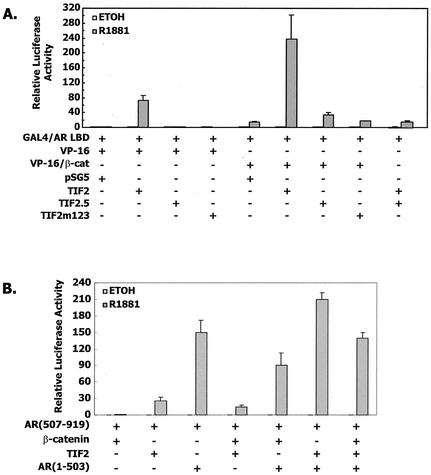
FIG. 3.
(A) Combined effect of VP16/β-catenin and TIF2 on androgen receptor AF-2. CV-1 cells were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, 50 ng of VP16/β-catenin, or 30 ng of VP16 expression vector, 50 ng of pSG5-TIF2, TIF2.5, or TIF2m1,2,3, or an equimolar amount of the pSG5 expression vector. (B) Combined effect of β-catenin, TIF2, and the androgen receptor NTD on androgen receptor-dependent transcription. CV-1 cells were cotransfected with 10 ng of pCMVhAR507-919, 100 ng of MMTV-LUC, 50 ng of β-catenin, 10 ng of pSG5-TIF2, 10 ng of AR(1-503), or an equimolar amount of the pSG5 expression vector.
TIF2.5 had a dominant negative effect on the interaction of TIF2 and the androgen receptor LBD on GAL4-mediated transcription, consistent with the interpretation that TIF2.5 bound to the androgen receptor but was unable to mediate formation of the transcriptional complex (Fig. 3, far right). However, neither TIF2.5 nor TIF2m1,2,3 blocked the effect of VP16/β-catenin. This result suggests that β-catenin bound to the androgen receptor independently of TIF2 and was not affected by the dominant negative TIF2.5 mutant.
The interaction of β-catenin with TIF2 and the androgen receptor NTD in affecting androgen receptor-dependent transcription is shown in Fig. 3B. In contrast to previous findings that β-catenin and SRC-1 had similar stimulatory effects on androgen receptor-dependent transcription, TIF2 was more potent at stimulating the MMTV-driven reporter construct. β-Catenin modulated TIF2 activation of androgen receptor-dependent transcription and enhanced the effect of the androgen receptor NTD. However, the effects of β-catenin on androgen receptor-dependent transcription in the presence of TIF2, the androgen receptor NTD, or both were small compared to the binding interactions that were assayed by GAL4-dependent transcription.
Androgen receptor, β-catenin, and TIF2 form a physical complex.
To demonstrate that androgen receptor, TIF2, and β-catenin formed a physical complex, we transfected COS-7 cells with expression plasmids for the three proteins and performed coimmunoprecipitation experiments. Figure 4 shows the results of these immunoprecipitation/Western blot experiments. Antibody to androgen receptor was able to immunoprecipitate TIF2 in the absence of ligand, but in a manner that was enhanced by the presence of R1881 (Fig. 4A). This is in agreement with reports that p160 coactivators such as TIF2 can interact with the conserved glutamine-rich region of the androgen receptor NTD in a ligand-independent manner (5). Antibody to androgen receptor also coimmunoprecipitated β-catenin in an androgen-dependent association, as we had previously published (52) (Fig. 4B).
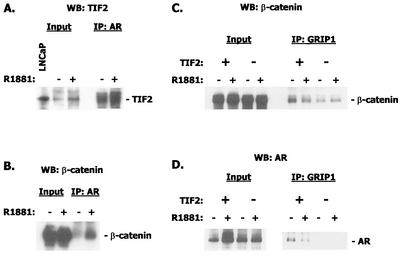
FIG. 4.
Immunoprecipitation/Western blotting of TIF2, androgen receptor, and β-catenin. (A) Immunoprecipitation (IP) with androgen receptor antibody and Western blotting (WB) with antibody to TIF2. The left panels show the input material. LNCaP cell extracts were included as an additional standard. (B) Immunoprecipitation with antibody to β-catenin and Western blot with androgen receptor antibody. (C) Immunoprecipitation with antibody to murine GRIP-1 fragment and Western blot with β-catenin. Note that the presence of exogenous TIF2 enhances the signal generated with endogenous TIF2. (D) Same as C but Western blot with androgen receptor antibody.
GRIP-1 antibody cross-reacts with TIF2 and pulls down β-catenin. Because of the abundance of TIF2 in COS-7 cells, we performed the experiment in the presence and absence of exogenous TIF2 expression vector to demonstrate specificity. The association of these two, presumably via binding to the androgen receptor, was not hormone dependent (Fig. 4C). We were also able to detect androgen receptor in the complex precipitated by GRIP-1 antibody. Similar to the association of GRIP-1 and β-catenin, this association was not androgen dependent but was augmented by exogenous TIF2 expression (Fig. 4D).
β-Catenin binding to helix 12.
The AF-2 domain of the androgen receptor LBD is formed after a ligand agonist binds to the LBD and helix 12 forms a surface with helices 3, 4, and 5 to which coactivator molecules and the androgen receptor NTD bind (31, 41). We examined the effects of individual amino acid alterations in helix 12 in order to develop a fingerprint of helix 12 amino acids that affect androgen receptor LBD binding and transcriptional activation when combined with VP16/β-catenin, TIF2, and the androgen receptor NTD.
Mutations in the androgen receptor C terminus were chosen based on known changes that caused androgen insensitivity syndromes (V889M, P892L, M895T, V903M, P904S, L907F, and G909R), as referenced in the androgen receptor database (http://ww2.mcgill.ca/androgendb/). Q902R is a mutation identified in prostate cancer (48). M894D is an androgen insensitivity mutation (49). Other helix 12 amino acids were each changed to alanine. The mutations were introduced into GAL4/AR LBD plasmids for the mammalian two-hybrid assay and into AR DBD+LBD(507-919) for the transactivation assay with an MMTV LTR reporter construct.
The constructs were assayed for their ligand binding activity to address concerns about the effect of reduced ligand binding on the interaction of the androgen receptor LBD with binding partners. The Kd of the wild-type androgen receptor construct was 0.083 nM, in agreement with a previously reported result with the same construct of 0.11 nM (19). Ligand binding of the different helix 12 mutants as reflected by the Kd ranged from 0.08 nM to 0.739 nM. Because ligand-binding affinity varied approximately 10-fold, binding and transcription studies with these constructs were done in the presence of 100 nM R1881. For each construct, we assayed either GAL4-dependent transcription or androgen receptor-dependent transcription in the presence of VP16/β-catenin, TIF2, or the androgen receptor NTD. Results of binding and transcription experiments are shown in Table 1.
TABLE 1.
Binding activity and transcriptional activity of helix 12 mutant androgen receptor constructsa
| Mutation | R1881 binding
|
Binding to GAL4/AR LBD (% of wild-type binding)
|
MMTV LTR transcriptional activity (% of wild-type activity)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Kd (nM) | Bmax (dpm/0.09 ml of cells) | VP16/β-catenin | TIF2 | VP16/AR (1-660) | VP16/β-catenin | TIF2 | AR (1-503) | |
| None | 0.0832 | 2,630 | ||||||
| V889M | 0.222 | 1,779 | −0.14 ± 0.18 | 15.76 ± 0.43 | 2.94 ± 0.36 | 2.09 ± 0.96 | 43.03 ± 4.96 | 9.24 ± 1.56 |
| F891A | 0.739 | 1,201 | 0.2 ± 0.10 | 0.05 ± 0.23 | 0.06 ± 0.07 | −0.01 ± 0.10 | −0.04 ± 0.04 | 0.24 ± 0.09 |
| P892L | 0.333 | 762 | −0.03 ± 0.02 | 0.05 ± 0.10 | 0 ± 0.05 | 0.17 ± 0.06 | 0.26 ± 0.05 | 0.45 ± 0.16 |
| E893A | 0.0512 | 2,120 | 10.26 ± 3.8 | 58.1 ± 7.36 | 9.68 ± 1.98 | 5.19 ± 1.35 | 27.31 ± 4.07 | 27.51 ± 7.47 |
| M894A | 0.133 | 1,893 | 31.15 ± 6.8 | 0.15 ± 0.13 | 34.6 ± 8.47 | 23.82 ± 3.27 | 2.16 ± 1.44 | 52.04 ± 7.89 |
| M894D | 0.166 | 2,326 | 17.38 ± 4.64 | −0.02 ± 0.26 | 4.13 ± 0.08 | 42.41 ± 0.24 | 0.16 ± 0.20 | 7.22 ± 4.49 |
| M895T | 0.222 | 1,454 | 9.63 ± 1.52 | 3.4 ± 1.02 | 16.09 ± 3.45 | 30.98 ± 4.98 | 31.2 ± 5.32 | 47.37 ± 4.58 |
| A896V | 0.166 | 1,944 | 114 ± 23.08 | 29.77 ± 9.23 | 43.43 ± 10.3 | 47.81 ± 7.05 | 40.79 ± 10.62 | 69.44 ± 8.94 |
| E897V | 0.111 | 2,170 | 207.6 ± 60.15 | 2.47 ± 1.97 | 0.13 ± 0.05 | 84.5 ± 13.7 | 0.38 ± 0.35 | 0.23 ± 0.04 |
| I898A | 0.166 | 2,149 | 18.09 ± 1.31 | 43.91 ± 6.61 | 0.68 ± 0.31 | 43.78 ± 13.57 | 73.45 ± 17.39 | 26.1 ± 5.31 |
| I899A | 0.222 | 2,163 | 15.7 ± 6.02 | 45.33 ± 5.02 | 15.98 ± 7.01 | 59.6 ± 30.4 | 85.8 ± 16.3 | 69.04 ± 1.2 |
| Q902R | 0.0739 | 2,691 | 74.31 ± 21.43 | 0.06 ± 0.30 | 1.03 ± 0.11 | 27.45 ± 1.53 | 0.44 ± 0.25 | 6.75 ± 1.79 |
| V903M | 0.0832 | 2,146 | 85.76 ± 29.0 | 26.04 ± 11.92 | 34.71 ± 5.37 | 66.99 ± 13.58 | 27.67 ± 4.56 | 64.64 ± 11.04 |
| P904S | 0.665 | 3,014 | 0.87 ± 0.37 | 1.64 ± 1.14 | 8.06 ± 0.73 | 6.4 ± 0.75 | 14.54 ± 0.50 | 46.44 ± 2.69 |
| K905A | 0.111 | 1,838 | 65.67 ± 25.25 | 46.27 ± 7.87 | 45.61 ± 9.68 | 66.8 ± 4.77 | 1.02 ± 0.14 | 13.61 ± 2.13 |
| L907F | 0.222 | 2,420 | 13.61 ± 7.21 | 40.32 ± 11.63 | 19.89 ± 4.4 | 22.54 ± 5.80 | 47.07 ± 6.49 | 52.67 ± 10.18 |
| G909R | 0.333 | 2,266 | 59.29 ± 9.41 | 17.24 ± 0.56 | 17.38 ± 1.04 | 31.02 ± 4.41 | 17.7 ± 2.12 | 48.08 ± 8.73 |
Two-hybrid interaction was determined with 10 ng of wild-type GAL4/AR LBD or the indicated point mutants and 50 ng of either VP16/β-catenin, TIF2, or VP16/AR(1-660), together with 100 ng of pFR-LUC and 10 ng of Renilla null luciferase reporters. Transactivation assay was determined by transfecting 10 ng of wild-type pCMVhAR507-919 or the indicated mutants, 100 ng of MMTV-LUC, and 50 ng of VP16/β-catenin, TIF2, or pCMVhAR1-503, together with 100 ng of pFR-LUC and 10 ng of Renilla null luciferase reporters. Cells were treated with 100 nM R1881 in phenol red-free medium for 24 h and assayed for firefly and Renilla luciferase activities as described in Materials and Methods. The induction by R1881 was determined for each mutant in the presence of individual factors, and the induction of wild-type GAL4/AR LBD and pCMVhAR507-919 were set at 100%, The percent binding activity to GAL/AR LBD mutants or percent transcriptional activity mediated by pCMVhAR507-919 mutants was then calculated as (induction of mutant − 1)/(induction of wild type − 1) × 100. Negative values indicate that the induction of the mutant by R1881 was less than 1 (the ethanol control value taken as 1).
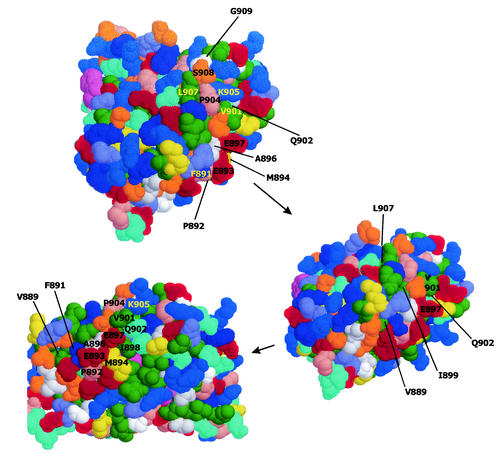
The locations of mutated amino acids are shown on the contour drawing of the androgen receptor in Fig. 5. Mutations at amino acids 891 and 892 abrogated all interactions of the androgen receptor LBD with VP16/β-catenin, TIF2, and the androgen receptor NTD. Adjacent to those two amino acids, mutation of V889 and E893 reduced binding and activity but more so for VP16/β-catenin and the androgen receptor NTD than for TIF2. M894 was also important for the interactions. M894A reduced the interaction with TIF2 more than with VP16/β-catenin and the androgen receptor NTD. However, M894D affected androgen receptor LBD interactions with both TIF2 and the androgen receptor NTD. Note in Fig. 5 that M894 is immediately adjacent to P892, a critical determinant of androgen receptor interaction with TIF2, VP16/β-catenin, and the androgen receptor NTD. M895 is barely exposed to the surface, lying beneath E893 and M894, and therefore was not labeled in Fig. 5. The M895T mutation reduced binding but preserved transcriptional activity in interactions with TIF2, VP16/β-catenin, and the androgen receptor NTD.
FIG. 5.
Space-filled model with Van der Waals radii of androgen receptor LBD viewed with CHIME according to the structure published by Matias et al. (31). The surface of the molecule shown at the tip was rotated upward and to the left to show the progressive views, as shown by the arrows. Amino acids mutated for the functional studies are indicated.
Other mutations were found to selectively affect interactions with one or another of VP16/β-catenin, TIF2, and the androgen receptor NTD. For example, E897V disrupted the interaction with TIF2 and the androgen receptor NTD but preserved the interaction with VP16/β-catenin. It is noteworthy that mutation of both amino acids 897 and 902 preserved the interaction with VP16/β-catenin but abrogated the interactions with TIF2 and the androgen receptor NTD. However, we found no mutation that abrogated the interaction of the androgen receptor LBD with VP16/β-catenin but preserved the interaction with TIF2 and the androgen receptor NTD, although E893A approximated this phenotype. Lastly, P904S, which appears on the model in Fig. 5 to be separated from the critical region around amino acids 891 and 892, also had a substantial effect on binding of TIF2, VP16/β-catenin, and the androgen receptor NTD and an effect on the transcriptional interaction of the androgen receptor LBD with VP16/β-catenin and TIF2. The expression levels of all the helix 12 mutant androgen receptor constructs were found to be similar by Western blotting (data not shown). Therefore, differences in activity were not due to differences in expression.
Analysis of androgen receptor-binding regions of β-catenin.
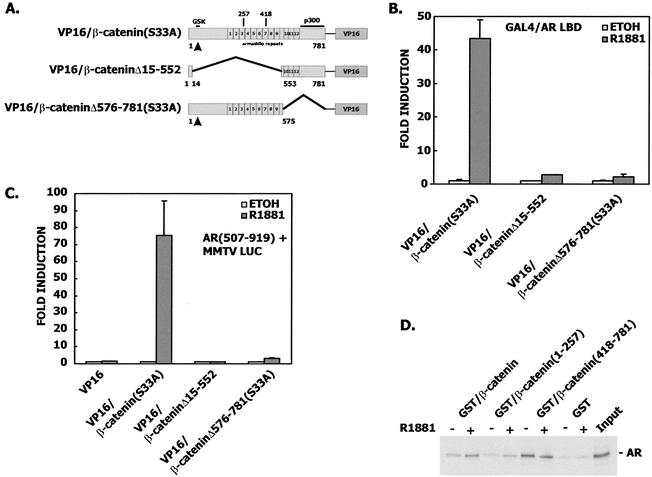
We also examined regions of β-catenin that were required for binding to the androgen receptor. Yang has recently shown that the armadillo repeats are required for yeast two-hybrid interaction between the androgen receptor and β-catenin (62). β-Catenin constructs comparing an S33A mutant full-length expression vector with constructs that removed either the N- or C-terminal armadillo repeats showed that either of the two large deletions nearly abrogated binding (Fig. 6A and B) and transactivation (Fig. 6C). The S33A mutant β-catenin was used as a positive control in these experiments because the Δ15-552 construct is deleted of the N-terminal phosphorylation sites that target β-catenin for degradation.
FIG. 6.
(A) Structural arrangement of VP16/β-catenin constructs in this experiment, demonstrating the location of armadillo repeats. (B) CV-1 cells in a mammalian two-hybrid assay were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, 10 ng of Renilla null luciferase reporter, and 50 ng of VP16/β-catenin(S33A), VP16/β-cateninΔN, or VP16/β-cateninΔC(S33A). (C) CV-1 cells in a transactivation assay were cotransfected with 10 ng of pCMVhAR507-919, 100 ng of MMTV-LUC, 50 ng of VP16/β-catenin(S33A), VP16/β-cateninΔN, VP16/β-cateninΔC(S33A), or equimolar amounts of VP16 empty vector. (D) In vitro interaction of the androgen receptor LBD with β-catenin. [35S]AR LBD was synthesized in vitro and incubated in the presence or absence of 1 μM R1881, with glutathione-Sepharose beads bound with GST-β-catenin, GST-β-catenin(1-257), or GST-β-catenin(418-781). Control beads contained GST alone. Beads were washed, and bound 35S-labeled proteins were eluted and analyzed by SDS-PAGE and autoradiography. The input lane contained 20% of the total radioactivity added for each reaction.
Mulholland also examined the physical interaction of androgen receptor and β-catenin by GST pulldown, showing that androgen receptor-GST interacted most strongly with the β-catenin armadillo repeats (35). Because androgen receptor-GST constructs often do not show ligand dependence (44), we employed β-catenin-GST constructs and showed that ligand-dependent androgen receptor pulldown occurred with full-length and C-terminal fragment of β-catenin (that included armadillo repeats 8 to 12) but to a lesser degree with the N-terminal region that included armadillo repeats 1 to 3 (Fig. 6D). The association of androgen receptor with GST-β-catenin and GST-β-catenin(1-257) was ligand responsive. The association of androgen receptor with GST-β-catenin(418-781) showed no ligand responsiveness. Therefore, there was demonstrable physical association of androgen receptor and portions of β-catenin even though these associations were inert for transcriptional activation. The binding of full-length GST-β-catenin to the androgen receptor that was enhanced by the presence of R1881 is quite similar to the coimmunoprecipitation of β-catenin and the androgen receptor that occurred in the absence of androgen but was enhanced by the presence of androgen (52).
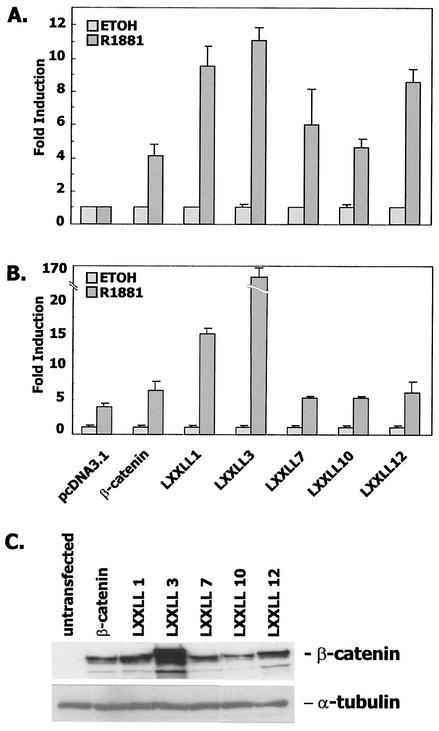
Nuclear receptor coactivators bind to the AF-2 region of androgen receptor via peptide motifs called NR boxes with the consensus sequence LXXLL. β-Catenin contains five such domains in armadillo repeats 1, 3, 7, 10, and 12. To determine the role, if any, of these repeats, we mutated each individually to AXXAA and then used the full-length β-catenin construct in binding (Fig. 7A) and transactivation (Fig. 7B) assays. Mutation of the LXXLL motifs in armadillo repeats 1 and 3 actually stimulated both binding to and transactivation of androgen receptor, although the effect may have been due to a large degree to overexpression of the LXXLL3 construct (Fig. 7C). Mutations of the other three LXXLL domains had very little effect on binding of VP16/β-catenin to GAL4/AR LBD or on the transcriptional interaction of β-catenin with androgen receptor.
FIG. 7.
LXXLL motifs in armadillo repeats of β-catenin are not required for the interaction with androgen receptor LBD. (A) CV-1 cells in a mammalian two-hybrid assay were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, 100 ng of wild-type β-catenin, individual LXXLL→AXXAA mutants, or an equimolar amount of pcDNA3.1 (B) CV-1 cells in a transactivation assay were cotransfected as described for panel A except that pCMVhAR507-919 and MMTV-LUC were used. (C) Western blot analysis of wild-type β-catenin and its mutants in SKBR3 cells, chosen for this analysis because they have very low endogenous expression of β-catenin.
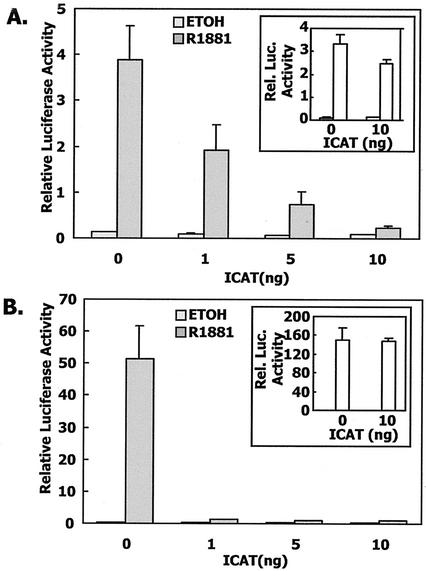
β-Catenin has a common mode of binding to diverse molecules such as E-cadherin and the adenomatous polyposis coli (APC) protein. These proteins bind to a groove formed by the armadillo repeats and are anchored to two lysine residues at positions 312 and 435 (13). The inhibitor of β-catenin and T-cell factor (TCF) 4 (ICAT) protein interferes with TCF4/β-catenin interaction by binding to the same region of the β-catenin armadillo repeats that interacts with TCF4 (W. Weis, personal communication) (46). We found that ICAT expression completely abrogated binding of β-catenin to the androgen receptor LBD in a mammalian two-hybrid assay and blocked the β-catenin-induced enhancement of androgen receptor transcriptional activation (Fig. 8). However, ICAT had no effect on the interaction of TIF2 and the androgen receptor LBD (Fig. 8, insets).
FIG. 8.
ICAT can block the functional and physical interaction of androgen receptor LBD and β-catenin. (A) CV-1 cells in a mammalian two-hybrid assay were cotransfected with 10 ng of GAL4/AR LBD, 100 ng of pFR-LUC, and 50 ng of VP16/β-catenin, together with increasing amounts of ICAT (0 to 10 ng/well). Equimolar amounts of empty vector were also included. As controls, triplicate cultures of CV-1 cells were also cotransfected as described above except that 50 ng of TIF2 was added instead of β-catenin (inset). (B) Transactivation assays. As in panel A except that pCMVhAR507-919 and MMTV-LUC were used in the transfection.
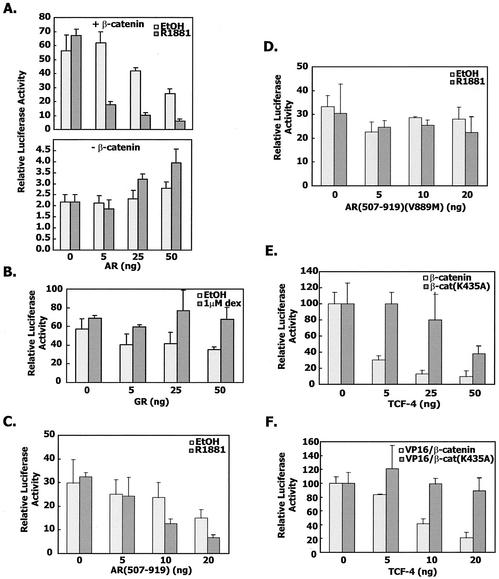
We also examined the degree of cross talk between TCF-dependent and androgen-dependent transcription. Androgen receptor interfered with the transcription from a TCF-responsive promoter in a manner that was to some degree hormone dependent. In the absence of β-catenin, activation of the TCF-responsive reporter was very low and not affected by the presence of androgen receptor (Fig. 9A) (35, 38). In contrast, the glucocorticoid receptor did not influence the activation of a TCF-responsive reporter by TCF-4 and β-catenin (Fig. 9B) (35). We also showed that the androgen receptor LBD was able to interfere with TCF-dependent transcription. However, when the AR(507-919)V889M mutant was titrated in the reporter assay, no effect on TCF-dependent transcription was seen (Fig. 9C and D and Table 1).
FIG. 9.
Cross talk between androgen receptor-dependent and TCF-dependent transcription. (A) CV-1 cells were cotransfected with increasing amounts of pCMVhAR, 10 ng of TCF4, and 100 ng of OT-LUC, together with or without 100 ng of β-catenin expression plasmid. Sixteen hours after transfection, cells were cultured in phenol red-free medium and treated with 1 nM R1881. At 24 h after induction, cells were harvested and assayed for firefly and Renilla luciferase activities as described in Materials and Methods. The average values were normalized for cotransfected Renilla, and the error bars represented the standard deviation within a given experiment. (B) CV-1 cells were cotransfected with increasing amounts of pSVLGR, 10 ng of TCF-4, 100 ng of OT-LUC, and 100 ng of β-catenin. Cells were treated as in A but with 100 nM dexamethasone instead of R1881. (C) TCF-4-dependent transcription from the OT reporter construct in the presence of the β-catenin expression plasmid and AR(507-919) in the presence or absence of R1881. (D) Same as in panel C except the AR(507-919) construct contained the V889M mutation that disrupted the interaction with β-catenin. (E) CV-1 cells were transfected with the MMTV-LUC reporter, AR(507-919), expression vector for β-catenin or β-catenin(K435A), and increasing amounts of the expression vector for TCF-4. (F) The experiment was carried out as in panel E except for the use of VP16/β-catenin fusion constructs instead of β-catenin.
Lastly, TCF-4 expression also interfered with β-catenin (Fig. 9E) and VP16/β-catenin (Fig. 9F) enhancement of androgen receptor-dependent transcription from the MMTV LTR. We also used the β-catenin mutant construct K435A as a negative control in these cross talk experiments. K435 is one of the residues critical for the interaction between β-catenin and molecules like TCF, E-cadherin, and APC (16, 57). We have found that the β-catenin K435A mutation preserves interaction with the androgen receptor even though it diminishes interaction with TCF-4 by more than 80% (L.-N. Song and E. P. Gelmann, unpublished observations). Therefore, the β-catenin K435A expression vector blunted the inhibitory effect of TCF-4 on androgen receptor-dependent transcription (Fig. 9E and F).
DISCUSSION
Our data show that β-catenin binds to the androgen receptor, activates transcription to some degree, and modulates the effects of TIF2 and the androgen receptor NTD. Of interest is the finding that the androgen receptor binding site for β-catenin is adjacent but not identical to the AF-2 binding sites for TIF2 and the androgen receptor NTD. Moreover, β-catenin binding to the androgen receptor is absolutely dependent on the presence of a ligand agonist, suggesting that the conformation induced by binding of a ligand agonist is required for β-catenin binding. p160 coactivator binding to nuclear receptors occurs at a specific groove that is created by the juxtaposition of helices 3, 4, 5, and 12 that is formed by swiveling of helix 12 over the ligand binding pocket (11, 32, 37, 42, 61). When a receptor antagonist is bound, helix 12 assumes a position unfavorable for coactivator binding. Mutational analysis has confirmed that helices 3, 4, 5, and 12 are the sites of activating factor 2 (AF-2) activity, which is important for p160 coactivator binding (42). Three-dimensional analysis of the androgen receptor LBD has shown that the androgen receptor has a similar structure and undergoes similar conformational changes in response to ligand binding as was demonstrated for other nuclear hormone receptors (31, 41).
β-Catenin appears to complex with the androgen receptor in proximity to the androgen receptor NTD or TIF2. β-Catenin binding depends on the integrity of K720, which also affects TIF2 binding to some degree, while the androgen receptor NTD and TIF2 require V716 (19). Both residues are in helix 3. Moreover, β-catenin depends on a subset of amino acids in helix 12 that are required for the androgen receptor NTD and TIF2 interactions. Two amino acids are required for androgen receptor LBD interactions with the androgen receptor NTD and TIF2 but have no effect on interactions with VP16/β-catenin. β-Catenin enhanced the binding and activity of the androgen receptor complex with TIF2 and the androgen receptor NTD. This implies but does not prove that β-catenin can bind to the androgen receptor at the same time that either of these other proteins binds to the androgen receptor.
Importantly, the dominant negative TIF2.5 mutant, which contains only the nuclear receptor interacting domain, completely abrogated TIF2 coactivation but had no effect on β-catenin interaction with the androgen receptor. The mechanism of β-catenin's interaction with these moieties is not yet clear. It may be that β-catenin acts to stabilize binding and formation of a transcriptional complex. Alternatively, β-catenin may recruit additional transcriptional machinery to the complex and thereby enhance the degree of transcriptional activation. Other explanations include stabilization of hormone binding and interference with putative binding of corepressors similar to those identified for the progesterone receptor (22). Taken together, the data suggested that ligand-bound androgen receptor forms a binding groove with helices 3, 4, 5, and 12 that binds to the armadillo repeats of β-catenin and allows β-catenin to bind in such a way that it can complement the transcriptional activity of both TIF2 and the androgen receptor NTD.
Interactions between the androgen receptor and WNT signaling pathways have important implications for our understanding of prostate cancer. We had initially found that a small fraction of prostate cancer specimens contain 5′ dominant activating mutations affecting the phosphorylation sites of β-catenin (9, 56). Since prostate cancers generally do not have a high rate of APC mutations and have not been found to harbor disruptions in WNT signaling components, the significance of the β-catenin mutations across the spectrum of prostate cancers was unclear (60). Recent findings have now provided strong evidence for cross talk between the WNT and androgen receptor signaling pathways.
Androgen can induce nuclear localization of β-catenin, and β-catenin can enhance androgen receptor signaling (35, 38, 52, 62). However, whereas β-catenin is a moderate activator of androgen receptor activity, in reporter assays ligand-bound androgen receptor decreased TCF-4 transcriptional activity, presumably by competing for β-catenin binding to TCF-4. Because β-catenin has functions in the cytoplasm as well as the nucleus, ligand binding of androgen receptor has the potential to alter the intracellular distribution of β-catenin and thereby either affect cell surface signals from E-cadherin or exert subtle effects on WNT signaling.
β-Catenin also interacts with the retinoic acid receptor-α. In the right cellular milieu, retinoids can increase β-catenin stability and localization to the cell surface (7). In other contexts, β-catenin can enhance transcriptional activation by retinoic acid receptor α and can compete with TCF for binding to β-catenin (12). β-Catenin does not interact with all nuclear hormone receptors and, specifically, has failed to show interaction with the retinoid X receptor, peroxisome proliferator-activated receptor, estrogen receptor, and glucocorticoid receptor (12, 35, 52).
Coactivator molecules contain a consensus peptide of the motif LXXLL, where L is leucine and X is any amino acid, in a region called the NR box, which binds to the groove in steroid hormone receptors (15, 55). Many of these proteins belong in the steroid receptor coactivator (SRC) family and bind to the AF-2 region in the C terminus of ligand-bound receptors such as the estrogen receptor. Other proteins, such as CREB-binding protein, complex with the androgen receptor near the DBD to participate in a multiprotein transcriptional complex (1, 22, 29). Hormone-dependent binding of p160 coactivators through the LXXLL motifs results in the activation of histone acetyltransferase activity, which plays a role in chromatin remodeling to allow active transcription of DNA (6, 14).
Elimination of any one LXXLL motif in β-catenin had no inhibitory effect on the interaction with androgen receptor. In fact, mutation of the two proximal LXXLL motifs stimulated both binding and coactivation of androgen receptor. Even though we cannot exclude the possibility that more than one LXXLL motif might be required for the interaction, we still predict that these LXXLL motifs are not crucial for the interaction with androgen receptor because our ongoing studies suggested that several residues outside armadillo repeats 1, 3, 7, 10, and 12 are required for the interaction (data not shown). Furthermore, Yang has argued that LXXLL motifs 7, 10, and 12 are not likely to be exposed on the surface of β-catenin and are probably not available for binding (62). Moreover, he has shown that the armadillo repeats, in particular repeat 6, are essential for androgen receptor binding.
The β-catenin armadillo repeats form a rigid contour of α-helices formed by the armadillo repeats. APC, TCF-4, and ICAT bind to β-catenin over a common region of the molecule's surface and anchor at two lysine residues called charge buttons (23, 24; W. Weis, personal communication). Since ICAT blocked androgen receptor binding to β-catenin, we propose that the AF-2 region of the androgen receptor can form a similar surface to interact with β-catenin even when it is bound to the androgen receptor NTD or TIF2. Further studies will be needed to elucidate the structural details of androgen receptor binding to β-catenin.
Acknowledgments
This work was supported by Public Health Service grants CA87855 and CA96854 from the NCI to E.P.G.
REFERENCES
- 1.Aarnisalo, P., J. J. Palvimo, and O. A. Janne. 1998. CREB-binding protein in androgen receptor-mediated signaling. Proc. Natl. Acad. Sci. USA 95:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, M., R. Takayanagi, A. Tomura, K. Imasaki, S. Kato, K. Goto, T. Yanase, S. Ikuyama, and H. Nawata. 2000. Androgen-insensitivity syndrome as a possible coactivator disease. N. Engl. J. Med. 343:856-862. [DOI] [PubMed] [Google Scholar]
- 3.Barker, N., P. J. Morin, and H. Clevers. 2000. The yin-yang of TCF/beta-catenin signaling. Adv. Cancer Res. 77:1-24. [DOI] [PubMed] [Google Scholar]
- 4.Barrack, E. R. 1996. Androgen receptor mutations in prostate cancer. Mt. Sinai J. Med. 63:403-412. [PubMed] [Google Scholar]
- 5.Bevan, C. L., S. Hoare, F. Claessens, D. M. Heery, and M. G. Parker. 1999. The AF1 and AF-2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 19:8383-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorklund, S., G. Almouzni, I. Davidson, K. P. Nightingale, and K. Weiss. 1999. Global transcription regulators of eukaryotes. Cell 96:759-767. [DOI] [PubMed] [Google Scholar]
- 7.Byers, S., M. Pishvaian, C. Crockett, C. Peer, A. Tozeren, M. Sporn, M. Anzano, and R. Lechleider. 1996. Retinoids increase cell-cell adhesion strength, beta-catenin protein stability, and localization to the cell membrane in a breast cancer cell line: a role for serine kinase activity. Endocrinology 137:3265-3273. [DOI] [PubMed] [Google Scholar]
- 8.Castagnaro, M., D. W. Yandell, B. Dockhorn-Dworniczak, H. J. Wolfe, and C. Poremba. 1993. Androgen receptor gene mutations and p53 gene analysis in advanced prostate cancer. Verh. Dtsch. Ges. Pathol. 77:119-123. (In German.) [PubMed]
- 9.Chesire, D. R., C. M. Ewing, J. Sauvageot, G. S. Bova, and W. B. Isaacs. 2000. Detection and analysis of beta-catenin mutations in prostate cancer. Prostate 45:323-334. [DOI] [PubMed] [Google Scholar]
- 10.Culig, Z., A. Hobisch, A. Hittmair, M. V. Cronauer, C. Radmayr, G. Bartsch, and H. Klocker. 1997. Androgen receptor gene mutations in prostate cancer. Implications for disease progression and therapy. Drugs Aging 10:50-58. [DOI] [PubMed] [Google Scholar]
- 11.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Easwaran, V., M. Pishvaian, Salimuddin, and S. Byers. 1999. Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr. Biol. 9:1415-1418. [DOI] [PubMed] [Google Scholar]
- 13.Eklof, S. K., S. G. Fridman, and W. I. Weis. 2001. Molecular mechanisms of beta-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC-beta-catenin complex. EMBO J. 20:6203-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman, L. P. 1999. Increasing the complexity of coactivation in nuclear receptor signaling. Cell 97:5-8. [DOI] [PubMed] [Google Scholar]
- 15.Glass, C. K., D. W. Rose, and M. G. Rosenfeld. 1997. Nuclear receptor coactivators. Curr. Opin. Cell Biol. 9:222-232. [DOI] [PubMed] [Google Scholar]
- 16.Graham, T. A., C. Weaver, F. Mao, D. Kimelman, and W. Xu. 2000. Crystal structure of a beta-catenin/Tcf complex. Cell 103:885-896. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, C. W., B. He, R. T. Johnson, O. H. Ford, J. L. Mohler, F. S. French, and E. M. Wilson. 2001. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 61:4315-4319. [PubMed] [Google Scholar]
- 18.Gregory, C. W., R. T. Johnson, Jr., J. L. Mohler, F. S. French, and E. M. Wilson. 2001. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 61:2892-2898. [PubMed] [Google Scholar]
- 19.He, B., J. A. Kemppainen, J. J. Voegel, H. Gronemeyer, and E. M. Wilson. 1999. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 274:37219-37225. [DOI] [PubMed] [Google Scholar]
- 20.He, B., J. A. Kemppainen, and E. M. Wilson. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275:22986-22994. [DOI] [PubMed] [Google Scholar]
- 21.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz, K. B., T. A. Jackson, D. L. Bain, J. K. Richer, G. S. Takimoto, and L. Tung. 1996. Nuclear receptor coactivators and corepressors. Mol. Endocrinol. 10:1167-1177. [DOI] [PubMed] [Google Scholar]
- 23.Huber, A. H., W. J. Nelson, and W. I. Weis. 1997. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90:871-882. [DOI] [PubMed] [Google Scholar]
- 24.Huber, A. H., and W. I. Weis. 2001. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105:391-402. [DOI] [PubMed] [Google Scholar]
- 25.Huggins, C., and C. V. Hodges. 1941. Studies on prostatic cancer; effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res. 1:293-297. [DOI] [PubMed] [Google Scholar]
- 26.Huggins, C., R. E. Stevens, and C. L. Hodges. 1941. Studies on prostatic cancer II. The effect of castration on clinical patients with carcinoma of the prostate. Arch. Surg. 43:209. [Google Scholar]
- 27.Koivisto, P., T. Visakorpi, and O. P. Kallioniemi. 1996. Androgen receptor gene amplification: a novel molecular mechanism for endocrine therapy resistance in human prostate cancer. Scand. J. Clin. Lab. Investig. Suppl. 226:57-63. [PubMed] [Google Scholar]
- 28.Langley, E., Z. X. Zhou, and E. M. Wilson. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270:29983-29990. [DOI] [PubMed] [Google Scholar]
- 29.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, and P. Chambon. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcelli, M., M. Ittmann, S. Mariani, R. Sutherland, R. Nigam, L. Murthy, Y. Zhao, D. DiConcini, E. Puxeddu, A. Esen, J. Eastham, N. L. Weigel, and D. J. Lamb. 2000. Androgen receptor mutations in prostate cancer. Cancer Res. 60:944-949. [PubMed] [Google Scholar]
- 31.Matias, P. M., P. Donner, R. Coelho, M. Thomaz, C. Peixoto, S. Macedo, N. Otto, S. Joschko, P. Scholz, A. Wegg, S. Basler, M. Schafer, U. Egner, and M. A. Carrondo. 2000. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J. Biol. Chem. 275:26164-26171. [DOI] [PubMed] [Google Scholar]
- 32.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi, Y., H. Uemura, K. Fujinami, K. Mikata, M. Harada, H. Kitamura, Y. Koizumi, and Y. Kubota. 2000. Fluorescence in situ hybridization evaluation of c-myc and androgen receptor gene amplification and chromosomal anomalies in prostate cancer in Japanese patients. Prostate 43:225-232. [DOI] [PubMed] [Google Scholar]
- 34.Morin, P. J. 1999. Beta-catenin signaling and cancer. Bioessays 21:1021-1030. [DOI] [PubMed] [Google Scholar]
- 35.Mulholland, D. J., H. Cheng, K. Reid, P. S. Rennie, and C. C. Nelson. 2002. The androgen receptor can promote beta-catenin nuclear translocation independently of adenomatous polyposis coli. J. Biol. Chem. 277:17933-17943. [DOI] [PubMed] [Google Scholar]
- 36.Newmark, J. R., D. O. Hardy, D. C. Tonb, B. S. Carter, J. I. Epstein, W. B. Isaacs, T. R. Brown, and E. R. Barrack. 1992. Androgen receptor gene mutations in human prostate cancer. Proc. Natl. Acad. Sci. USA 89:6319-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 38.Pawlowski, J. E., J. R. Ertel, M. P. Allen, M. Xu, C. Butler, E. M. Wilson, and M. E. Wierman. 2002. Liganded androgen receptor interaction with beta-catenin: nuclear co-localization and modulation of transcriptional activity in neuronal cells. J. Biol. Chem. 277:20702-20710. [DOI] [PubMed] [Google Scholar]
- 39.Quigley, C. A., A. De Bellis, K. B. Marschke, M. K. el Awady, E. M. Wilson, and F. S. French. 1995. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr. Rev. 16:271-321. [DOI] [PubMed] [Google Scholar]
- 40.Rimm, D. L., E. R. Koslov, P. Kebriaei, C. D. Cianci, and J. S. Morrow. 1995. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA 92:8813-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sack, J. S., K. F. Kish, C. Wang, R. M. Attar, S. E. Kiefer, Y. An, G. Y. Wu, J. E. Scheffler, M. E. Salvati, S. R. Krystek, Jr., R. Weinmann, and H. M. Einspahr. 2001. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc. Natl. Acad. Sci. USA 98:4904-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937. [DOI] [PubMed] [Google Scholar]
- 43.Simental, J. A., M. Sar, M. V. Lane, F. S. French, and E. M. Wilson. 1991. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 266:510-518. [PubMed] [Google Scholar]
- 44.Sun, Z., J. Pan, and S. P. Balk. 1997. Androgen receptor-associated protein complex binds upstream of the androgen-responsive elements in the promoters of human prostate-specific antigen and kallikrein 2 genes. Nucleic Acids Res. 25:3318-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, H., N. Sato, Y. Watabe, M. Masai, S. Seino, and J. Shimazaki. 1993. Androgen receptor gene mutations in human prostate cancer. J. Steroid Biochem. Mol. Biol. 46:759-765. [DOI] [PubMed] [Google Scholar]
- 46.Tago, K., T. Nakamura, M. Nishita, J. Hyodo, S. Nagai, Y. Murata, S. Adachi, S. Ohwada, Y. Morishita, H. Shibuya, and T. Akiyama. 2000. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 14:1741-1749. [PMC free article] [PubMed] [Google Scholar]
- 47.Taplin, M. E., G. J. Bubley, Y. J. Ko, E. J. Small, M. Upton, B. Rajeshkumar, and S. P. Balk. 1999. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 59:2511-2515. [PubMed] [Google Scholar]
- 48.Taplin, M. E., G. J. Bubley, T. D. Shuster, M. E. Frantz, A. E. Spooner, G. K. Ogata, H. N. Keer, and S. P. Balk. 1995. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N. Engl. J. Med. 332:1393-1398. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J., F. Saatcioglu, O. A. Janne, and J. J. Palvimo. 2001. Disrupted amino- and carboxyl-terminal interactions of the androgen receptor are linked to androgen insensitivity. Mol. Endocrinol. 15:923-935. [DOI] [PubMed] [Google Scholar]
- 50.Tilley, W. D., G. Buchanan, T. E. Hickey, and J. M. Bentel. 1996. Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin. Cancer Res. 2:277-285. [PubMed] [Google Scholar]
- 51.Tovar, A., A. Sanchez-Capelo, J. D. Galindo, A. Cremades, and R. Penafiel. 1997. Antiandrogenic effect of RU-486 in the mouse kidney. Int. J. Biochem. Cell Biol. 29:361-366. [DOI] [PubMed] [Google Scholar]
- 52.Truica, C. I., S. Byers, and E. P. Gelmann. 2000. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 60:4709-4713. [PubMed] [Google Scholar]
- 53.Visakorpi, T., E. Hyytinen, P. Koivisto, M. Tanner, R. Keinanen, C. Palmberg, A. Palotie, T. Tammela, J. Isola, and O. P. Kallioniemi. 1995. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 9:401-406. [DOI] [PubMed] [Google Scholar]
- 54.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 56.Voeller, H. J., C. I. Truica, and E. P. Gelmann. 1998. Beta-catenin mutations in human prostate cancer. Cancer Res. 58:2520-2523. [PubMed] [Google Scholar]
- 57.von Kries, J. P., G. Winbeck, C. Asbrand, T. Schwarz-Romond, N. Sochnikova, A. Dell'Oro, J. Behrens, and W. Birchmeier. 2000. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol. 7:800-807. [DOI] [PubMed] [Google Scholar]
- 58.Wallen, M. J., M. Linja, K. Kaartinen, J. Schleutker, and T. Visakorpi. 1999. Androgen receptor gene mutations in hormone-refractory prostate cancer. J. Pathol. 189:559-563. [DOI] [PubMed] [Google Scholar]
- 59.Wang, C., and T. Uchida. 1997. Androgen receptor gene mutations in prostate cancer. Nippon Hinyokika Gakkai Zasshi 88:550-556. (In Japanese.) [DOI] [PubMed]
- 60.Watanabe, M., H. Kakiuchi, H. Kato, T. Shiraishi, R. Yatani, T. Sugimura, and M. Nagao. 1996. APC gene mutations in human prostate cancer. Jpn. J. Clin. Oncol. 26:77-81. [DOI] [PubMed] [Google Scholar]
- 61.Westin, S., R. Kurokawa, R. T. Nolte, G. B. Wisely, E. M. McInerney, D. W. Rose, M. V. Milburn, M. G. Rosenfeld, and C. K. Glass. 1998. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature 395:199-202. [DOI] [PubMed] [Google Scholar]
- 62.Yang, F., X. Li, M. Sharma, C. Y. Sasaki, D. L. Longo, B. Lim, and Z. Sun. 2002. Linking beta-catenin to androgen-signaling pathway. J. Biol. Chem. 277:11336-11344. [DOI] [PubMed] [Google Scholar]
- 63.Yarbrough, W. G., V. E. Quarmby, J. A. Simental, D. R. Joseph, M. Sar, D. B. Lubahn, K. L. Olsen, F. S. French, and E. M. Wilson. 1990. A single base mutation in the androgen receptor gene causes androgen insensitivity in the testicular feminized rat. J. Biol. Chem. 265:8893-8900. [PubMed] [Google Scholar]