Abstract
We have designed a modified version of the Dam identification technique and used it to probe higher-order chromatin structure in Saccharomyces cerevisiae. We fused the bacterial DNA methyltransferase Dam to the DNA-binding domain of TetR and targeted the resulting chimera to Tet operators inserted in the yeast genome at the repressed locus HML. We then monitored the methylation status of HML and other sequences by a quantitative technique combining methylation-sensitive restriction and real-time PCR. As expected, we found that TetR-Dam efficiently methylated HML in cis. More strikingly, when TetR-Dam was present at HML, we observed increased methylation in the III-L subtelomeric region but not in intervening sequences. This effect was lost when the HML silencers were inactivated by mutations. When the HM silencers and the Tet operators were transferred to a plasmid, strong methylation was clearly observed not only in the III-L subtelomeric region but also at other telomeres. These data indicate that HM silencers can specifically associate with telomeres, even those located on different chromosomes.
Eukaryotic genomes are divided into functional compartments in which transcription is potentially active (euchromatin) or repressed (heterochromatin), and these compartments correspond to distinct physical domains of the nucleus. Consequently, there is a correlation between the position of a gene in the nucleus and its transcriptional activity. Relocation into a heterochromatic domain usually correlates with transcriptional repression (this has been well described for the Drosophila gene brown, for instance [10, 13]). In contrast, enhancers, which stimulate gene expression, seem to act in part by excluding genes from heterochromatin (18). What determines the spatial positioning of genes within the nucleus and the functional consequences of heterochromatin proximity are therefore important questions to address.
Saccharomyces cerevisiae has proved to be a powerful tool with which to study the role of heterochromatin in gene repression. Extensive work in S. cerevisiae has shown that the mating type loci HML and HMR are kept transcriptionally repressed by cis-acting elements called silencers (24). The silencers act by recruiting a complex of proteins called Sir (silent information regulator) proteins (35). This complex then spreads along the nucleosomal fiber through multiple interactions with histone tails (21, 22).
The telomeres play an important role in silencer activity in S. cerevisiae (40). Mutations that disrupt telomere structure and/or function greatly impair silencer function (15, 26, 31-33). Silencers also become less active when moved away from the telomeres along the same chromosome (30). These results, together with microscopy analyses, have led to a “reservoir” model in which telomere clusters are thought to constitute a subnuclear compartment that sequesters limiting silencing factors, including the Sir proteins (19, 30, 36). In accordance with this model, tethering of a weak silencer to the nuclear periphery, in which the telomeres are clustered, facilitates its repression (2). Relocation to the peripheral compartment probably does not cause repression per se (43). Rather, it appears to provide a high local concentration of silencing factors (2) and/or to prevent the switch to an active state (15). One prediction of the model is that silencers may have to associate with the telomeric compartment, at least transiently, to establish silencing. However, direct telomere-silencer interactions have not yet been evidenced.
In this work we asked whether silencers can physically interact with telomeres. This question cannot be addressed by microscopy because the distance between the mating type silencers and their proximal telomeres is below the resolution limit of this technique. We therefore designed an in vivo system in which a DNA methyltransferase is targeted to a silencer and the methylation of telomeric sites is precisely measured. Our data show that HML silencers preferentially associate with their proximal telomere, III-L. However, when the same silencers are removed from their natural location and placed on a plasmid, this specificity is lost, and the silencers can associate with any telomere. Therefore, silencers have the inherent capacity to interact in trans with telomeres, but intrachromosomal constraints appear to restrain their association mainly to their proximal telomere. We propose that telomeres play an active role in silencer-mediated silencing by addressing silencers into a nuclear repressive compartment.
MATERIALS AND METHODS
Plasmid constructions.
Molecular biology manipulations were performed as described previously (39). The S. cerevisiae SURE2 strain (Stratagene), grown at 30°C, was used for plasmids containing TetO112. All constructs were verified by DNA sequencing.
The TetR open reading frame was PCR amplified off plasmid p6501 (a generous gift of F. Feuerbach and U. Nehrbass). NotI and EcoRI restriction site were inserted at the end of the upstream and downstream primers TetRa (AATTCGCGGCCGCGCCCTTGCTCACCATGGACC) and TetRb (AGTTGGAATTCAGATCTCGCTCTAGAACTAGTGGA) (in all sequences, restriction sites designed in oligonucleotides for cloning purposes are shown in boldface) (Table 1). The PCR product was then cloned into EcoRI- and NotI-cut pCmycDam (45). Then a BglII-SalI fragment containing the genes for TetR, Myc, and Dam was inserted after the GAL1/10 promoter at the BamHI and XhoI sites of pESC-HIS (Stratagene), resulting in plasmid pTetRDam.
TABLE 1.
Coordinates of the primers used for quantitative PCR
| GATC no. | Upstream primer (5′→3′) | Downstream primer (5′→3′) |
|---|---|---|
| 1 | TATGTGATGATTCGCTTGGAAGGG | GCAATTTCATCTACAGGCTTGGAGG |
| 2 | CGAGAAATTCGGTGACTCTAAGGC | GCAGAGAACAGTTGCTTATGCG |
| 3 | GCTTAAACGAAGAATACCAGAAGCACG | CATAGTCAGGAATCGCGACACTAGC |
| 4 | CTTACTTGTTGCTGCTCCCTCTCC | CTTGAAGGGAGAGAGTGATGTCTCCG |
| 5 | CTTATCAGAGCATAGTTGGTCAGC | GCATCACAATATACAGTTAATGCCACCTG |
| 6 | TATTGTGATGCATCTCATGGAGC | GTATTCTGCCTCAGTAGATGG |
| 7 | CCTTGTCTATTAGTTCCGGGTC | CTCATGAAGGTGTTATCGCTGC |
| 8 | TCACTGCTCTTTTCTGTGTTCC | GACCAAACTTACGATCTTTGG |
| 9 | GCATATATATAATTAAGCGGGAGC | CAATATACTTACAGAGACCTC |
| 10 | GAAACCGTCTTCCTCGGATACG | CAGATTGAGCGACTTAGAAGGTGCTGG |
| 11 | GTTGTATCCTTGATAGCTCCTTATCCG | GTACGGCTACTGACCTAGATACTCAGG |
| 12 | ATGAGGATTGAAATTGTTCTTGG | AACAGAAAGAAAAGGAGCACGAGGC |
| 13 | CCAAATCAACCTTTCTAGGC | CTTGCCCATGTTTAAGAAGAGG |
| 14 | CTCTGGCTTTCAAAATGATAGCG | GAGTTTAAGGAAGAGGATAACGCATCCG |
| 15 | CGAGGAGCTGATCAAGGACCAGG | AATATGCAGTACCATTCCGCTC |
| 16 | CTGAGGTGAACACACCCACGCC | GCAGATTAACTTTGCTACGAGAGGG |
| 17 | GTAAGGTAGAGAGCCCTTCCG | GCCTTCGATTGAACATCCTGCCAG |
| 18 | TCCAATTCCAAATTCTAGGGACG | GTCAATGAGTAGTAGATAGTAAAGCC |
| 19 | TCAGTGCCCAACTCAGCTTCCG | GTGGCCTTCCTTCCTTTGGTGGAGC |
| 20 | CTATACTCCAGCAGAGGAACCC | CTTTCTGAGCATTTCCTAACACG |
| Myo5 | GAAGGCCACCTACAGCAGGC | CCTGCTCTTCTGATACGCACG |
| Spt15 | GAATCGAGATGGTACAAAACCAGC | CTCTAATACGCATGATGACAGCAGC |
| Y′ | GCGCAGATCTGAAAGTTGGAGTTTTTCAGCG | GCGGGATCCAACCACACCTCCGAAATCTGC |
| KAN | GGTCAGACTAAACTGGCTGACGG | CCATGAGTGACGACTGAATCCG |
HML derivatives with LexA binding sites were all constructed as follows. A TRP1 fragment was amplified from pFL39 (4) with primers TRPa (AGTTGAAGCTTACTAGTGGTCGAAAAAAGAAAAGGAGAGGGCC) and TRPb(AATTCAAGCTTGGCAAGTGCACAAACAATACTTAAATAAATACTAC). It was inserted downstream of the HML-I sequence at the unique HindIII site. During this cloning, an SpeI site was created immediately downstream of HML-I. The region flanking HML-I to the right between positions 15220 and 15915 was amplified with primers HDa (AGTTGAGATCTCGGAACACATTCTTATAAATCTATAGG) and HDb (AATTCAGATCTGCGCGCCCTCTAATACTATAAAGGACTTGG). It was inserted at the unique BglII site to serve as a target during homologous recombination. The downstream primer also contains at its end a BssHII site. This procedure was carried out in parallel on the wild-type HML sequence and on the IΔ242 mutant form (29). It yielded plasmids pITRPHD and piTRPHD, respectively.
The HML-E silencer and URA3 reporter gene were amplified from a previously described plasmid (27) with primers E+URAa (AAGCTGGAGCTCGCGCGCCGGTTGATGACATGATTTTGTATCGTC) and E+URAb (AATTCGAGCTCTCATTACGACCGAGATTCCCG) and inserted upstream of the I sequence at the unique SacI site. The upstream primer also contains a BssHII restriction site. As above, this was done with the wild-type and mutant HML to give pEURA3I and pEURA3i, respectively. A PCR fragment containing four binding sites for LexA was obtained by PCR amplification of plasmid pSH18-34 (Stratagene) with primers LexAa (AGTTGACTAGTCCATATCTAATCTTACCTC) and LexAb (AATTCACTAGTCGCATTATCATCCCTC). It was cloned at a unique SpeI site, resulting in plasmids pEURA3ILEXA and pEURA3iLEXA.
Plasmids used to introduce TetO112-containing constructs at HML were derived from plasmids pITRPHD and piTRPHD. First, a plasmid harboring the kanMX4 gene adjacent to 112 TetO2 operators was built. A 5.6-kb SalI-BamHI TetO2 fragment obtained by excision from p306tetO2x112 (from the Nasmyth laboratory) was inserted between the SalI and BglII sites of pFA6a-KanMX4 (47), resulting in plasmid pTETKAN. A 180-bp SpeI-XbaI fragment from plasmid pITRPHD or piTRPHD removing the ATG of TRP1 was replaced by the 6.7-kb SpeI TETKAN fragment of pTETKAN, resulting in plasmids pITETKAN and piTETKAN, respectively.
pSILTet was derived from pITETKAN by insertion of the HML-E silencer and URA3 at a unique KpnI site upstream of HML-I. PCR amplification was performed with primers E+URA-KpnIa (AAGCTGCCAGGTACCTGGCGGTTGATGACATGATTTTGTATCGTC) and E+URA-KpnIb (AATTCCCAGGTACCTGGTCATTACGACCGAGATTCCCG). These primers each contain a BstXI site (boldface) engineered to create KpnI-compatible ends after BstXI digestion of the PCR products. The 1.5-kb NotI kanMX fragment of pFA6a-KanMX4 was cloned into the NotI site of pEURA3ILexA, resulting in plasmid pSIL.
pCENTet is a LEU2-CEN-ARS plasmid containing the 6.7-kb SpeI fragment from pTETKAN cloned at the SpeI site of pRS315 (41). pCEN is also a pRS315-based plasmid, in which the 1.5-kb NotI kanMX fragment of pFA6a-KanMX4 was cloned into the NotI site of pRS315.
Yeast strains, media, and methods.
Manipulations of S. cerevisiae were performed as described previously (38). The S. cerevisiae strains used in this study are all derivatives of S150-2B (MATa leu2-3,112 URA3-52 TRP1-289 his3Δ gal2 gal4::LEU2). All gene replacements were confirmed by Southern blot analysis.
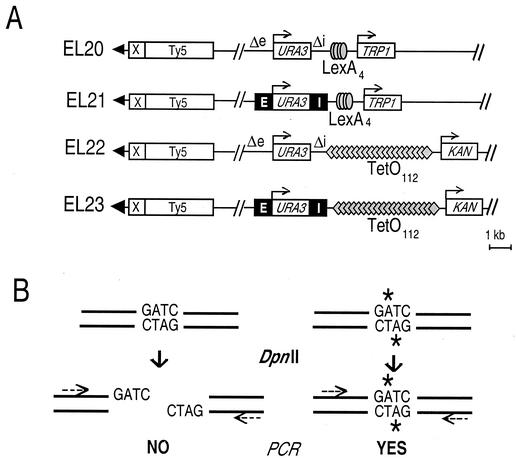
To insert LexA binding sites at HML, we transformed EG42 (5) with BssHII-digested, LexA4-containing HML constructs. We then screened for loss of the LEU2′-′lacZ marker by a filter-based β-galactosidase color assay. This resulted in strains EL20 and EL21 (see Fig. 1A). To integrate the TetO112 array at HML, we transformed the PvuII-linearized plasmids pITETKAN and piTETKAN into strains EL20 and EL21. This gave rise to strains EL22 and EL23, respectively (see Fig. 1A).
FIG. 1.
Outline of the experimental system. (A) Schematic representation of the left end of chromosome III in the test strains. The HML-E and HML-I silencers are drawn as solid boxes (E and I). A fragment containing either four LexA sites or a cluster of 112 TetO sites was inserted next to HML-I, together with the adjacent TRP1 or KAN resistance gene, respectively, used for selection purposes. The subtelomeric III-L region was left unaltered and carries an X subtelomeric element and a Ty5 retrotransposon known to be subject to telomere-driven silencing. Arrowheads, telomeric repeats. (B) Principle of the PCR-based quantification assay. Primer pairs are designed so that each one brackets a single GATC. Unmethylated sites are cut by DpnII and fail to be amplified. In contrast, sites that have been methylated by Dam (stars) become resistant to DpnII and can yield a PCR fragment. This permits quantification of the methylation level for a given GATC.
Analysis of URA3 expression.
The expression of URA3 was monitored essentially as described by Fourel et al. (17) by spotting 10 μl of serial dilutions of overnight culture onto appropriate selective synthetic medium with or without 5-fluoroorotic acid (1 g/liter).
Quantitative PCR.
Cells carrying the TetR-Dam plasmid were grown overnight in selective medium with 2% raffinose. Genomic DNA was prepared, and equal amounts of DNA were incubated for 16 h at 37°C with DpnII. The enzyme was heat inactivated for 20 min at 65°C, and the mixture was ethanol precipitated. Samples were then assayed by quantitative PCR with a LightCycler (Roche Molecular Biochemicals) according to the manufacturer's recommendations. A standard dilution series of nondigested genomic DNA was included in every experiment to allow relative quantification of each sample. Each sample was assayed in triplicate, revealing a standard error of about 5% for the quantitation of the DNA sample. Figures 3 to 5 only show the results of one set of experiments for better clarity. Each of the experiments was repeated at least twice, with independent DNA preparations. Identical patterns of methylation profile were obtained, although some variation in the absolute quantity of methylated GATC sequences was observed, perhaps as the result of varying expression of the methyltransferase (data not shown).
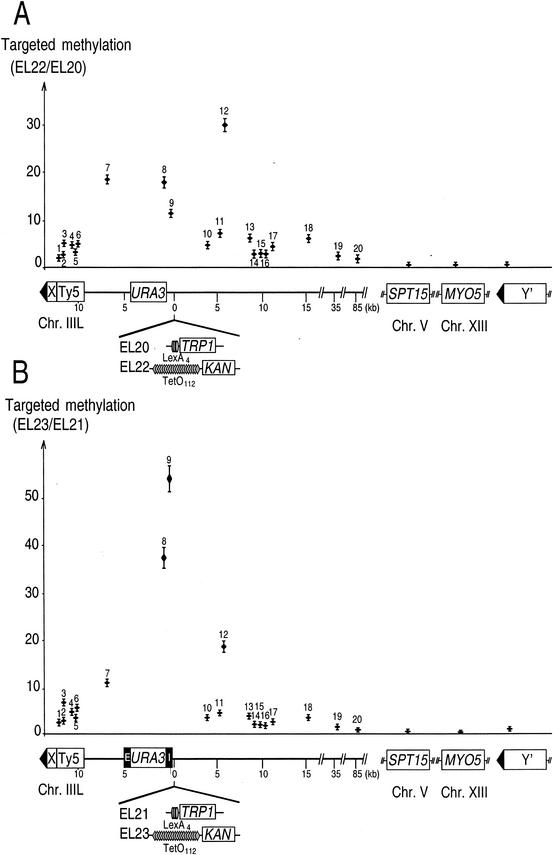
FIG.3.
Targeted methylation profiles of silenced and nonsilenced HML regions and other loci. (A) TetR-Dam targeted to HML via TetO112 specifically methylates a broad region of the left arm of chromosome III. Targeted methylation is the ratio of the methylation frequency for a given GATC in a TetO112-containing strain to that in a LexA4-containing strain. The number above each point refers to the relative position of the probed GATC along the chromosome. A schematic representation of the left arm of chromosome III is depicted below the graph, together with the right-side insertions at HML that differed between the strains compared. Other GATCs located on chromosomes other than chromosome III were also assayed. Two are located in the coding sequence of MYO5 and SPT15 on chromosomes XIII and V, respectively, and one is present in the Y′ subtelomeric repeats, which are found next to approximately half of the S. cerevisiae telomeres. The latter GATC is located 400 bp away from the telomeric TG1-3 repeats, in a conserved region. (B) Targeted methylation profile of strains carrying functional HML silencers. Legend is otherwise the same as for Fig. 1.
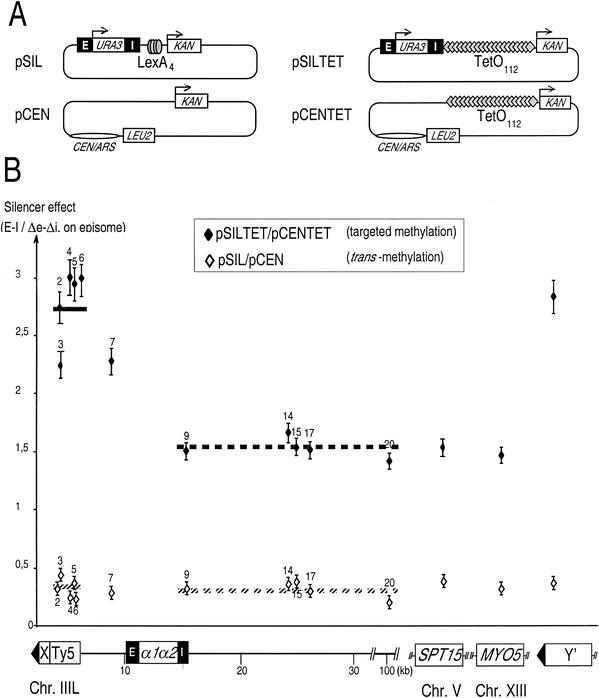
FIG. 5.
Plasmid-borne HML associates with III-L subtelomere and Y′ chromosome ends. (A) Scheme of the plasmids used in this study. The thin lines indicate bacterial pUC vector sequences. pSIL and pSILTet derive from the same construct used to modify the HML locus, described in Fig. 1A. pCENTet includes the TetO112 cluster and the KAN gene inserted in pRS315, a CEN-ARS vector. pCEN is identical to pCENTet except that it lacks the TetO112 cluster. (B) Ratio of methylation frequency with the SIL to the CEN plasmids containing TetO112 or not. A diagram of the left arm of chromosome III is shown below the graph, with the positions of the HML silencers and the two α1 and α2 mating type genes. Note that in that case, in which plasmids were transformed in the original S150-2B strain and selected for resistance to kanamycin, the chromosomal HML locus is not modified. Legend is otherwise the same as for Fig. 4 except that fewer sites were analyzed and reference and subtelomeric silencer effect levels are the averages of values obtained for sites 9, 14, 15, 17, 20, and 2 to 6.
RESULTS
System to target and detect methylation in S. cerevisiae.
The Dam identification technique is based on the fusion between the bacterial methyltransferase Dam and a DNA-binding protein (45). One can then identify the binding sites for the protein of interest by virtue of their cis methylation. Our goal was to examine physical interactions between loci in the yeast S. cerevisiae. In particular, we wanted to address whether the silenced mating type locus HML interacts with telomeres in a repressive nuclear compartment. We reasoned that we could adapt the technique to suit our purposes. Our rationale was that if Dam was tethered to HML, then loci that interacted with HML would become preferentially methylated. One important aspect of this system is that, methylation being a covalent modification, a trace of the interaction would remain even if the interaction was transient.
For this method to work satisfactorily, two preliminary conditions had to be met. First, the recruitment of Dam to HML and the cis-methylation of this locus had to be efficient. Second, the amount of unbound Dam free to diffuse in the nucleus and cause background methylation had to be minimal. In order to find these optimal conditions, we tried two different targeting systems. In a first series of experiments, we expressed a LexA-Dam hybrid protein in strains containing four LexA sites inserted into HML loci with or without silencers (Fig. 1A, EL20 and EL21). Methylation was examined by Southern blotting after restriction of genomic DNA with the methylation-sensitive enzymes DpnI and DpnII. We found methylation of the HML region to be no greater than that of a control locus on another chromosome, indicating inefficient targeting or high background (data not shown).
We therefore turned to another system in which Dam was fused to TetR and an array of 112 TetO sites (hereafter called TetO112) was inserted at HML loci with and without functional silencers (Fig. 1A, EL22 and EL23). By Southern analysis (see above), we observed increased methylation of the HML region in comparison to other loci (data not shown). The effect was maximal when the expression of TetR-Dam driven by the GAL1 promoter was kept low by growing the cells in the presence of raffinose, not galactose (data not shown). In all the experiments presented hereafter, the cells were transformed with TetR-Dam and raffinose was used as the carbon source.
We then devised a system to quantify the amount of methylation at individual GATC sites accurately and sensitively. Its principle, outlined in Fig. 1B, relies on quantitative PCR. Genomic DNA is overdigested with DpnII, which only cuts nonmethylated GATC sequences. There is no endogenous GATC methylation in S. cerevisiae, so in the absence of Dam methylation, no GATC is methylated and no amplification product can be formed. In contrast, a fragment encompassing a GATC site that has been methylated by Dam can be amplified by PCR. Therefore, the amount of PCR product directly reflects the methylation level of the GATC site examined.
To evaluate this assay, we sought conditions in which binding of TetR-Dam should be high and easily detectable. We therefore carried out our pilot experiments in strain EL22, in which HML is tagged with TetO sites and bears mutations that prevent silencing (Fig. 1A). We verified that these mutations incapacitated HML silencing, as indeed EL22 did not grow on 5-fluoroorotic acid-containing medium (Fig. 2). In these conditions, the chromatin at and around HML should be open and permissive for TetR-Dam binding. We extracted genomic DNA from strain EL22 expressing TetR-Dam, digested it with DpnII, and then performed quantitative PCR measurements with primer pairs that spanned chromosome III. Primer pairs within SPT15 on chromosome V and MYO5 on chromosome XIII and in Y′ subtelomeric repeats (present at several chromosome ends, but not III-L in our strain) were used as controls. Each primer pair brackets only one GATC site.
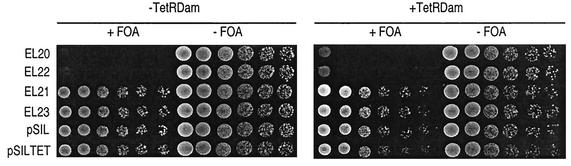
FIG. 2.
HML silencing is not affected by insertion of TetO sites and expression of TetR-Dam. Silencing is measured by the ability of cells to grow in the presence of 5-fluoroorotic acid (FOA), a drug that kills cells expressing URA3. A representative experiment is shown, and identical results were obtained with independent isolates from each strain. From left to right: nondiluted culture, three successive 10-fold dilutions, then two threefold dilutions. Growth on 5-fluoroorotic acid, and therefore silencing, depended on the presence of the E and I silencers flanking the URA3 reporter gene (EL21, EL23, pSIL, and pSILTet). It was not affected by the presence of flanking insertions (LexA or TetO sites) or the expression of TetR-Dam (− or + TetR-Dam). It was also properly silenced when placed on a plasmid (EL21 and EL23 versus pSIL and pSILTet).
We were concerned that different sites of the genome may display inherently different susceptibility to methylation because of their sequence or the local DNA structure. To take this variability into account, we normalized our data as follows. We first measured the methylation in strain EL20, which is identical to EL22 except that it contains LexA binding sites instead of TetO112 at HML (Fig. 1A). Consequently, TetR-Dam is not bound to HML in this strain but diffuses within the nucleus. This first methylation measurement therefore quantifies differences in accessibility and/or methylation efficiency between different sequences. We then measured methylation in strain EL22 (Fig. 1A). For each primer pair, this second value was divided by the value obtained in strain EL20, and the ratio was named targeted methylation. The results of this experiment are shown in Fig. 3A.
The most salient finding was that the targeted methylation was highest, up to a value of 30, for GATC sites located in the HML region (sites 7 to 12). In other words, sites around HML were methylated up to 30-fold more efficiently when TetR-Dam was recruited to HML than when it diffused freely. In contrast, for sites outside of chromosome III, the values of targeted methylation were not significantly different from 1. These two results reflect efficient targeting of TetR-Dam to TetO112 and demonstrate the feasibility of our approach.
The highly methylated region around HML spanned about 15 kb. It displayed local variations in methylation (compare site 12 to 10 and 11), which might be explained by local differences in chromatin structure. The extent of methylation dropped off sharply for sites further than about 7.5 kb from the TetO112 sites. This distance is similar to that observed in Drosophila melanogaster, in which targeted methylation extended about 5 kb in either direction from the binding site. Importantly, we noticed that all the sites on the left arm of chromosome III, even those most distant from TetO112, were more methylated than any of the sites on other chromosomes. Sites 19 and 20, for instance, 38 and 88 kb away from TetO112, respectively, displayed about 50% more targeted methylation than SPT15, MYO5, and Y′ (Fig. 3A). A possible explanation is that chromosome III is folded into a given domain of the yeast nucleus and that HML is more likely to interact with sequences on III than on other chromosomes, which localize to other nuclear regions. Previous work in S. cerevisiae (8, 12), as well as findings reported in mammalian cells (6, 9), seem to support this idea.
We were also concerned that silencing may limit access of TetR-Dam to TetO112. We therefore examined the targeted methylation pattern obtained with TetR-Dam targeted to a wild-type silenced HML locus (Fig. 1A, strain EL23). We first tested whether the presence of TetO112 or the expression of TetR-Dam would interfere with HML silencing. As shown in Fig. 2, we observed that neither modification altered the capacity of HML silencers to silence a URA3 reporter gene. These results are consistent with previous studies showing that DNA methylation does not interfere with silencing (20) and that binding of TetR does not modify the higher-order organization of chromatin (3, 15, 34).
As in the pilot experiment above, we used a reference strain to normalize our measurements. In this case it was EL21, a strain related to EL23 that contains LexA binding sites in the place of TetO112. The targeted methylation value was determined by dividing the methylation obtained in strain EL23 by that obtained in EL21. The pattern that we observed was overall similar to that obtained in the absence of functional HML silencers (compare Fig. 3B and Fig. 3A). Methylation was elevated in a 15-kb region centered on TetO112 and declined progressively beyond this limit. For sites outside of chromosome III, the methylation levels were again very low (Fig. 3B, SPT15, MYO5, and Y′). Thus, silencing at HML did not prevent targeting of TetR-Dam to TetO112 and ensuing DNA methylation.
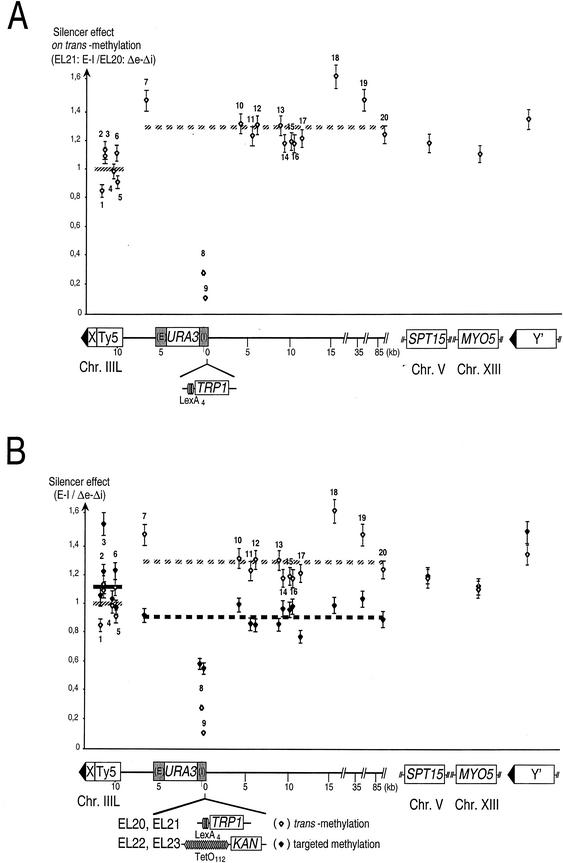
Finally, we were concerned that the presence of silencing at HML may have an effect on the pattern of methylation at other genomic sites independent of targeting. To evaluate this possibility, we now plotted methylation values obtained with EL21 divided by those obtained with EL20. None harbor TetO112 sites, and EL20 lacks silencers at HML, whereas they are present in EL21. The resulting value was referred to as the silencer effect on trans methylation (Fig. 4A). For any given site, a silencer effect greater than 1 means that the site is more methylated when silencers are present at HML than in their absence. A silencer effect smaller than 1 means the opposite. Three major phenomena became apparent when we did this calculation (Fig. 4A).
FIG.4.
Silencer effects on trans methylation and on targeted methylation. The specific influence of silencers on TetR-Dam-mediated methylation can be inferred from processing the same raw methylation data obtained for strains EL20 to EL23 through calculating a different type of ratio from that presented in Fig. 3. Thus, silencer effect is the ratio of the methylation frequency for a given GATC in a strain carrying intact E and I silencers at HML to one in which these have been mutated, and this in either of two settings: upon targeting of TetR-Dam to HML via TetO112 (targeted methylation, solid diamonds), or upon expression of TetR-Dam without any Tet operators to which it may bind (trans methylation, open diamonds). For the sake of clarity, trans methylation is presented by itself in panel A and targeted methylation is presented together with trans methylation in panel B to facilitate comparisons. Sites along chromosome III can be grouped into three classes displaying distinctive behaviors. Sites 8 and 9, within HML-I, are in a region where both trans and targeted methylation are hampered due to the presence of silenced chromatin. Sites 7 and 10 to 20 yield an average reference level of silencer effect along chromosome III (broken line) that differs between trans and targeted methylation settings (hatched and dark lines, respectively). Sites 1 to 6 all appear similarly influenced by their proximity to telomere III-L with regard to both trans- and targeted TetR-Dam-mediated methylation and yield a subtelomeric average silencer effect level (solid lines).
The strongest effect was observed for sites 8 and 9, which were about fivefold less accessible to the enzyme in the presence of active silencers. This agrees with previous reports showing that silenced regions are less accessible to the modifying enzymes (20). Second, sites 7 and 10 to 20 as well as SPT15, MYO5, and Y′ sites were slightly but significantly more methylated by the enzyme in the presence of silencers at HML than in their absence (Student's t test: P < 0.0009). Although we have no definitive explanation for this observation, it may result from variations in the level of expression of TetR-Dam, which might be slightly higher in EL21 than in EL20. Finally, sites 1 to 6 were clearly less accessible to the enzyme than the latter set of sites (compare solid and broken lines, which indicate the average silencer effect for each group). This shows that the mere presence of a silenced HML locus influences chromatin structure in the III-L subtelomeric region. Although it was not investigated further, we presume that this decreased accessibility to TetR-Dam in fact reflects an increase in III-L telomeric silencing dependent on the HML-E and -I silencers. This unanticipated result therefore strongly suggests that the HML-E and -I silencers reinforce silencing emanating from telomere III-L.
Silencing of HML permits interaction with telomere III-L.
To visualize the specific contribution of the silencers to the targeted methylation profile, we plotted the ratio of targeted methylation in the silenced versus the nonsilenced context (silencer effect on targeted methylation) (Fig. 4B, solid diamonds). The silencer effect on trans methylation is represented again to facilitate comparison (open diamonds). The methylation of sites closest to HML (sites 8 and 9) was not as deeply affected in the silenced context upon targeting of TetR-Dam compared to trans-methylation, suggesting that targeting of TetR-Dam at HML compensates in part for the loss of accessibility due to the presence of silenced chromatin. Second, methylation of reference sites along chromosome III (sites 7 and 10 to 20) was reduced upon TetR-Dam targeting to HML (compare hatched and dark broken lines), which was not the case for sites located internally on other chromosomes (SPT15 and MYO5). This phenomenon might be accounted for by a locally reduced availability of TetR-Dam within chromosome III nuclear territory upon its trapping to multiple sites at a single locus of this chromosome. TetR-Dam is indeed known to be expressed in limiting amounts in our system.
By contrast, and most importantly, the methylation of the subtelomeric element Ty5 (sites 1 to 6) increased in the silenced context compared to reference sites of chromosome III (compare solid and broken dark lines). The differences between the cluster formed by sites 7 and 10 to 20 and the one formed by sites 1 to 6 are highly significant (Student's t test: P < 0.000001). This finding is all the more striking because this increased methylation at sites 1 to 6 contrasts with the reduced accessibility of sites 1 to 6 to trans methylation upon silencing at HML (see Fig. 4A). One model which is consistent with these results is that the presence of silencers at HML increases HML-telomere III-L interactions, thereby allowing specific methylation of Ty5 sequences upon targeting of TetR-Dam to HML, which, as at HML, compensates for the reduced accessibility to a trans-acting enzyme associated with reinforced silencing.
Finally, the presence of silencers at HML also seemed to increase methylation in the subtelomeric regions of other chromosomes upon targeting of TetR-Dam at HML (Fig. 4B, Y′ site, compare solid and open diamonds). Although this result is a first hint that HML silencers may be endowed with the capacity to interact with all telomeres, the effect was moderate (Student's t test: 0.01 < P < 0.05), and this hypothesis therefore required independent confirmation. Altogether, these data strongly suggest that HML silencers preferentially associate with their proximal telomere.
Plasmid-borne silencers interact with the III-L Ty5 subtelomeric region and also with Y′ telomeres.
The above results suggested a physical contact between the silencers at HML and Ty5 at telomere III-L as well as, to a lesser extent, with subtelomeric regions of other chromosomes. However, the magnitude of the observed effects was rather limited, and interpretation was further complicated by a cis effect of HML silencers on TetR-Dam-mediated methylation independent of its targeting to HML (see Fig. 4A). We therefore decided to investigate this model further by asking whether plasmid-borne silencers can interact with telomeric regions.
We targeted TetR-Dam to a plasmid and assessed whether the presence of plasmid-borne silencers would lead to increased methylation of telomeric regions. For this purpose, we created a series of plasmids (Fig. 5A). pSILTet and pSIL both contain the HML silencers (SIL). The first plasmid contains TetO112, but the second does not. HML provides the ARS and CEN functions in these plasmids (1, 25, 28). Plasmid pCENTet contains TetO112 but no silencer, while plasmid pCEN has neither. We determined that the plasmid copy number in cells grown under selective conditions was sixfold higher for pSIL than for pSILTet and for pCEN than for pCENTet and fivefold higher for the SIL compared to the corresponding CEN plasmids (data not shown). As expected, the flanking silencers repressed the expression of the URA3 gene carried by the plasmids whether or not TetR-Dam was expressed (Fig. 2). Importantly, the presence of pSIL, pCEN, pSILTet, or pCENTet did not interfere with chromosomal silencing (data not shown). The KAN gene present on pSILTet and pCENTet was strongly methylated upon TetR-Dam expression (data not shown). Altogether, these controls validate the use of the plasmid system to study the effects of targeted methylation.
We measured methylation in the presence of TetR-Dam and of plasmid pSILTet. Again, for normalization purposes, these values were divided by those obtained in the presence of pCENTet, which does not contain a silencer. This ratio is plotted in Fig. 5B (solid diamonds). The value obtained for the control sites MYO5 and SPT15 was about 1.5. This means that even the control sites received about 50% more methylation when the cells contained pSILTet than when they contained pCENTet. The most likely explanation for this fact is that TetR-Dam is slightly more expressed in the former situation, for an unknown reason. We found that sites 9, 14, 15, 17, and 20 behaved like the control loci, with ratios between 1.45 and 1.65. In contrast, two regions clearly behaved differently: first, sites within and around Ty5, and second, the site within Y′. In both cases, the ratio was about 3, a value significantly different from that of the controls (Student's t test: P < 0.0000006). This shows that Ty5 and the Y′ elements are more methylated in the presence of pSILTet. This could have one of two causes. First, pSILTet, and its tethered TetR-Dam, could interact with these regions more than with other genomic loci. Alternatively, these regions could become inherently more accessible to background methylation just because of the presence of the silencers on a plasmid.
We tested this by using plasmids pSIL, which contains the silencers but not TetO112, and pCEN, which contains neither. We measured methylation in the presence of pSIL and divided that value by that obtained with pCEN. This ratio is plotted in Fig. 5B (open diamonds). Its value was about 0.4 for the control sites. Again, why the ratio deviates from 1 could be due to slight variations in TetR-Dam expression between strains. At any rate, the relevant result is that all sites behave like the control sites. In other words, no site becomes more or less accessible to TetR-Dam in the presence of the plasmid-borne silencers. This rules out our second hypothesis, and we conclude that HML, when present on a plasmid, can interact with the Ty5 element present at telomere III-L and also with the Y′ sequences present on other telomeres.
DISCUSSION
Many genetic arguments have suggested functional interactions between silencers and telomeres (5, 16, 30), but until now direct proof for physical contacts was lacking. We have adapted a DNA methyltransferase targeting assay to S. cerevisiae and coupled it to a quantitative method of methylation detection. This allowed us to investigate the interactions of the HML silencers with other loci. We show that, in its natural context, HML interacts with the proximal subtelomeric element Ty5 and to a lesser extent with Y′ elements present at other telomeres. This interaction specifically depends on the presence of functional silencers at HML. Furthermore, although not addressed here, it is presumably transient and may occur at specific stages of the cell cycle.
Our results further suggest that the physical interaction between HML silencers and the III-L subtelomeric region contribute to efficient silencing of the Ty5 retrotransposon (46). Given that a GATC site located between telomere III-L and HML is not affected by the presence of HML silencers (Fig. 4A, site 7), it seems unlikely that the putative cooperation between Ty5 silencing and HML silencers would derive from a continuous propagation of silent chromatin emanating from HML. This is consistent with previous results demonstrating that repression at HML is not affected by the expression of a URA3 reporter gene located between HML and telomeres (30).
Many proteins, including Rap1, Sir2, Sir3, and Sir4, are known to associate both with silencers and with telomeres and could potentially bridge these two compartments. Interestingly, a Sir-dependent telomere folding in the immediate subtelomeric region has been described (11, 42). Although this looping occurs over much smaller distances than the interactions described here, both phenomena could possibly rely on the same mechanisms.
In light of the physical associations between telomere and silencer described here, the role of telomeres in silencer-mediated repression certainly needs to be reconsidered. In its simplest form, the reservoir model assumes that silencers can function autonomously as long as the local concentration of silencing factors is sufficient. However, a situation in which a silent locus is located away from the nuclear periphery has never been reported (43), and the establishment of silencing appears to require an intact telomeric compartment (15). The existence of specific telomere-silencer associations points to a more direct role of the telomeric compartment than just as a storage space for silencing factors. The fact that overexpression or delocalization of the Sir proteins can rescue HML silencing at nontelomeric sites has been put forward as a strong argument in favor of the reservoir model (30, 32). However, this may also be explained by an improved association of the silencers with telomeres, which would emancipate them from the chromosomal context and mimic the situation observed with silencer-containing plasmids (Fig. 5).
Physical associations might also explain the functional cooperation observed between various types of silencing elements (5, 17, 27). These interactions could involve many of the factors present both at telomeres and at silencer elements. Although the association of silencers with telomeres might be essential for their silencing function, one may imagine that certain silencers may be less telomere dependent than others, requiring less frequent or less prolonged association. This might be the case with HMR-E, which appears to serve as a dominant repressor of expression (7, 14, 44, 48). Overall, we propose that telomeres form a platform that facilitates the coalescence of silencers and consequently organize the yeast silencing compartment. Such long-range interactions are likely to play a key role in the establishment and/or maintenance of silenced domains in other organisms like Drosophila melanogaster (37).
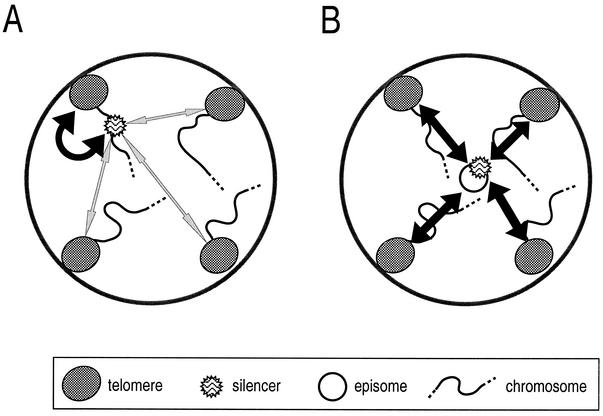
When HML silencers were placed on a plasmid, we observed that these now interacted not only with the Ty5 III-L region but also with other telomeric regions. It is unlikely that the silencers in an episomal state lack some chromatin structure necessary to direct interaction with the III-L telomere, since these can still interact with various telomeres and appear functional in imposing silencing on a reporter gene. Rather, the episome might lack some intrachromosomal constraint that causes HML to interact preferentially with its proximal telomere (Fig. 6A). For instance, when carried on a plasmid, the movements of the HML region are likely to be less constrained in the nucleoplasm. The sequence would then be free to interact with several, if not all, chromosome ends (Fig. 6B). This is in full agreement with the fact that plasmid-borne silencers are more active than those inserted far away from a telomere (30). Therefore, the dynamic properties of a chromosomal segment determine its capacity to be silenced. In agreement with this idea, it was recently demonstrated that telomeres provide strong constraints on chromosome movements that contrast with the ability of other parts of the genome to diffuse rapidly in G1 phase (23).
FIG. 6.
Model for physical associations between silencers and telomeres. (A) A chromosomal silencer can interact with telomeres (bidirectional arrow) but displays strong preference for a proximal partner. (B) In contrast, a plasmid-borne silencer associates equally with all chromosome ends, and this difference likely reflects its free movement in the nucleoplasm.
By using a methyltransferase targeting assay, we showed the existence of direct interactions between HML silencers and telomeres. This finding closes the circle of evidence suggesting that functional cooperation between silencing elements in S. cerevisiae relies at least in part on direct physical interactions. We have brought direct evidence for the existence of silencing compartments that contain telomeres and HM silencers.
Acknowledgments
We thank F. Feuerbach, U. Nehrbass, B. van Steensel, and K. Nasmyth for the gift of reagents and advice as well as K. Bystricky and S. Gasser for critical reading of the manuscript.
This work was supported by La Ligue Nationale contre le Cancer.
REFERENCES
- 1.Abraham, J., K. A. Nasmyth, J. N. Srathern, A. J. Klar, and J. B. Hicks. 1984. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J. Mol. Biol. 176:307-331. [DOI] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., A. M. Neiman, D. C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394:592-595. [DOI] [PubMed] [Google Scholar]
- 3.Aragon-Alcaide, L., and A. V. Strunnikov. 2000. Functional dissection of in vivo interchromosome association in Saccharomyces cerevisiae. Nat. Cell Biol. 2:812-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonneaud, N., O. Ozier-Kalogeropoulos, G. Y. Li, M. Labouesse, L. Minvielle-Sebastia, and F. Lacroute. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7:609-615. [DOI] [PubMed] [Google Scholar]
- 5.Boscheron, C., L. Maillet, S. Marcand, M. Tsai-Pflugfelder, S. M. Gasser, and E. Gilson. 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 15:2184-2195. [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, S., S. Gilchrist, J. M. Bridger, N. L. Mahy, J. A. Ellis, and W. A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211-219. [DOI] [PubMed] [Google Scholar]
- 7.Buck, S. W., and D. Shore. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9:370-384. [DOI] [PubMed] [Google Scholar]
- 8.Burgess, S. M., and N. Kleckner. 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 13:1871-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croft, J. A., J. M. Bridger, S. Boyle, P. Perry, P. Teague, and W. A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csink, A. K., and S. Henikoff. 1996. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381:529-531. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin, D., S. M. Kantrow, R. A. Liberatore, and V. A. Zakian. 2000. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol. 20:7991-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 13.Dernburg, A. F., K. W. Broman, J. C. Fung, W. F. Marshall, J. Philips, D. A. Agard, and J. W. Sedat. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85:745-759. [DOI] [PubMed] [Google Scholar]
- 14.Ehrenhofer-Murray, A. E., D. H. Rivier, and J. Rine. 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier, E. Gilson, J. C. Olivo-Marin, H. Scherthan, and U. Nehrbass. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4:214-221. [DOI] [PubMed] [Google Scholar]
- 16.Fourel, G., C. Boscheron, E. Revardel, E. Lebrun, Y. F. Hu, K. C. Simmen, K. Muller, R. Li, N. Mermod, and E. Gilson. 2001. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fourel, G., E. Revardel, C. E. Koering, and E. Gilson. 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18:2522-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francastel, C., M. C. Walters, M. Groudine, and D. I. Martin. 1999. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell 99:259-269. [DOI] [PubMed] [Google Scholar]
- 19.Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S. M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottschling, D. E. 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA 89:4062-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 22.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383:92-96. [DOI] [PubMed] [Google Scholar]
- 23.Heun, P., T. Laroche, K. Shimada, P. Furrer, and S. M. Gasser. 2001. Chromosome dynamics in the yeast interphase nucleus. Science 294:2181-2186. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, S. G., and J. R. Broach. 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10:1021-1032. [DOI] [PubMed] [Google Scholar]
- 25.Kimmerly, W. J., and J. Rine. 1987. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol. Cell. Biol. 7:4225-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde, E. J. Louis, and S. M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653-656. [DOI] [PubMed] [Google Scholar]
- 27.Lebrun, E., E. Revardel, C. Boscheron, R. Li, E. Gilson, and G. Fourel. 2001. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics 158:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine, M. S., S. Enomoto, S. L. Finstad, and J. Berman. 1992. Yeast telomere repeat sequence (TRS) improves circular plasmid segregation, and TRS plasmid segregation involves the RAP1 gene product. Mol. Cell. Biol. 12:1997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahoney, D. J., and J. R. Broach. 1989. The HML mating type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9:4621-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maillet, L., C. Boscheron, M. Gotta, S. Marcand, E. Gilson, and S. M. Gasser. 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10:1796-1811. [DOI] [PubMed] [Google Scholar]
- 31.Maillet, L., F. Gaden, V. Brevet, G. Fourel, S. G. Martin, K. Dubrana, S. M. Gasser, and E. Gilson. 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcand, S., S. W. Buck, P. Moretti, E. Gilson, and D. Shore. 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 10:1297-1309. [DOI] [PubMed] [Google Scholar]
- 33.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97:621-633. [DOI] [PubMed] [Google Scholar]
- 34.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 35.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8:2257-2269. [DOI] [PubMed] [Google Scholar]
- 36.Palladino, F., T. Laroche, E. Gilson, A. Axelrod, L. Pillus, and S. M. Gasser. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75:543-555. [DOI] [PubMed] [Google Scholar]
- 37.Pirrotta, V. 1997. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 13:314-318. [DOI] [PubMed] [Google Scholar]
- 38.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Shore, D. 2001. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11:189-198. [DOI] [PubMed] [Google Scholar]
- 41.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 43.Tham, W. H., J. S. Wyithe, P. K. Ferrigno, P. A. Silver, and V. A. Zakian. 2001. Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell 8:189-199. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. S., L. M. Johnson, and M. Grunstein. 1994. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol. Cell. Biol. 14:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Steensel, B., and S. Henikoff. 2000. Identification of in vivo DNA targets of chromatin proteins with tethered dam methyltransferase. Nat. Biotechnol. 18:424-428. [DOI] [PubMed] [Google Scholar]
- 46.Vega-Palas, M. A., S. Venditti, and E. Di Mauro. 1997. Telomeric transcriptional silencing in a natural context. Nat. Genet. 15:232-233. [DOI] [PubMed] [Google Scholar]
- 47.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 48.Xu, E. Y., S. Kim, and D. H. Rivier. 1999. SAS4 and SAS5 are locus-specific regulators of silencing in Saccharomyces cerevisiae. Genetics 153:25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]