Abstract
The cyclin D-dependent kinase is a critical mediator of mitogen-dependent G1 phase progression in mammalian cells. Given the high incidence of cyclin D1 overexpression in human neoplasias, the nature and complexity of cyclin D complexes in vivo have been subjects of intense interest. Besides its catalytic partner, the nature and complexity of cyclin D complexes in vivo remain ambiguous. To address this issue, we purified native cyclin D1 complexes from proliferating mouse fibroblasts by affinity chromatography and began to identify and functionally characterize the associated proteins. In this report, we describe the identification of Hsc70 and its functional importance for cyclin D1 and cyclin D1-dependent kinase maturation. We demonstrate that Hsc70 associates with newly synthesized cyclin D1 and is a component of a mature, catalytically active cyclin D1/CDK4 holoenzyme complex. Our data suggest that Hsc70 promotes stabilization of newly synthesized cyclin D1, thereby increasing its availability for assembly with CDK4. In addition, our data demonstrate that Hsc70 remains bound to cyclin D1 following its assembly with CDK4 and Cip/Kip proteins, where it ensures the formation of a catalytically active complex.
Cell cycle transitions require the sequential and ordered activation of the cyclin-dependent kinases (CDKs) and inactivation of CDK inhibitors. The D-type cyclins, the regulatory subunits of the CDK4/6 kinase, are induced through mitogen-triggered signaling events during the G1 phase of the cell cycle. Mitogenic stimuli trigger the accumulation of active cyclin D1/CDK4 complexes through both increased expression and decreased proteolysis of cyclin D1 and through the promotion of cyclin D1/CDK4 assembly (46). Expression of cyclin D1 depends upon activation of a signal transduction cascade involving Ras, Raf-1, and the extracellular signal-regulated protein kinases (ERK1/2) (2, 3, 10, 27, 30, 53, 54). Accumulation of cyclin D1 during G1 also relies upon mitogen-dependent inhibition of glycogen synthase kinase 3β via activation of phosphatidylinositol 3-kinase (14). Inhibition of glycogen synthase kinase 3β results in decreased cyclin D1 proteolysis (14) and nuclear accumulation of the cyclin D1/CDK4 kinase (4, 5, 14).
Expression of CDK4/6, unlike that of the D-type cyclins, does not strictly depend upon mitogens and can thus be detected in quiescent cells. Upon CDK4/6 translation, they are recruited into high-molecular-weight, cytoplasmic, Hsp90-containing complexes via CDC37 (32, 50). Hsp90 and CDC37 serve to stabilize CDK4 via their capacity to direct proper CDK4 folding. Upon proper folding, CDK4 is released from this cytoplasmic complex and is competent for assembly with a D-type cyclin or is shunted into dead-end inhibitor complexes with a member of the Ink4 family of CDK inhibitors (40).
Although CDK4 is present in quiescent cells, it does not assemble with D-type cyclins even in the presence of nonlimiting, ectopically expressed cyclin, suggesting the existence of a mitogen-regulated “assembly factor” that serves to bring cyclin D1 and CDK4 together (10, 26). Recent evidence suggests that members of the Cip/Kip family may serve as this elusive assembly factor. Both p21Cip1 and p27Kip1 can promote assembly of cyclin D/CDK4 complexes in vitro (28) and are components of the active cyclin D holoenzyme in vivo (28). Additional support comes from experiments demonstrating that cells deficient for Cip/Kip proteins do not assemble cyclin D/CDK complexes (9, 37). These results suggest that the minimal complexity of the cyclin D holoenzyme would be a ternary complex composed of a D-type cyclin, CDK4 (CDK6), and p21Cip1 (or p27Kip1). The predicted molecular mass of this complex, approximately 90 to 100 kDa, is smaller than the size described for the cyclin D holoenzyme in vivo. Several groups have characterized the active cyclin D holoenzyme as a complex with a molecular mass ranging from 150 to 200 kDa (32, 34, 40). At face value, these results indicate that there is at least one more unidentified component in the cyclin D holoenzyme.
In order to identify critical regulatory components of the active cyclin D/CDK4 holoenzyme, we purified cyclin D1 complexes from proliferating mammalian cells. Here we report the identification of Hsc70 as a novel cyclin D1 binding protein. We demonstrate that Hsc70 is present in high-molecular-weight complexes containing newly synthesized cyclin D1 and is a component of an active 158-kDa cyclin D1/CDK4 holoenzyme. We provide evidence that Hsc70 functions in the maturation of cyclin D1, thereby facilitating the assembly of an active cyclin D1/CDK4 holoenzyme.
MATERIALS AND METHODS
Cell culture conditions and transfections.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium containing glutamine and supplemented with antibiotics (Cellgro) and 10% fetal calf serum (BioWhittaker). Insect Sf9 cells were grown in Grace's medium supplemented with 10% heat-inactivated fetal calf serum. Baculoviruses encoding Flag-D1, CDK4, and p21Cip1 were described previously (5, 16). Standard protocols for baculovirus manipulation were followed (51). Derivatives of NIH 3T3 cells engineered to overexpress Flag-tagged cyclin D1 and Flag-D1-T286A were described previously (14).
The Hsc70 cDNA was subcloned into pcDNA3 (Invitrogen) for expression in mammalian cells or pVL1393 for production of baculovirus (Pharmingen). Transient expression in NIH 3T3 cells was achieved by transfection of the indicated expression plasmids with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions.
Affinity purification and analysis.
Protein complexes were purified from whole-cell extracts prepared in Tween 20 buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, 20 U of aprotinin per ml, 5 μg of leupeptin per ml, 0.4 mM NaVO4, and 0.4 mM NaF) with M2-agarose (Sigma). Following extensive washes with Tween 20 buffer, complexes were eluted with excess Flag peptide or with 0.1 M glycine, pH 3.5. Purified complexes were digested with trypsin and directly analyzed by tandem mass spectroscopy or resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) prior to preparation for tandem mass spectroscopy analysis. Where indicated, affinity-purified complexes were loaded directly onto a Superdex 200 HR 10/30 column and fractionated with the Biologic chromatography system (Bio-Rad) prior to analysis. Molecular mass standards were as follows: thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa). The void volume of each column was 9 ml, and fraction collection commenced at 10 ml.
Tandem mass spectroscopy analysis of the samples was performed by microcapillary electrospray liquid chromatography-tandem mass spectrometry. Briefly, the protein gel bands, visualized by Coomassie brilliant blue, were digested with sequencing grade trypsin (Roche) in situ, and the peptides were extracted as described previously (47). The recovered peptides were fractionated on a 7.5-cm (100 μm inner diameter) reverse-phase C18 capillary column attached inline to a ThermoFinnigan LCQ-Deca ion trap mass spectrometer. The entire digested sample was loaded as described previously (19), and the peptides were eluted by ramping a linear gradient from 2 to 60% solvent B in 90 min. Solvent A consisted of 5% acetonitrile, 0.5% acetic acid, and 0.02% heptafluorobutyric acid, and solvent B consisted of 80:20 acetonitrile-water containing 0.5% acetic acid and 0.02% heptafluorobutyric acid. The flow rate at the tip of the needle was set to 300 nl/min by programming the high-pressure liquid chromatography (HPLC) pump and use of a split line.
The mass spectrometer cycled through four scans as the gradient progressed. The first was a full mass scan, followed by three tandem mass scans of the successive three most intense ions. A dynamic exclusion list was used to limit collection of tandem mass spectra for peptides that eluted over a long period of time. All tandem mass spectra were searched with the SEQUEST computer algorithm against the National Center for Biotechnology Information nonredundant protein database (June 2000). Each high-scoring peptide sequence was manually compared with the corresponding tandem mass spectrum to ensure that the match was correct.
Immunoblotting, immunoprecipitation, and protein kinase assays.
For detection of cyclin D1 complexes, cellular lysates prepared in Tween 20 buffer were resolved on denaturing polyacrylamide gels, transferred to nitrocellulose membranes (Millipore), and blotted with antibodies specific for total cyclin D1 (D1-17-13G), CDK4 (Santa Cruz), Hsc70 (StressGen), or p21Cip1 (Santa Cruz) or the 9E10 antibody directed against Myc-tagged cyclin E. Sites of antibody binding were visualized with protein A-conjugated horseradish peroxidase (EY Laboratories). For detection of cyclin D1-dependent kinase activity, cells were harvested in Tween 20 immunoprecipitation buffer, and following precipitation with the cyclin D1 monoclonal antibody, protein kinase assays with 1 μg of recombinant glutathione S-transferase (GST)-retinoblastoma protein (Rb) were performed as previously described (43).
For immunoprecipitation analysis of associated proteins, protein complexes were precipitated from either affinity-purified fractions or total cellular lysates prepared in Tween 20 buffer as described above. Immune complexes with the indicated antibodies were collected with protein A-Sepharose. Precipitated proteins were resolved by SDS-PAGE, detected by immunoblot analysis, and visualized by enhanced chemiluminescence (New England Nuclear).
Immunofluorescence.
NIH 3T3 cells seeded on glass coverslips were transfected with expression vectors encoding the indicated DNAs. Cells were fixed 48 h following transfection with methanol-acetone (1:1). For visualization of cyclin D1- and bromodeoxyuridine-positive cells, coverslips were first stained with the Flag-specific M2 monoclonal antibody (Sigma). Secondary antibody staining was performed for 30 min with biotinylated anti-mouse immunoglobulin followed by streptavidin-conjugated Texas Red (Vector Laboratories). Following a brief treatment with 1.4 N HCl, coverslips were incubated with antibromodeoxyuridine antibodies (Research Diagnostics Inc.) and fluorescein isothiocyanate-conjugated anti-sheep immunoglobulin antibodies (Vector). In all cases, DNA was visualized with Hoechst 33258 dye at a 1:500 dilution. Coverslips were mounted on glass slides with Vectashield mounting medium (Vector).
RESULTS
Identification of Hsc70 as a cyclin D1 binding protein.
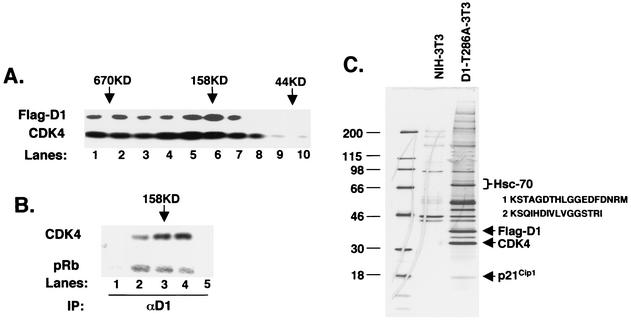
To identify proteins that physically associate with cyclin D1 in vivo, we set out to immunopurify cyclin D1 complexes from proliferating mouse fibroblasts. To circumvent difficulties associated with purification of low-abundance proteins, we wished to use NIH 3T3 fibroblasts which overexpress either epitope-tagged cyclin D1 (D1-3T3) or a stable, constitutively nuclear cyclin D1 mutant, cyclin D1-T286A (D1-T286A) (14). These cells have been characterized previously (14), and they overexpress Flag-D1 by approximately 10-fold (4). Enrichment of endogenous cyclin D1 was observed in nuclear, catalytically active 150- to 200-kDa complexes (34, 40). Thus, prior to purification of cyclin D1 complexes from these cells, we first confirmed that epitope-tagged cyclin molecules were incorporated into the active 150- to 200-kDa complexes.
Lysates prepared from D1-3T3 (Fig. 1) or D1-T286A-3T3 (data not shown) were loaded onto a Superdex 200 gel filtration column. Fractions were collected beginning at the void volume, and samples from each fraction were subjected to Western blot analysis with either cyclin D1- or CDK4-specific antibodies. We chose to use the cyclin D1 antibody rather than the M2 antibody due to its increased sensitivity for Western analysis and because Flag-D1 migrates with an altered mobility relative to endogenous cyclin D1, thereby allowing us to distinguish between ectopic and endogenous proteins (16). While a peak of Flag-D1 eluted between 150 and 200 kDa (Fig. 1A, lanes 5 to 7), it could also be readily detected in high-molecular-weight fractions (Fig. 1A, lanes 1 to 4). Endogenous cyclin D1 is not apparent at this exposure. CDK4 was present in most fractions coincident with a molecular mass ranging from 600 kDa to 100 kDa (Fig. 1A, lower panel). As reported previously, very little CDK4 was present in complexes, consistent with a molecular mass of a heterodimer of either CDK4-cyclin D1 or CDK4-INK (50 to 70 kDa) (40). Similar results were observed with D1-T286A (data not shown).
FIG. 1.
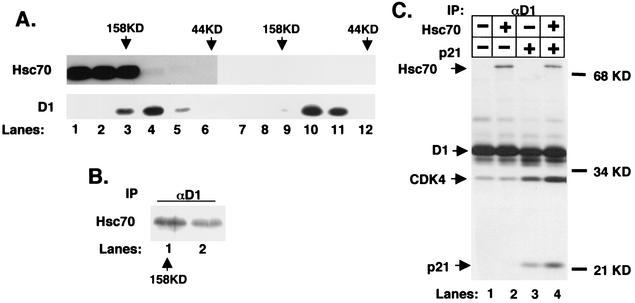
Hsc70 copurifies with cyclin D1. (A) Lysates were fractionated by gel filtration chromatography, and 5% of each fraction was resolved by SDS-PAGE; elution of cyclin D1 (top panel) and CDK4 (bottom panel) was visualized by immunoblot analysis. The positions of eluting molecular weight standards are indicated at the top. (B) Fractions corresponding to lanes 4 to 8 in panel A were immunoprecipitated (IP) with a monoclonal antibody specific for cyclin D1 and either blotted for associated CDK4 (top) or assayed for their ability to phosphorylate recombinant GST-Rb (bottom). (C) Detection of cyclin D1 and cyclin D1-associated proteins by silver stain. Lane 1 contains molecular weight markers (sizes shown in kilodaltons), lane 2 contains proteins that nonspecifically bind to M2 beads from control NIH 3T3 lysates, and lane 3 contains cyclin D1 complexes isolated from FlagD1-T286A-3T3 lysates (an essentially identical pattern was recovered from Flag-D1-3T3 cells; data not shown). Proteins were eluted with excess Flag peptide. Positions of Flag-D1, CDK4, and p21Cip1 are indicated to the right, as is the position of Hsc70, along with peptides identified by mass spectrometry.
To determine if fractions eluting with a molecular mass of 150 to 200 kDa contained catalytically active cyclin D1/CDK4 complexes, these were subjected to precipitation with a cyclin D1-specific antibody and either assayed for their capacity to phosphorylate recombinant retinoblastoma protein (pRb) in vitro or subjected to immunoblot analysis for associated CDK4. Catalytically active cyclin D1/CDK4 complexes were only detected in fractions that eluted between 150 and 200 kDa (Fig. 1B, lanes 2 to 4, corresponding to lanes 5 to 7 of Fig. 1A). Although cyclin D1 was present in higher-molecular-weight fractions, there was no detectable association of cyclin D1 with CDK4 in these fractions (Fig. 1B, lane 1; data not shown), nor was the cyclin D1 in these fractions catalytically active.
To identify cyclin D1-associated proteins, Flag-tagged cyclin D1-T286A (Fig. 1C) was immunoaffinity purified on M2-agarose beads from whole-cell extracts under conditions that preserved the catalytically active cyclin D1 holoenzyme (33). Cyclin D1 and cyclin D1-copurifying proteins were resolved on a denaturing polyacrylamide gel and visualized by silver staining (Fig. 1C). Several proteins copurified with cyclin D1 but were absent in a control purification from parental NIH 3T3 cells (Fig. 1C). A similar pattern of associated proteins was observed when wild-type Flag-D1 was purified (data not shown).
Bands were excised, digested with trypsin, and subjected to microcapillary electrospray liquid chromatography-tandem mass spectrometry (17). As expected, mass spectra revealed the presence of CDK4 and p21Cip1 as prominent copurifying proteins (Fig. 1C). Here we describe the identification of the 70-kDa protein that copurifies with a stoichiometry approaching that of CDK4 and p21Cip1 (other copurifying proteins will be described elsewhere). Good mass spectra were obtained for two tryptic peptides that were identical with the published sequence of murine Hsc70. While the sequence of peptide 2 (inset, Fig. 1C) is found in both Hsp70 (heat shock-inducible protein 70) and Hsc70 (heat shock cognate or constitutive protein 70), peptide 1 is specific for Hsc70, suggesting that this protein is Hsc70 rather than Hsp70.
Growth factor-dependent regulation of cyclin D1-Hsc70 association.
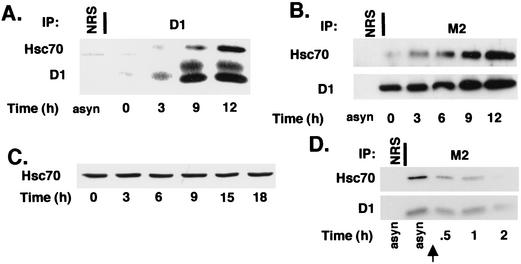
We next assessed binding of Hsc70 to cyclin D1 and determined if binding is mitogen dependent. NIH 3T3 cells arrested in G0 by serum deprivation for 36 h were stimulated to reenter the cell cycle by addition of serum-derived growth factors and harvested at various intervals thereafter. Lysates were prepared from these cells and, following precipitation with a cyclin D1-specific antibody, were resolved on a denaturing polyacrylamide gel. Growth factor-dependent cyclin D1 induction was detectable by 3 h and increased through 12 h, which corresponds to the G1/S boundary, as determined by pulse-labeling with bromodeoxyuridine (data not shown) (42). Coprecipitating Hsc70 was detectable by 9 h and increased through 12 h (Fig. 2A, top panel). CDK4 binding to cyclin D1 was also detectable by 9 h (data not shown).
FIG. 2.
Mitogen-dependent regulation of cyclin D1-Hsc70 binding. (A) Wild-type NIH 3T3 cells were used as an asynchronous culture (asyn) or synchronized by serum deprivation for 36 h and stimulated to reenter the cell cycle by addition of 10% fetal calf serum. Cell lysates were prepared at the indicated intervals (bottom) and immunoprecipitated (IP) with normal rabbit serum (NRS) or a cyclin D1-specific monoclonal antibody (D1). Precipitated proteins were then subjected to immunoblot analysis with antibodies specific for Hsc70 (top) or cyclin D1 (bottom). (B) D1-3T3 cells were synchronized as in panel A. Cell lysates were prepared at the indicated intervals (bottom) and precipitated with normal rabbit serum (NRS) or the M2 monoclonal antibody and subjected to immunoblot analysis with antibodies specific for Hsc70 (top panel) or cyclin D1 (bottom panel). (C) Cell lysates prepared as for panel A were subjected to direct Western analysis with an Hsc70-specific antibody. (D) Whole-cell lysates prepared from asynchronous D1-3T3 cells or D1-3T3 cells cultured in medium containing 0.1% fetal calf serum for the indicated intervals were precipitated with normal rabbit antiserum (NRS) or with the M2 monoclonal antibody. Precipitates were immunoblotted with either Hsc70 (top panel) or cyclin D1 (lower panel) antibodies. The arrow indicates the point at which cells were placed in medium containing 0.1% serum.
While these data suggest that Hsc70-cyclin D1 interactions might be regulated by mitogens, we could not rule out that this increased association during cell cycle reentry reflected increased synthesis of cyclin D1 or Hsc70 during this interval. Therefore, we assessed cyclin D1-Hsc70 binding in cells (D1-3T3) that constitutively overexpress cyclin D1 independently of mitogens. D1-3T3 cells were rendered quiescent by serum deprivation for 36 h and stimulated to reenter the cell cycle with serum-derived growth factors. Flag-D1 was precipitated from cell lysates prepared at various intervals following serum stimulation. Levels of cyclin D1 and cyclin D1-associated Hsc70 were determined by subsequent immunoblot analysis.
A modest increase in Flag-D1 was detectable during this time course (Fig. 2B, bottom panel). As ectopic cyclin D1 expression is not mitogen dependent, this increase likely results from decreased cyclin proteolysis during G1 phase (see Fig. 4) (14). While cyclin D1 is expressed at high levels in quiescent D1-3T3 cells, no Hsc70 binding was detectable in arrested cells (Fig. 2B, top panel, lane 2). Binding of Hsc70 to cyclin D1 increased coincident with serum-derived mitogens and continued to increase as cells progressed through G1 phase (Fig. 2B, lanes 3 to 6). We failed to detect Hsp90 in any cyclin D1 precipitates, consistent with previous work demonstrating that this chaperone interacts primarily with CDK4 prior to its assembly with cyclin D1 (50). Hsc70 protein levels remained constant throughout the course of this experiment (Fig. 2C), demonstrating that the lack of Hsc70 binding does not reflect the absence of Hsc70 protein in quiescent cells.
FIG. 4.
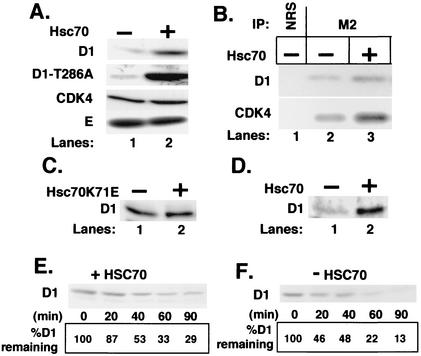
Hsc70 decreases cyclin D1 proteolysis. (A) NIH 3T3 cells were cotransfected with (lane 2) or without (lane 1) a vector encoding Hsc70 along with vectors encoding either cyclin D1 and CDK4 (top and bottom panels), cyclin D1-T286A and CDK4 (second panel), or Myc-tagged cyclin E and CDK2 (bottom panel). Levels of the indicated proteins were determined by direct Western blot analysis of total cell lysates prepared from cells transfected with the indicated vectors. (B) Whole-cell extracts were prepared from NIH 3T3 cells cotransfected with either Flag-D1 and CDK4 (lanes 1 and 2) or Flag-D1, CDK4, and Hsc70 and precipitated with either normal rabbit serum (lane 1) or the M2 monoclonal antibody (lanes 2 and 3). Cyclin D1 and coprecipitating CDK4 were detected by immunoblot analysis with antigen-specific antibodies. (C) NIH 3T3 cells were cotransfected with a plasmid encoding cyclin D1 with (lane 2) or without (lane 1) a vector encoding Hsc70K71E. Levels of cyclin D1 were monitored by immunoblot analysis. (D) p21/p27−/− MEFs were transfected with a plasmid encoding cyclin D1 without (lane 1) or with (lane 2) a plasmid encoding Hsc70. Levels of cyclin D1 were monitored by immunoblot analysis. (E and F) NIH 3T3 cells transfected with vectors encoding either Flag-D1 and CDK4 or Flag-D1, CDK4, and Hsc70 were treated with 50 μg of cycloheximide per ml, and lysates prepared from these cells at the intervals indicated at the bottom of each panel were subjected to immunoblot analysis for cyclin D1. The percentage of cyclin D1 remaining at each time point is indicated at the bottom of each lane.
If cyclin D1 and Hsc70 associate in a growth factor-dependent fashion, removal of serum should result in dissociation of this complex. To test this hypothesis, whole-cell lysates were prepared from asynchronously proliferating D1-3T3 cells or from D1-3T3 cells that had been cultured in medium containing 0.1% fetal calf serum for 0.5, 1, or 2 h. Cyclin D1 complexes were collected with the M2 monoclonal antibody, and levels of cyclin D1 and D1-associated Hsc70 were assessed by immunoblotting. As shown above, high levels of cyclin D1-associated Hsc70 were apparent in proliferating cells (Fig. 2D, lane 2). However, a significant decrease in coprecipitating Hsc70 was apparent 30 min following serum removal (Fig. 2D, lane 3), and by 2 h little or no coprecipitating Hsc70 was detected (Fig. 2D, lane 5). While levels of cyclin D1 also decreased during serum starvation due to increased protein degradation, loss of Hsc70 binding occurred prior to loss of cyclin D1 (Fig. 2D, compare upper and lower panels, lane 3). These results demonstrate that Hsc70 binds to cyclin D1 in vivo and that association is growth factor dependent.
Hsc70 is present in high-molecular-weight complexes and the active 158- to 200-kDa cyclin D1 complex.
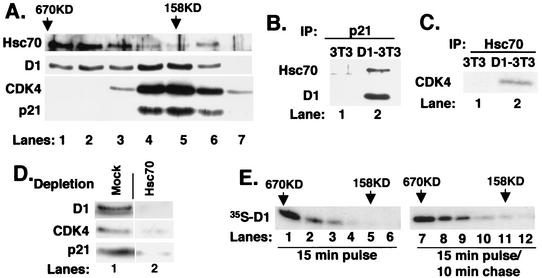
To determine the approximate size of the cyclin D1-Hsc70 complexes, Flag-D1 was purified by M2 affinity chromatography and subsequently resolved by gel filtration chromatography. Fractions collected following gel filtration were analyzed for cyclin D1, Hsc70, p21Cip1, and CDK4. While cyclin D1 was enriched in complexes of 158 to 200 kDa (Fig. 3A, lanes 4 to 5), D1 was also detected in complexes with a mass of 400 to 600 kDa (lane 1). As noted previously, p21Cip1 and CDK4 were associated with cyclin D1 in complexes within the 158- to 200-kDa range (Fig. 3A, lanes 4 to 6). Strikingly, while a majority of Hsc70 was associated with cyclin D1 in the larger complexes (Fig. 3A, lanes 1 to 3), Hsc70 was also associated with cyclin D1 in complexes with an apparent mass of 158 to 200 kDa (Fig. 3A, lanes 4 to 6).
FIG. 3.
Cyclin D1 and Hsc70 are components of multiple complexes. (A) Affinity-purified cyclin D1 complexes were resolved by gel filtration. Ten percent of each fraction was subjected to immunoblot analysis with antibodies specific for Hsc70 (top panel), cyclin D1 (second panel), CDK4 (bottom panel), or p21Cip1 (bottom panel). (B and C) The fraction corresponding to lane 5 in panel A was precipitated with a p21-specific antiserum (B) or an Hsc70 monoclonal antibody (C). Associated proteins were monitored by immunoblot analysis with antibodies specific for the proteins indicated to the left of each panel. (D) The fraction corresponding to lane 5 of part A (158 kDa) was subjected to two rounds of immunodepletion with either normal rabbit antiserum (mock) or the Hsc70-specific monoclonal antibody. Codepletion of cyclin D1 (top panel), CDK4 (middle panel), and p21Cip1 (lower panel) was monitored by immunoblot analysis with the respective antibodies. (E) Asynchronous NIH 3T3 cells were pulse-labeled with [35S]methionine for 15 min (lanes 1 to 6) or pulse-labeled and subsequently chased for 10 min with excess unlabeled methionine for 10 min (lanes 7 to 12). Lysates prepared from these cells were resolved by gel filtration chromatography, and cyclin D1 was precipitated from fractions ranging from 670 kDa to 100 kDa (indicated at the top). Proteins were then analyzed by SDS-PAGE, and cyclin D1 was visualized by autoradiography.
Because chaperones regulate folding of newly synthesized protein, we considered the possibility that high-molecular-weight cyclin D1-Hsc70 complexes (>400 kDa) might represent newly synthesized cyclin D1 associated with Hsc70. The absence of cyclin D1-associated CDK4 in these fractions (Fig. 3A, lanes 1 to 3) is consistent with this notion. To address this, we either pulse-labeled NIH 3T3 cells with [35S]methionine for 15 min or pulsed for 15 min and chased with excess unlabeled methionine for 10 min. Lysates were prepared from these cells, complexes were resolved by gel filtration chromatography, and cyclin D1 was isolated by immunoprecipitation.
In cells pulse-labeled for 15 min with no chase period, a majority of newly synthesized cyclin D1 was present in large complexes of >600 kDa (Fig. 3E, lanes 1 to 2), with none detectable in the 158-kDa fraction (Fig. 3E, lanes 4 to 6). In contrast, following a 10-min chase with unlabeled methionine, radiolabeled cyclin D1 could be detected in fractions that corresponded to a molecular mass of 158 to 200 kDa (Fig. 3D, lanes 10 to 12). These data indicate that the large (>600-kDa) cyclin D1-Hsc70 complexes represent newly synthesized cyclin D1 and imply that Hsc70 functions in the regulation of newly translated cyclin D1.
The above data suggest that Hsc70 associates with newly synthesized cyclin D1 in the absence of CDK4 in proliferating cells. The presence of Hsc70 in 158- to 200-kDa complexes (Fig. 3A, lanes 4 to 6) indicates that Hsc70 remains bound to cyclin D1 following its assembly with CDK4 and is a component of a quaternary complex composed of D1, CDK4, p21Cip1, and Hsc70.
To determine if p21Cip1, cyclin D1, and Hsc70 are indeed present in the same complex, affinity-purified cyclin D1 that fractionated at 158 kDa was reprecipitated with a p21Cip1-specific antiserum and subsequently immunoblotted with either the Hsc70-specific monoclonal antibody or a cyclin D1-specific antibody. Both cyclin D1 and Hsc70 were detected in the p21Cip1 precipitate (Fig. 3B, lane 2). Precipitation of the 158-kDa fraction with the Hsc70 antibody revealed the presence of CDK4 (Fig. 3C).
The data above strongly supported the existence of a tetrameric D1-CDK4-Hsc70-p21Cip1 complex. To provide conclusive support for the existence of this complex, we subjected the fraction corresponding to the affinity-purified 158-kDa cyclin D1 complex (lane 5 of Fig. 3A) to immunodepletion with the Hsc70 monoclonal antibody or with a control antibody. Immunoblot analysis of the depleted extracts revealed that removal of Hsc70 resulted in a corresponding depletion of cyclin D1, CDK4, and p21Cip1 (Fig. 3D, lane 2, upper, middle, and lower panels, respectively). Mock depletion failed to remove cyclin D1, CDK4, or p21Cip1 (lane 1). These data demonstrate that while Hsc70 associates with cyclin D1 in high-molecular-weight complexes devoid of CDK4 and p21Cip1, it remains associated with the catalytically active 158-kDa cyclin D1-CDK4-p21Cip1 complex.
Stabilization of cyclin D1 by Hsc70.
The above results demonstrate that Hsc70 is present in both high-molecular-weight complexes (>400 kDa) and the active 158- to 200-kDa holoenzyme. CDK4 also cofractionates in high-molecular-weight complexes containing the Hsp90-CDC37 chaperone complex that regulates CDK4 folding and stabilization (32, 50). Cyclin E maturation is also regulated by a chaperonin complex, the chaperone containing t-complex (CCT), wherein cyclin E is first cycled through high-molecular-weight complexes containing CCT prior to its association with CDK2 (55). We thus considered whether Hsc70 might regulate maturation of cyclin D1 and thereby determine cyclin D1 protein stability, as was demonstrated for CDK4-chaperone and cyclin E-chaperonin complexes. If Hsc70 regulates cyclin D1 maturation, we reasoned that expression of Hsc70 would promote increased cyclin D1 protein levels and increased binding to its catalytic partner CDK4.
Cyclin D1 and CDK4 were transiently overexpressed in NIH 3T3 cells with or without Hsc70. Direct immunoblot analysis with a cyclin D1 antibody revealed a significant increase in cyclin D1 in cells cotransfected with Hsc70 (Fig. 4A, top panel). In contrast, levels of CDK4 (Fig. 4A, bottom panel) or Myc-tagged cyclin E (Fig. 4A, bottom panel) were not affected by expression of Hsc70, demonstrating the specificity of the targeted interaction. Additionally, increased levels of CDK4 were detected in cyclin D1 precipitates (Fig. 4B, lane 2 to 3), consistent with Hsc70-dependent promotion of cyclin D1 maturation. The binding and release cycle of Hsc70 with folding intermediates is dependent upon Hsc70 ATPase activity (8). This activity can be inhibited by mutation of lysine 71 (39). To determine if Hsc70 “chaperone” activity is required for cyclin D1 stabilization, we transfected NIH 3T3 cells with plasmids encoding Hsc70K71E along with cyclin D1 and CDK4. Direct immunoblot analysis with a cyclin D1 antibody revealed no increase in cyclin D1 levels in cells cotransfected with Hsc70K71E (Fig. 4C).
Previous work demonstrated a role for Cip/Kip proteins in cyclin D1 stabilization via regulation of cyclin nuclear export (5, 20). To determine whether increased cyclin D1 levels detected in cells overexpressing Hsc70 were dependent upon Cip/Kip, we tested the ability of Hsc70 to promote increased levels of cyclin D1 in p21/p27 double null (p21/p27−/−) murine embryonic fibroblasts (MEFs). Cyclin D1 was transiently overexpressed in p21/p27−/− MEFs with or without wild-type Hsc70. Immunoblot analysis revealed a significant elevation in cyclin D1 levels in p21/p27−/− MEFs overexpressing Hsc70 (Fig. 4D).
These data suggest that in the absence of ectopic Hsc70, overexpressed cyclin D1 could not be properly folded and incorporated into CDK4 complexes and was likely shunted off for proteolysis. To test this, we measured cyclin D1 turnover in the absence and presence of ectopic Hsc70. We utilized the protein synthesis inhibitor cycloheximide (50 μg/ml) to block new protein synthesis and monitored the rate of cyclin D1 decay by direct Western blot analysis with a cyclin D1 monoclonal antibody. Cycloheximide was added to asynchronously proliferating NIH 3T3 cells transfected with vectors encoding either cyclin D1 and CDK4 or cyclin D1, CDK4, and Hsc70, and cells were harvested at the indicated intervals. By 20 min, greater than 50% of cyclin D1 had been degraded in the absence of ectopic Hsc70 (Fig. 4F). In the presence of ectopic Hsc70, greater than 50% of cyclin D1 still remained at 40 min (Fig. 4E). These results demonstrate that Hsc70 can stabilize cyclin D1.
We have previously shown that proteolysis of mature cyclin D1 depends on phosphorylation of threonine 286 (16). This residue is targeted by the protein kinase glycogen synthase kinase 3β, preferentially in the context of a cyclin D1/CDK4 complex (14). We reasoned that if increased cyclin D1 turnover in the absence of Hsc70 resulted from improper folding, phosphorylation would not be required for proteolysis and would thus occur independently of Thr-286 phosphorylation. Consistent with this hypothesis, coexpression of Hsc70 effectively promoted increased stabilization of the nonphosphorylatable D1-T286A mutant (Fig. 4A, second panel). In addition, immunoblotting with a phospho-Thr-286-specific antibody revealed that overexpression of Hsc70 did not affect turnover of phosphorylated cyclin D1 (data not shown). These data demonstrate that overexpression Hsc70 stabilizes cyclin D1, likely through the promotion of cyclin D1 protein maturation.
Reconstitution of a 158-kDa cyclin D1 holoenzyme with Hsc70.
Active cyclin D complexes can be reconstituted in insect Sf9 cells by coinfection with only a D-type cyclin and CDK4 (7, 33). These complexes have an apparent size of 70 kDa (7), approximately 100 kDa smaller than that of the native cyclin D1 holoenzyme. Both p21Cip1 and p27Kip1 have been identified as components of the active 150- to 200-kDa cyclin D/CDK complex (7, 9, 28, 49). However, a ternary complex composed of cyclin D1, CDK4, and p21 (p27) is predicted to elute with a mass of approximately 100 kDa, smaller than that of the native cyclin D1 holoenzyme. Copurification of Hsc70 with the 158-kDa cyclin D1 complex suggested that Hsc70 might represent the missing 50 to 80 kDa. Therefore, we determined if coexpression of Hsc70 along with cyclin D1, CDK4, and p21Cip1 is sufficient to reconstitute a 158-kDa cyclin D1 complex.
Lysates prepared from Sf9 cells infected with cyclin D1, CDK4, and p21Cip1 with and without Hsc70 were resolved by gel filtration chromatography. Fractions were subjected to immunoblot analysis with either an Hsc70 antibody or a cyclin D1 monoclonal antibody. In the absence of ectopic Hsc70, cyclin D1 complexes ranged from 70 to 130 kDa (Fig. 5A, lanes 10 to 11), consistent with a ternary complex composed of cyclin D1 (36 kDa), CDK4 (34 kDa), and p21Cip1(21 kDa). In the presence of ectopic Hsc70, a subset of cyclin D1 complexes eluted at 158 kDa (Fig. 5A, lane 3), consistent with the formation of a D1-CDK4-p21-Hsc70 quaternary complex. Immunoblot analysis confirmed the presence of Hsc70 in the 158-kDa fraction with cyclin D1 (Fig. 5A, top panel, lane 3). In addition, Hsc70 was found to coprecipitate with cyclin D1 from fractions corresponding to 158 kDa (Fig. 5B).
FIG. 5.
Reconstitution of a 158-kDa cyclin-CDK complex. (A) Sf9 insect cells were infected with baculoviruses encoding cyclin D1, CDK4, and p21Cip1 with (lanes 1 to 6) or without (lanes 7 to 12) Hsc70. Lysates prepared from these cells were resolved by gel filtration chromatography and subjected to immunoblot analysis for Hsc70 (top panel) or cyclin D1 (bottom panel). The elution positions of molecular weight standards are indicated at the top of each panel. (B) Fractions corresponding to lanes 3 and 4 of part A were precipitated with the cyclin D1 monoclonal antibody. Coprecipitation of Hsc70 was monitored by immunoblotting with the Hsc70 monoclonal antibody. (C) Lysates prepared from Sf9 insect cells infected with baculoviruses encoding either cyclin D1 and CDK4 (lane 1), cyclin D1, CDK4, and Hsc70 (lane 2), cyclin D1, CDK4, and p21Cip1 (lane 3), or all four (lane 4) and metabolically labeled with [35S]methionine were precipitated with the cyclin D1 monoclonal antibody. Positions of Hsc70, cyclin D1, CDK4, and p21Cip1 are indicated to the left of the panel and were verified independently by immunoprecipitation with antigen-specific antibodies (data not shown). Labeled proteins were visualized by autoradiography.
Given the presence of Hsc70 in the active cyclin D1/CDK4/p21 complex, we also considered whether Hsc70-dependent folding of cyclin D1 might be critical for assembly of cyclin D1 with CDK4. Insect Sf9 cells make an ideal system to explore the regulation of cyclin D1/CDK4 assembly. Sf9 cells express the necessary machinery to support production of an active cyclin D1/CDK4 kinase but assembly remains inefficient, suggesting that under conditions of chronic cyclin D1-CDK4 overexpression, there are rate-limiting components (Fig. 5C) (50). Expression of Hsc70 alone did not promote a reproducible increase in cyclin D1-CDK4 association (Fig. 5C, lanes 1 to 2), while expression of p21Cip1 resulted in a significant increase in cyclin-CDK binding (Fig. 5C, compare lanes 2 and 3). Expression of p21Cip1 did not affect Hsc70-cyclin association (Fig. 5C, compare lanes 2 and 4). Thus, while p21Cip1 may be necessary for highly efficient cyclin D1/CDK4 assembly, cyclin D1-Hsc70 interactions occur independently of p21Cip1. These data suggest that Hsc70 is not an assembly factor for the cyclin D1/CDK4 holoenzyme.
While p21Cip1 is required for cyclin D1/CDK4 assembly, it is also an effective inhibitor of cyclin D1-dependent cell cycle progression when ectopically expressed at high levels (15, 21). One explanation for this observed cell cycle inhibition is that under conditions of transient transfection of only cyclin D1-CDK4-p21Cip1, Hsc70 becomes a limiting component. The ability of ectopic Hsc70 to promote stabilization of transiently overexpressed cyclin D1 is consistent with this notion. We therefore considered the possibility that ectopic Hsc70 might compensate for p21Cip1 overexpression via increasing cyclin D1 stability and thereby promote the formation of catalytically active cyclin D1 holoenzyme.
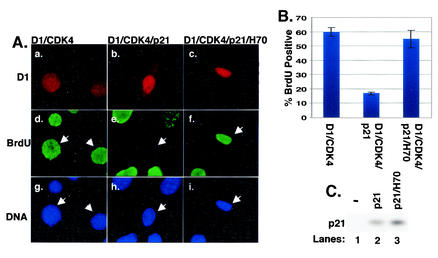
To address this notion NIH 3T3 cells were transiently transfected with vectors encoding cyclin D1 and CDK4, cyclin D1, CDK4, and p21Cip1, or cyclin D1, CDK4, p21Cip1, and Hsc70. Twenty-four hours posttransfection, cells were labeled with bromodeoxyuridine for an additional 20 h and subsequently fixed and stained with antibodies specific for ectopic cyclin D1 (M2 monoclonal, specific for an N-terminal Flag epitope tag; red) and bromodeoxyuridine (green). A majority of cells expressing only cyclin D1 and CDK4 were bromodeoxyuridine positive (Fig. 6A, panels a and d; quantitation in Fig. 6B). Cotransfection of an equivalent concentration of vector encoding p21Cip1 resulted in a dramatic reduction in cells that stained positively for both cyclin D1 and bromodeoxyuridine (Fig. 6A, panels b and e; Fig. 6B), consistent with p21Cip1-dependent cell cycle arrest. Cotransfection of Hsc70 was sufficient to restore cell cycle progression in the presence of ectopic p21Cip1 (Fig. 6A, panels c and f; Fig. 6B). The failure of cells cotransfected with both Hsc70 and p21Cip1 to undergo cell cycle arrest was not due to reduced p21Cip1 expression in these cells (Fig. 6C, lanes 2 to 3). These data demonstrate that Hsc70 association with cyclin D1/CDK4 complexes attenuates the inhibitory activity of p21Cip1 without compromising the ability of p21Cip1 to promote cyclin D1/CDK4 assembly.
FIG. 6.
Hsc70 maintains cyclin D1-dependent cell cycle progression in the presence of inhibitory levels of p21Cip1. (A) NIH 3T3 cells were transfected with vectors encoding Flag-D1 and CDK4, Flag-D1, CDK4, and p21Cip1, or Flag-D1, CDK4, p21Cip1, and Hsc70; 24 h posttransfection, cells were labeled with bromodeoxyuridine (BrdU) for 20 h. Cells were fixed and stained with monoclonal M2 antibody (red), bromodeoxyuridine (green), and Hoechst dye (blue). (B) Quantitation of panel A, representing three independent experiments; error bars represent standard errors between experiments. (C) Whole-cell lysates prepared from cells transfected as for panel A were resolved by SDS-PAGE, and levels of p21Cip1 were monitored by immunoblot analysis with a p21-specific antibody.
DISCUSSION
Identification of Hsc70 as a cyclin D1 binding protein in vivo.
The cyclin D1-dependent kinase is a critical mediator of G1 phase progression, deregulation of which contributes to a variety of cancers in both mouse model systems and humans (13, 45). As such, characterization of cyclin D1 and cyclin D1 complexes in vivo is critical for elucidation of its growth-regulatory functions. The D-type cyclins, and cyclin D1 in particular, can bind to a variety of proteins, including those involved in cell cycle regulation, Rb, CDK4/6, and Cip/Kip family members (46), DNA-binding transcription factors such as nuclear hormone receptors (38, 41, 56), STAT family members (6), v-Myb (18), the Myb-like protein DMP1 (24), and the DNA-modifying enzyme p300/CREB-binding protein-associated protein (35). Many of these interactions have been documented by reconstitution and likely occur at substoichiometric levels.
We purified cyclin D1 complexes under native conditions that maintained the integrity of the cyclin D1-dependent kinase. Visualization of the purified cyclin D1 complex by silver staining revealed several polypeptides that specifically copurified with cyclin D1. Tandem mass spectroscopy analysis of the 70-kDa copurifying protein revealed two peptides that were identical to murine Hsc70, the constitutively expressed member of the Hsp70 family of chaperones (1). Immunoblot analysis with a monoclonal antibody that recognizes Hsc70 but not other Hsp70 family members confirmed that Hsc70 does indeed associate with cyclin D1 and cyclin D1 complexes. Hsc70 copurified with cyclin D1 in both high-molecular-weight complexes (>400 kDa) and fractions containing the active cyclin D1/CDK4/p21Cip1 holoenzyme (150 to 200 kDa), suggesting that it may perform multiple regulatory functions with regard to cyclin D1.
Hsp70/Hsc70 chaperones localize to both nuclear and cytosolic compartments, where they regulate protein maturation and function (1). In the cytosol, they bind newly synthesized proteins and maintain them in a folding-competent conformation (1, 8). The Hsp70/Hsc70 family also regulates protein degradation via associated cochaperones such as CHIP (36) and controls the activity of certain nuclear DNA-binding transcription factors (11, 22).
Considering the role of chaperones in the maturation of other cell cycle proteins such as CDK4 and cyclin E along with the documented role of protein degradation in the regulation of cyclin D levels, Hsc70 function could either promote or attenuate cyclin D1 protein accumulation. Consistent with the former, we found that ectopic expression of Hsc70 reduces the rate of cyclin D1 turnover, thereby increasing cyclin D1 steady-state levels. Increased cyclin D1 levels coincided with its availability to associate with and activate its catalytic partner CDK4. We also noted that Hsc70 could promote accumulation of cyclin D2, suggesting that Hsc70 likely functions in the maturation of all D-type cyclins (data not shown). In addition, an Hsc70 mutant that lacks ATPase activity, Hsc70K71E, failed to stabilize cyclin D1, demonstrating that Hsc70 chaperone function is required for cyclin D1 maturation.
The fact that a majority of copurifying Hsc70 is found in high-molecular-weight fractions, where newly synthesized cyclin D1 is preferentially localized, suggests that Hsc70 initially facilitates folding of monomeric cyclin D1 prior to its assembly into complexes with CDK4. The necessity for coexpression of Hsc70 along with cyclin D1 implies that Hsc70 becomes a limiting factor when cyclin D1 is overexpressed. In turn, the absence of sufficient Hsc70 results in improper D1 folding and increased cyclin proteolysis. Similar conclusions were reached in the examination of cyclin E maturation by the chaperonin complex CCT (55). Newly synthesized cyclin E is found to preferentially cofractionate in high-molecular-weight complexes with CCT, and only after brief chase intervals is it found in lower-molecular-weight fractions containing its catalytic partner CDK2. Similar to the Hsc70-D1 relationship, CCT functions to stabilize newly synthesized cyclin E, and thus, in its absence cyclin E is rapidly degraded via the 26S proteasome.
Whether Hsc70 promotes protein maturation versus protein triage may largely depend upon associated cochaperones such as CHIP (11, 36) or Bag1 (31, 48, 52). Thus, it is possible that Hsc70-dependent regulation of cyclin D1 depends upon an associated cochaperone. Newly synthesized cyclin D1 is enriched in high-molecular-weight complexes with Hsc70. Associated regulatory cochaperones could partially account for the large size of these complexes. Alternatively, the relative mass of these “folding” complexes may also reflect the capacity of multiple Hsc70 molecules to bind to unfolded and extended cyclin D1. Hsc70 regulates protein folding via stabilization of unfolded polypeptides (1). It preferentially recognizes seven-residue peptides containing a hydrophobic core (23, 44); such motifs are predicted to occur approximately every 36 residues (8). Based on this, the Hsc70-D1 ratio could approach 7:1, resulting in a putative 530-kDa complex. Based on this, we anticipate that the stoichiometry of Hsc70 to D1(newly synthesized) would be greater than 1, and as cyclin D1 is folded, Hsc70 will be released and the Hsc70-D1 ratio would approach 1:1.
The cyclin D1 holoenzyme has been characterized as a 158- to 200-kDa entity that contains a D-type cyclin, CDK4, and a Cip/Kip family member that acts to assemble the cyclin D/CDK4 complex and maintain it in the nucleus (9, 28, 40). The predicted size of such a complex is approximately 90 to 100 kDa, suggesting the existence of an additional component. Our data suggest that a complex composed of cyclin D1, CDK4, p21Cip1, and Hsc70 constitutes at least one of the active 158- to 200-kDa cyclin D1 complexes. While a majority of the Hsc70 that copurifies with cyclin D1 fractionates in high-molecular-weight complexes, Hsc70 is also present in the 158-kDa cyclin D1/CDK4/p21Cip1 complex (Fig. 3). This is in contrast to CDK4-associated chaperone complexes, which do not stably interact with cyclin-bound CDK4 (12, 29, 50).
If Hsc70 regulates proper folding of cyclin D1 prior to association with CDK4, what is its role in the mature quaternary complex? One possibility is that Hsc70 promotes mitogen-dependent assembly of cyclin D1 with CDK4. The ability of mitogens to promote association of Hsc70 with cyclin D1 is temporally consistent with this idea. A second possibility is that the continued presence of Hsc70 promotes “productive” assembly of the quaternary holoenzyme complex, thereby ensuring cyclin D1/CDK4 catalytic activity in the presence of Cip/Kip molecules. Three observations suggest that the latter is the most likely function. First, expression of Hsc70, unlike that of p21Cip1 (28), is not sufficient to promote D1/CDK4 assembly in an Sf9 system, arguing that Hsc70 is not an assembly factor. Second, we found that transfection of mouse fibroblasts with equimolar concentrations of plasmids encoding p21Cip1, cyclin D1, and CDK4 resulted in cell cycle arrest. In contrast, coexpression of Hsc70 prevented cell cycle arrest, consistent with its ability to maintain catalytically active D1/CDK4/p21Cip1 complexes. Finally, preliminary work suggests that expression of Hsc70 is sufficient to promote increased cyclin D1-dependent kinase activity in the presence of high concentrations of p21Cip1 or p27Kip1 (data not shown).
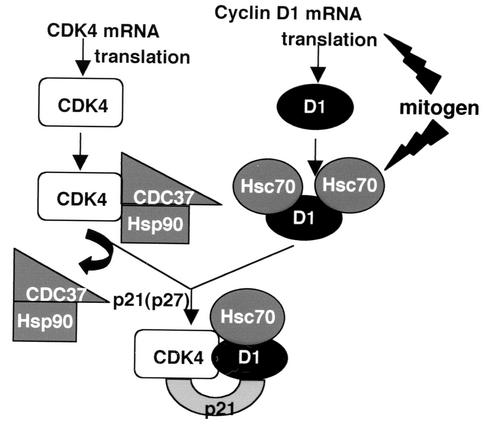
The previous demonstration that Hsp90 and CDC37 determine the stability of newly synthesized CDK4, along with our demonstration that Hsc70 interacts with cyclin D1 and cyclin D1/CDK4 complexes to facilitate cyclin stabilization and complex activation, indicates that chaperones are critical regulators of cyclin/CDK maturation and function. Collectively, these data are consistent with the model illustrated in Fig. 7. Maturation and stabilization of constitutively synthesized CDK4 are regulated by Hsp90 via the CDC37 targeting subunit (50). In response to mitogens, cyclin D1 is produced and associates with Hsc70. In the absence of mitogen-activated Hsc70, cyclin D1 is unstable due to its propensity for misfolding and aggregation. Misfolded cyclin D1 is rapidly degraded and is unavailable for assembly with CDK4. Binding of Hsc70 in response to mitogen stimulation functions to fold cyclin D1, thereby increasing its stability. Cyclin D1 stabilization during protein maturation is independent of p21Cip1 or p27Kip1 binding (Fig. 4C). Following chaperone-mediated subunit maturation, cyclin D1-CDK4 assembly is promoted by a Cip/Kip family protein (28). In contrast to Hsp90 and CDC37, which appear to dissociate from CDK4 prior to its assembly with a D-type cyclin, Hsc70 remains a component of the holoenzyme, where it functions to maintain cyclin-CDK activity in the presence of a potentially inhibitory Cip/Kip subunit.
FIG. 7.
Model depicting the participation of chaperones in the regulation of cyclin D1/CDK4 complex maturation (details in text).
Our data suggest that the association of Hsc70 with cyclin D1 constitutes yet one more potentially rate-limiting, mitogen-regulated step in the activation of the cyclin D/CDK enzyme. It is not clear how mitogens regulate Hsc70 function with regard to cyclin D1. While Hsc70 abundance does not fluctuate significantly during the cell cycle, Hsc70 gene expression does increase during G1 phase (25). It is therefore possible that it is newly synthesized Hsc70 that regulates cyclin D1. Alternatively, Hsc70 may be subject to mitogen-regulated posttranslational modifications. Consistent with this notion, we noted that copurified Hsc70 migrates as multiple bands on silver-stained gels. In addition, our preliminary experiments suggest that activated MEK1 is sufficient to trigger cyclin D1-Hsc70 association (data not shown). This is consistent with the ability of MEK1 to drive both cyclin D1 accumulation and its assembly with CDK4 (10). Further efforts are required to elucidate how growth factor-dependent signaling pathways regulate cyclin D1 maturation via modulation of Hsc70 activity.
Acknowledgments
We thank Richard Morimoto for providing the Hsc70 cDNA, David Smith for the Hsc70K71E cDNA, Greg Enders for providing the CDK2 cDNA, and Bruce Clurman for Myc-tagged cyclin E.
We acknowledge support from the American Cancer Society (grant RPG-00-303-01), National Institutes of Health (grant CA93237), the V Foundation (J.A.D.), and the National Science and Engineering Research Council of Canada (A.E.).
REFERENCES
- 1.Agshe, V. R., and F.-U. Hartl. 2000. Roles of molecular chaperones in cytoplasmic protein folding. Cell Dev. Biol. 11:15-25. [DOI] [PubMed] [Google Scholar]
- 2.Aktas, H., H. Cai, and G. M. Cooper. 1997. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol. Cell. Biol. 17:3850-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 4.Alt, J. R., J. L. Cleveland, M. Hannink, and J. A. Diehl. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 14:3102-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alt, J. R., A. B. Gladden, and J. A. Diehl. 2002. p21Cip1 promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J. Biol. Chem. 277:8517-8523. [DOI] [PubMed] [Google Scholar]
- 6.Bienvenu, F., H. Gascan, and O. Coqueret. 2001. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J. Biol. Chem. 276:16840-16847. [DOI] [PubMed] [Google Scholar]
- 7.Blain, S. W., E. Montalvo, and J. Massague. 1997. Differential interaction of the cyclin-dependent kinase (CDK) inhibitor p27Kip1 with cyclin A-CDK2 and cyclin D2-CDK4. J. Biol. Chem. 272:25863-25872. [DOI] [PubMed] [Google Scholar]
- 8.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, C., P. Olivier, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21Cip1 and p27Kip1 CDK ‘inhibitors' are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18:1571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, M., V. Sexl, C. J. Sherr, and M. F. Roussel. 1998. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1). Proc. Natl. Acad. Sci. USA 95:1091-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93-96. [DOI] [PubMed] [Google Scholar]
- 12.Dai, K., R. Kobayashi, and D. Beach. 1996. Physical interaction of the mammalian CDC37 with CDK4. J. Biol. Chem. 271:22030-22032. [DOI] [PubMed] [Google Scholar]
- 13.Diehl, J. A. 2002. Cyclin to cancer with cyclin D1. Cancer Biol. Ther. 1:226-231. [DOI] [PubMed] [Google Scholar]
- 14.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl, J. A., and C. J. Sherr. 1997. A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol. Cell. Biol. 17:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehl, J. A., F. Zindy, and C. J. Sherr. 1997. Inhibition of cyclin D phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11:957-972. [DOI] [PubMed] [Google Scholar]
- 17.Emili, A., D. M. Schieltz, J. R. Yates III, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 18.Ganter, B., S. Fu, and J. S. Lipsick. 1998. D-type cyclins repress transcriptional activation by the v-Myb but not the c-Myb DNA-binding domain. EMBO J. 17:255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatlin, C. L., G. R. Kleemann, L. G. Hays, A. J. Link, and J. R. Yates III. 1998. Protein identification at the low femtomole level from silver-stained gels with a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263:93-101. [DOI] [PubMed] [Google Scholar]
- 20.Gerace, L. 1995. Nuclear export signals and the fast track to the cytoplasm. Cell 82:341-344. [DOI] [PubMed] [Google Scholar]
- 21.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 cdk-interacting protein cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 22.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-580. [DOI] [PubMed] [Google Scholar]
- 23.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 24.Hirai, H., and C. J. Sherr. 1996. Interaction of D-type cyclins with a novel Myb-like transcription factor, DMP1. Mol. Cell. Biol. 16:6457-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt, C. R., A. J. Parsian, P. C. Goswami, and C. A. Kozak. 1999. Characterization and expression of the mouse Hsc70 gene. Biochim. Biophys. Acta 1444:315-325. [DOI] [PubMed] [Google Scholar]
- 26.Kato, J.-Y., M. Matsuoka, D. K. Strom, and C. J. Sherr. 1994. Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol. Cell. Biol. 14:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkhoff, E., and U. R. Rapp. 1997. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Mol. Cell. Biol. 17:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 29.Lamphere, L., F. Fiore, X. Xu, L. Brizuela, S. Keezer, C. Sardet, G. F. Draetta, and J. Gyuris. 1997. Interaction between Cdc37 and Cdk4 in human cells. Oncogene 14:1999-2004. [DOI] [PubMed] [Google Scholar]
- 30.Lavoie, J. N., G. L'Allemain, A. Brunet, R. Muller, and J. Pouyssegur. 1996. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 271:20608-20616. [DOI] [PubMed] [Google Scholar]
- 31.Luders, J., J. Demand, and J. Hohfeld. 2000. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275:4613-4617. [DOI] [PubMed] [Google Scholar]
- 32.Mahony, D., D. A. Parry, and E. Lees. 1998. Active cdk6 complexes are predominantly nuclear and represent only a minority of the cdk6 in T cells. Oncogene 16:603-611. [DOI] [PubMed] [Google Scholar]
- 33.Matsushime, H., D. E. Quelle, S. A. Shurtleff, M. Shibuya, C. J. Sherr, and J.-Y. Kato. 1994. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 14:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McConnell, B. B., F. J. Gregory, F. J. Stott, E. Hara, and G. Peters. 1999. Induced expression of p16INK4a inhibits both CDK4- and CDK2-associated kinase activity by reassortment of cyclin-CDK-inhibitor complexes. Mol. Cell. Biol. 19:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon, C., T. Suthiphongchai, J. DiRenzo, and M. E. Ewen. 1999. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc. Natl. Acad. Sci. USA 96:5382-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 37.Muraoka, R. S., A. E. G. Lenferink, B. Law, E. Hamilton, D. M. Brantley, L. R. Roebuck, and C. L. Arteaga. 2002. ErbB2/Neu-induced, cyclin D1-dependent transformation is accelerated in p27-haploinsufficient mammary epithelial cells but impaired in p27-null cells. Mol. Cell. Biol. 22:2204-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuman, E., M. H. Ladha, N. Lin, T. M. Upton, S. J. Miller, J. DiRenzo, R. G. Pestell, P. W. Hinds, S. F. Dowdy, M. Brown, and M. E. Ewen. 1997. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol. Cell. Biol. 17:5338-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Brien, M. C., K. M. Flaherty, and D. B. McKay. 1996. Lysine 71 of the chaperone protein Hsc70 is essential for ATP hydrolysis. J. Biol. Chem. 271:15874-15877.8. [DOI] [PubMed] [Google Scholar]
- 40.Parry, D., D. Mahony, K. Wills, and E. Lees. 1999. Cyclin D-CDK subunit arrangement is dependent on the availability of competing INK4 and p21 class inhibitors. Mol. Cell. Biol. 19:1775-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petre, C. E., Y. B. Wetherill, M. Danielsen, and K. E. Knudsen. 2002. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J. Biol. Chem. 277:2207-2215. [DOI] [PubMed] [Google Scholar]
- 42.Quelle, D. E., R. A. Ashmun, S. E. Shurtleff, J. Y. Kato, D. Bar-Sagi, M. F. Roussel, and C. J. Sherr. 1993. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7:1559-1571. [DOI] [PubMed] [Google Scholar]
- 43.Rimerman, R. A., A. Gellert-Randleman, and J. A. Diehl. 2000. Wnt1 and MEK1 cooperate to promote cyclin D1 accumulation and cellular transformation. J. Biol. Chem. 275:14736-14742. [DOI] [PubMed] [Google Scholar]
- 44.Rudiger, S., L. Germeroth, J. Schneider-Mergener, and B. Bukau. 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16:1501-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 46.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 47.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 48.Song, J., M. Takeda, and R. I. Morimoto. 2001. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 49.Soos, T. J., H. Kiyokawa, J. S. Yan, M. S. Rubin, A. Giordano, A. DeBlasio, S. Bottega, B. Wong, J. Medelsohn, and A. Koff. 1996. Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ. 7:135-146. [PubMed] [Google Scholar]
- 50.Stepanova, L., X. Leng, S. B. Parker, and J. W. Harper. 1997. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 10:1491-1502. [DOI] [PubMed] [Google Scholar]
- 51.Summers, M. D., and G. E. Smith. 1987. A manual of methods for baculovirus vectors and insect culture procedures. Texas Agricultural Experiment Station Bulletin 1555. Texas Agricultural Experiment Station, Amarillo.
- 52.Takayama, S., T. Sato, S. Krajewski, K. Kochel, S. Irie, J. A. Millan, and J. C. Reed. 1996. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell 80:279-284. [DOI] [PubMed] [Google Scholar]
- 53.Treinies, I., J. F. Paterson, S. Hooper, R. Wilson, and C. J. Marshall. 1999. Activated MEK stimulates expression of AP1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol. Cell. Biol. 19:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber, J. D., D. M. Raben, P. J. Phillips, and J. J. Baldassare. 1997. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D in G1 phase. Biochem. J. 326:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Won, K.-A., R. J. Schumacher, G. W. Farr, A. L. Horwich, and S. I. Reed. 1998. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol. Cell. Biol. 18:7584-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwijsen, R. M., E. Wientjens, R. Klompmaker, J. van der Sman, R. Bernards, and R. J. Michalides. 1997. CDK-independent activation of estrogen receptor by cyclin D1. Cell 88:405-415. [DOI] [PubMed] [Google Scholar]