Abstract
Two closely related p21-activated kinases from Saccharomyces cerevisiae, Ste20 and Cla4, interact with and are regulated by Cdc42, a small Rho-like GTPase. These kinases are argued to perform a common essential function, based on the observation that the single mutants are viable whereas the double mutant is inviable. Despite having a common upstream regulator and at least one common function, these molecules also have many distinct cellular signaling roles. Ste20 signals upstream of several mitogen-activated protein kinase cascades (e.g., pheromone response, filamentous growth, and high osmolarity), and Cla4 signals during budding and cytokinesis. In order to investigate how these kinases are directed to distinct functions, we sought to identify specificity determinants within Ste20 and Cla4. To this end, we constructed both chimeric fusions and point mutants and tested their ability to perform unique and shared cellular roles. Specificity determinants for both kinases were mapped to the C-terminal kinase domains. Remarkably, the substitution of a single amino acid, threonine 818, from Ste20 into an otherwise wild-type Cla4, Cla4D772T, conferred the ability to perform many Ste20-specific functions.
Protein kinases of the PAK (p21-activated kinase) family are found in all eukaryotic species examined (1, 5, 23, 46). This family is defined by sequence similarity in the kinase domains and by the occurrence of a domain that can be bound by Cdc42, a p21 GTPase of the RAS superfamily (23). Cdc42 is thought to activate and thereby contribute to the regulation of these protein kinases (27, 45).
Saccharomyces cerevisiae cells contain three members of this family, Ste20, Cla4, and Skm1, each of which appears to perform a distinct set of cellular roles. Ste20, the founding member of the PAK family, was identified by its role in the pheromone response pathway and was subsequently shown to participate in two other signal transduction pathways, the haploid invasive growth pathway and the osmosensing high-osmolarity glycerol (HOG) pathway (16, 17, 26, 28, 31, 32, 34). Cla4 was identified by its requirement for viability in the absence of the G1 cyclins Cln1 and Cln2 (9). Cla4 promotes normal septin function and the subsequent regulation of polarized growth (2, 4, 10, 21, 42, 44). In addition to these unique roles, it appears that Cla4 and Ste20 share at least one function, a possibility that follows from the observation that their simultaneous loss is lethal (9). Skm1 was identified by sequence similarity to Ste20 and Cla4 and has not yet had a function ascribed to it (24). Mutations in SKM1 confer no observable phenotype, nor do they show synthetic genetic interactions with CLA4 or STE20 (24).
In an effort to understand how Ste20 and Cla4 can carry out both unique and shared functions, we sought to identify regions of each protein that were responsible for directing functional specificity by constructing chimeric proteins. The large N-terminal regions of Ste20 and Cla4 contain the PAK domains. This N-terminal region of Cla4, but not Ste20, also contains a pleckstrin homology (PH) domain. Aside from the PAK domains, which exhibit 42% identity, the N-terminal domains of Cla4 and Ste20 show little sequence similarity. In contrast, the C-terminal regions, which contain the 11 subdomains that characterize all protein kinases, are highly similar (54% identical over the kinase domains) except at the extreme C terminus, where each protein has a unique, relatively short sequence. In the case of Ste20, this extreme C-terminal segment has been argued elsewhere to be a protein-protein interaction domain required for binding to Ste4, the β subunit of the pheromone response pathway heterotrimeric G protein (18). Whether the Cla4 C-terminal segment has an analogous function is not known; no targets have been identified.
We constructed an initial set of chimeras based on the apparent domain structure summarized above. The functional properties of these chimeras argued that the N-terminal domains were interchangeable, and we therefore constructed a set of chimeras in which regions within the C-terminal kinase domain and protein-protein interaction domain were swapped. Analysis of this set of chimeras suggests that specificity determinants can be mapped to the C-terminal regions of Cla4 and Ste20. Specifically, the presence of kinase subdomains III through X from Cla4 is sufficient to provide the morphological function of Cla4 to the chimeras. Conversely, the presence of Ste20 sequence spanning kinase subdomain IX to the C terminus is sufficient to direct chimeric fusions to function in the pheromone response and HOG pathways. Strikingly, a single amino acid substitution in an otherwise wild-type Cla4 (Cla4D772T) allows it to carry out many Ste20-specific functions.
MATERIALS AND METHODS
Yeast manipulations.
Strains used in this study are listed in Table 1. Standard media and yeast manipulations were used (35, 37).
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| SY3357a | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ | D. Mitchell and G. Sprague |
| SY3360a | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ cla4::TRP1 | D. Mitchell and G. Sprague |
| SY3748a | MATα leu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ cla4::TRP1 ste20::HIS3 〈pRS316ADE8CLA4〉 | This study |
| SY3749a | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ ste20::HIS3 | This study |
| SY3750a | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ ste20::HIS3 bar1::kanamycin | This study |
| SY3751a | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ cla4::TRP1 cln1::HIS3 cln2::HIS3 〈pRS316ADE8CLA4〉 | This study |
| SY3752b | MATα Σ ura3-52 ste20::ura3− (made ura3− on 5-FOA plate) leu2::URA3 | April Goehring |
| SY3753a | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ ste20::HIS3 ssk1::URA3 | This study |
| SY3970a | MATa/α leu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ ste20::HIS3 | April Goehring |
| SY3971a | MATα leu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ ste20::TRP1 | David Mitchell |
| 227 | MATα lys1 cry1 | Ira Herskowitz |
These yeast strains are derivatives of YPH499 and YPH500 (S288C) (39).
This yeast strain is a derivative of HY334 (of the Σ1278 background) provided by G. Fink, Whitehead Institute for Biomedical Research, Cambridge, Mass.
Construction of plasmids.
The expression of all chimeric constructs, as well as the full-length STE20 and CLA4 constructs, was driven by 446 bp of STE20 upstream sequence (coordinates −1 to −446 with respect to the STE20 translation initiation codon).
(i) pSTE20-HA (pSL2731) contains PCR-amplified STE20 (with primersMK55:5′-CCG-CCT-TCT-AGT-AGG-GCC-CCT-GCC-C-3′ and MK56:5′-CGA-GAG-AGG-ATG-TTA-CCG-CGG-TAG-GAA-3′), which was cut at the flanking ApaI and SacII restriction sites and then ligated into the corresponding sites of pRS315 (39). A DNA fragment encoding a C-terminal triple hemagglutinin (HA) epitope was fused to STE20 to make pSTE20-HA by recombination-based subcloning (20). The PCR fragment used for this subcloning contained DNA encoding the HA epitope flanked on one side by 40 bp that are homologous to STE20 sequence and on the other side by 40 bp homologous to pRS315 sequence. The primers used to amplify the triple HA epitope were MK77 (5′-GAA-ACC-GTA-AAT-TTG-GAC-GTA-ACT-GAA-GAT-GAT-AAA-CAA-AAG-ATG-TCG-CGA-TAC-CCA-TAC-GAT-3′) and MK78 (5′-CCA-GTG-AAT-TGT-AAT-ACG-ACT-CAC-TAT-AGG-GCG-AAT-TGG-AGC-TAG-TTA-ACA-GCA-GCG-TAA-TC-3′).
(ii) pCLA4-HA (pSL2732) contains PCR-amplified CLA4 (with primers MK57:5′-CTG-GCT-CTC-TAA-AGG-CCA-GGC and MK58:5′-CGT-AGG-ATA-AAC-CGC-GGT-ATT-AAA-CG-3′). The PCR product was cut with MluI, which cleaves at position −344 with respect to the ATG of CLA4, and treated with T4 DNA polymerase to make the ends blunt. The resulting product was cut at the SacII site within primer MK57 and ligated to SphI (made blunt with T4 DNA polymerase) and SacII-cut pSTE20. As for pSTE20-HA, a DNA fragment encoding a C-terminal triple HA epitope was fused to CLA4 by recombination-based subcloning to obtain pCLA4-HA. The primers used to amplify this triple HA epitope were MK78 and MK79 (5′-GAT-CCA-AAG-GAT-TTG-ACA-TCA-CTG-TTG-GAG-TGG-AAG-GAA-ATG-TCG-CGA-TAC-CCA-TAC-GAT-3′).
(iii) pCS1 (pSL2740) contains CLA4 coding sequence 1 to 1526 fused to STE20 coding sequence 1695 to 2820. This chimera contains the N-terminal, presumptive regulatory region from Cla4 and the C-terminal kinase and protein interaction domains of Ste20. A PCR fragment from pCLA4 was amplified by using the primers MK57 and MK60 (5′-GGC-CAC-ACC-AGC-CGG-CTG-TGC-GAC-3′). This fragment was cut with NaeI and ligated to the 2.2-kb NaeI fragment from pSTE20. The ligation product containing the CS1 chimera was amplified with MK56 and MK57, cut with ApaI and SacII, and ligated into the corresponding sites in pRS315. A C-terminal triple HA epitope was introduced into pCS1 as described for pSTE20-HA.
(iv) pSC1 (pSL2733) contains STE20 coding sequence 1 to 1697 fused to CLA4 coding sequence 1530 to 2529. This chimera contains the N-terminal, presumptive regulatory region of Ste20 and the C-terminal kinase and presumptive protein interaction domains of Cla4. The pCLA4 NciI/SspI fragment was ligated into the pSTE20 NaeI/Ecl136III sites. A C-terminal triple HA epitope was fused to SC1 as described for pCLA4-HA.
An additional group of chimeras having fusion points not dictated by available restriction sites was made by recombination in vivo between DNA fragments synthesized by PCR and designed to have homologous ends (36). The primers contained 40 bases from one gene and 20 bases from the other. Following transformation of yeast with such PCR-generated fragments and a linearized vector, intact plasmids were recovered and the presence of the designed chimera was verified by DNA sequencing. Each construct contains a C-terminal triple HA epitope fusion. The numbers on this set of chimeras denote the kinase subdomain in which the fusion was made. For example, the CS3 chimera contains the N-terminal region of Cla4 fused to the C-terminal region of Ste20 within kinase subdomain III.
(v) pCS3 (pSL2741) contains CLA4 coding sequence 1 to 1836 fused to STE20 coding sequence 2002 to 2820.
(vi) pSC3 (pSL2734) contains STE20 coding sequence 1 to 2001 fused to CLA4 coding sequence 1837 to 2529.
(vii) pCS7 (pSL2742) contains CLA4 coding sequence 1 to 2133 fused to STE20 coding sequence 2272 to 2820.
(viii) pSC7 (pSL2735) contains STE20 coding sequence 1 to 2271 fused to CLA4 coding sequence 2134 to 2529.
(ix) pCS8 (pSL2743) contains CLA4 coding sequence 1 to 2202 fused to STE20 coding sequence 2338 to 2820.
(x) pSC8 (pSL2736) contains STE20 coding sequence 1 to 2337 fused to CLA4 coding sequence 2203 to 2529.
(xi) pCS9 (pSL2744) contains CLA4 coding sequence 1 to 2298 fused to STE20 coding sequence 2437 to 2820.
(xii) pSC9 (pSL2737) contains STE20 coding sequence 1 to 2436 fused to CLA4 coding sequence 2299 to 2529.
(xiii) pCS10 (pSL2745) contains CLA4 coding sequence 1 to 2355 fused to STE20 coding sequence 2494 to 2820.
(xiv) pSC10 (pSL2738) contains STE20 coding sequence 1 to 2493 fused to CLA4 coding sequence 2356 to 2529.
(xv) pCS11 (pSL2746) contains CLA4 coding sequence 1 to 2418 fused to STE20 coding sequence 2557 to 2820.
(xvi) pSC11 (pSL2739) contains STE20 coding sequence 1 to 2556 fused to CLA4 coding sequence 2419 to 2529.
(xvii) pCS10D772T (pSL2747) contains CLA4 coding sequence 1 to 2355 fused to STE20 coding sequence 2494 to 2820, except that the Cla4 aspartic acid 772 is replaced with a threonine.
Versions of full-length STE20 and CLA4 as well as the chimeras were also cloned into the BamHI site of the high-copy-number vector YEp351 by in vivo recombination. The high-copy-number constructs are designated pSL2752 to pSL2768 in the following order: STE20, CLA4, SC1 to SC10, CS1 to CS10, and CS10D772T.
The contribution of single amino acids to the functional specificity of Ste20 and Cla4 was tested by making point mutations in the full-length genes. These mutants were made by PCR-based recombination in yeast as described above for the chimeras. STE20T818D(pSL2751) was made with the primers MK85 (5′-GAA-ATG-ATC-GAG-GGG-GAG-CCT-CCA-TAT-TTA-AAT-GAA-GAT-CCG-CTA-AGA-GCA-CTG-TAT-TTA-3′) and MK78. CLA4D772T(pSL2749) was made with the primers MK83 (5′-GGT-GAA-CCA-CCA-TAT-TTA-AAT-GAA-ACT-CCA-CTA-AAG-GCG-3′) and MK78. Mutations were confirmed by DNA sequencing.
Two plasmids were constructed specifically for kinase assays. First, pHA (pSL2771) was constructed by PCR-based recombination (contains the same sequence as pSTE20-HA except that the STE20 coding sequence has been deleted). In addition, a catalytically inactive form of glutathione S-transferase (GST)-Ste11 was prepared by subcloning a 1.0-kb HindIII/NcoI fragment from plasmid pSL2560 (30) into the HindIII/NcoI sites of plasmid pGA1970 (generously provided by B. Errede and G. Ammer). The resulting plasmid (pSL2772) contains pGEX-2T carrying a GST fusion of the Myc epitope-tagged STE11Δ/K444R,T596I truncation allele which contains a truncation of 341 amino acids at the amino terminus as well as replacements of lysine residue 444 by arginine and threonine residue 596 by isoleucine. Constructs were confirmed by DNA sequencing.
Western analysis.
All constructs used in this study contain a C-terminal triple HA epitope tag. To evaluate the level of expression of the chimeras, extracts were prepared and used for Western analysis. Specifically, SY3357 strains containing individual constructs were grown to mid-log phase, pelleted, and resuspended in buffer (8 M urea, 5% sodium dodecyl sulfate, 40 mM Tris [pH 6.8], 0.1 mM EDTA, 0.4 mg of bromophenol blue/ml, and 1% β-mercaptoethanol). Samples were vortexed for 5 min, boiled for 2 min, and centrifuged for 2 min, and the resulting extracts were then subjected to electrophoresis through a 10% polyacrylamide gel and transferred to nitrocellulose. To detect the chimeric proteins, the Western blots were probed with a 1:1,000 dilution of monoclonal HA antibody (12CAS.16.4) and then with a 1:3,000 dilution of Bio-Rad goat anti-mouse immunoglobulin G-horseradish peroxidase conjugate. To detect Pgk1, which serves as a loading control, the Western blots were probed with a 1:10,000 dilution of monoclonal Pgk1 antibody and subsequently with a 1:3,000 dilution of Bio-Rad goat anti-mouse antibodies. Proteins were visualized with ECL (Pierce).
Morphological assays.
For the routine assessment of cellular morphology, mid-log cultures grown in selective medium were fixed with 3.7% formaldehyde. A minimum of 200 cells was observed with a Zeiss Axioplan 2 microscope (100× oil-immersion objective). All assays were performed in triplicate. To assess the morphological response to α-factor, mid-log-phase cells were pelleted and resuspended in fresh medium, and synthetic α-factor was added (final concentration of 6 μM). Samples were collected after 4 h of α-factor exposure and fixed with 3.7% formaldehyde, and shmoo formation was assessed with a Zeiss Axioplan 2 microscope (100× oil-immersion objective). To observe bud scar patterns, SY3970 cells (ste20Δ/ste20Δ) were grown to mid-log phase, fixed with 3.7% formaldehyde, and stained with 0.1 mg of calcofluor white/ml for 15 min as described previously (6). At least 50 cells were observed per sample; all samples were assayed in triplicate.
β-Galactosidase assays.
Cells were prepared and assayed as described previously (12). Samples were assayed in triplicate.
Halo and quantitative mating assays.
Halo assays were done as described previously (41). Suspensions of fresh MATa bar1 cells (SY3750) were spread on the surface of selective plate medium and allowed to dry. A sterile solution of synthetic α-factor dissolved in water was applied to filter disks, which were placed on the plates. Quantitative mating assays were done with SY3749 strains containing individual chimeras and 227 as a tester strain as described previously (41). Bilateral mating assays were performed exactly like the quantitative mating assays, except that SY3749 strains containing the chimeras were crossed to SY3971 strains containing the matching chimera.
Kinase assays.
GST-Ste11Δ/K444R,T596I was expressed in Escherichia coli and purified over glutathione-Sepharose (40). Ste20, Cla4, Ste20T818D, and Cla4D772T were immunopurified from yeast extracts as described previously (47), except that 200 ml of yeast culture was prepared per kinase assay sample, the log-phase yeast cultures were treated for 90 min with 6 μM α-factor, and monoclonal HA antibody (12CAS.16.4) was utilized to form antibody-antigen complexes. Yeast extracts were prepared from ste20Δbar1Δ cells (SY3750) containing wild-type or mutant kinases. Prior to the carrying out of phosphorylation reactions, protein kinase immune complexes were incubated in kinase reaction buffer for 20 min at 30°C (50 mM Tris-HCl [pH 7.5], 40 mM magnesium chloride, 1 mM dithiothreitol, 0.5 mM sodium orthovanadate, 5 μg of aprotinin/ml, and 5 μg of leupeptin/ml) that was supplemented with 400 mM ATP in order to stimulate autophosphorylation. The immune complexes were subsequently washed twice with kinase reaction buffer. Kinase assays were then carried out in 30 μl of kinase buffer containing 1 μM GST-Ste11Δ/K444R,T596I or 1 μM myelin basic protein (New England Biolabs) and ATP (1,000 Ci/mol) for 20 min at 30°C. These reactions were terminated with 30 μl of 2× Laemmli buffer followed by 5 min of boiling. Terminated reaction mixtures were then subjected to electrophoresis through an 8% polyacrylamide gel and transferred to nitrocellulose. Ste11Δ/K444R,T596I phosphorylation was detected from the blots with a Storm 860 phosphodetector system (Amersham Biosciences, Piscataway, N.J.) and quantified with ImageQuant V1.11 (Molecular Dynamics). Blots were then subjected to Western analysis as described above; monoclonal Myc antibody (9E10 serum) was used at a 1:50 dilution to detect Ste11 protein. Blots were developed with ECL Plus (Amersham, Arlington Heights, Ill.), detected with the Storm 860 scanner, and quantified with ImageQuant V1.11 software.
Salt sensitivity assay.
Single colonies of the SY3753 strain containing the indicated plasmid were patched onto selective medium. These patches were replica plated to rich medium supplemented with 1.2 M NaCl and were incubated at 30°C.
Invasive growth assay.
Fresh cultures grown in selective medium were diluted in water and spotted onto yeast extract-peptone-dextrose plates. Plates were incubated at 30°C for 3 days. After this, cells were washed from the surface of these plates under a stream of water to reveal the presence or absence of invasive growth (34).
RESULTS
Chimeric fusions were made between CLA4 and STE20.
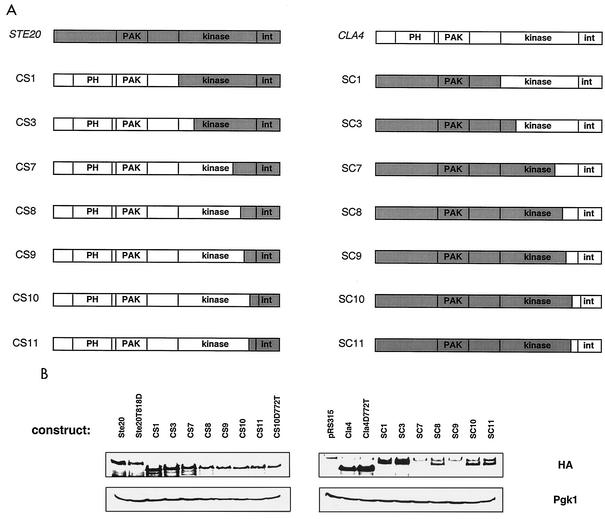
Cla4 and Ste20 perform both overlapping and distinct cellular functions. As one means to identify regions of these proteins that direct their activity to specific cellular roles, we have constructed a series of chimeric gene fusions between CLA4 and STE20. The chimeras that form the basis of this report are represented schematically in Fig. 1, and their construction is detailed in Materials and Methods. In brief, each construct contains a 447-bp fragment of the upstream region of STE20 to drive expression and a fragment capable of encoding a triple HA epitope fusion at the C terminus. To provide controls, constructs with these features, but containing wild-type STE20 and CLA4, were also prepared.
FIG. 1.
Structure and expression of chimeric proteins. (A) Schematic representation of the encoded chimeric fusion proteins. Abbreviations: PAK, PAK domain; PH, pleckstrin homology domain; int, protein-protein interaction domain. (B) Each chimera contains a C-terminal triple HA epitope fusion that was utilized to assess protein expression as described in Materials and Methods. Pgk1 served as a loading control. The band found above all samples in the Cla4-containing blot (probed with monoclonal HA antibody) is a background band commonly observed when using this antibody.
In order to assess chimeric protein expression and function, each construct was transformed into wild-type, cla4Δ, and ste20Δ strains listed in Table 1. Protein expression was assessed by Western analysis of yeast cell extracts (Fig. 1). Although there was some variation in expression levels and although all proteins containing the Cla4 C terminus were expressed at a lower level than were proteins with the Ste20 C terminus, the biological activities of the chimeras did not correlate with their expression levels. In an attempt to compensate for this expression discrepancy, all constructs were tested for function when expressed from a high-copy-number plasmid (YEp351) as well as from a low-copy-number plasmid (pRS315). In almost every instance, altering the expression level of the constructs had no effect on the observed function; exceptions are noted when appropriate.
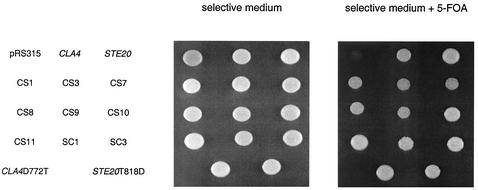
The ability of each chimera to function was assessed initially by measuring the 5-fluoroorotic acid (5-FOA) resistance of a cla4Δ ste20Δ strain that contained a CLA4-URA3 plasmid and a chimera on a LEU2-based plasmid. Survival on 5-FOA requires the loss of the CLA4-URA3 plasmid; this loss is lethal if the chimera does not provide the shared essential function. Nine chimeras allowed for viability on 5-FOA and thus were able to perform the shared essential function (Fig. 2). By this same criterion, some of the chimeric proteins were nonfunctional (data not shown). In addition to the shared essential function, chimeras were tested for the ability to perform Cla4- and Ste20-specific functions. As detailed below, all functional chimeras were able to perform not only the shared essential role but also a subset of Cla4- and/or Ste20-specific functions. In addition, all chimeras that were unable to perform the shared essential function were also nonfunctional in all Cla4-specific and Ste20-specific assays (data not shown).
FIG. 2.
Ability of chimeric fusions to perform the essential function shared by CLA4 and STE20. Strain SY3748 (ste20Δ cla4Δ 〈pRS316ADE8CLA4〉) was transformed with pRS315-derived plasmids carrying full-length CLA4, full-length STE20, or a chimeric gene fusion. The resultant strains were grown to saturation in selective medium and spotted onto either selective medium or selective medium supplemented with 0.1% 5-FOA. Each spot contains 7.5 × 106 cells. The ability to grow on 5-FOA indicates the ability to lose the CLA4-URA3 plasmid and thus provide the shared essential function.
A C-terminal segment of Ste20 directs chimeric proteins to function in the pheromone response pathway.
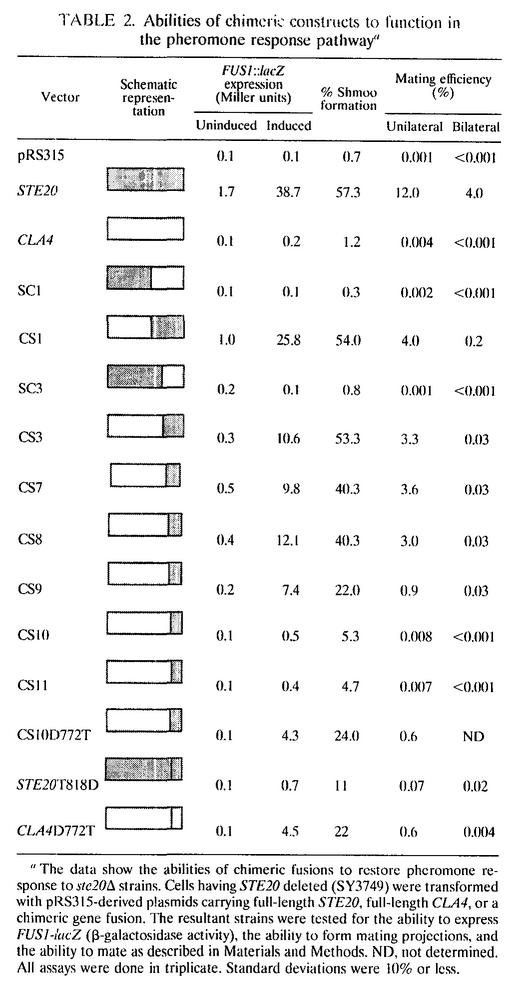
Ste20 is thought to function upstream of several mitogen-activated protein (MAP) kinase cascades found in the pheromone response pathway, the haploid invasive growth pathway, and the osmolarity (HOG) pathway. We tested the abilities of the chimeric proteins to function in each of these pathways. To assess the abilities of the chimeras to restore normal pheromone response pathway activity, low-copy-number plasmids encoding the chimeras were transformed into ste20Δ strains and the transformants were tested for pheromone-mediated transcriptional induction of pheromone-responsive genes, shmoo formation, and cell cycle arrest (Table 2 and Fig. 3). In addition, unilateral and bilateral mating efficiencies were measured (Table 2).
TABLE 2.
Abilities of chimeric constructs to function in the pheromone response pathwaya
The data show the abilities of chimeric fusions to restore pheromone response to ste20Δ strains. Cells having STE20 deleted (SY3749) were transformed with pRS315-derived plasmids carrying full-length STE20, full-length CLA4, or a chimeric gene fusion. The resultant strains were tested for the ability to express FUS1-lacZ (β-galactosidase activity), the ability to form mating projections, and the ability to mate as described in Materials and Methods. ND, not determined. All assays were done in triplicate. Standard deviations were 10% or less.
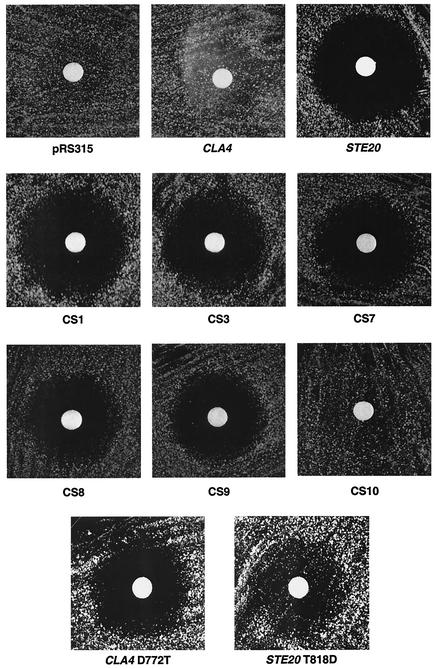
FIG. 3.
Ability of chimeric fusions to restore G1 arrest to ste20Δ bar1Δ strains. Cells having STE20 deleted (SY3750) were transformed with pRS315-derived plasmids carrying full-length STE20, full-length CLA4, or a chimeric gene fusion. The resultant strains were spread on the surface of selective plate medium and allowed to dry. A sterile solution of synthetic α-factor dissolved in water was applied to filter disks, which were placed on the plates.
The CS1, CS3, CS7, CS8, and CS9 chimeras restored pheromone response activity to substantial levels. The CS10 and CS11 chimeras conferred an evident but low activity on the pheromone response pathway. For example, they conferred a 100-fold-lower mating efficiency than did the CS9 chimera, even though these three chimeric proteins are expressed at indistinguishable levels (Fig. 1). Therefore, chimeric studies map the determinants that confer Ste20 specificity to a C-terminal region containing kinase subdomains IX, X, and XI and the protein-protein interaction domain.
The data presented above indicate that the CS1, CS3, CS7, CS8, and CS9 chimeras exhibit substantial Ste20 function. As a final and stringent test of their ability to carry out all Ste20 activities, we performed bilateral mating tests in which the two mating partners rely on the same chimera to provide Ste20 function. By this test, the chimeric proteins (CS1, CS3, CS7, CS8, and CS9) were considerably enfeebled compared to wild-type Ste20 (Table 2). One interpretation of all these data is that these chimeras are competent to activate the signal transduction pathway that leads through the MAP kinase cascade but are not competent to carry out the polarity reorientation function of Ste20, as evidenced by a decrease in bilateral mating. In this view, it appears that the N terminus of Ste20 is important for bilateral mating, potentially by instilling the ability to direct cell polarity.
A single amino acid substitution in the Cla4 kinase domain confers the ability to perform Ste20-specific functions.
The CS9 and CS10 chimeras differ in sequence at only two amino acids, suggesting that one or both of these residues might be an important specificity determinant. CS9 contains a threonine (T818 of Ste20) and an arginine (R821 of Ste20) instead of an aspartate (D772 of Cla4) and a lysine (K775 of Cla4). To test whether these residues confer specificity, we performed several experiments. First, we asked whether single amino acid substitutions in the CS10 chimera would confer the ability to carry out Ste20 functions. The CS10K775R substitution functioned as the CS10 chimera (data not shown), but the CS10D772T substitution functioned as the CS9 chimera (Table 2). That is, CS10D772T conferred nearly the same mating efficiency as the CS9 chimera did.
Prompted by this latter finding, we tested whether the D772T substitution in an otherwise wild-type Cla4 would have altered specificity. Strikingly, this Cla4 mutant restored transcriptional induction and unilateral mating competence to ste20Δ cells to the same extent as the CS10D772T chimera did (Table 2). Conversely, the equivalent substitution in an otherwise wild-type Ste20, T818D, substantially reduced its ability to function in the pheromone response pathway (Table 2). Therefore, replacement of aspartate 772 in Cla4 by the Ste20 residue occupying the equivalent position in the kinase subdomain structure, threonine 818, is sufficient to confer Ste20-like activity on Cla4. Moreover, T818 of Ste20 is crucial for Ste20 function, as the Ste20T818D mutant is impaired in pheromone response pathway activity (Table 2).
The presence of residual activity of Ste20T818D in the pheromone response pathway, greater than that seen for Cla4, implies that other regions of Ste20 also contribute to functional specificity. This interpretation is supported by the finding that the Cla4D772 mutant was defective in bilateral mating whereas the Ste20D818T mutant allowed for unilateral and bilateral mating equally well, normalized to wild-type Ste20 activity in these assays. Therefore, it appears that Ste20D818T may retain the competence to establish cell polarity.
The Cla4D772T mutant protein is expressed at levels identical to those of wild-type Cla4 (Fig. 1), implying that the ability of the Cla4D772T mutant to function like Ste20 is a gain in activity and not an increase in the protein expression. Interestingly, we found that high-copy-number wild-type CLA4 did allow for a low level of transcriptional induction in response to pheromone (37-fold less than the wild type) but did not restore shmoo formation, G1 arrest, or mating efficiency (data not shown). Hence, high-copy-number CLA4 cannot compensate for the loss of STE20 in almost every instance. However, altering a single amino acid in Cla4 (without altering the expression level) confers the ability to function like Ste20 in the pheromone response pathway.
A single amino acid substitution in Cla4 leads to a gain in the ability to phosphorylate Ste11, a known Ste20 substrate.
A single amino acid substitution in an otherwise wild-type Cla4 at position 772 confers the ability to function in the pheromone response pathway. The Cla4D772T mutant protein is expressed at the same level as is wild-type Cla4 (Fig. 1). Previous studies (11, 15) have failed to discern a substantial difference in the localization of Ste20 and Cla4. Both are located at sites of polarized growth (small bud tips and shmoo tips). Nonetheless, we examined the localization of wild-type and mutant Cla4 under pheromone-inducing conditions but failed to detect differences (data not shown). Therefore, expression levels and cellular localization cannot explain the observed functional difference between wild-type Cla4 and the Cla4D772T mutant.
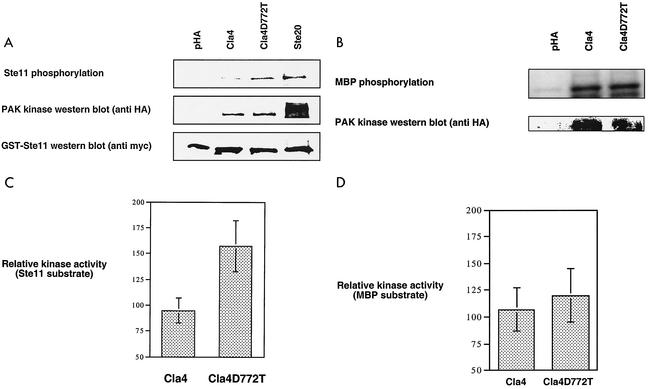
Another mechanism that could explain this functional difference is an increase in the ability of the Cla4D772T kinase to phosphorylate a Ste20 target in the pheromone response pathway. To test this, we performed kinase assays with immunopurified kinase complexes and bacterially purified Ste11, a known Ste20 substrate that functions in the pheromone response pathway. We found that the Cla4D772T mutant phosphorylates Ste11 nearly twice as well (1.7-fold) as does wild-type Cla4 (Fig. 4). This increase is of the same magnitude as the change in in vitro activity seen for a number of regulated protein kinases. For example, the myosin light chain kinase 1 isoform is activated twofold by Src in vitro, and the specificity of JNK2 versus JNK1 for the substrate c-Jun is about threefold (3, 13). More important, the magnitude of increased phosphorylation of Ste11 by the Cla4D772T mutant in vitro is comparable to the 1.9-fold increase in Ste7 hyperphosphorylation (another pheromone response MAP kinase component) seen in vivo following pheromone treatment (48). The two isoforms of Cla4 showed no difference in the ability to phosphorylate myelin basic protein (Fig. 4). Therefore, the increase in Cla4D772T kinase activity is substrate specific. The increased ability of the Cla4D772T mutant to phosphorylate Ste11 appears sufficient to increase the ability to act in the pheromone response pathway.
FIG. 4.
Ability of immunopurified kinases to phosphorylate Ste11. Phosphorylation reactions were performed with immunopurified PAK complexes from ste20Δ bar1Δ pheromone-treated yeast extracts (prepared as described in Materials and Methods) with either bacterially expressed and purified GST-Ste11Δ/K444R,T596I (at a 1 μM final concentration) (A) or 1 μM myelin basic protein (MBP) (B). Relative phosphorylation levels were determined by measuring the radioactivity of the labeled protein bands directly from blots and dividing it by the quantified amount of Cla4 or Cla4D772T kinase as described in Materials and Methods (C and D). The data presented are derived from seven independent trials.
Threonine 772 substitution is sufficient to direct otherwise wild-type Cla4 to function in the HOG pathway.
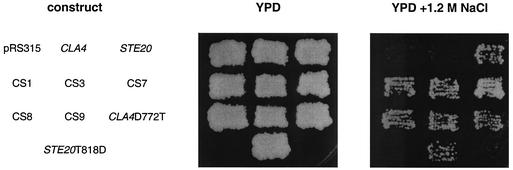
A high salt concentration in the growth medium leads to the activation of the HOG MAP kinase cascade by two independent upstream mechanisms, one requiring the Sho1 receptor and the other requiring the Sln1 receptor. Activation of either branch is sufficient to activate the HOG pathway and confer the ability to grow on medium containing high salt concentrations (29). Cells lacking both Ste20 and Ssk1 (a kinase required for the function of the Sln1 branch of the HOG pathway) are salt sensitive whereas the single mutants are substantially less salt sensitive (28). This observation led to the proposal that Ste20 functions downstream of Sho1 in the HOG pathway. The salt sensitivity of the ste20Δ ssk1Δ strain was utilized to test the ability of the chimeras to function in the HOG pathway. Plasmids carrying the chimeras were transformed into a ste20Δ ssk1Δ strain (SY3753), and the resultant strains were tested for the ability to grow on medium containing a high salt concentration. Expression of the chimeras CS1, CS3, CS7, CS8, and CS9, the same chimeras that conferred pheromone response activity, conferred the ability to grow on 1.2 M NaCl to the same extent as did the full-length STE20 plasmid (Fig. 5). In addition, the CS10D772T, Cla4D772T, and Ste20T818D constructs also provided growth on high-salt media. Thus, in addition to conferring the ability to function in the pheromone response pathway, the threonine substitution is also sufficient to allow otherwise wild-type Cla4 to function in the HOG pathway.
FIG. 5.
Ability of chimeric fusions to function in the HOG pathway. Cells having STE20 and SSK1 deleted (SY3753) were transformed with pRS315-derived plasmids carrying full-length STE20, full-length CLA4, or a chimeric fusion. The resultant strains were grown as patches on selective medium and were then replica plated to yeast extract-peptone-dextrose (YPD) (rich medium) or yeast extract-peptone-dextrose plus 1.2 M NaCl.
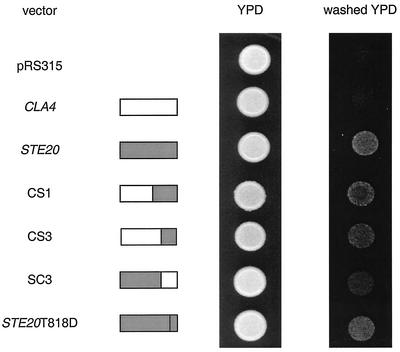
Haploid invasive growth is restored to ste20Δ cells by two distinct subsets of chimeras.
Filamentous growth is normally evaluated in the Σ strain background. Cells from this background lacking STE20 (SY3752) are defective for agar invasion as determined by the plate washing assay (34). By this assay, three of the chimeras (CS1, CS3, and SC3) restored invasion to ste20Δ cells to various extents (Fig. 6). In addition, the SC1 chimera restored agar invasion to the ste20Δ strain when expressed on a high-copy-number plasmid (data not shown). The CS3 and SC3 chimeras harbor nonoverlapping Ste20 sequences, raising the possibility that the distinct regions of Ste20 confer different facets of Ste20 function during haploid invasive growth, facets that alone can restore invasion. In support of this notion, the CS1 and CS3 chimeras, but not the SC3 chimera, restored cell elongation during haploid invasive growth as assessed by the single cell assay (data not shown) (8). The Ste20T818D mutant, which functions poorly in the pheromone response pathway, did allow for haploid invasive growth in the ste20Δ background. Perhaps this construct provides the same subset of haploid invasive growth activity as does the SC3 chimera.
FIG. 6.
Ability of chimeric fusions to restore haploid invasive growth to ste20Δ strains. Cells of the filamentous strain background having STE20 deleted (SY3752) were transformed with pRS315-derived plasmids carrying full-length STE20, full-length CLA4, or a chimeric gene fusion. The resultant strains were grown to saturation in selective medium and spotted onto yeast extract-peptone-dextrose (YPD) plates. Each spot contains 7.5 × 106 cells. The plates were incubated at 30°C for 3 days and were then washed as described in Materials and Methods.
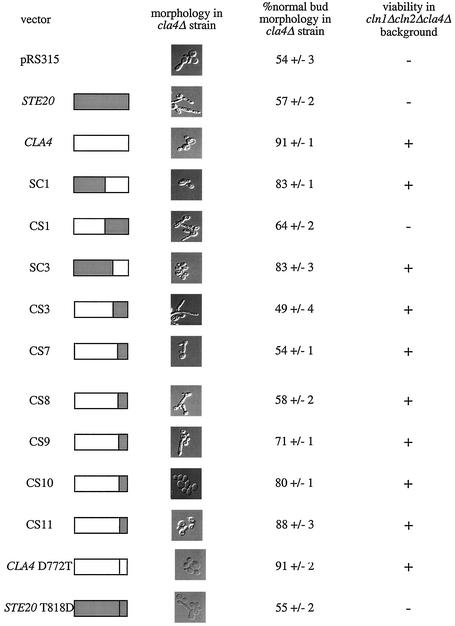
Distinct sets of chimeras perform the Cla4-specific morphological and G1 functions.
Each chimeric fusion was tested for the ability to perform two Cla4-specific functions, a morphological function and a presumed G1 function. Cells lacking Cla4 activity have elongated buds due to a G2 cell cycle delay (4, 10, 25, 33, 44). The ability of chimeras to restore normal bud morphology to cla4Δ strains was quantified in mid-log-phase cultures (Fig. 7). Four chimeras (SC1, SC3, CS10, and CS11) restored normal morphology to cla4Δ cells. The structures of these chimeras suggest that the N-terminal regions of Cla4 and Ste20 are functionally interchangeable and further suggest that the C-terminal region of Cla4 from kinase subdomain III through kinase subdomain X is sufficient to allow execution of the Cla4 morphological function.
FIG. 7.
Ability of chimeric fusions to restore Cla4-specific functions to cla4Δ strains. Cells having CLA4 deleted (SY3360) were transformed with pRS315-derived plasmids carrying full-length CLA4, full-length STE20, or a chimeric gene fusion. These strains were grown to mid-log phase in selective medium, fixed with 3.7% formaldehyde, and visualized with Nomarski optics. The morphologies that were observed were quantified as described in Materials and Methods. Chimeric fusions were also tested for the ability to restore viability to the SY3751 strain (cln1Δ cln2Δ cla4Δ 〈pRS316ADE8CLA4〉) on medium containing 0.1% 5-FOA. Growth on 5-FOA indicates the ability to lose the CLA4 plasmid and thus restore viability to the triple mutant background.
Cla4 was originally identified by its requirement for viability in strains lacking the G1 cyclins Cln1 and Cln2, suggesting a G1 function for Cla4 (9). Chimeras were tested for the ability to carry out this function by determining whether their presence allowed for the loss of a wild-type CLA4-URA3 plasmid from a cln1Δ cln2Δ cla4Δ strain. As indicated by growth on 5-FOA, eight of the chimeric fusions were able to restore viability to the cln1Δ cln2Δ cla4Δ strain (Fig. 7). These eight included the four that were able to perform the Cla4 morphological function (SC1, SC3, CS10, and CS11) and four others (CS3, CS7, CS8, and CS9). Interestingly, seemingly nonoverlapping regions of Cla4 are able to bypass the requirement for Cln1 and Cln2. These results suggest that the G1 function of Cla4 may be restored by more than one mechanism and further suggest that the G1 and morphological functions of Cla4 are distinct, because distinct sets of chimeras are capable of performing these cellular roles.
The Ste20 N terminus is involved in bipolar bud site selection.
Ste20 is required for normal bipolar budding in diploid cells (38). Each chimeric fusion was tested for the ability to restore this function to diploid cells lacking Ste20. We found that low-copy-number STE20 did not restore bipolar budding to an appreciable level (data not shown). High-copy-number STE20 (YEp351 based) restored bipolar budding to 33%. The high-copy-number SC1 and SC3 chimeras also restored a high degree of bipolar budding to diploids (Table 3). Therefore, the N-terminal region of Ste20 appears to be important for allowing bipolar bud site selection in diploid yeast.
TABLE 3.
Abilities of chimeric fusions to restore diploid bud site selection to diploid yeast lacking STE20a
| Construct | % Bipolar budding |
|---|---|
| YEp351 | 3 ± 1 |
| CLA4 | 2 ± 1 |
| STE20 | 33 ± 4 |
| CS1 | 9 ± 3 |
| CS3 | 9 ± 4 |
| CS7 | 9 ± 3 |
| CS8 | 9 ± 2 |
| CS9 | 9 ± 3 |
| CS10 | 9 ± 3 |
| CS11 | 6 ± 1 |
| SC1 | 25 ± 6 |
| SC3 | 24 ± 4 |
Diploid cells having STE20 deleted (SY3970) were transformed with YEp351-derived plasmids carrying full-length STE20, full-length CLA4, or a chimeric gene fusion. These strains were grown to mid-log phase in selective medium, fixed with 3.7% formaldehyde, stained with calcofluor, and visualized with Nomarski optics. The bud scar staining observed was quantified as described in Materials and Methods.
DISCUSSION
The ability of protein kinase homologues to function redundantly is a recurring theme throughout cell biology (7, 14, 22, 43). The yeast PAK kinases Cla4 and Ste20 appear to provide an example of this phenomenon: loss of both kinases is lethal whereas the loss of a single one is not. These kinases have unique cellular roles as well, as mutations in CLA4 and STE20 confer strikingly different phenotypes. The intriguing ability of Cla4 and Ste20 to carry out a shared activity as well as unique activities prompted the construction of chimeric proteins to identify regions of these kinases that direct functional specificity. Here, 10 functional chimeric proteins and two point mutants were investigated. Each of these was able to perform the shared essential function, suggesting that regions of these kinases can be readily interchanged.
Functional specificity determinants map to the C-terminal regions of Cla4 and Ste20.
Four chimeras (SC1, SC3, CS10, and CS11) as well as the Cla4D772T mutant (data not shown) restored the Cla4 morphological function. These chimeras share the sequence extending from the Cla4 kinase subdomain III to subdomain X. Because subdomains III to X are thought to dictate substrate specificity for protein kinases, we infer that these chimeras may be able to phosphorylate Cla4-specific targets. The observation that the CS10 and CS11 chimeras restore normal bud morphology even though they lack the presumptive Cla4 protein-protein interaction domain argues that this domain may not be required for the Cla4 morphological function. In a complementary manner, six chimeras containing C-terminal regions of Ste20 restored Ste20-specific functions, albeit to various degrees (CS1, CS3, CS7, CS8, and CS9). Hence, a Ste20 specificity determinant appears to map to a C-terminal region, from kinase subdomain IX through to the end of the protein interaction domain.
A threonine substitution at residue 772 is able to direct an otherwise wild-type Cla4 to function in the pheromone response pathway and the HOG pathway.
The ability of the Cla4D772T point mutant to function in the pheromone response and HOG pathways suggests that the kinase domains of Ste20 and Cla4 are largely interchangeable and that threonine 818 of Ste20 is crucial for determining Ste20 specificity. The Cla4D772T mutant protein is expressed at levels identical to those of wild-type Cla4. Therefore, it appears that the Cla4D772T mutant has gained a Ste20-specific activity, potentially the ability to phosphorylate a Ste20-specific substrate. Several lines of evidence support this possibility. First, by analogy to the mammalian PAK1 crystal structure, threonine 818 is found within a docking site that interfaces with the PAK domain in an autoinhibited state (19). Upon activation, this region is likely to be solvent exposed and potentially serve as a substrate docking site. Second, Kallunki et al. identified a stretch of 23 amino acids corresponding to the equivalent kinase domain position that determines the ability of JNK2, a MAP kinase, to bind and phosphorylate c-Jun, a specific target (13). Finally, we performed kinase assays with immunopurified Cla4 and Cla4D772T complexes by using either Ste11 (a Ste20 substrate in the pheromone response and HOG pathways) or myelin basic protein (a nonspecific substrate). We found that the Cla4D772T mutant phosphorylates Ste11 nearly twice as well as it does wild-type Cla4 but phosphorylates myelin basic protein equally as well as it does wild-type Cla4. Therefore, the mechanism by which Cla4D772T attains altered specificity appears to be in part due to an increase in the ability to specifically phosphorylate Ste11.
A distinct putative Ste20 functional specificity determinant was previously identified by Leeuw et al. (18), who demonstrated a physical interaction between the extreme C-terminal domain of Ste20 and Ste4 (the β subunit of the heterotrimeric G protein in the pheromone response pathway). They further demonstrated a requirement for this region for Ste20 function in the pheromone response pathway (18). Interestingly, the absence of this region from the Cla4D772T mutant appears to severely decrease the bilateral mating efficiency (Table 2). Therefore, the very C-terminal region may be crucial in polarization events involved in the mating process. In addition, the N terminus of Ste20 appears to be important for bilateral mating. Chimeras containing the Cla4 N terminus are severely compromised for bilateral mating compared to mating with a wild-type strain (Table 2). In further support of the idea that the Ste20 N terminus is important for bilateral mating, the Ste20T818D mutant shows essentially the same mating efficiency in bilateral and unilateral tests. Thus, Ste20 appears to contain two regions that are important for bilateral mating, the N-terminal region and the very C-terminal region that is known to interact with Ste4. We suggest that these regions of Ste20 are important for reorienting the polarity of the actin cytoskeleton.
Two sets of chimeras allow for haploid invasive growth in ste20Δ cells.
Ste20 contributes to several physiological changes that filamentous cells undergo during haploid invasive growth, including cell elongation and bud site selection. Three chimeras restored agar invasion to ste20Δ cells but appeared to do so by different means. CS1 and CS3 restored cell elongation to ste20Δ cells, but SC3 did not. The nonoverlapping sequences found in CS3 and SC3 suggest that the N-terminal and C-terminal regions of Ste20 may impart distinct Ste20 activities during haploid invasive growth; each activity is sufficient to restore invasion.
Two distinct sets of chimeras allow for viability in the cla4Δ cln1Δ cln2Δ background.
Cla4 was identified by its requirement for viability in cells lacking the G1 cyclins Cln1 and Cln2. Two sets of chimeras restored viability to the cla4Δ cln1Δ cln2Δ strain, the chimeras that restore normal bud morphology to cla4Δ cells (SC1, SC3, CS10, and CS11) and four chimeras shown to perform Ste20-specific functions (CS3, CS7, CS8, and CS9). Chimeras SC3 (from the first set) and CS3 (from the second set) do not have overlapping sequences, and so it is not possible to ascribe function in the cln1Δ cln2Δ cla4Δ strain to a particular region of Cla4. Rather, perhaps this result suggests that either Ste20 or Cla4 kinase activity is able to provide the essential function in the triple mutant but that wild-type Ste20 is regulated in a way that precludes it from carrying out that function. For example, wild-type Ste20 may be subject to a negative regulatory input. If so, the observation that CS1 cannot carry out the essential function whereas CS3 can suggests that the site of regulation may be found within kinase subdomain I, II, or III. An appealing possibility is that Cln1 and Cln2 are required to activate Ste20. In this view, the lethality of cln1Δ cln2Δ cla4Δ and ste20Δ cla4Δ strains would have at least one common basis.
Ste20 and Cla4 functional specificity maps to several regions within these kinases.
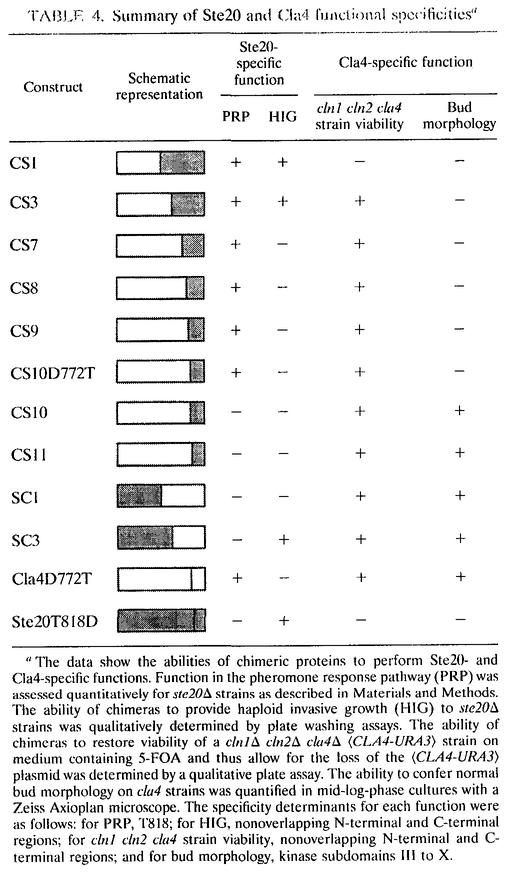
The chimeric molecules and the point mutants containing regions of Ste20 and Cla4 allowed us to map several specificity determinants, as summarized in Table 4. These determinants map to distinct regions of the kinases depending on the cellular role being investigated. For example, threonine 818 directs Ste20 function to the pheromone response pathway and the HOG pathway, whereas two distinct, nonoverlapping segments of Ste20 promote invasive growth. In a similar manner, kinase subdomains III to X allow execution of the Cla4 morphological function, whereas chimeras with nonoverlapping sequences allow for viability in a cln1Δ cln2Δ cla4Δ strain. These results suggest that subsets of Ste20 and Cla4 function are separable and are potentially governed by different factors.
TABLE 4.
Summary of Ste20 and Cla4 functional specificitiesa
The data show the abilities of chimeric proteins to perform Ste20- and Cla4-specific functions. Function in the pheromone response pathway (PRP) was assessed quantitatively for ste20Δ strains as described in Materials and Methods. The ability of chimeras to provide haploid invasive growth (HIG) to ste20Δ strains was qualitatively determined by plate washing assays. The ability of chimeras to restore viability of a cln1Δ cln2Δ cla4Δ 〈CLA4-URA3〉 strain on medium containing 5-FOA and thus allow for the loss of the 〈CLA4-URA3〉 plasmid was determined by a qualitative plate assay. The ability to confer normal bud morphology on cla4 strains was quantified in mid-log-phase cultures with a Zeiss Axioplan microscope. The specificity determinants for each function were as follows: for PRP, T818; for HIG, nonoverlapping N-terminal and C-terminal regions; for cln1 cln2 cla4 strain viability, nonoverlapping N-terminal and C-terminal regions; and for bud morphology, kinase subdomains III to X.
Acknowledgments
We thank David Mitchell, April Goehring, Phil Kinsey, Scott Given, Lucia Liverio, Greg Smith, David Rivers, Paul Cullen, Hilary Kemp, Monique Dail, Karen Sprague, Tom Stevens, Bruce Bowerman, Judith Eisen, and Ira Herskowitz for providing advice, strains, and/or plasmids.
This work was supported by research (GM-30027) and training (GM07413) grants from the NIH.
REFERENCES
- 1.Bagrodia, S., S. J. Taylor, C. L. Creasy, J. Chernoff, and R. A. Cerione. 1995. Identification of a mouse p21Cdc42/Rac activated kinase. J. Biol. Chem. 270:22731-22737. [DOI] [PubMed] [Google Scholar]
- 2.Benton, B. K., A. Tinkelenberg, I. Gonzalez, and F. R. Cross. 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 17:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birukov, K. G., C. Csortos, L. Marzilli, S. Dudek, S. F. Ma, A. R. Bresnick, A. D. Verin, R. J. Cotter, and J. G. Garcia. 2001. Differential regulation of alternatively spliced endothelial cell myosin light chain kinase isoforms by p60(Src). J. Biol. Chem. 276:8567-8573. [DOI] [PubMed] [Google Scholar]
- 4.Bose, I., J. E. Irazoqui, J. J. Moskow, E. S. Bardes, T. R. Zyla, and D. J. Lew. 2001. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 276:7176-7186. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. L., L. Stowers, M. Baer, J. Trejo, S. Coughlin, and J. Chant. 1996. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr. Biol. 6:598-605. [DOI] [PubMed] [Google Scholar]
- 6.Chant, J., and J. R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 8.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 10.Gulli, M., M. Jaquenoud, Y. Shimada, G. Niederhauser, P. Wiget, and M. Peter. 2000. Phosphorylation of the cdc42 exchange factor cdc24 by the PAK-like kinase cla4 may regulate polarized growth in yeast. Mol. Cell 6:1155-1167. [DOI] [PubMed] [Google Scholar]
- 11.Holly, S. P., and K. J. Blumer. 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147:845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis, E. E., D. C. Hagen, and G. F. Sprague, Jr. 1988. Identification of a DNA segment that is necessary and sufficient for α-specific gene control in Saccharomyces cerevisiae: implications for regulation of α-specific and a-specific genes. Mol. Cell. Biol. 8:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallunki, T., B. Su, I. Tsigelny, H. K. Sluss, B. Derijard, G. Moore, R. Davis, and M. Karin. 1994. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 8:2996-3007. [DOI] [PubMed] [Google Scholar]
- 14.Klinghoffer, R. A., C. Sachsenmaier, J. A. Cooper, and P. Soriano. 1999. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 18:2459-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leberer, E., W. Cunle, T. Leeuw, A. Fourest-Lieuvin, J. Segall, and D. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leberer, E., D. Dignard, D. Harcus, L. Hougan, M. Whiteway, and D. Y. Thomas. 1993. Cloning of Saccharomyces cerevisiae STE5 as a suppressor of a Ste20 protein kinase mutant: structural and functional similarity of Ste5 to Far1. Mol. Gen. Genet. 241:241-254. [DOI] [PubMed] [Google Scholar]
- 17.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391:191-195. [DOI] [PubMed] [Google Scholar]
- 19.Lei, M., W. Lu, W. Meng, M. C. Parrini, M. J. Eck, B. J. Mayer, and S. C. Harrison. 2000. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102:387-397. [DOI] [PubMed] [Google Scholar]
- 20.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 21.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. J. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 23.Manser, E., T. Leung, H. Salihuddin, Z. S. Zhao, and L. Lim. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367:40-46. [DOI] [PubMed] [Google Scholar]
- 24.Martin, H., A. Mendoza, J. M. Rodriguez-Pachon, M. Molina, and C. Nombela. 1997. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol. Microbiol. 23:431-444. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell, D. A., and G. F. Sprague, Jr. 2001. The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G2/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:488-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moskow, J. J., A. S. Gladfelter, R. E. Lamson, P. M. Pryciak, and D. J. Lew. 2000. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7559-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posas, F., E. A. Witten, and H. Saito. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18:5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Printen, J. A., and G. F. Sprague, Jr. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 1999. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 274:16861-16870. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 35.Rose, M. D., F. Winston, and P. Hieter (ed.). 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Santos, R. C., N. C. Waters, C. L. Creasy, and L. W. Bergman. 1995. Structure-function relationships of the yeast cyclin-dependent kinase Pho85. Mol. Cell. Biol. 15:5482-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman, F., G. R. Fink, and J. B. Hicks (ed.). 1982. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sheu, Y. J., Y. Barral, and M. Snyder. 2000. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5235-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 41.Sprague, G. F. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 42.Sreenivasan, A., and D. Kellogg. 1999. The elm1 kinase functions in a mitotic signaling network in budding yeast. Mol. Cell. Biol. 19:7983-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi, T. 1995. Cytokine signaling through nonreceptor protein tyrosine kinases. Science 268:251-255. [DOI] [PubMed] [Google Scholar]
- 44.Tjandra, H., J. Compton, and D. Kellogg. 1998. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr. Biol. 8:991-1000. [DOI] [PubMed] [Google Scholar]
- 45.Tu, H., and M. Wigler. 1999. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ushiro, H., T. Tsutsumi, K. Suzuki, T. Kayahara, and K. Nakano. 1998. Molecular cloning and characterization of a novel Ste20-related protein kinase enriched in neurons and transporting epithelia. Arch. Biochem. Biophys. 355:233-240. [DOI] [PubMed] [Google Scholar]
- 47.Wu, C., M. Whiteway, D. Thomas, and E. Lebere. 1995. Molecular characterization of Ste20, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984-15992. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, Z., A. Gartner, R. Cade, G. Ammerer, and B. Errede. 1993. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell. Biol. 13:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]