Abstract
During the course of a study aimed at identifying inner ear-specific transcripts, a 1,906-bp murine cDNA predicted to encode a secreted 469-aa protein with two domains of homology with the secreted phospholipases A2 was isolated. This transcript is specifically expressed in the inner ear from embryonic day 9.5. The encoded 95-kDa glycoprotein is the major protein of the utricular and saccular otoconia and thus was named otoconin-95. By immunohistofluorescence, otoconin-95 also was detected in the cupulae of the semicircular canals and in previously undescribed transient granular structures of the cochlea. Otoconin-95 was found to be synthesized by various nonsensory cell types, but not by the supporting cells of the sensory epithelia, which produce the otoconial precursor vesicles. In addition, multiple isoforms generated by differential splicing were observed in different combinations during development. Based on the present results, we propose a model for the formation of the otoconia.
The mammalian inner ear is composed of two sensory organs: the cochlea, which is the auditory component, and the vestibule, responsible for the control of balance. The latter consists of the saccule and the utricle, which detect linear acceleration, and the three semicircular canals sensitive to angular acceleration. Each sensory epithelium is covered by an acellular gelatinous membrane, namely the tectorial membrane in the cochlea, the cupulae in the ampullae of the semicircular canals, and the otoconial membranes in the saccule and the utricle. The displacement of the acellular membrane relative to the sensory epithelia leads to the deflection of the stereocilia of sensory hair cells, which in turn opens the mechanotransduction channels, leading to cell depolarization.
The otoconial membranes of the saccule and utricle are overlayed with dense biominerals made of a filamentous organic matrix and calcium carbonate (CaCO3). These biominerals are found as large deposits (otoliths) in most fish and as numerous small crystals (otoconia) in all other vertebrates. Otoliths and otoconia have been classified in three groups according to the crystalline form of CaCO3, i.e., vaterite, aragonite, or calcite. In mammals, otoconia are calcitic, whereas in amphibians they are calcitic in the utricle and aragonitic in the saccule (1–4). Each type of otoconia is characterized by a distinct set of proteins collectively named otoconins (3). The major protein of aragonitic otoconia has been purified from Xenopus laevis saccule and sequenced (5). In contrast, none of the otoconins of the calcitic otoconia have been characterized so far. In mice, the rate of otoconia production is highest between embryonic days 15 and 17 (E15–E17), although the biominerals continue to grow until postnatal day 7 (P7) (6, 7). Different hypotheses for the genesis of otoconia have been proposed (reviewed in ref. 8). It is now generally accepted that otoconia are produced from protrusions of the saccular and utricular supporting cells that form vesicular structures above the maculae (6, 7, 9, 10). These noncrystalline structures, here referred to as preotoconia, are 3–10 μm in diameter in the mouse (11), are surrounded by a membrane with characteristics of intracellular organelles (11), and have a high calcium content (7, 9, 11–13). The mechanism of subsequent crystal formation is unknown.
With the aim of identifying proteins specific to the inner ear, we used a PCR amplification-based approach to generate a subtracted mouse cochlear cDNA library. This process led us to identify the major protein of murine otoconia, which was named otoconin-95. Based on the biochemical characterization of otoconin-95 and the identification of the cells producing the protein during development, we propose a model for the biogenesis of mammalian otoconia.
MATERIALS AND METHODS
Construction of a Subtracted Cochlear cDNA Library.
Extraction of total RNA from cochleae, dorsal root ganglia, and cartilage of P2, P15, and E20 BALB/c mice, respectively, purification of poly(A)+ RNA, and generation of double-stranded cDNA were carried out as described (14). Sau 3AI-digested cochlear cDNA fragments were subjected to three rounds of PCR-coupled subtraction by using the representational difference analysis method (15). The subtracting cDNA mixture comprised amplified Sau 3AI-digested cDNA fragments from cartilage and dorsal root ganglia (20 μg each) and seven Sau 3AI-digested clones (2.5 μg each) isolated from a previous subtracted cochlear cDNA library (14). The final differential products were cloned into pBluescript KS+ (Stratagene).
Whole-Mount in Situ Hybridization and Northern Blot Analysis.
Whole E8.5–E12 Rj:Swiss mouse embryos were treated and hybridized as described (16). The sense and antisense riboprobes were derived from the pC3 cDNA clone (nucleotide positions 669–942). One microgram of each poly(A)+ RNA preparation was fractionated, blotted, and hybridized with a probe derived from pC3, following standard procedures (17).
Cloning of the Full-Length Ocn-95 cDNA and Nucleotide Sequence Analysis.
The 274-bp pC3 sequence was extended by 5′ and 3′ rapid amplification of cDNA ends (RACE)–PCR. The amplifications were performed on oligo(dT)-primed double-stranded P2 cochlear cDNA ligated to adaptors, as described (14). The amplified 5′ and 3′ RACE-PCR products, obtained with primer 5′-901-TCTGCTCTCACCTCACCAGCCACTT and primer 5′-695-GTGTGACAAGGCTGCTGTGGAGTGC, respectively, were cloned into the pCR 2.1 TA cloning vector (Invitrogen). DNA sequencing was carried out as described (14), and sequence analysis was performed by using the Genetics Computer Group Package (18).
Alternative Splicing Analysis.
Reverse transcription–PCR was performed on total RNA extracted from E11.5, P0, P2, P15, and adult inner ears by using the GeneAmp RNA PCR kit (Perkin–Elmer) and several primer pairs. All of the alternative regions were localized in the DNA fragment amplified by using primer 5′-695-GTGTGACAAGGCTGCTGTGGAGTGC and primer 5′-1482-CTCCGTCTGGTGACTTGAGGCTTTGA; the corresponding amplification products were purified and sequenced. Partial gene structure was determined by PCR amplifications carried out on mouse genomic DNA by using the primer pairs 5′-1100-TGACAGATTGGCCTTCGTGC and 5′-1231-CTTCCACAGTAGCAGCC, and 5′-999-GCCCTAGCCAAAGGTACAACCC and 5′-1144-GTCATGCTGTCCCCATCACCCA; the intron/region C junction was sequenced.
Gene Mapping.
A length polymorphism was detected between the C57BL/6 and SEG strains in the 3′ untranslated region of Ocn-95 by PCR amplification using the primer pair 5′-1623-CCTCCTGGTCCCTTGGGGGCCAGA and 5′-1818-TGCTGACAAGTTTCAGGAGGCTCGTG. Using this polymorphism, segregation of Ocn-95 was compared with that of marker loci from chromosome 15, which previously were typed in 91 DNA samples derived from backcross progeny between the C57BL/6 and SEG strains of the European Collaborative Interspecific Backcross panel (19). The results were processed with the help of genelink software (20).
Anti-Otoconin-95 Sera and Immunohistofluorescence Studies.
A cDNA fragment from positions T775 to A1137 was amplified with primers containing a BamHI (forward primer) or KpnI (reverse primer) restriction site in their 5′ termini. The digested PCR product was cloned into the expression vector pQE30 (Qiagen, Chatsworth, CA). The encoded 6× His-tagged recombinant polypeptide was overexpressed in Escherichia coli and purified according to the QIAexpressionist protocol (Qiagen). Five hundred micrograms of either the purified recombinant protein or one of two keyhole limpet hemocyanin-coupled synthetic peptides corresponding to the N-terminal region (NH2-LDTPNPQELPPGLSK-COOH) or C-terminal end (NH2-KPPGPLGARPLGGK-COOH) of the protein was used to prime New Zealand White rabbits. The animals received four boosts (500 μg) at 4-week intervals. Tissue preparation and immunolabeling were performed as described (14).
Protein Extraction and Analysis.
Tissue samples were subjected to three successive extraction steps (3). (i) They were first homogenized as described (14); after centrifugation, the supernatant (fraction 1) was collected. (ii) The pellet was sonicated for 5 min in 0.2% SDS/100 mM sodium acetate, pH 7.4 and centrifuged at 13,000 g for 5 min. This extraction step was repeated three times, and the three supernatants were pooled (fraction 2). (iii) The final pellet was incubated overnight at 4°C in 20% EDTA, pH 7.4 (fraction 3). Saccular and utricular maculae from seven adult mice were microdissected in artificial endolymph (21) under a stereomicroscope, and the otoconial complexes were harvested with a siliconized pulled Pasteur pipette. After incubation in 500 μl of 0.025% SDS/100 mM sodium acetate, pH 7.4 for 5 min and centrifugation, the supernatant was collected, and the pellet was decalcified in 8 μl of 20% EDTA, pH 7.4. Proteins were separated on 7.5% reducing SDS/polyacrylamide gels and transferred onto nitrocellulose membranes (Bio-Rad) following standard procedures (22). Immunodetection using the ECL kit (Amersham) with a 1/10,000 serum dilution and colloidal gold staining using AuroDye forte (Amersham) were performed according to the manufacturer’s instructions. N-glycosidase-F treatment on cochlear fraction 1 and vestibular fraction 2 extracted at P2 was performed as described (14).
RESULTS
Identification of an Inner Ear-Specific Transcript Encoding a Phospholipase A2 (PLA2)-Related Protein.
As a preliminary test of the subtracted mouse cochlear cDNA library, 34 clones were randomly selected and sequenced. Among the 19 different sequences identified, one (pC3 for partial C3, 274 bp long) showed 77% nucleotide sequence identity with a human cDNA clone, AF5, isolated from a teratocarcinoma cell line (23). Because AF5 encoded a protein with similarity to PLA2 enzymes, the corresponding gene, whose expression had not been detected in any of the normal human tissues tested, had been named PLA2L for PLA2-like gene.
By in situ hybridization, performed on whole-mount E8.5–E12.5 embryos (Fig. 1) and tissue sections at P2 (not shown), the C3 transcript was detected exclusively in the inner ear from E9.5 onward. Reverse transcription–PCR amplifications, carried out on total RNA extracted from various P2 or adult tissues, confirmed the inner ear specificity of C3 (not shown).
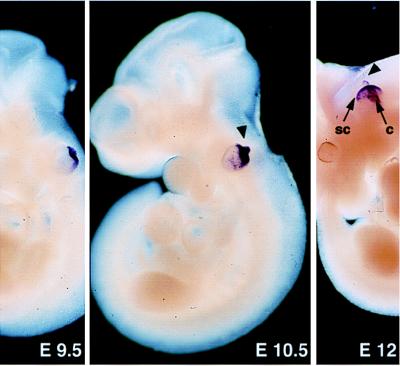
Figure 1.
Expression of Ocn-95 at early embryonic stages. At E9.5–E10.5, the growing endolymphatic duct (arrowhead) and the dorsal part of the otic vesicle are strongly labeled. At E12, the endolymphatic duct, the forming semicircular canals (sc), and the cochlea (c) are stained. Magnification: ×25 (E9.5), ×17 (E10.5), and ×6 (E12).
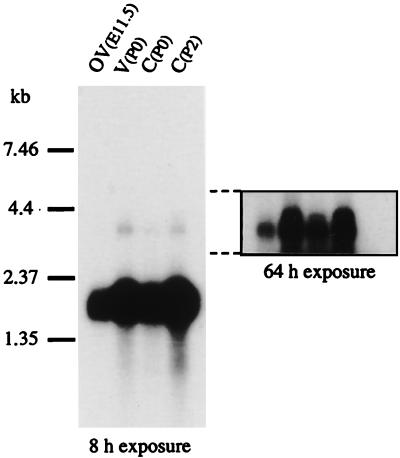
Northern blot analysis of poly(A)+ RNA from the otic vesicle and the cochlea or vestibule collected at E11.5 and P0 or P2, respectively, by using a pC3-derived probe, detected a highly expressed 1.9-kb transcript (Fig. 2) and two minor transcripts of ≈3 and 3.5 kb, which are likely to correspond to nuclear precursors because they were shown to contain one or two introns introducing an in-frame stop codon. In the inner ear from P15 and adult mice, a faint 1.9-kb band could be observed only after a long exposure (data not shown).
Figure 2.
Northern blot analysis of mouse poly(A)+ RNA at E11.5, P0, and P2. OV, otic vesicle; V, vestibule; C, cochlea.
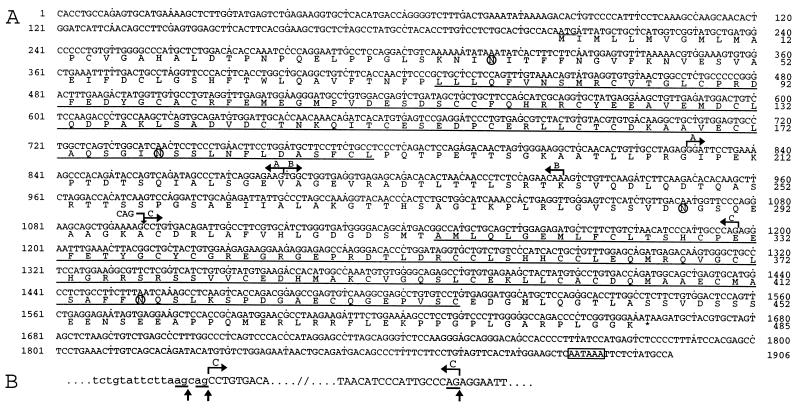
A 1,906-bp C3 cDNA was reconstituted by using 5′ and 3′ rapid amplification of cDNA ends–PCR. The translation initiation site was identified at position 207 by the presence of an adequate Kozak consensus sequence (GCCACAATGA; ref. 24) preceded by three stop codons (positions 33, 39, and 69). The initiation codon is followed by a 1,458-bp ORF and a 242-bp 3′ untranslated region containing a polyadenylation signal (Fig. 3A). The deduced protein begins with a 16-aa sequence that fits the predictions for a signal sequence (25). Its cleavage would leave a mature 469-aa protein, with a predicted molecular mass of 50.8 kDa. Four potential N-glycosylation sites at positions 37, 178, 288, and 417 were detected (Fig. 3A). Because it subsequently was shown to be a 95-kDa component of the otoconia (see below), the C3-encoded protein hereafter is referred to as otoconin-95 and the corresponding gene is referred to as Ocn-95.
Figure 3.
(A) Ocn-95 cDNA and deduced amino acid sequences. The translation initiation codon is underlined, and a polyadenylation signal is boxed. The alternative regions (A, B, C, and CAG insertion) are indicated by broken arrows. The CAG insertion at position 1096 is predicted to result in the insertion of an alanine at position 298. Loss of A, (A + B), and C is predicted to result in in-frame deletions of 17 (G208-E224), 33 (G208-T240), and 34 (A297-P330) aa, respectively. Loss of (A+B) is also responsible for an amino acid substitution (K241D). Regions A and B are, according to the structure of PLA2L (28), encoded by two adjacent exons. The two sPLA2-like domains are underlined, and four potential N-glycosylation sites are circled. (B) Alternative splicing at the intron/region C junction. Lowercase and uppercase letters denote intronic and exonic sequences, respectively. The alternative region C is indicated between broken arrows. The three acceptor splice sites (canonical AG underlined) are highlighted by vertical arrows. Region C corresponds to the 5′ part of a 172-bp human exon (28) and its 3′ end fits well with an acceptor splice site (29). Amplification on mouse genomic DNA of a 132-bp fragment between T1100 and G1231 confirmed that the 102-bp region C corresponds to the 5′ part of a longer exon. Sequence analysis of the intron-region C junction revealed two overlapping acceptor splice sites. The use of the most 5′ one results in the CAG insertion at position 1096.
Analysis of the deduced amino acid sequence demonstrated homology with the secreted PLA2s (sPLA2s), which constitute one of the two major classes of PLA2s (26). Two sPLA2-like domains of 117 and 122 aa were detected (Fig. 3A). These domains are only 37% identical, suggesting ancient duplication, and each sPLA2-like domain displays 30–37% identity with the numerous sPLA2s. The strong sequence homology with the putative protein encoded by the human PLA2L gene was found to extend throughout the protein, leading us to consider PLA2L and Ocn-95 as orthologs. During computer-aided alignment of otoconin-95 with the sPLA2-related proteins, an homology with otoconin-22 was noted. This 127-aa protein, extracted from the X. laevis saccular otoconia (5) consists of a single sPLA2-like domain that displays ≈31% identity with the sPLA2-like domains of otoconin-95.
The sPLA2s are 115- to 140-aa Ca2+-dependent enzymes with seven intramolecular disulfide bonds. They have been classified into group I and group II, based on their primary sequences (27). The sPLA2-like domains of murine otoconin-95, human PLA2L (23), and Xenopus otoconin-22 (5) all possess some residues that are characteristic of group I sPLA2s and others that are group II specific. In addition, these proteins lack several conserved residues of sPLA2s, which are involved in the catalysis, suggesting that they do not have a PLA2 activity.
Differential Splicing of Ocn-95 Transcripts During Mouse Development.
Analysis of Ocn-95 expression by reverse transcription–PCR demonstrated the existence of different combinations of various transcripts during development. At E11.5, six transcripts encoding putative isoforms differing in the length of the interdomain linking the two sPLA2-like domains were identified. These transcripts result from the presence/absence of a CAG insertion at position 1096, combined with the presence/absence of a 51-bp (region A) or a 99-bp (region A plus region B) sequence (Fig. 3A). At this stage, most of the transcripts lack the CAG insertion, and the major amplified product is deleted for region A. At P0/P2, only transcripts containing the complete 1,906-bp sequence were observed, with the CAG insertion in a minority of them. At P15, these two transcripts also were detected, but the one with the CAG insertion was by far predominant. In addition, another transcript with a 102-bp deletion (region C, Fig. 3A) was amplified. This transcript encodes a protein with incomplete inter and second sPLA2-like domains. In adult mice, the transcripts observed were the same as those detected at E11.5 but those containing region A were amplified at the same level as those without it. According to the partial structure of human PLA2L (28) as well as our analysis of Ocn-95 (see Fig. 3 legend), we could conclude that all of the transcripts observed result from alternative splicing. In particular, we established that inclusion/exclusion of region C is determined by the use of three alternative acceptor splice sites, whereas regions A and B represent cassette exons (Fig. 3 A and B and legend).
Cells Synthesizing Otoconin-95 During Inner Ear Formation.
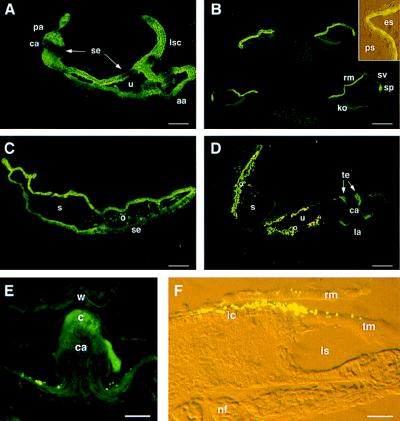
The specificity of the antisera raised against three distinct regions of otoconin-95 was ascertained by both immunoblot and immunofluorescence analysis of tissues at P0. All three antisera gave the same staining pattern, and no signal was observed with the preimmune sera. The results illustrated hereafter have been obtained with the 046CP4 serum raised against the C-terminal peptide. At E10.5, otoconin-95 was detected in the epithelium of the dorsal wall of the otic vesicle and in the emerging endolymphatic duct (not shown). At E14.5, in the vestibule, otoconin-95 was detected in the nonsensory epithelia of the semicircular canals and in the walls of the utriculo-saccular complex (Fig. 4A). In contrast, no immunostaining of the future sensory patches of the semicircular canals (the cristae ampullaris), the utricle, and the saccule (utricular and saccular maculae) was observed. At the same stage, in the cochlea, otoconin-95 was observed in the epithelial cells of the roof of the cochlear duct, but not in Kölliker’s organ, the future sensory organ of Corti (not shown). At birth, in the cochlea (Fig. 4B), the labeling was restricted to the spiral prominence and Reissner’s membrane (only the cell layer facing the endolymphatic space was labeled). In the vestibule, a signal was observed in the transitional cells and in the epithelial cells of the roof of the saccule (Fig. 4C) and utricle. At P4, the transitional cells adjacent to the cristae (Fig. 4D) were labeled while the staining of the other epithelial cells of the labyrinth began to fade. At P10, cellular immunostaining was no longer observed in the inner ear and only specific acellular structures were labeled.
Figure 4.
Distribution of otoconin-95 in the developing inner ear. (A) Section through the three ampullae and part of the utriculo-saccular complex at E14.5. (B) Four sections of the cochlear duct at P0. Note that cochlear cell differentiation proceeds with a base-to-apex gradient of maturation. (Inset) A detailed view of the two cell layers of the Reissner’s membrane. (C) Saccule at P0. (D) Vestibule at P4. (E) Posterior ampulla at P21. The punctate signals above the epithelium on each side of the cristae were not reproducible; they may correspond to displaced utricular otoconia. (F) Cochlea at P8. Inset and F micrographs were taken under both daylight and fluorescent light with Nomarski optics. The 046CP4 serum was used at 1:1,000 dilution. aa, anterior ampulla; c, cupula; ca, crista ampullaris; es, endolymphatic space; ic, interdental cells; is, inner sulcus; ko, Kölliker’s organ; la, lateral ampulla; lsc, lateral semicircular canal; nf, nerve fiber; o, otoconia; pa, posterior ampulla; ps, perilymphatic space; rm, Reissner’s membrane; s, saccule; se, sensory epithelium; sp, spiral prominence; sv, stria vascularis; te, transitory epithelium; tm, tectorial membrane; u, utricle; w, wall of the ampulla. (Scale bars: A, B, and D, 50 μm; C and E, 30 μm; F, 15 μm.)
Otoconin-95 in Acellular Structures of the Inner Ear.
Immunostaining of the forming otoconia first was detected at E15.5 (not shown). After birth, in the vestibule, the strongest signal was associated with the saccular and utricular otoconia (Fig. 4D). A staining of the cupulae also was observed from P8 onward (Fig. 4E). In the cochlea, a bright punctate staining was observed above the tectorial membrane in its limbal region (Fig. 4F). This reproducible staining was transient; it first was detected at E20, was strongest at P8, and had disappeared at P10. The labeled granules, observed all along the length of the cochlea, were 3- to 4-fold smaller in diameter than the otoconia, regardless of the stage.
Biochemical Characterization of Otoconin-95.
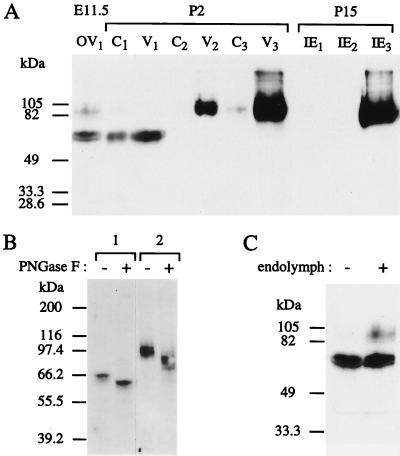
Immunoblot analysis detected several bands in the inner ear protein extracts (Fig. 5A). At E11.5, two bands of ≈65 kDa and several bands ranging from 85 to 105 kDa were obtained from the otic vesicles. At P0/P2, both the cochlear and the vestibular “soluble protein fractions” (fraction 1, see Materials and Methods) contained only the ≈65-kDa proteins. However, considering that from E15.5 onward the otoconia are forming, the proteins present in the “insoluble fraction” sequentially were extracted by sonication in the presence of SDS (fraction 2) and by decalcification with EDTA (fraction 3) (3). The 85- to 105-kDa proteins were detected in the vestibular fraction 2, but the bulk of these forms were present in fraction 3. Therefore, otoconia only contain the 85- to 105-kDa isoforms of otoconin-95. A faint band at ≈95 kDa also was observed in the cochlear fraction 3. At P15, the ≈65-kDa forms were no longer detected in the inner ear, and the 85- to 105-kDa proteins were detected only in the decalcified fraction. Because the highest molecular mass of the proteins predicted by the various Ocn-95 transcripts is ≈51 kDa, it was hypothesized that the protein backbone was subjected to posttranslational modifications. Given that four potential N-glycosylation sites had been detected within the otoconin-95 amino acid sequence (Fig. 3A), the presence of N-linked sugars was tested by N-glycosidase F (PNGase F) digestion (Fig. 5B). After PNGase F treatment, the 65-kDa bands shifted to 60 kDa, and the 85- to 105-kDa ones to 75–95 kDa, therefore showing that all isoforms are N-glycosylated. However, the bulk of the difference between the apparent molecular masses of 65 and 85–105 kDa remained unexplained. Considering that the 65-kDa forms were detected only when a cellular immunostaining was observed, we investigated whether the ≈65-kDa forms might represent the intracellular precursors of the secreted 85- to 105-kDa ones. Proteins were extracted from E11.5 otic vesicles either intact or slitted and extensively washed in PBS to remove endolymph. The 65-kDa bands were observed in both conditions whereas the 85- to 105-kDa bands were detected only when the endolymph was preserved (Fig. 5C), demonstrating that the 65-kDa forms are indeed intracellular precursors of the 85- to 105-kDa ones, which are rapidly exported to the endolymph upon the final maturation step.
Figure 5.
Immunoblot analysis of otoconin-95. (A) Differential extraction of otoconin-95 isoforms. Soluble proteins were extracted from otic vesicle at E11.5 (OV1), cochlea (C1), and vestibule (V1) at P2, and inner ear (IE1) at P15. Proteins of the remaining insoluble fractions were sequentially extracted by sonication in the presence of 0.2% SDS (C2, V2, and IE2, respectively) and decalcification in the presence of 20% EDTA (C3, V3, and IE3, respectively). (B) N-glycosydase F sensitivity of the 65-kDa (lane 1) and 85- to 105-kDa (lane 2) forms. Fractions C1 and V2 were incubated either with (+) or without (−) N-glycosydase F (PNGase F). (C) Identification of the intracellular and secreted isoforms of otoconin-95. Proteins were extracted from E11.5 otic vesicles in which endolymph had been retained (+) or removed (−). Molecular mass standards are indicated on the left.
Otoconin-95 Is the Major Protein of Murine Otoconia.
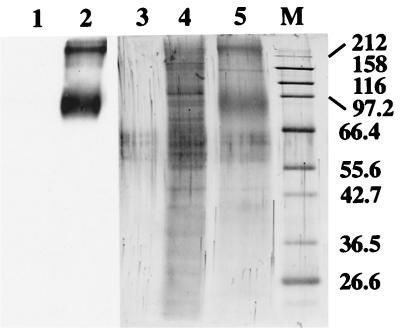
As otoconin-95 appeared to be expressed at high levels in the developing inner ear and to be mostly incorporated into the otoconia, it was hypothesized that it could be identical to the 90- to 100-kDa protein that was reported as the major protein of the rat otoconia (4). To test this hypothesis, saccular and utricular otoconial complexes (otoconia + otoconial membrane) from adult mice were microdissected. The proteins from the otoconial membranes were extracted and the pelleted otoconia were decalcified. The solubilization of the otoconial membrane and the concomitant purification of the otoconia were monitored both by light microscopy and immunodetection of otogelin (14), a marker of the otoconial membranes (not shown). Many proteins could be detected in the otoconial membrane extract after colloidal gold staining (Fig. 6, lane 4), whereas a major broad band of 85–105 kDa and another strong band at the top of the gel were detected from the otoconia (lane 5). Immunoblot analysis using the 046CP4 antiserum failed to detect any signal from the otoconial membranes (lane 1), whereas, in the otoconia, a strong 85- to 105-kDa band and a band at the top of the gel (which thus may represent aggregates of otoconin-95) could be seen (lane 2). Therefore, otoconin-95 is the major protein of murine otoconia.
Figure 6.
Protein content of the otoconial complex. Immunodetection of otoconin-95 (1/15 of the sample, lanes 1 and 2) and colloidal gold staining (14/15 of the sample, lanes 4 and 5) of proteins extracted from the otoconial membranes (lanes 1 and 4) and otoconia (lanes 2 and 5) of adult mice; lane 3, control lane for colloidal gold staining loaded without protein added. Molecular masses (in kDa) of the marker proteins in lane M are indicated on the right. Note that the bands, migrating between the 55.6- and 66.4-kDa markers, observed in each lane (including lane 3) are artifacts seen only in the presence of reducing agents.
Otoconin-95 and Genetic Inner Ear Defects.
At least three mouse mutants with isolated otoconial defects (30–33), and more than 20 mouse mutants with vestibular dysfunction and deafness (ref. 34; 1998 Mouse Genome Database), have been reported. To test whether an Ocn-95 defect could underlie one of these mutants, the chromosomal mapping of this gene was undertaken. Given that PLA2L already had been mapped to 8q24 (28), which is homologous to a region of mouse chromosome 15, linkage analysis with polymorphic chromosome 15 markers was undertaken. Ocn-95 was assigned to between D15Mit65 and D15Mit144, located at positions 29 and 31.2, respectively (1998 Mouse Genome Database). None of the aforementioned mutations have been mapped to this region. Likewise, none of the reported human loci for isolated inner ear dysfunction have been localized to the PLA2L region.
DISCUSSION
To date, three inner ear-specific proteins have been reported in mammals (14, 35), all of them being components of the acellular membranes. In the present study, we have characterized another inner ear-specific glycoprotein, otoconin-95, which is the major protein of murine calcitic otoconia. In addition, the human PLA2L orphan gene (23) was identified as the Ocn-95 ortholog.
The Major Proteins of Otoconia Are Related to sPLA2.
In both the rat otoconia and the Xenopus utricular (calcitic) otoconia, the presence of a major 90-to 100-kDa protein of unknown sequence has been reported (3). Therefore, calcitic otoconia probably contain a similar 90- to 100-kDa major protein, regardless of the species. In contrast, the Xenopus saccular (aragonitic) otoconia contain a major 22-kDa protein (otoconin-22) (5), which is a sPLA2-related 127-aa glycoprotein with two N-glycosylation sites. Otoconin-95 is also a N-glycosylated sPLA2-related protein. However, whereas otoconin-22 consists of a single sPLA2-like domain, otoconin-95 has two such domains, linked by an interdomain of variable length (because of alternative splicing), and N- and C-terminal extensions. The present data indicate that posttranslational modifications other than N-glycosylations occur before secretion of otoconin-95. In this respect, it is noteworthy that six potential O-glycosylation sites, all outside of the sPLA2-like domains (T22 in the N-terminal end, T203, S252, T254, and T255 in the interdomain, and S456 in the C-terminal end), were predicted by the netoglyc software (36). Although highly divergent, the sPLA2-like domains of otoconin-95 and otoconin-22 share two common features. (i) Several amino acids required for the PLA2 activity are substituted, making it unlikely that otoconin-95 and otoconin-22 have such an activity. Along this line, attempts to detect a PLA2 activity for otoconin-22 have been unsuccessful (5). (ii) The domains have characteristics of both group I and group II sPLA2s, suggesting that Ocn-22 and Ocn-95 derived from a gene ancestral to the divergence of these two sPLA2s groups. Accordingly, it is reasonable to assume that the major 40- to 50-kDa protein of vateritic otoliths found in chondrostean fish (3) also belongs to the same class of sPLA2s-related glycoproteins.
Insights into the Formation of the Otoconia.
We found that otoconin-95 is synthesized by numerous nonsensory epithelial cell types of the membranous labyrinth, but not by the supporting cells of the saccular and utricular maculae, which release the preotoconia. Otoconin-95 is produced from E9.5 onward and seems to accumulate within the endolymph several days before the appearance of preotoconia, between E14.5 and E15.5 (6, 7, 10, 13). Based on these results, we propose the following model for the formation of the otoconia: despite the absence of PLA2 activity, the two sPLA2-like domains of otoconin-95 are able to interact specifically with the membrane of the preotoconia. A possible resorption of this membrane would allow the interaction of otoconin-95 with the Ca2+-rich vesicular content. Otoconin-95 thus may be involved in the initiation and/or growth of the crystal via Ca2+ or CaCO3 binding. This proposal is supported by the results of Pote and Ross (3), who demonstrated that a 90- to 100-kDa protein of rat otoconia is able to bind calcium. Given the low pI of the extracellular forms of otoconin-95 (major band at pI ≤ 4.3, data not shown), we suggest that negative charges, carried by the protein backbone and oligosaccharides (possibly glycosaminoglycan chains), may bind Ca2+. Therefore, the production by separate cell populations of the Ca2+-rich substance and otoconin-95 allows crystallization to occur only when these two components come into contact, upon the release of the preotoconia above the maculae. Six different protein backbones, which differ by the length of the interdomain, were found at E11.5, when otoconin-95 accumulates in the endolymph. Some of them thus may be involved in the initiation of crystallization. The two transcripts encoding the longest isoforms of the protein (which are the only ones detected at birth) could be involved in the growth of the biomineral. In contrast, the isoform with a truncated second sPLA2-like domain, whose transcript was detected at P15, might inhibit the growth of the crystal.
The question of whether otoconia form only during inner ear development or are renewed throughout life is still debated. In adults, low levels of Ocn-95 transcripts could be detected by Northern blot and reverse transcription–PCR analyses. Interestingly, the same combination of Ocn-95 transcripts was found at E11.5 and in adult mice. Two E11.5 splice variants, no longer detected at P0/P2 or P15, thus are re-expressed at low levels in adult mice, therefore supporting the idea that neogenesis of otoconia occurs during adult life.
Otoconin-95 in Other Acellular Structures of the Inner Ear.
Otoconin-95 was detected in the cupulae of the cristae ampullaris from P8 onward. It is noteworthy that these gelatinous extracellular matrices have an affinity for calcium (37) and can be calcified in pathological conditions (38, 39). We also reproducibly detected otoconin-95 in transient granular structures overlaying the tectorial membrane in its limbal portion from E20 to P8. The cellular staining that was observed in the cochlea up to P8 supports the proposal that these “tectorial granules” are indeed acellular structures specific to the cochlea. Their location immediately underneath the Reissner’s membrane (Fig. 4F) suggests a role in the positioning of the forming tectorial membrane at the surface of the spiral limbus, by preventing the tectorial membrane from adhering to the Reissner’s membrane. These transient granules have not been reported before. However, it is noteworthy that spherical structures have been described in the same location in adult guinea pigs treated with a cholinomimetic agent (40).
Acknowledgments
We thank J.-P. Hardelin, M. Tosi, and V. Kalatzis for critical reading of the manuscript, F. Crozet for helpful discussions, and S. Blanchard and F. Levi-Acobas for technical help. We are grateful to D. Demêmes and M. T. Nicolas for help with microdissection and to D. Simon and J. Jaubert for help with gene mapping. This work was supported by grants from the Association Française contre les Myopathies (5870 MG-1997) and the European Economic Community (PL95–1324). E.V. was supported by a fellowship from the Caisse Nationale d’Assurance Maladie des Professions Indépendantes (CANAM).
ABBREVIATIONS
- E(n)
embryonic day
- P(n)
postnatal day
- PLA2
phospholipase A2
- SPLA2
secreted PLA2
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF093591).
References
- 1.Carlström D. Biol Bull. 1963;125:441–463. [Google Scholar]
- 2.Marmo F, Balsamo G, Franco E. Cell Tissue Res. 1983;233:35–43. doi: 10.1007/BF00222230. [DOI] [PubMed] [Google Scholar]
- 3.Pote K G, Ross M D. Comp Biochem Physiol. 1991;98:287–295. doi: 10.1016/0305-0491(91)90181-c. [DOI] [PubMed] [Google Scholar]
- 4.Pote K G, Ross M D. J Ultrastruct Mol Struct Res. 1986;95:61–70. doi: 10.1016/0889-1605(86)90029-7. [DOI] [PubMed] [Google Scholar]
- 5.Pote K G, Hauer III C R, Michel H, Shabanowitz J, Hunt D F, Kretsinger R H. Biochemistry. 1993;32:5017–5024. doi: 10.1021/bi00070a007. [DOI] [PubMed] [Google Scholar]
- 6.Anniko M. Am J Otolaryngol. 1980;1:400–410. doi: 10.1016/s0196-0709(80)80021-4. [DOI] [PubMed] [Google Scholar]
- 7.Takumida M, Harada Y. Arch Oto-Rhino-Laryngol. 1984;241:9–15. doi: 10.1007/BF00457911. [DOI] [PubMed] [Google Scholar]
- 8.Lim D J. In: Ultrastructural Atlas of the Inner Ear. Friedman I, Ballantyne J, editors. London: Butterworth; 1984. pp. 245–269. [Google Scholar]
- 9.Harada Y. Scanning Electron Microsc. 1979;III:963–966. [PubMed] [Google Scholar]
- 10.Nakahara H, Bevelander G. Anat Rec. 1979;193:233–241. doi: 10.1002/ar.1091930205. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H, Ikeda K, Takasaka T. Hearing Res. 1995;90:212–218. doi: 10.1016/0378-5955(95)00168-7. [DOI] [PubMed] [Google Scholar]
- 12.Anniko M, Wikstrom S O, Wroblewski R. Acta Oto-Laryngol. 1987;104:285–289. doi: 10.3109/00016488709107330. [DOI] [PubMed] [Google Scholar]
- 13.Harada Y. The Vestibular Organs: SEM Atlas of the Inner Ear. Amsterdam: Kugler & Ghedini; 1988. [Google Scholar]
- 14.Cohen-Salmon M, El-Amraoui A, Leibovici M, Petit C. Proc Natl Acad Sci USA. 1997;94:14450–14455. doi: 10.1073/pnas.94.26.14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Nature (London) 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breen M, Deakin L, MacDonald B, Miller S, Sibson R, Tartelin E, Avner P, Bourgade F, Guénet J-L, Montagutelli X, et al. Hum Mol Genet. 1994;3:621–627. [Google Scholar]
- 20.Montagutelli X. J Hered. 1990;81:490–491. doi: 10.1093/oxfordjournals.jhered.a111033. [DOI] [PubMed] [Google Scholar]
- 21.Ross M D, Pote K G, Cloke P L, Corson C. Physiologist. 1980;23:S129–S130. [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 23.Feuchter-Murthy A E, Freeman J D, Mager D L. Nucleic Acids Res. 1993;21:135–143. doi: 10.1093/nar/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis E A. J Biol Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 27.Davidson F F, Dennis E A. J Mol Evol. 1990;31:228–238. doi: 10.1007/BF02109500. [DOI] [PubMed] [Google Scholar]
- 28.Kowalski P E, Freeman J D, Nelson D T, Mager D L. Genomics. 1997;39:38–46. doi: 10.1006/geno.1996.4471. [DOI] [PubMed] [Google Scholar]
- 29.Senapathy P, Shapiro M B, Harris N L. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 30.Lane P. Mouse News Lett. 1986;75:28. [Google Scholar]
- 31.Ornitz D M, Bohne B A, Thalmann I, Harding G W, Thalmann R. Hearing Res. 1998;122:60–70. doi: 10.1016/s0378-5955(98)00080-x. [DOI] [PubMed] [Google Scholar]
- 32.Peterson A, Biddle F. Mouse News Lett. 1970;43:19. [Google Scholar]
- 33.Sweet H O. Mouse News Lett. 1980;63:19. [Google Scholar]
- 34.Steel K P. Annu Rev Genet. 1995;29:675–701. doi: 10.1146/annurev.ge.29.120195.003331. [DOI] [PubMed] [Google Scholar]
- 35.Legan P K, Rau A, Keen J N, Richardson G P. J Biol Chem. 1997;272:8791–8801. doi: 10.1074/jbc.272.13.8791. [DOI] [PubMed] [Google Scholar]
- 36.Hansen J E, Lund O, Engelbrecht J, Bohr H, Nielsen J O, Hansen J-E S, Brunak S. Biochem J. 1995;308:801–813. doi: 10.1042/bj3080801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamata S. Arch Histol Cytol. 1991;54:173–180. doi: 10.1679/aohc.54.173. [DOI] [PubMed] [Google Scholar]
- 38.Johnsson L G, Rouse R C, Hawkins J E, Jr, Kingsley T C, Wright C G. Am J Otolaryngol. 1981;2:284–298. doi: 10.1016/s0196-0709(81)80038-5. [DOI] [PubMed] [Google Scholar]
- 39.Johnsson L G, Rouse R C, Wright C G, Henry P J, Hawkins J E., Jr Am J Otolaryngol. 1982;3:77–90. doi: 10.1016/s0196-0709(82)80037-9. [DOI] [PubMed] [Google Scholar]
- 40.Prieto J J, Rueda J, Rubio M E, Merchan J A. Hearing Res. 1991;54:59–66. doi: 10.1016/0378-5955(91)90136-w. [DOI] [PubMed] [Google Scholar]