Abstract
Eukaryotic initiation factor 4E (eIF4E) binds the mRNA cap structure and forms eIF4F complexes that recruit 40S subunits to the mRNA. Formation of eIF4F is blocked by eIF4E-binding proteins such as 4E-BP1, which interacts with eIF4E via a motif in the center of its 118-residue sequence. 4E-BP1 plays key roles in cell proliferation, growth, and survival. Binding of 4E-BP1 to eIF4E is regulated by hierarchical multisite phosphorylation. Here we demonstrate that three different features in the C terminus of 4E-BP1 play distinct roles in regulating its phosphorylation and function. Firstly, we identify a new phosphorylation site in its C terminus (S101). A serine or glutamate at this position is required for efficient phosphorylation at Ser65. A second C-terminal site, S112, directly affects binding of 4E-BP1 to eIF4E without influencing phosphorylation of other sites. Thirdly, a conserved C-terminal motif influences phosphorylation of multiple residues, including rapamycin-insensitive sites. These relatively long-range effects are surprising given the reportedly unstructured nature of 4E-BP1 and may imply that phosphorylation of 4E-BP1 and/or binding to eIF4E induces a more-ordered structure. 4E-BP2 and -3 lack phosphorylatable residues corresponding to both S101 and S112. However, in 4E-BP3, replacement of the alanine at the position corresponding to S112 by serine or glutamate did not confer the ability to be released from eIF4E in response to insulin.
Eukaryotic initiation factor 4E (eIF4E) binds to the 5′-cap structure of eukaryotic mRNAs and forms complexes containing other initiation factors, which facilitate the attachment of the 40S subunit to the mRNA and the identification of the AUG start codon. eIF4E interacts directly with the modular scaffold protein eIF4G, which then interacts with other proteins such as the helicase eIF4A, the multisubunit factor eIF3, and the poly(A)-binding protein PABP (22). eIF4E is thought to play a key role in the regulation of mRNA translation and cellular physiology. For example, artificial overexpression of eIF4E can lead to cell transformation or dysregulation of cell growth (7, 25; reviewed in reference 15), and eIF4E levels are markedly elevated in many human tumors (6). Overexpression of eIF4E enhances the translation of mRNAs with structured 5′-untranslated regions and also increases the transport of cyclin D1 mRNA into the cytoplasm (reviewed in reference 15).
The availability of eIF4E is regulated by small heat-stable inhibitor proteins, termed eIF4E-binding proteins (4E-BPs), that bind to a similar site on eIF4E as eIF4G does, and thus block the formation of complexes between eIF4E and eIF4G (15). They thus act to repress cap-dependent translation. By far the best understood of the 4E-BPs is 4E-BP1, which has been the subject of extensive investigation. Human 4E-BP1 contains 118 amino acids, with the eIF4E-binding motif located between residues 54 and 60 (15). Association of 4E-BP1 with eIF4E is regulated by a range of stimuli: for example, insulin, which activates mRNA translation, induces the phosphorylation of 4E-BP1 and its release from eIF4E, allowing the protein to bind eIF4G to form initiation factor complexes (15). Increased phosphorylation of 4E-BP1 caused by any stimulus so far tested is blocked by rapamycin, indicating a role for the mammalian target of rapamycin (mTOR) signaling pathway in this effect. A second, well-characterized target for mTOR signaling is the 70-kDa protein kinase that phosphorylates ribosomal protein S6 (S6K1) (2). Phosphorylation of S6K1 (which activates it) is also blocked by rapamycin, but it is believed to lie on a different branch of the mTOR pathway from 4E-BP1 (43). Very recent data suggest that a binding partner for mTOR, termed raptor, may act as a scaffold protein to facilitate regulation of 4E-BP1 by mTOR (18, 24).
There is a substantial level of interest in the 4E-BPs, stimulated by a range of recent findings. For example, overexpression of 4E-BP1 or -2 reverses the transformed phenotype of cells overexpressing eIF4E, indicating that 4E-BP1 and -2 act as tumor suppressors (36). Chromosomal knockout of 4E-BP1 affects the development of adipose tissue in mice (41). Recent data show that, in common with S6K1, 4E-BP1 regulates animal growth and/or cell size (11, 28). Indeed, a number of lines of evidence indicate that, in animals, TOR signaling serves to link nutritional and hormonal signals to the regulation of cell size (16, 48). The observation that rapamycin reverses the growth of tumors linked to dysregulation of signaling through phosphatidylinositide (PI) 3-kinase also points to a key role for mTOR signaling in cell proliferation (1, 31). Furthermore, 4E-BP1 appears to be important in cell survival and apoptosis (26, 40).
Phosphorylation of 4E-BP1 is complex: early studies identified at least five sites of phosphorylation in 4E-BP1 which play differing roles in modulating the binding of 4E-BP1 to eIF4E or in permitting the phosphorylation of other sites in the protein (i.e., phosphorylation of 4E-BP1 is hierarchical) (14, 17, 29). Two N-terminal threonines (T37 and T46 in the human protein) are required for phosphorylation of T70, which in turn is required for phosphorylation of S65. Phosphorylation at T70 appears to be of major importance in bringing about the release of 4E-BP1 from eIF4E, while it has been suggested that phosphorylation of S65 prevents the reassociation of eIF4E and 4E-BP1 (23, 27): 4E-BP1 phosphorylated at this site is not found in association with eIF4E (29). S65 and T70 are relatively close in the primary sequence to the eIF4E-binding motif. In vivo, phosphorylation of S65 and T70 is sensitive to rapamycin (17, 29). A fifth site is S83, which appears to be of minor importance in the control of 4E-BP1 in vivo. Although mTOR phosphorylates some sites in 4E-BP1 in vitro, it is far from clear that it is a physiologically relevant kinase, and the kinases acting on 4E-BP1 in vivo remain to be identified. Bacterially expressed 4E-BP1 has little if any folded structure, consistent with its stability to heat and acids (12). Most attention has been focused on the roles in controlling 4E-BP1 of features (e.g., phosphorylation sites) near its N terminus or adjacent to the 4E-BP1 binding motif (14, 17, 23, 29, 30). Indeed, Tee and Proud recently identified a novel motif near its N terminus that is required for phosphorylation of 4E-BP1 in response to insulin (40). In contrast, little attention has been paid to the role of features in the C terminus in regulating its phosphorylation or function.
Here we identify a novel regulatory phosphorylation site in the C terminus of 4E-BP1 that is required for phosphorylation of S65 in vivo. We show that phosphorylation at a second C-terminal site even further from the eIF4E-binding motif is directly required for the release of 4E-BP1 from eIF4E, independently of any effect on the phosphorylation at other sites. We also demonstrate that a motif at the extreme C terminus of 4E-BP1 plays a key role in the phosphorylation of several sites and in facilitating release of 4E-BP1 from eIF4E in response to insulin. These findings provide new insights into the complex control of 4E-BP1 and provoke a reassessment of the hierarchical regulation of 4E-BP1.
MATERIALS AND METHODS
Methods.
Unless otherwise indicated, all reagents were from Sigma or Merck. Asp-N was from Roche. The antiserum for eIF4E was as described previously (44), and anti-eIF4G was kindly provided by Simon Morley (Sussex, United Kingdom). The phosphospecific antisera for 4E-BP1 were from Cell Signaling. Anti-Myc (9E10) was from Sigma.
Cells.
HEK293 cells were grown and transfected as described previously (46). In most experiments, 5 μg of DNA was used per 10-cm-diameter dish. For experiments in which association of 4E-BP1 with eIF4E was to be studied, we used less DNA (<1 μg/dish), since previous experience has shown that it is hard to observe release if very high levels of 4E-BP1 are expressed (presumably because a sufficient proportion of the protein remains in hypophosphorylated states that still bind eIF4E). Before insulin treatment, cells were starved of serum overnight. Cell extracts were either processed directly or (where stated) subjected to m7GTP-Sepharose chromatography or Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen). Cell lysates (400 μg of protein) were applied to m7GTP-Sepharose (packed volume, 20 μl) and mixed for 1 h at 4°C. After the beads were washed twice in lysis buffer (1 ml), the bound proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. For isolation on NTA-agarose, 200 μg of protein was applied to 10 μl (packed volume) of beads. Blots were visualized by enhanced chemiluminescence (ECL).
Gel electrophoresis and immunoblotting.
Gel electrophoresis and immunoblotting were performed as described previously (35). In most cases gels containing 12.5% acrylamide were used, but to display the mobility shifts of differently phosphorylated forms of 4E-BP1, gels containing 13.5% acrylamide and 0.36% methylene bis-acrylamide were used (33).
Production of antisera.
Antisera designed to recognize human 4E-BP1 when phosphorylated at S112 were raised against the synthetic peptide AGGEESpQFEMDC (Sp denotes phosphoserine). The C-terminal cysteine is present to facilitate coupling of the peptide to the carrier and to the resin for the antibody purification procedure. For immunization, peptides were conjugated to keyhole limpet hemocyanin. All immunization procedures were performed at Diagnostics Scotland (Edinburgh). Phosphospecific antisera were purified over columns comprising immobilized phosphopeptide conjugated to CH-activated Sepharose. To ensure that the phosphospecific antiserum did not contain antibodies that reacted with the nonphosphorylated protein, such antibodies were removed by passing the preparation over a column of the nonphosphorylated peptide, and the unbound material was used for the immunoblot analyses.
Expression vectors.
Human 4E-BP1 and its mutants were expressed as His-Myc fusion proteins from vectors similar to those described previously for the rat protein (40), except that the tags are present at the N terminus of the 4E-BP1 polypeptide. Point mutations were introduced using the QuikChange system (Stratagene).
In vitro or in vivo labeling and peptide mapping.
In vitro or in vivo labeling and peptide mapping were performed exactly as described previously (46) except that His- and Myc-tagged 4E-BP1 was purified by chromatography on NTA-agarose while endogenous 4E-BP1 was immunoprecipitated from extracts of HEK293 cells that had been metabolically labeled using [32P]orthophosphate. In vitro labeling of bacterially expressed 4E-BP1 proteins was performed using DYRK2 prepared as described previously (3). In some experiments, 4E-BP1 was phosphorylated by activated recombinant Erk or CK2, which were obtained from the Division of Signal Transduction Therapy, University of Dundee, or Promega (Southampton, United Kingdom), respectively.
RESULTS
S101 is a novel phosphorylation site in 4E-BP1.
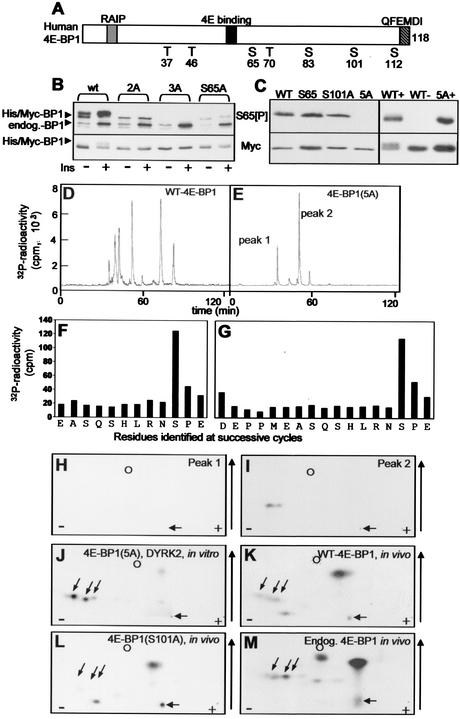
HEK293 cells were transfected with a vector encoding human 4E-BP1 (with hexahistidine and Myc tags) (shown schematically in Fig. 1A) and subsequently treated with insulin. A pronounced signal was observed in Western blots by use of an antibody designed to recognize 4E-BP1 when 4E-BP1 is phosphorylated at S65 (Fig. 1B). As has frequently been described previously, insulin induces a shift in the mobility of 4E-BP1 toward slower-migrating, more highly phosphorylated species. These slower-migrating species likely correspond to forms phosphorylated at T70 and, especially, S65 (33, 45). We were surprised when using this antibody to observe a signal from samples derived from cells transfected with vectors encoding mutants of 4E-BP1 in which S65 is converted to alanine (S65A or S65A T70A [2A] mutants) (Fig. 1B). The strength of this signal was not affected by insulin. This suggested that 4E-BP1 contains a further site of phosphorylation that is recognized by the anti-S65[P] antibody. Inspection of the amino acid sequence of human 4E-BP1 reveals that S101 is in a sequence context similar to that of S65 (RNS65PV and RNS101PE). The weaker signal for the S65A versus the 2A mutant likely reflects the slightly lower level of expression of this mutant. To determine whether the signal observed for the S65A or 2A mutants with the anti-S65[P] antibody was due to S101, this residue was also mutated to alanine (to give the 3A mutant). This resulted in loss of the signal seen with this phosphospecific antibody (Fig. 1B), strongly suggesting that the S65[P]-antibody also recognizes 4E-BP1 phosphorylated at S101 and showing that there are no other sites in 4E-BP1 that are recognized by this antibody (as expected from inspection of the sequence). The ability of antibodies that had been raised to recognize the phosphorylation site at S65 to also recognize that at S101 may vary between sources or batches and may therefore not have been observed in other studies (e.g., reference 17). It is formally possible that the anti-S65[P] antibody did not actually recognize S65[P] but only Ser101. To check this, we made use of the fact that Erk phosphorylates S65 very efficiently (19, 23) but acts much less efficiently at other sites. Recombinant wild-type 4E-BP1, and mutants in which T36, T37, T70, and S83, or these four residues plus S65, are mutated to alanine [4E-BP1(S65) and 4E-BP1(5A), respectively] or in which S101 was mutated to alanine (S101A) were incubated in vitro with Erk. Samples were then analyzed by SDS-PAGE and Western blotting using the S65[P] antibody. As can be seen from Fig. 1C, strong signals were observed for the wild type and the S65 and S101A proteins after treatment with Erk. No signal at all was seen for the 5A variant: since this contains S101, it seems that Erk does not phosphorylate this site, consistent with earlier data (23). Since the wild-type and S65 proteins contain both S65 and S101, in these cases the antibody could be picking up either in their phosphorylated forms. However, a strong signal was still observed for the S101A mutant, in which no reaction with S101[P] is possible (Fig. 1C). These data indicate that the so-called S65[P] antibody does indeed recognize 4E-BP1 phosphorylated at S65. It thus recognizes both this site and phosphoserine 101.
FIG. 1.
Identification of a novel in vivo phosphorylation site in 4E-BP1. (A) Schematic illustration of wild-type human 4E-BP1, showing the sites of phosphorylation, the eIF4E-binding site, the RAIP and QFEMDI motifs, and the tags present in the fusion proteins employed in these studies. (B) HEK293 cells were transfected with vectors encoding His-Myc-tagged-versions of wild-type human 4E-BP1 or mutants in which either S65 alone (S65A), S65 and T70 (2A), or S65, T70, and S101 (3A) were mutated to A. Where indicated, cells were stimulated with insulin (100 nM; 30 min). Extracts were analyzed by SDS-PAGE and Western blotting with the anti-S65[P] antibody (top gel) or anti-Myc (bottom gel). Positions of recombinant (His/Myc) and endogenous 4E-BP1 polypeptides are shown. (C) (Left) Recombinant wild-type His-Myc-tagged 4E-BP1 or mutants in which either T37, T46, T70, and S83 are mutated to A (S65), S101 is mutated to A (S101A), or T37, T46, T70, S65, and S83 are mutated to alanine (5A) were incubated with activated Erk and unlabeled ATP. (Right) Recombinant wild-type His-Myc-tagged 4E-BP1 or the 5A mutant was incubated with DYRK2 and unlabeled ATP (+) or with ATP alone (−). Samples were then analyzed by SDS-PAGE and Western blotting using the anti-S65[P] antibody or anti-Myc, as a loading control. (D and E) Wild-type 4E-BP1 or the 5A mutant (in which T37, T46, T70, S65, and S83 are mutated to A) was radiolabeled using DYRK2 in vitro. Following cleavage by Asp-N, peptides were resolved by reverse-phase HPLC (C18 column, with in-line detection of 32P radioactivity, as described previously [46]). The two main radioactive peaks observed for 4E-BP1(5A) areindicated. (F and G) Peak 1 (F) or 2 (G) from the HPLC run for which results are shown in panel E was subjected to solid-phase Edman degradation, and the radioactivity released at each cycle was monitored. The residues identified at each cycle are also shown. (H to J) Two-dimensional phosphopeptide maps of peaks 1 (H) and 2 (I) from the HPLC run for which results are shown in panel E or of 4E-BP1(5A) phosphorylated in vitro by DYRK2 and digested with Asp-N (J). (K to M) Two-dimensional phosphopeptide maps of His-Myc-tagged 4E-BP1 (wild type [K] or S101A [L]) or endogenous 4E-BP1 (M) phosphorylated in HEK293 cells were generated by Asp-N digestion of the indicated protein that had been radiolabeled in vivo. In panels H to M, the positions of the origin (horizontal arrow), marker (dinitrophenyl-lysine) (circle), and peptides corresponding to peptides containing S101 (diagonal arrows) are shown to facilitate comparisons between maps, as are the direction of chromatography (vertical arrow) and the polarity of electrophoresis (plus and minus signs).
It was important to establish by a second independent method that S101 is indeed phosphorylated in vivo. The best approach would be to metabolically label 4E-BP1 in vivo and perform protein chemical analyses to establish whether S101 is indeed labeled. To achieve this, it is crucial to be able to identify unambiguously the phosphopeptide(s) corresponding to Ser101. The sequence around S101 closely resembles the recently reported consensus for phosphorylation by the kinases DYRK2 and DYRK3 (3): there is an arginyl residue at −2 and a prolyl residue at +1. This is, however, also true of S65. Therefore, to study whether S101 was phosphorylated by these kinases in vitro, we used recombinant 4E-BP1, made in Escherichia coli, in which S65 (and indeed four other sites of phosphorylation, T36, T47, T70, and S83) was mutated to alanine. The resulting 4E-BP1(5A) protein, or wild-type 4E-BP1, was incubated in vitro with recombinant DYRK2 (also made in E. coli) in the presence of [γ-32P]ATP and MgCl2. The phosphorylated proteins were digested with Asp-N (1 μg/100 μl), and the resulting phosphopeptides were analyzed by reverse-phase high-pressure liquid chromatography (HPLC) (as described previously [46]). Five to six main labeled peaks were observed for wild-type 4E-BP1 (Fig. 1D), while the 5A mutant gave only two main peaks, indicated as peak 1 and peak 2 in Fig. 1E. The peaks that are present in Fig. 1D but absent from Fig. 1E likely correspond to residues that are absent from the 5A mutant, in particular S65, which, as already noted, lies in a good consensus for phosphorylation by DYRK2. Other data (not shown) confirm that DYRK2 also phosphorylates S65 in wild-type 4E-BP1. The material in the fractions corresponding to peaks 1 and 2 from the 5A mutant was subjected to analysis by mass spectrometry and solid-phase Edman degradation. Peak 1 contained a peptide with m/z 1435.0280: this matches the predicted mass of peptide EASQSHLRNSPE, containing a phosphate group (residues 92 to 103 of human 4E-BP1). Solid-phase Edman analysis confirmed the sequence and revealed that the radioactivity was released at cycle 10, corresponding to Ser101 (Fig. 1F). This peptide arises by miscleavage by Asp-N at the N-terminal side of E92, which can occur at low efficiency.
Peak 2 contained two peptides (m/z 2003.7540 and 2019.7625). These both correspond to the peptide DEPPMEASQSHLRNSPE containing a single phosphate group (residues 87 to 103 of human 4E-BP1). The peptide with the higher mass represents a species in which M91 has undergone oxidation to the sulfone (hence increasing the mass by 16). Solid-phase Edman analysis again confirmed the sequence and showed that radioactivity was released at cycle 15, once more corresponding to S101 (Fig. 1G). Peptide mapping showed that the material in peak 1 ran as a single radiolabeled peptide (Fig. 1H), while peak 2 resolved into two labeled peptides upon electrophoresis (Fig. 1I). These presumably represent the variants of peptide 87-103 containing either methionine or methionine sulfone, which are not resolved by reversed-phase HPLC (Fig. 1E). The relative amounts of these two species varied markedly between experiments, presumably reflecting different extents of oxidation of M91. The three peptides seen for peaks 1 and 2 correspond to those observed when an Asp-N digest of 4E-BP1(5A) phosphorylated by DYRK2 is analyzed by 2-dimensional mapping (Fig. 1J).
To demonstrate directly that S101 is phosphorylated in vivo, HEK293 cells were transfected with vectors encoding wild-type 4E-BP1(His/Myc) or the S101A(His/Myc) mutant. Forty-eight hours later, cells were metabolically labeled with [32P]orthophosphate and then lysed, and the tagged 4E-BP1 was purified on Ni-agarose. The isolated protein was digested with Asp-N, and 2-dimensional peptide maps were prepared. Several radiolabeled peptides were observed in the map from wild-type 4E-BP1 (Fig. 1K); two (highly positively charged) peptides were absent from the map derived from the S101A mutant (Fig. 1L). Their positions on 2-dimensional maps correspond exactly to those of the two phosphopeptides derived from 4E-BP1(5A) labeled by DYRK2, which contain phosphoserine 101 (Fig. 1H to J). This suggested that S101 was indeed a site of phosphorylation in vivo. To verify that labeling of S101 was not an artifact of overexpression of 4E-BP1 in HEK293 cells, we also analyzed the phosphorylation of the endogenous 4E-BP1. After labeling of cells with [32P]orthophosphate and lysis, 4E-BP1 was immunoprecipitated with an anti-4E-BP1 antiserum and digested with Asp-N. Two-dimensional peptide mapping revealed radiolabeled peptides in precisely the positions of the peptides containing S101 (Fig. 1M). Thus, S101 is phosphorylated in endogenous 4E-BP1.
S101 is constitutively phosphorylated in vivo.
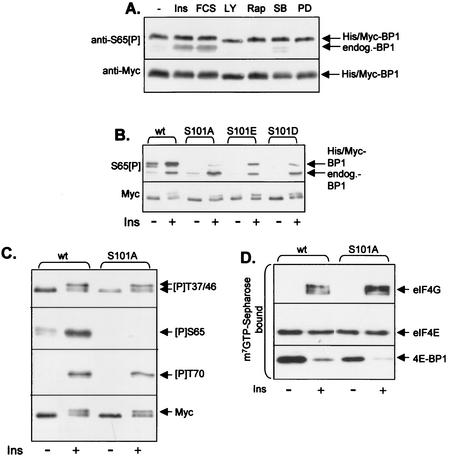
To assess whether phosphorylation of S101 was subject to regulation, we wished to exploit the observation that, as described above, the anti-S65[P] antibody cross-reacts with S101[P]. To confirm this cross-reactivity, we incubated wild-type 4E-BP1 or the 5A mutant with DYRK2 in vitro. The anti-S65[P] antibody recognized both the wild-type protein and the 5A mutant after treatment with DYRK2 but gave no signal if DYRK2 was omitted from the reaction (Fig. 1C). As described above (Fig. 1D to G), DYRK2 phosphorylates the 5A mutant only at S101, demonstrating that the anti-S65[P] antibody does indeed recognize 4E-BP1 phosphorylated at this site, in addition to S65[P] itself. To examine the possible regulation of the phosphorylation of S101 in vivo, we used 4E-BP1(S65A) to eliminate the signal from phosphorylation at S65. This mutant was expressed in HEK293 cells which were subsequently treated with insulin or fetal calf serum (as stimuli) or with inhibitors of PI 3-kinase (LY294002), mTOR (rapamycin), p38 mitogen-activated protein kinase (α/β isoforms) (SB202190), or MEK (PD184352). Samples of the resulting cell lysates were analyzed by SDS-PAGE and immunoblotting using anti-S65[P]. As shown in Fig. 2A, phosphorylation of S101 was not affected by any of these stimuli or signaling inhibitors. Similar data were obtained for another cell line of human origin, HeLa cells (data not shown). The existence of this additional basal phosphorylation site that is detected by the anti-S65[P] antibody means that caution must be exercised in interpreting data obtained by this antibody, since the signal observed reflects the sum of the signals due to phosphorylation at S65 and S101 (e.g., 4E-BP1 that is bound to eIF4E may appear to be phosphorylated at S65 due to the signal from S101).
FIG. 2.
S101 is constitutively phosphorylated in vivo, and this affects phosphorylation at S65. (A) HEK293 cells were transfected with a vector encoding 4E-BP1(S65A) and 48 h later were treated (for 30 min) with either insulin (100 nM) (Ins), fetal calf serum (10% [vol/vol]) (FCS), LY294002 (30 μM) (LY), rapamycin (100 nM) (Rap), SB202190 (10 μM) (SB), or PD184352 (10 μM) (PD). The signal for the lower band, which corresponds to the endogenous 4E-BP1, is mainly due to phosphorylation of this protein at S65 and is accordingly increased by insulin and serum treatment. (B) HEK293 cells were transfected with wt 4E-BP1 or mutants in which S101 was mutated to A or to the acidic residue D or E. Where indicated, cells were stimulated with insulin (100 nM; 30 min). Extracts were analyzed by SDS-PAGE and Western blotting with the anti-S65[P] antibody (upper panel) or anti-Myc (lower panel). (C) Conditions were as described for panel B, but only the wild-type (wt) and S101A vectors were used. Blots were developed with anti-Myc or the indicated phosphospecific antisera for 4E-BP1. The signals for the endogenous 4E-BP1 in these extracts are too faint to be seen at this exposure of the immunoblots. (D) Conditions were as described for panel B, but extracts were subjected to affinity chromatography on m7GTP-Sepharose and SDS-PAGE followed by immunoblotting with the indicated antisera.
As discussed above, the phosphorylation of 4E-BP1 involves a complex interplay between multiple sites of phosphorylation, which play distinct roles in modulating the phosphorylation of other sites in 4E-BP1 or its release from eIF4E (14, 17, 29, 30). To assess whether phosphorylation at S101 played a role in modulating the function of 4E-BP1, we first assessed whether mutation of S101 to alanine affected phosphorylation at other sites in this protein. Insulin brought about a marked increase in the signal observed with the S65[P] antibody (Fig. 2B). Since insulin does not affect phosphorylation at S101 (Fig. 2A), this reflects increased phosphorylation at S65. In contrast, very little increase in the signal was observed with the S101A mutant. Thus, S101 appears to be required for phosphorylation at S65, a site whose phosphorylation apparently prevents the binding of 4E-BP1 to eIF4E (21), since 4E-BP1 phosphorylated at S65 does not bind efficiently to eIF4E (23, 27, 29, 34). If phosphorylation, and thus a negative charge, at S101 is required for phosphorylation at S65, mutation of S101 to an acidic residue might restore the ability of insulin to bring about phosphorylation at S65. As shown in Fig. 2B, insulin did elicit some phosphorylation of S65 when S101 was mutated to Glu or (less efficiently) Asp. Thus, a negative charge at S101 is sufficient for phosphorylation of S65 in response to insulin in vivo.
Phosphorylation of S65 has previously been shown to require prior phosphorylation of 4E-BP1 at T37, T46, and T70 (14, 17, 29). To assess whether the effect of loss of S101 on phosphorylation of S65 was due to an effect on phosphorylation at these sites, we made use of appropriate phosphospecific antisera. The signals seen with these antisera were similar for the wild type and S101A mutants, in the absence or presence of insulin (Fig. 2C), indicating that S101 probably affects phosphorylation at S65 directly rather than through an effect on any of the sites previously shown to influence phosphorylation at S65 (14, 17, 29, 30).
Although phosphorylation of S65 appears to act to prevent (re)association of 4E-BP1 with eIF4E, it does not appear to be required for the release of 4E-BP1 from eIF4E (17, 30). Consistent with this, both the wild-type and S101A mutant 4E-BP1 proteins were released from eIF4E in response to insulin, allowing insulin to promote assembly of eIF4F complexes, observed as increased binding of eIF4G to eIF4E (Fig. 2D).
S112 is directly required for release of 4E-BP1 from eIF4E.
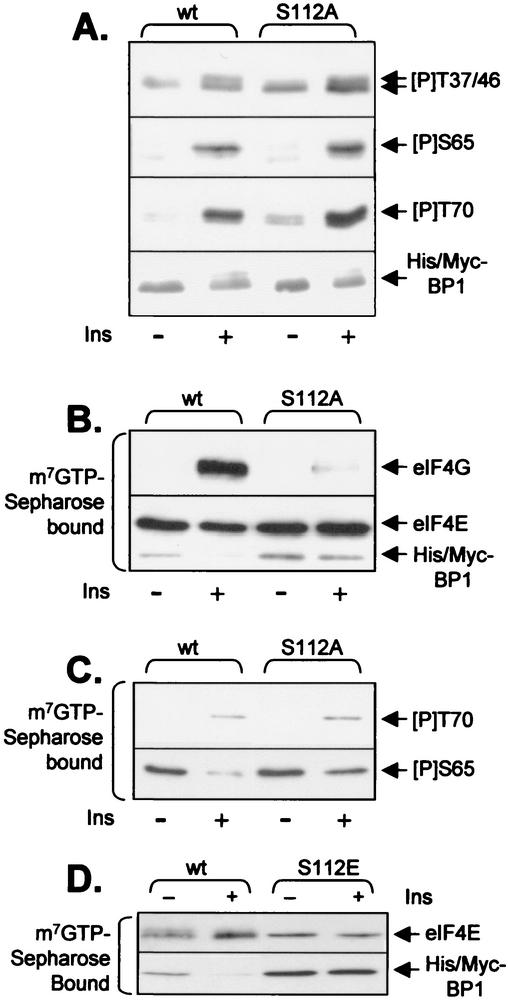
The data described above show that S101, a phosphorylation site at the C terminus of 4E-BP1, unexpectedly plays an important role in modulating the phosphorylation of a site (S65) in its central region near the eIF4E-binding site. Other reports have suggested that S112 in human 4E-BP1 is required for insulin-induced release of 4E-BP1 from eIF4E (47) and that the corresponding residue in rat 4E-BP1 (S111) is phosphorylated in response to insulin in fat cells (20). Taken together with the data reported here for S101, it was possible that S112 in human 4E-BP1 also acted to prime phosphorylation at other sites in 4E-BP1. To assess this, we again used the phosphospecific antisera described above. Mutation of S112 to alanine had no detectable effect on basal or insulin-stimulated phosphorylation at T37, T46, S65, or T70 (Fig. 3A). However, it is clear, as reported earlier (47), that this mutation does impair the ability of insulin to elicit release of 4E-BP1 from eIF4E: whereas wild-type 4E-BP1 was released almost completely in response to insulin, the S112A mutant remained bound (Fig. 3B). Consistent with this, insulin failed to promote binding of eIF4G to eIF4E in cells expressing 4E-BP1(S112A) (Fig. 3B). Thus, S112 influences the release of 4E-BP1 by a direct effect on binding to eIF4E rather than via effects on the phosphorylation of other sites, especially T70, the site thought to play the major role in release.
FIG. 3.
S112 directly modulates release of 4E-BP1 from eIF4E. HEK293 cells were transfected with a vector encoding His-Myc-tagged wild-type (wt) 4E-BP1 or the S112A or S112E mutant. (A and B) Samples were processed exactly as for Fig. 2C and D. (C) Samples were processed as for Fig. 2D but were subjected to immunoblotting using the anti-T70[P] or anti-S65[P] antisera as indicated. (D) HEK293 cells were transfected with a vector encoding His-Myc-tagged wild-type 4E-BP1 or the S112E mutant. Forty-eight hours later, after overnight serum starvation, some plates were treated with insulin, as indicated. After lysis, equal amounts of cell protein were subjected to affinity chromatography on m7GTP-Sepharose, and the bound material was analyzed by SDS-PAGE and Western blotting using anti-eIF4E or anti-Myc (as indicated).
m7GTP pulldowns followed by Western blotting with the phosphospecific antisera revealed that T70 is phosphorylated in the 4E-BP1(S112A) protein bound to eIF4E in insulin-treated cells (Fig. 3C). This confirms that the S112A mutation does not prevent phosphorylation of T70 and, most significantly, demonstrates that phosphorylation of T70 does not suffice to bring about release of 4E-BP1 from eIF4E, since phosphorylation at S112 is also needed for this. In contrast, similar analysis for the anti-S65[P] antibody revealed no increase in signal following insulin treatment for the S112A 4E-BP1 protein bound to eIF4E, despite the fact that insulin increases phosphorylation of S65 in the bulk of the 4E-BP1(S112A, Fig. 3A). One interpretation of this finding is that S65 is phosphorylated only following release of 4E-BP1 from eIF4E, perhaps because it is occluded in the eIF4E-4E-BP1 complex. Indeed, Erk can phosphorylate S65 in free 4E-BP1 but not when it is complexed with eIF4E (9).
To assess whether replacement of S112 by an acidic residue would mimic the effects of phosphorylation of this residue, we prepared a mutant in which S112 was replaced by Glu (this was chosen in preference to Asp, as the latter was almost ineffective at position 101 [Fig. 2A]). This mutant was expressed in HEK293 cells, and we studied whether it was released from eIF4E in response to insulin, like wild-type 4E-BP1. As shown in Fig. 3D, this S112E variant was not released from eIF4E in response to insulin treatment. This suggested that replacement of serine with glutamate at this position does not mimic phosphorylation. However, an alternative explanation is that this serine is not actually phosphorylated in HEK293 cells, but that it is important, for other reasons, that there is a serine at this position. The first point would be in line with a report indicating that the corresponding residue (S111) is not phosphorylated in 4E-BP1 in rat adipocytes (10).
S112 is constitutively phosphorylated in HEK293 cells and is not affected by rapamycin or wortmannin.
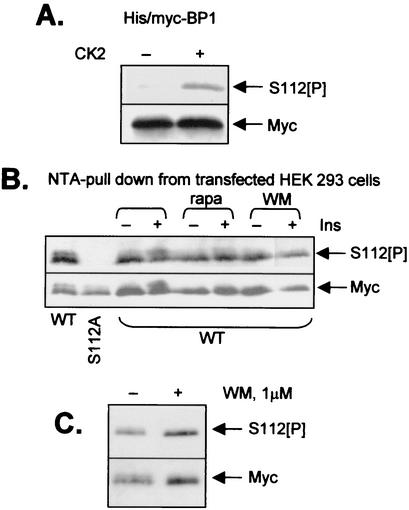
To study whether S112 is indeed phosphorylated in HEK293 cells, we raised a phosphospecific antibody using a synthetic peptide corresponding to the amino acid sequence around S112 in human 4E-BP1 (which included a phosphoserine at the position equivalent to S112 [see Materials and Methods]). This antibody was purified successively over affinity columns of immobilized phosphopeptide (to purify antibodies that react with the peptide containing S112[P]) and then over a dephosphopeptide column (i.e., one on which is immobilized a peptide of identical sequence but with serine instead of phosphoserine). Only the unbound material from the second column (which removes unwanted antibodies that react with the nonphosphorylated peptide) was used in the immunoblot experiments described here. To verify that the resulting antibody was indeed phosphospecific, we made use of the fact that CK2 can phosphorylate S112 in 4E-BP1 (20). As shown in Fig. 4A, the antibody failed to recognize recombinant human 4E-BP1 expressed in bacteria, while a clear signal was observed for 4E-BP1 after treatment with CK2 and ATP/MgCl2. This confirms the specificity of the antibody for 4E-BP1 phosphorylated at S112.
FIG. 4.
Ser112 is constitutively phosphorylated in a wortmannin-insensitive manner. (A) Recombinant 4E-BP1 (expressed in E. coli with His and Myc tags) was incubated with or without CK2 and ATP-MgCl2, as indicated. Samples were then analyzed by SDS-PAGE and Western blotting using either anti-Myc (loading control) or an antibody raised against a peptide corresponding to the sequence around Ser112 of 4E-BP1 and containing a phosphoseryl residue as the position equivalent to Ser112 (designated S112[P]). (B) HEK293 cells were transfected with vectors encoding Myc-His-tagged forms of wild-type 4E-BP1 or an S112A mutant (as indicated). Thirty-six hours later, cells were starved of serum overnight. Where shown, cells were pretreated with rapamycin (100 nM; 30 min) (rapa) or wortmannin (100 nM; 30 min) (WM) prior to addition of insulin (where indicated; 15 min; 100 nM). Samples were subjected to affinity chromatography on NTA-agarose (to isolate the His-tagged 4E-BP1), and the bound material was analyzed as for panel A. (C) Conditions were the same as for panel B, but cells expressing wild-type 4E-BP1 were treated with wortmannin at 1 μM for 45 min prior to lysis (where indicated) and analysis by SDS-PAGE and Western blotting using anti-S112[P] or anti-Myc (loading control), as indicated.
However, the titer of this antiserum was poor. Thus, to analyze the phosphorylation of S112 4E-BP1 in vivo, we overexpressed His-Myc-tagged 4E-BP1 in HEK293 cells and then purified the protein on NTA-agarose from a large amount (500 μg) of cell protein. Following immunoblotting, the anti-S112[P] antibody gave a weak but clear signal (Fig. 4B). As expected, no signal was observed for the S112A mutant. Pretreatment of cells with insulin did not alter the strength of the signal, indicating that S112 is constitutively phosphorylated, at least in HEK293 cells. Treatment of cells with rapamycin did not decrease the signal, indicating that basal phosphorylation at S112 is not maintained by the mTOR pathway. An inhibitor of PI 3-kinase (wortmannin [42]) also had no effect on phosphorylation at S112. This was true both at 100 nM, which entirely inhibits PI 3-kinase and has also been shown to completely inhibit the phosphorylation of p53 by ATM in vivo (37), and at 1 μM (Fig. 4C), a concentration known to completely inhibit ATM activity in vitro (4, 38). These data conflict with an earlier report for primary rat adipocytes which showed both that insulin increased phosphorylation at S112 and that this was sensitive to inhibition of PI 3-kinase (20). Our data also suggest that S112 is not a target for phosphorylation by ATM, contrary to the conclusions of an earlier study (47), at least in HEK293 cells. Thus, S112, like S101, is a constitutive phosphorylation site that plays a key role in the regulation of 4E-BP1, in this case because it is directly required for release of 4E-BP1 from eIF4E. Thus, taken with other data (27, 34), the release of 4E-BP1 from eIF4E seems to require phosphorylation of at least two sites (T70, an insulin-regulated site, and S112, a constitutive site) which together contribute to weakening its affinity for eIF4E. A glutamate at 112 clearly does not mimic phosphoserine at this position.
The C-terminal tail of 4E-BP1 plays a key role in the phosphorylation of multiple sites but is not essential for inputs from mTOR.
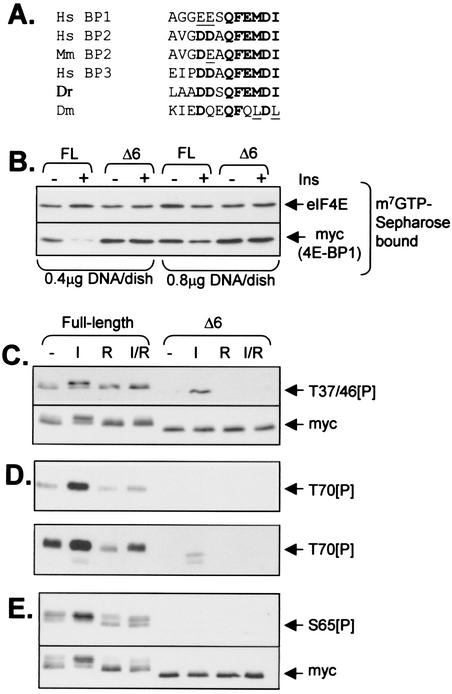
In view of the fact that Ser112 is far from the eIF4E-binding region in the unfolded structure of 4E-BP1, its effect on the interaction between eIF4E and 4E-BP1 is surprising. Immediately C-terminal to this residue is a sequence (QFEMDI) that is found in all known mammalian 4E-BPs, including human 4E-BP2 and 4E-BP3 (Fig. 5A). It is found in all the available mammalian 4E-BP sequences and in that of the only other vertebrate homolog known, from zebra fish (Danio rerio). A related sequence is found in the Drosophila homolog (Fig. 5A). It was conceivable that this common feature played a role in the binding of 4E-BP1 to eIF4E and that phosphorylation at S112 might interfere with this interaction. To test whether this motif was required for binding to eIF4E in vivo, we created a Δ6 mutant in which it was deleted. Analysis of m7GTP-Sepharose pulldown material from cells expressing the Δ6 mutant revealed that this mutant can bind to eIF4E (Fig. 5B). Indeed, when one takes into account the similar levels of expression of the wild-type and Δ6 mutant proteins (see Myc blot, Fig. 5C, lower panel), it is clear that a rather larger amount of the 4E-BP1 bound to eIF4E is the Δ6 mutant compared to the wild type. Thus, the QFEMDI motif is not required for binding, and in fact its removal may actually favor binding in vivo. While our studies were in progress, Schalm and Blenis (39) reported data which they interpreted as indicating that the FEMDI motif in 4E-BP1 is required for signaling from mTOR to 4E-BP1. However, their work did not address why this motif was important—e.g., whether it affected the phosphorylation of 4E-BP1, especially at sites known to be modulated via mTOR, or its biological function (sequestering eIF4E). We therefore considered it important to assess the effect of the Δ6 mutation on the level of phosphorylation of specific sites in 4E-BP1.
FIG. 5.
The C-terminal tail of 4E-BP1 is dispensable for binding to eIF4E but plays a key role in the phosphorylation of the protein at multiple sites in vivo. (A) Sequence alignments of the extreme C termini of 4E-BPs from Homo sapiens (Hs), Mus musculus (Mm), Danio rerio (Dr) (zebra fish), and Drosophila melanogaster (Dm). Corresponding sequences in rat and mouse 4E-BP1 are identical to the human sequence shown. Residues conserved in all, or almost all, sequences are boldfaced. Conservative replacements are underlined. (B to E) Full-length 4E-BP1 and the Δ6 mutant were expressed in HEK293 cells as His-Myc-tagged polypeptides. Cells were starved of serum for 16 h prior to further analysis. In some cases (I), cells were treated with insulin (100 nM; 60 min). Where indicated (R), cells were pretreated with rapamycin (100 μM; 30 min) prior to addition of insulin and/or lysis. (B) Equal amounts of cell lysate protein were subjected to affinity chromatography on m7GTP-Sepharose, and the bound material was analyzed by SDS-PAGE and Western blotting using antibodies for eIF4E or Myc as indicated; cells were transfected with 0.4 or 0.8 μg of DNA (as indicated). These data exemplify the importance of low-level transfection in order to be able to observe release of wild-type 4E-BP1 in response to insulin (it is clear that little release is seen at the higher transfection/expression levels). (C to E) Amounts of cell extract containing equal amounts of recombinant wild-type and Δ6 mutant protein (see Myc blot, panel C, lower section) were analyzed directly (C and D) by SDS-PAGE and Western blotting using anti-T37/46[P] (C) or anti-T70[P] (D) phosphospecific antisera or were subjected to affinity chromatography on NTA-agarose (E) prior to analysis by SDS-PAGE and Western blotting using the anti-S65[P] phosphospecific antibody. The lower part of panel D shows a longer exposure of the same immunoblot. The signal for the endogenous 4E-BP1 is now also visible as faint bands below those due to the overexpressed His-Myc-tagged 4E-BP1. The lower part of panel E shows the results of reprobing the blot shown in the upper part with anti-Myc.
As shown in Fig. 5C, in full-length 4E-BP1, T37/46 are already substantially phosphorylated in serum-starved cells, and only a small increase, if any, was seen after insulin treatment. Rapamycin had no effect on the basal phosphorylation of these sites. The Δ6 truncation drastically reduced the phosphorylation of T37/46, indicating an important role for the QFEMDI motif in their phosphorylation. Surprisingly, insulin increased the phosphorylation of T37/46 in the Δ6 mutant, and this effect was blocked by rapamycin. For wild-type 4E-BP1, basal phosphorylation at T70 was low but increased markedly in response to insulin, an effect that was blocked by rapamycin (Fig. 5D). In the Δ6 variant, phosphorylation at T70 was greatly reduced from that in the wild-type protein. Insulin did induce a small, rapamycin-sensitive increase in the signal for T70[P], but this was much weaker than that for the full-length protein and was visible only after extended exposure of the blot (Fig. 5D, lower panel). In the case of blotting performed using the S65 phosphospecific antibody, the signal was far weaker than that for the wild-type protein (Fig. 5E). After long exposure times, a weak signal was observed, which did not change in response to insulin or rapamycin (data not shown). Given that this antibody reacts with phosphorylated forms of both S65 and S101, this weak signal likely represents phosphorylation at S101, indicating that the QFEMDI motif is not required for phosphorylation of this site. Deletion of the QFEMDI motif therefore decreases the phosphorylation of at least four sites in 4E-BP1, two of which are sensitive to rapamycin (S65 and T70) and two of which are not (T37 and T46).
Conversion of A84 to Ser or Glu does not suffice to allow insulin to release 4E-BP3 from eIF4E.
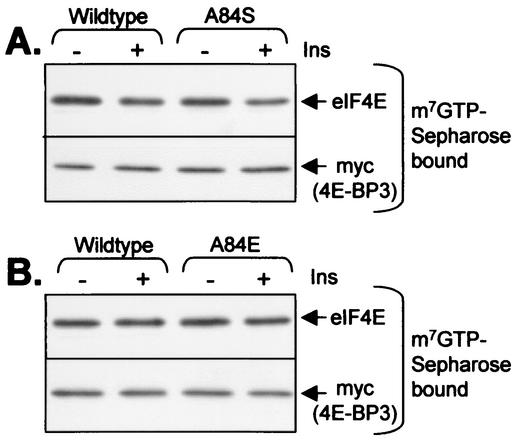
Tee and Proud have previously shown that insulin does not bring about the release of 4E-BP3 from eIF4E (40). Given the importance of S112 for the release of 4E-BP1 from eIF4E, the fact that 4E-BP3 possesses an alanyl residue at this position (A84, Fig. 5A) offered a potential explanation for this. To study this further, we generated mutants in which A84 was replaced by serine or glutamate. The sequence around A84 in 4E-BP3 is similar to that around S112 in 4E-BP1 (Fig. 5A), so one might expect that the kinase that acts on this site in 4E-BP1 might also phosphorylate S84 in this 4E-BP3 variant. Both variants were expressed in HEK293 cells, and the ability of insulin to induce their release was analyzed by affinity chromatography on m7GTP-Sepharose. As shown in Fig. 6, neither the A84S nor the A84E mutant was released in response to insulin, i.e., they both behaved like the wild-type protein. It therefore seems that a phosphorylatable residue at this position is not sufficient to allow insulin to bring about release of 4E-BP3 from eIF4E.
FIG. 6.
Effect of mutating A84 on the behavior of 4E-BP3. Mutants were created in which A84 in 4E-BP3 was replaced by serine (A84S, panel A) or glutamate (A84E, panel B). These variants were expressed in HEK293 cells in parallel with the wild-type protein. Cells were starved of serum overnight and in some cases were treated for 60 min with 100 nM insulin. Extracts were subjected to affinity chromatography on m7GTP-Sepharose followed by analysis of the bound material by SDS-PAGE and immunoblotting with the indicated antibodies.
DISCUSSION
The present data reveal differing roles for three important regulatory features located in the hitherto poorly studied C terminus of 4E-BP1. Firstly, we identify a novel in vivo phosphorylation site at S101 and show that this site plays a key role in the insulin-induced phosphorylation of S65, a site near the eIF4E-binding motif of 4E-BP1 that affects the affinity of 4E-BP1 for eIF4E. Secondly, we demonstrate that S112 is required for insulin-induced release of 4E-BP1 from eIF4E and that this is due to a direct effect on eIF4E-4E-BP1 binding rather than to an influence of this site on phosphorylation of other residues in 4E-BP1. Since S112 is constitutively phosphorylated, and a mutant with alanine at this position is not released efficiently in response to insulin, we infer that phosphorylation at this site is required for release, although other explanations are formally possible. Thirdly, we show for the first time that the extreme C terminus, which contains the TOR signaling (TOS) motif reported by Schalm and Blenis (39), is critical for phosphorylation of several sites in 4E-BP1. However, this input is not strictly linked to inputs from mTOR, as discussed in more detail below. These findings point to key roles for the C-terminal region of 4E-BP1 in its regulation. Given that recombinant 4E-BP1 has little, if any, folded structure, it is surprising that sites far from the 4E-binding motif have effects on its association with eIF4E or the phosphorylation of other sites distant in its primary sequence, and this issue is discussed further below.
It is thus now clear that phosphorylation of S65 requires at least two priming events—phosphorylation at S101 as well as at T70. Certain other protein kinases such as glycogen synthase kinase 3 and casein kinase 1 also require priming phosphorylation events for their action, but in these cases priming phosphorylation occurs only a few residues C- or N-terminal (respectively) from their sites of action. In the former case, structural studies have revealed that this priming mechanism involves a phosphate-binding site that interacts with the priming phosphoserine in the +4 position relative to the target residue (5, 13). The kinase acting at S65 is clearly dependent on a phosphorylation event much further away than this (at S101). Although S101 in human 4E-BP1 is phosphorylated by DYRK isoforms in vitro, these kinases may not be responsible for phosphorylating this site in vivo. Indeed, although the 4E-BP1 proteins from the rat and mouse each have a serine at this position, these residues do not lie in a consensus for phosphorylation by DYRK, suggesting that other kinases act on these sites. It is important to note that the sequence around S101 and its equivalents in rodent 4E-BP1 is generally highly conserved: it is identical at 16 out of 20 residues around this serine. In fact the following 10 residues are identical in rat, mouse, and human 4E-BP1.
Phosphorylation at S65 also requires the C-terminal QFEMDI motif, but this probably reflects the fact that this feature is needed for phosphorylation at the corresponding priming site, T70. The fact that two widely separated phosphorylation sites (T70 and S101) are required for phosphorylation at S65 could imply either that the kinase acting at this site has a docking site that recognizes both phosphoseryl residues individually or that phosphorylation of the otherwise unfolded 4E-BP1 polypeptide induces structure which is recognized by the S65 kinase. Extensive work will be required to investigate these possibilities.
Our data show that phosphorylation at S112 influences the binding of 4E-BP1 to eIF4E without affecting its phosphorylation at other sites, i.e., directly, despite the fact that S112 also lies quite far from the eIF4E-binding site. This may again imply the existence either of long-range interactions within the 4E-BP1 molecule or the fact that when phosphorylated, or when associated with eIF4E, 4E-BP1 may adopt a more-structured conformation in which, e.g., S112 is adjacent to eIF4E. These new data indicating a direct role for phosphorylation of S112 in modulating the binding of 4E-BP1 are interesting in the light of recent findings that phosphorylation of S65 alone or at S65 and T70 did not suffice to effect release of 4E-BP1-based peptides from eIF4E (29, 32). These authors interpreted this effect as indicating a role for phosphorylation at T37 and T46 in release, and the present data indicate that phosphorylation at S112 also plays an important role in modulating eIF4E-4E-BP1 binding, thus helping to explain their data. Thus, release of 4E-BP1 from eIF4E appears to require its phosphorylation at S65 and T70, which is enhanced by insulin, and at S112, which is constitutively phosphorylated in HEK293 cells.
An important related point is that the sequences of 4E-BP2 and -3 do not have a serine at the position corresponding to S112, even though in other respects their C termini are very similar to that of 4E-BP1—in both proteins, alanine is present at this position (Fig. 5A). Furthermore, neither protein contains a residue equivalent to S101. These differences raise important questions about the regulation of these 4E-BPs. Interestingly, neither protein has so far been shown to dissociate from eIF4E in response to stimuli such as insulin. Recent data clearly show that insulin fails to induce release of 4E-BP3 from eIF4E under conditions where 4E-BP1 does dissociate (40). For almost all the phosphorylation sites in 4E-BP2 and -3, phosphospecific antisera are not available, and we have not explored the phosphorylation of these proteins in any detail. Converting alanine 84 in 4E-BP3 to serine or glutamate did not allow 4E-BP3 to be released from eIF4E in response to insulin. It therefore seems that a phosphorylatable residue at this position is insufficient to allow insulin to bring about release and that additional inputs are likely required. Another major difference between 4E-BP1 and 4E-BP3 is that the latter contains only one residue equivalent to T37 and T46. This may impair the phosphorylation of other sites in 4E-BP3 (e.g., those corresponding to S65 and T70 in 4E-BP1) and could also in part underlie its failure to be released in response to insulin.
To study the phosphorylation of S112 in 4E-BP1, we developed an appropriate phosphospecific antiserum. By use of this reagent, it was clear that S112 is constitutively phosphorylated in HEK293 cells. This contrasts with an earlier report for rat adipocytes (10) which indicated that this site was not phosphorylated. That study involved 32P labeling of the protein in vivo, and it may be that the rate of turnover of phosphate on this constitutively phosphorylated residue is so low that no significant labeling occurred. In our studies, we found that phosphorylation of S112 was not increased by insulin, in contrast to a second report for rat adipocytes, where an increase was observed (20). Again, this study employed in vivo labeling with [32P]orthophosphate, and it may be that insulin increases the turnover of phosphate on S112 without a change in its net level of phosphorylation. Contrary to the suggestion made by Heesom et al. (20), S112 does not act to “prime” phosphorylation of other sites in 4E-BP1. A further study suggested that S112 was a target for phosphorylation by ATM (47). This link is surprising given that DNA-damaging agents cause dephosphorylation of 4E-BP1 rather than inducing an increase (40) (although it should be noted that the conditions used by these workers do not activate ATM). The present study shows that the level of phosphorylation of S112 is not affected by wortmannin at concentrations reported to inhibit phosphorylation of p53 by ATM in vivo (37). Indeed, phosphorylation of p53 by ATM was almost completely inhibited by wortmannin concentrations 10 times lower than that used here. Nevertheless, some reports have indicated that (in vitro) ATM is inhibited only by higher concentrations of wortmannin (4, 38). However, even at 1 μM (which does completely inhibit ATM in vitro), wortmannin did not reduce the level of signal seen with the anti-S112[P] antibody. These data suggest that ATM is not responsible for the basal phosphorylation of S112 in HEK293 cells, implying the existence of another S112 kinase(s). ATM is presumably able to phosphorylate S112 in vitro because the following residue is Gln, creating a consensus site for phosphorylation by ATM. Our data also show that phosphorylation of S112 is also not influenced by the mTOR pathway, since phosphorylation of S112 was not affected by rapamycin.
Our analysis of the effect of deletion of the C-terminal QFEMDI motif confirms the initial observation of Schalm and Blenis (39) that a single point mutation within this motif eliminates the ability of insulin to induce the mobility shift in 4E-BP1 that normally accompanies its phosphorylation. These authors interpreted their data as indicating that this region was required for signaling from mTOR to 4E-BP1, and they called the FEMDI sequence a TOR signaling (TOS) motif. However, they did not establish whether, and how, it modulated the regulation of 4E-BP1. We demonstrate here that absence of the QFEMDI motif drastically affects the phosphorylation of 4E-BP1 at multiple sites (T37, T46, T70, and S65) and therefore its regulation. However, our data indicate that this is not specifically a motif required for mTOR signaling. Firstly, deletion of the motif affects the phosphorylation of two basal phosphorylation sites that are not sensitive to rapamycin in vivo, i.e., T37 and T46. Secondly, the increase in phosphorylation of T37, T46, and T70 in response to insulin observed in this Δ6 mutant is blocked by rapamycin, illustrating that mTOR can still regulate 4E-BP1 in the absence of this motif. Phosphorylation at T37 and T46 is actually more prone to inhibition by rapamycin in the Δ6 mutant (where it is eliminated by this drug) than in full-length 4E-BP1. It is possible, indeed likely, that S112 does not undergo phosphorylation in the Δ6 mutant (but we cannot easily check this because the Δ6 protein is not recognized by our anti-S112[P] antibody, due to the loss of the part of the epitope that it binds). However, lack of phosphorylation of S112 cannot be the cause of the effects of the Δ6 truncation on the phosphorylation of other sites, as they are still phosphorylated in the S112A mutant. Our data are consistent with the idea that the QFEMDI motif is needed for phosphorylation of basal sites in 4E-BP1, which are required for insulin-induced phosphorylation at other sites, rather than specifically for rapamycin-sensitive inputs from mTOR. In view of our data, further work to explore the precise role of the TOS motif in the S6 kinases (39) will be required.
It is now clear that the hierarchical phosphorylation of 4E-BP1 is even more complex than previously thought, with constitutive phosphorylation sites in the N (T37 and T46) and C (S101/112) termini playing crucial roles in its regulation. An increasing number of proteins are now known to be modulated by complex multisite phosphorylation. Two of many possible examples are p53 (37) and the SCFCdc4 ubiquitin ligase (8), both of which are, like 4E-BP1, key regulators of cell function. The existence of multiple phosphorylation sites in these proteins presumably facilitates their control by multiple inputs. Our data also identify a C-terminal motif required for phosphorylation of many sites in 4E-BP1. This is reminiscent of the N-terminal “RAIP” sequence which was recently shown to be required for phosphorylation of T37, T46, T70, and S65 in 4E-BP1 (40). The protein raptor (18, 24), which binds to mTOR, was recently identified as a potential scaffold that interacts with 4E-BP1and facilitates its phosphorylation. This raises the possibility that raptor may do so by binding the RAIP and/or QFEMDI motif. However, the findings that these motifs are required for phosphorylation of sites in 4E-BP1 that are not sensitive to rapamycin in vivo (40; this report) appear inconsistent with this idea.
The data presented here show that two phosphorylation sites near the C terminus of 4E-BP1 influence the functions or phosphorylation of the central region of 4E-BP1, which is surprising given that recombinant 4E-BP1 has no folded structure (12). It is possible that phosphorylation of 4E-BP1 induces a more-ordered structure: for example, as discussed above, phosphorylation at S101 and T70 may induce a structure that creates a docking site for the kinase that acts at S65, a residue whose phosphorylation in vivo requires prior phosphorylation at these two sites. The evidence that phosphorylation of S112 is required for release of 4E-BP1 from eIF4E even when T70 is phosphorylated suggests that this part of 4E-BP1 may contact eIF4E in the binary eIF4E-4E-BP1 complex, even though the interaction does not depend upon the C-terminal QFEMDI motif.
Acknowledgments
We are grateful to Gert Scheper and Nick Morrice (Dundee) for help with HPLC analysis and peptide chemistry, respectively. We thank Manjur Karim and John Hughes (UMIST) for recombinant 4E-BP1 and cDNAs, respectively.
This work was supported by grants (to C.G.P.) from the Biotechnology and Biological Sciences Research Council, The Wellcome Trust, and the Medical Research Council (G9901450).
REFERENCES
- 1.Aoki, M., E. Blazek, and P. K. Vogt. 2001. A role for the kinase mTOR in cellular transformation induced by the oncoproteins P3K and Akt. Proc. Natl. Acad. Sci. USA 98:136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avruch, J., C. Belham, Q. Weng, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26:115-154. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, L. E., and C. G. Proud. 2002. Differing substrate specificities of members of the DYRK family of arginine-directed protein kinases. FEBS Lett. 510:31-36. [DOI] [PubMed] [Google Scholar]
- 4.Chan, D. W., S. C. Son, W. Block, R. Ye, K. K. Khanna, M. S. Wold, P. Douglas, A. A. Goodarzi, J. Pelley, Y. Taya, M. F. Lavin, and S. P. Lees-Miller. 2000. Purification and characterization of ATM from human placenta. A manganese-dependent, wortmannin-sensitive serine/threonine protein kinase. J. Biol. Chem. 275:7803-7810. [DOI] [PubMed] [Google Scholar]
- 5.Dajani, R., E. Fraser, S. M. Roe, N. Young, V. Good, T. C. Dale, and L. H. Pearl. 2001. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105:721-732. [DOI] [PubMed] [Google Scholar]
- 6.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumours: its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 31:59-72. [DOI] [PubMed] [Google Scholar]
- 7.De Benedetti, A., and R. E. Rhoads. 1990. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc. Natl. Acad. Sci. USA 87:8212-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshaies, R. J., and J. E. Ferrell. 2001. Multisite phosphorylation and the countdown to S phase. Cell 107:819-822. [DOI] [PubMed] [Google Scholar]
- 9.Diggle, T. A., S. K. Moule, M. B. Avison, A. Flynn, E. J. Foulstone, C. G. Proud, and R. M. Denton. 1996. Both rapamycin-sensitive and -insensitive pathways are involved in the phosphorylation of the initiation factor 4E binding protein (4E-BP1) in response to insulin in rat epididymal fat cells. Biochem. J. 316:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fadden, P., T. A. Haystead, and J. C. Lawrence. 1998. Phosphorylation of the translational regulator, PHAS-I, by protein kinase CK2. FEBS Lett. 435:105-109. [DOI] [PubMed] [Google Scholar]
- 11.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher, C. M., A. M. McGuire, A.-C. Gingras, H. Li, H. Matsuo, N. Sonenberg, and G. Wagner. 1998. 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry 37:9-15. [DOI] [PubMed] [Google Scholar]
- 13.Frame, S., P. Cohen, and R. M. Biondi. 2001. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7:1321-1327. [DOI] [PubMed] [Google Scholar]
- 14.Gingras, A.-C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 translation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 16.Gingras, A.-C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 17.Gingras, A.-C., B. Raught, S. P. Gygi, A. Niedzwieka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieczyska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin, mTOR, mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 19.Haystead, T. A. J., C. M. M. Haystead, C. Hu, T. A. Lin, and J. C. Lawrence. 1994. Phosphorylation of PHAS-I by MAP kinase. Identification of a site phosphorylated by MAP kinase in vitro and in response to insulin in rat adipocytes. J. Biol. Chem. 269:23185-23191. [PubMed] [Google Scholar]
- 20.Heesom, K. J., M. B. Avison, T. A. Diggle, and R. M. Denton. 1998. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor 4E-binding protein 1 on the rapamycin-insensitive site (serine-111). Biochem. J. 336:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heesom, K. J., and R. M. Denton. 1999. Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E-BP1 by an mTOR-associated kinase. FEBS Lett. 457:489-493. [DOI] [PubMed] [Google Scholar]
- 22.Hentze, M. W. 1997. eIF4G: a multipurpose ribosome adapter? Science 275:500-501. [DOI] [PubMed] [Google Scholar]
- 23.Karim, M. M., J. M. X. Hughes, J. Warricker, G. C. Scheper, C. G. Proud, and J. E. G. McCarthy. 2001. A quantitative molecular mechanism for modulation of mammalian translation by the eIF4E-binding protein 1. J. Biol. Chem. 276:20750-20757. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 25.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 26.Li, S., N. Sonenberg, A.-C. Gingras, M. Peterson, S. Avdulov, V. A. Polunovsky, and P. B. Bitterman. 2002. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol. Cell. Biol. 22:2852-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, T.-A., X. Kong, T. A. J. Haystead, A. Pause, G. J. Belsham, N. Sonenberg, and J. C. Lawrence. 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653-656. [DOI] [PubMed] [Google Scholar]
- 28.Miron, M., J. Verdu, P. E. Lachance, M. J. Birnbaum, P. F. Lasko, and N. Sonenberg. 2002. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3:596-601. [DOI] [PubMed] [Google Scholar]
- 29.Mothe-Satney, I., G. J. Brunn, L. P. McMahon, C. T. Capaldo, R. T. Abraham, and J. C. Lawrence. 2000. Mammalian target of rapamycin-dependent phosphorylation of PHAS-1 in four (S/T)P sites detected by phospho-specific antibodies. J. Biol. Chem. 275:33836-33843. [DOI] [PubMed] [Google Scholar]
- 30.Mothe-Satney, I., D. Yang, P. Fadden, T. A. J. Haystead, and J. C. Lawrence. 2000. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol. Cell. Biol. 20:3558-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumours to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niedwiecka, A., J. Marcotrigiano, J. Stepinski, M. Jankowska-Anyszka, A. Wyslouch-Cieczyska, M. Dadlez, A.-C. Gingras, P. Mak, E. Darzynkiewicz, N. Sonenberg, S. K. Burley, and R. Stolarski. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319:615-635. [DOI] [PubMed] [Google Scholar]
- 33.Patel, J., X. Wang, and C. G. Proud. 2001. Glucose exerts a permissive effect on the regulation of the initiation factor 4E binding protein 4E-BP1. Biochem. J. 358:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pause, A., G. J. Belsham, A.-C. Gingras, O. Donzé, T. A. Lin, J. C. Lawrence, and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762-767. [DOI] [PubMed] [Google Scholar]
- 35.Price, N. T., S. F. Nakielny, S. J. Clark, and C. G. Proud. 1989. The two forms of the beta-subunit of initiation factor-2 from reticulocyte lysates arise from proteolytic degradation. Biochim. Biophys. Acta 1008:177-182. [DOI] [PubMed] [Google Scholar]
- 36.Rousseau, D., A.-C. Gingras, A. Pause, and N. Sonenberg. 1996. The eIF4E-binding protein-1 and protein-2 are negative regulators of cell growth. Oncogene 13:2415-2420. [PubMed] [Google Scholar]
- 37.Saito, S., A. A. Goodarzi, Y. Higashimoto, Y. Noda, S. P. Lees-Miller, E. Appella, and C. W. Anderson. 2002. ATM mediates phosphorylation at multiple p53 sites, including Ser46, in response to ionizing radiation. J. Biol. Chem. 277:12491-12494. [DOI] [PubMed] [Google Scholar]
- 38.Sarkaria, J. N., R. S. Tibbett, E. C. Busby, A. P. Kennedy, D. Hill, and R. T. Abraham. 1998. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 58:4375-4382. [PubMed] [Google Scholar]
- 39.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 40.Tee, A. R., and C. G. Proud. 2002. Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22:1674-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukiyama-Kohara, K., F. Poulin, M. Kohara, C. T. DeMaria, A. Cheng, Z. Wu, A. C. Gingras, A. Katsume, M. Elchebly, B. M. Spiegelman, M. E. Harper, M. L. Tremblay, and N. Sonenberg. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7:1128-1132. [DOI] [PubMed] [Google Scholar]
- 42.Vlahos, C. J., W. F. Matter, K. Y. Hui, and R. F. Brown. 1994. A specific inhibitor of phosphatidyl inositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 269:5241-5248. [PubMed] [Google Scholar]
- 43.Von Manteuffel, S. R., P. B. Dennis, N. Pullen, A.-C. Gingras, N. Sonenberg, and G. Thomas. 1997. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point upstream of p70S6k. Mol. Cell. Biol. 17:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, X., A. Flynn, A. J. Waskiewicz, B. L. J. Webb, R. G. Vries, I. A. Baines, J. Cooper, and C. G. Proud. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 273:9373-9377. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X., F. E. M. Paulin, L. E. Campbell, E. Gomez, K. O'Brien, N. Morrice, and C. G. Proud. 2001. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon subunit and their roles in vivo. EMBO J. 20:4349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, D.-Q. and, M. B. Kastan. 2000. Participation of ATM in insulin signalling through phosphorylation of eIF-4E-binding protein 1. Nat. Cell Biol. 2:893-898. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhart, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]