Abstract
Although phosphoinositide 3-kinase (PI 3-kinase) is essential for cell cycle progression, the molecular mechanisms that regulate its diverse biological effects are poorly understood. We demonstrate here that Rb, a key regulator of cell cycle progression, associates with p55 kDa (p55α and p55γ) regulatory subunits of PI 3-kinase in vivo and in vitro. Both confocal microscopy and biochemical analysis demonstrated the presence of p55γ in the nucleus. The 24-amino-acid N-terminal end of p55γ, which is unique among PI 3-kinase regulatory subunits, was sufficient to bind Rb. Addition of serum or growth factors to quiescent cells triggered the dissociation of Rb from p55. Ectopic expression of the 24-amino-acid N-terminal end of p55γ inhibited cell cycle progression, as evidenced by induction of cell growth arrest at the G0/G1 phase, inhibition of DNA synthesis, inhibition of cyclin D and cyclin E promoter activity, and changes in the expression of cell cycle-related proteins. The inhibitory effects of the N-terminal end of p55γ on cell cycle progression depended on the presence of functional Rb. These data demonstrate for the first time an association of p55γ with Rb and show that modification of this association can lead to cell cycle arrest.
Phosphatidylinositol 3-kinase (PI 3-kinase) is an essential component of growth factor-regulated pathways (21). After binding their cognate receptors, growth factors activate the class IA PI 3-kinases, which are composed of a regulatory subunit and a catalytic p110 subunit (19). This enzyme phosphorylates phosphatidylinositol bisphosphate [PI(4,5)P2], generating phosphatidylinositol (3,4,5) triphosphate (PIP3) and its derivative PI(3,4)P2 (21). Recent reports suggest that stimulation of the lipid kinase activity of PI 3-kinase leads to activation of c-AKT, PDK1, Rac, the protein kinase C-ɛ isoform, and p70S6 kinase (8). In addition, PI 3-kinase also possesses serine/threonine kinase activity (8). Signals mediated by the protein kinase activity of PI 3-kinase are different from the signals mediated by its lipid kinase activity (3, 7).
Several isoforms of both the p110 (α,β, δ) enzyme and its regulatory subunits (p85α and β, p55α and γ, and p50α and γ) have been characterized and cloned (21). These isoforms are key regulators of different PI 3-kinase-dependent responses. Although regulatory subunits have no enzymatic activity, they are needed to recruit p110 to specific cellular locations, thus regulating its catalytic activity. Regulatory subunits include p85α, p55α, and p50α, products of the p85α gene, and p85β and p55γ, encoded by the p85β and p55γ genes, respectively(8). p85α, the prototype regulatory subunit, contains two SH2 domains; an iSH2 domain, located between the two SH2 domains responsible for binding to p110; two proline-rich motifs; an SH3 domain; and a breakpoint cluster (bcr) gene homology region. While much is known about the roles of the SH2 domains of the regulatory subunits of PI 3-kinase in signaling, little is known about the roles of the other domains. In p55γ, a unique 24-amino-acid N-terminal domain replaces the SH3 and bcr domains. The specific function of this N-terminal domain of p55γ has not been found (14). Similarly, there is a 52% identity among the first 34 amino acids of p55α and p55γ (11).
The retinoblastoma tumor suppressor protein, Rb, and its family members p107 and p130, have been shown to be major modulators of the G1/S transition in mammalian cells (12, 23). Many of the functions of Rb depend on its association with other proteins at crucial phases in the cell cycle. When hypophosphorylated in the G0 and G1 phases of the cell cycle, Rb blocks cell cycle progression by binding to and suppressing the activities of transcription factors such as E2F family members (10). Phosphorylation of Rb, begun during late- to mid-G1 and continuing through S and G2/M, leads to inactivation of Rb and release of bound E2F proteins. E2F then activates genes involved in cell cycle progression (12). Several cell cycle-dependent serine/threonine protein kinases (CDKs) and cyclins have been found to phosphorylate Rb (18). In addition to the roles of Rb in cell cycle progression, the functions of Rb in cellular differentiation have begun to be explored (17, 26).
It has been reported that PI 3-kinase regulates the phosphorylation of Rb and that this process is crucial in PI 3-kinase-mediated signaling (5, 6, 20). Brennan et al. recently demonstrated that PI 3-kinase has a critical role in coupling the interleukin-2 (IL-2) receptor to E2F regulation through the induction of hyperphosphorylation of Rb via AKT-mediated pathways (5, 6). However, the molecular mechanisms regulating PI 3-kinase and Rb interactions remain to be defined. Here we report that Rb associated with PI 3-kinase through its 55-kDa regulatory subunits in cultured cells. Moreover, this association was regulated by the addition of growth factors. Expression of the unique N-terminal end of p55γ, which we found to be an Rb binding domain, strongly inhibited cell cycle progression in cells with wild-type Rb, but not in cells lacking functional Rb.
MATERIALS AND METHODS
Cell culture and transfections.
Cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2 in air. The cell lines, obtained from the American Type Culture Collection (Manassas, Va.), were routinely cultured either in Dulbecco's modified Eagle medium (DMEM)-F-12 medium supplemented with 5% fetal bovine serum (FBS) (COS7 cells) or 10% FBS (MCF-7, NIH 3T3, MDA-MB 468, and SAOS-2 cells) or in RPMI 1640 medium supplemented with 15% FBS (AU565 cells). Rb−/− mouse embryonic fibroblasts (MEFs) and MEFs from control littermates (Rb+/+) (gifts of T. Jacks) were cultured in DMEM supplemented with 10% heat-inactivated FBS (9). Cells were transfected using the Fugene-6 reagent (Boehringer Mannheim, Indianapolis, Ind.).
Cell cycle analysis and cell sorting.
For cell cycle analysis, the DNA and green fluorescent protein (GFP) contents of cells were determined by flow cytometry (FACScan; Becton-Dickinson) and the data were analyzed using a ModFit software package. GFP-positive cells were sorted on a FACSVantage (Becton-Dickinson). More than 75% of the sorted cells were GFP positive. Sorted cells were lysed in sodium dodecyl sulfate (SDS) sample buffer containing 10 mM dithiothreitol before analysis by Western blotting.
Immunoprecipitation, Western blot analysis, and immunostaining.
Total cell extracts, immunoprecipitation, and Western blot analysis were performed as described previously (24). Cell fractionations were carried out by the method of Ramsby and Makowski (15). Preparation and characterization of the antibody against His-p55γ have been described elsewhere (24). The purified antibody was directly conjugated to horseradish peroxidase (HRP) by using a kit from Pierce. The following antibodies were purchased from commercial sources: anti-Rb for immunoprecipitation (polyclonal, C-15; monoclonal, IF8; Santa Cruz); anti-Rb for immunoblotting (monoclonal G3-245; PharMingen); anti-bromodeoxyuridine (anti-BrdU; Oncogene); anti-FLAG (M2; Sigma); anti-hemagglutinin (anti-HA; Santa Cruz); anti-GFP (monoclonal [Promega] and polyclonal [Invitrogen]); anti-p27Kip1; and anti-cyclin D, anti-cyclin E, anti-p110α, anti-p110β, and anti-mitogen-activated protein kinase (anti-MAPK; Santa Cruz). Cells were cultured on coverslips incubated with BrdU (10 μg/ml) overnight 48 h after transfection. The coverslips were washed twice with phosphate-buffered saline and fixed in 4% paraformaldehyde for 15 min at room temperature. Double immunostaining with anti-GFP or anti-Rb and anti-BrdU or anti-FLAG antibodies was performed as described previously (25). Images were acquired by confocal laser scanning microscopy (LSM-410; Carl-Zeiss). The number of BrdU-positive GFP+ (transfected) cells was determined by counting 200 GFP+ cells transfected with the empty vector control or 200 GFP+ cells tranfected with the GFP fusion protein constructs. After immunostaining against BrdU and counterstaining of cell nuclei with propidium iodide, DNA synthesis was determined by counting the percentage of BrdU+ cells among transfected cells.
Preparation of plasmid constructs.
Construction of the cDNA encoding full-length p55γ has been described previously (24). The cDNA encoding the first N-terminal 24 amino acids of p55γ fused to the N-terminal end of GFP was obtained by digesting the cDNA encoding full-length p55γ with EcoRI and BamHI (internal site) and ligating it into EcoRI and BamHI sites of pEGFP-N1 (Clontech); the cDNA encoding the 24-amino-acid N-terminal end of p55γ fused to the C-terminal end of GFP was amplified by PCR (forward primer, 5′-TTTTGAATTCTATGGACCGCGATGACGCAGA; reverse primer, 5′-TTTTGGATCCATTTCAATATAAAATATCAGT) and cloned into the EcoRI and BamHI sites of pEGFP-C1 (Clontech) vectors. For expression of FLAG tagged p55 (F-p55), the cDNA encoding full-length p55γ was amplified by PCR and subcloned into EcoRI and SalI sites of the CMV2 plasmid (Kodak). The cDNA construct encoding only p55γ has been described elsewhere (24). The cDNA encoding a constitutively activated p110α (p110α*) of PI 3-kinase (a gift of J. Downward [16]) and the cDNA encoding a full-length HA-tagged p55α (a gift of C. R. Kahn [1]) were subcloned into pcDNA3. The sequence of cDNA inserts was confirmed by sequencing.
In vitro binding assays, in vitro kinase assay, and luciferase reporter assays.
Full-length Rb fused to glutathione S-transferase (GST-Rb1-928) (27), a gift from S. Mittnacht, and a GST-Rb379-928 fusion protein (a gift of R. Livingston) were expressed and purified. The preparation of GST-Rb proteins and in vitro binding experiments have been described previously (25).
The cyclin E promoter-luciferase reporter construct, a gift of T. Kouzarides (4), contains the major promoter region (−543 to +263) of the cyclin E gene cloned upstream of a luciferase reporter gene. The cyclin D promoter (−1745 to +1) reporter construct was from R. Pestell (22). Transfections and luciferase assays were done as previously described (25). A construct encoding β-galactosidase (β-Gal) was cotransfected, and expression of β-Gal was used as an internal control. Data were summarized from three independent experiments (means ± standard deviations [SD]).
RESULTS
The 55-kDa regulatory subunits of PI 3-kinase interact directly with pRb.
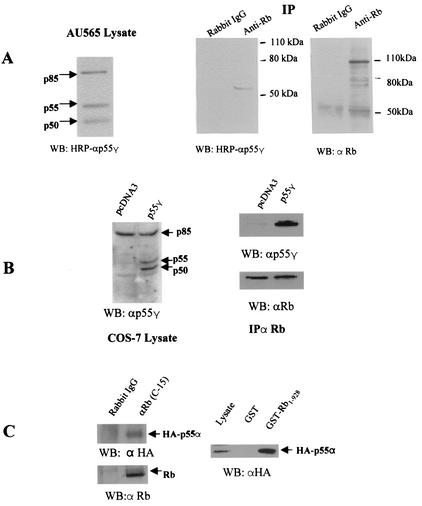
The anti-p55γ antibody used in Western blotting recognizes p85α, p55α, and p50α, due to the extensive homology among these regulatory subunits (24). We initially examined expression of these subunits in AU565 breast cancer cells by Western blotting. All three molecular weight forms were expressed in AU565 cells (Fig. 1A, left panel). In an effort to identify whether any regulatory subunit of PI 3-kinase physically interacts with Rb, we immunoprecipitated lysates of AU565 human breast cancer cells with an anti-Rb antibody. Proteins associated with Rb were analyzed by immunoblotting with a previously described HRP-conjugated anti-p55γ antibody (24). Only a 55-kDa immunoreactive band was detected in the Rb immunocomplex, but not in the immunocomplex precipitated by a nonspecific antibody (Fig. 1A, middle panel). Rb, as expected, was immunoprecipitated by the Rb antibody, but not by nonspecific immunoglobulin G (IgG) (Fig. 1A, right panel).
FIG. 1.
Rb associates with 55-kDa regulatory subunits of PI 3-kinase in vivo. (A) (Left) AU565 cell lysates were analyzed by Western blotting using an HRP-conjugated anti-p55γ antibody. Arrows indicate the position of the p85, p55, and p50 regulatory subunits of PI 3-kinase. (Center and right) AU565 cell lysates were immunoprecipitated with an anti-Rb antibody (C-15; Santa Cruz) or nonspecific rabbit IgG. Immunoprecipitated proteins were analyzed by Western blotting using an HRP-conjugated anti-p55γ antibody (center) or a monoclonal anti-Rb antibody (IF8; Santa Cruz) (right) as indicated. (B) (Left) COS7 cells were transfected with a construct encoding full-length p55γ or the pcDNA3 vector control. Due to the use of alternative translation initiation sites (24), both p55γ and p50γ are expressed. Cell lysates were analyzed by Western blotting using an HRP-conjugated anti-p55γ antibody. Arrows indicate the positions of p85, p55, and p50 subunits of PI 3-kinase. (Right) The cell lysates were also immunoprecipitated with an antibody to Rb, and immunoprecipitated proteins were analyzed by Western blotting using either HRP-conjugated anti-p55γ or anti-Rb antibodies as indicated. (C) (Left) A cDNA construct expressing a full-length C-terminal HA-tagged p55α was transfected into NIH 3T3 cells. Cell lysates were collected 48 h later and immunoprecipitated by an anti-Rb antibody or normal rabbit IgG. The immunocomplexes were analyzed using an anti-HA (upper panel) or anti-Rb antibody. (Right) COS7 cell lysates overexpressing p55α were incubated with purified GST or GST-Rb1-928. Bound proteins were analyzed by Western blotting using an anti-HA antibody.
Two 55-kDa subunit proteins of PI 3-kinase have been identified, p55α (11) and p55γ (14). As the antibody used could detect both proteins, the identity of the protein which bound Rb in the preceding experiments could not be determined. To first determine if p55γ could associate with Rb, a cDNA encoding full-length p55γ was introduced into COS7 cells, which, unlike AU565 cells, express very low levels of endogenous p55 (Fig. 1B, left panel). Cell lysates were immunoprecipitated using an anti-Rb antibody, and the interaction of Rb with p55γ was detected by Western blot analysis. The results showed that exogenous p55γ associated with Rb in vivo (Fig. 1B, right panel). Due to the alternative initiation of translation observed using this construct, p50γ is expressed as efficiently as p55γ (Fig. 1B, left panel). However, p50γ did not associate with Rb in COS7 cells.
We demonstrated that a 55-kDa protein that is recognized by an antibody to p55 γ was detected in Rb immunoprecipitates. p55α, a product of the p85α gene, has a structure similar to that of p55γ with 16 to 34 identical N-terminal amino acids (11). As our antibodies might also detect p55α, we tested if an exogenously expressed HA-tagged- p55α (1) could also bind Rb. We similarly found p55α in Rb immunoprecipitates (Fig. 1C, left panel). GST-Rb also bound the HA-tagged 55α (Fig. 1C, right panel).
Delineation of interaction regions in Rb and p55γ.
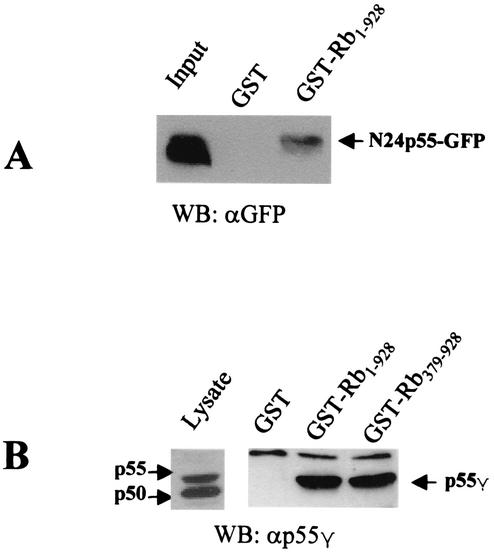
We then determined the region of p55γ that binds Rb. p55γ is identical to p50γ except for an additional 24 amino acids at the N-terminal end of p55γ (14). The fact that p50 did not associate with Rb suggested that the N-terminal 24-amino-acid sequence might encompass an Rb binding domain. Therefore, a fusion protein consisting of the 24-amino-acid N-terminal end of p55γ fused to GFP (N24p55-GFP) was overexpressed in COS7 cells. Cell lysates were incubated with GST-Rb fusion proteins. Results from Western blotting using an anti-GFP antibody showed that the N24p55-GFP fusion protein associated with GST-Rb but not with GST alone (Fig. 2A). GFP alone did not associate with Rb (data not shown). These data confirmed that the 24-amino-acid N-terminal end of p55γ was sufficient to bind Rb.
FIG. 2.
The N terminal 24 amino acids of p55 γ bind to Rb in vitro. (A) A cDNA construct encoding a chimeric protein composed of the N-terminal 24 amino acid residues of p55γ and GFP (N24p55-GFP) was transfected into COS7 cells. Cell lysates were incubated with purified GST-Rb1-928 or GST only. Blots were probed using an antibody to GFP. (B) (Left) COS7 cells were transfected with the cDNA construct expressing full-length p55γ. Cell lysates were analyzed by immunoblotting using an HRP-conjugated anti p55γ antibody. (Right) Cell lysates were also incubated with either purified GST-Rb1-928, a truncated GST-Rb379-928 fusion protein, or GST protein loaded onto glutathione-agarose beads. Associated proteins were analyzed by Western blotting using an anti-p55γ antibody.
Next, to begin to determine the domain of Rb that binds p55γ, COS7 cells were transfected with an expression construct encoding p55γ. The results shown in Fig. 2B demonstrate that two Rb fusion proteins (GST-Rb1-928 and GST-Rb379-928), but not GST alone, bound the 55γ subunit of PI 3-kinase. This result also suggested that the C-terminal end of Rb (amino acids 379 to 928), the so called big pocket domain of Rb, contained the binding domain for p55γ.
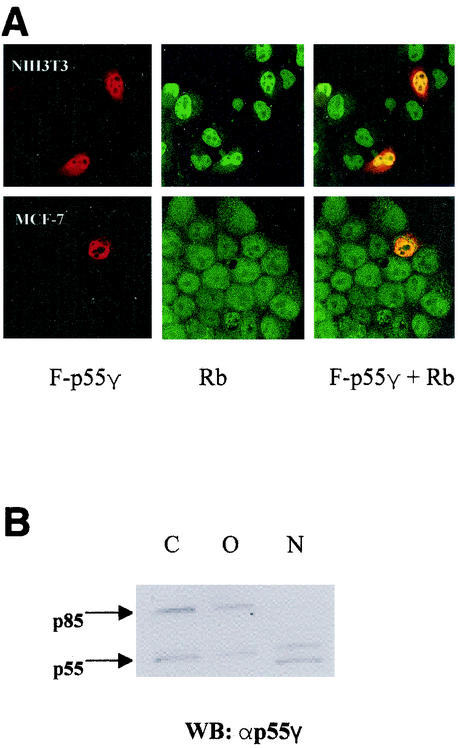
To further confirm the interaction of Rb with p55γ in living cells and to determine the subcellular localization of the interaction, we transfected cells (NIH 3T3 and MCF-7) with a construct expressing a FLAG-tagged full-length p55γ protein (F-p55). Immunostaining was conducted by using anti-Rb and anti-FLAG antibodies and fluorescein-labeled (αRb) and phycoerythrin-labeled (α FLAG) secondary antibodies. The data in Fig. 3A indicate a significant colocalization of Rb and p55γ in cultured cells. These data show that the p55γ regulatory subunit of PI 3-kinase associated with Rb in vivo and in vitro and that this interaction was localized to the nucleus.
FIG. 3.
Rb and p55 colocalize in the nucleus. (A) A cDNA construct encoding FLAG-tagged p55γ (F-p55) was transfected into NIH 3T3 (upper panels) and MCF-7 (lower panels) cells. The cells on coverslips were stained with an anti-FLAG (M-2) antibody and phycoerythrin-labeled rabbit anti-mouse IgG (red) and with an anti-Rb (C-15) antibody and fluorescein-labeled sheep anti-goat IgG (green). The images were taken on a laser-scanning confocal microscope as described in the text. Areas of colocalization appear yellow. (B) AU565 cells were fractionated into cytoplasmic (C), organelle (O), or nuclear (N) fractions as described in the text. Equal amounts of proteins were resolved by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting with antibodies to p55γ.
The localization of endogenous p55 proteins in log-phase AU565 cells was also determined by Western blot analysis of nuclear, cytoplasmic, and organelle fractions obtained by differential detergent extraction (15). The results of these experiments indicated that a subpopulation of p55 proteins was detected in the nuclei of AU565 cells. In addition, a band of around 50 kDa was observed. p85 was observed in the cytoplasm and organelles, but not in the nuclei, of AU565 cells.
Regulation of the interaction of Rb with PI 3-kinase in cultured cells by growth factors.
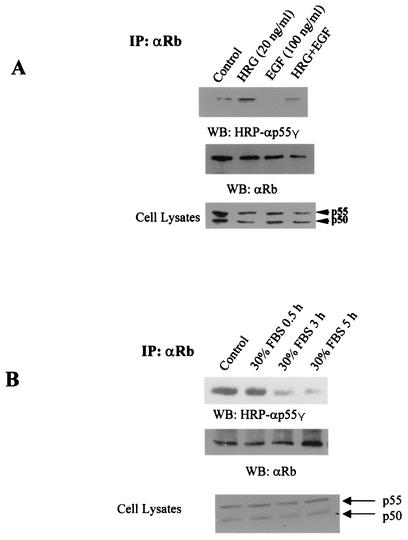
We next examined whether the interaction of Rb with p55γ could be regulated in cells under different growth conditions. In AU565 cells, heregulin induces differentiation and inhibits cell proliferation, while epidermal growth factor (EGF) stimulates cell proliferation (2, 13). AU565 cells were serum starved for 24 h (control). Cells were then treated for 20 h with the indicated concentrations of heregulin or EGF. Treatment of AU565 cells with heregulin at concentrations that inhibit cell growth increased the binding of p55γ to Rb (Fig. 4A). Conversely, treatment of cells with EGF at concentrations that stimulate growth inhibited the binding and also decreased the association induced by heregulin. Neither of the treatments changed the levels of p55γ and p50γ proteins (Fig. 4A, bottom panel).
FIG. 4.
Effect of growth factors on the interaction of Rb with p55γ. (A) AU565 cells were incubated in serum-free medium overnight and then treated with heregulin (HRG), EGF, or a combination for 20 h at the concentrations indicated. Cell extracts were immunoprecipitated with an anti-Rb antibody. Western blotting was performed using antibodies to p55γ or Rb as indicated. Aliquots of cell lysates were also analyzed using the HRP-conjugated anti-p55 antibody. (B) NIH 3T3 cells were cultured in serum-free DMEM-F-12 medium overnight (control). The medium was replaced by DMEM-F-12 medium containing 30% FBS, and cells were incubated for an additional 0.5, 3, or 5 h as indicated. Immunoprecipitation with an anti-Rb antibody and Western blotting with HRP-conjugated anti-p55 and anti-Rb were performed as described previously. Western blotting for p55γ of cell lysates was performed as described in the legend to panel A.
The regulation of the interaction of Rb and p55γ was also examined in NIH 3T3 cells. A strong association of Rb with p55 was noted in NIH 3T3 cells serum starved for 24 h (control). Addition of serum (30%) triggered the dissociation of p55γ from Rb as observed at 30 min. Binding was significantly decreased 3 h after serum stimulation (Fig. 4B). Western blot analysis determined that the level of p55γ was the same in all samples.
Expression of the N-terminal end of p55γ inhibits cell cycle progression in MCF-7 cells.
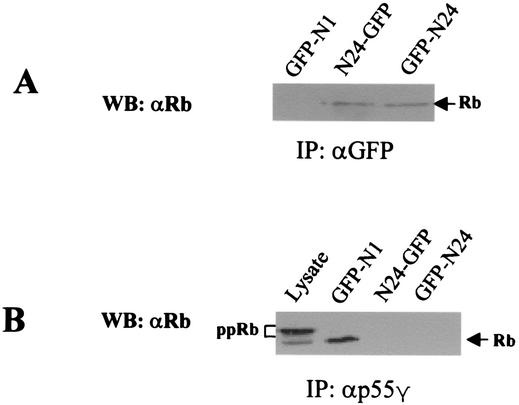
We next examined the effects of inhibiting the association of p55γ and Rb on cell cycle progression. It was reasoned that ectopic expression of the Rb-binding domain of p55γ could compete with endogenous p55γ for binding to Rb and block a p55-mediated signal transduction pathway that would lead to cell cycle progression. Therefore, we first examined whether the ectopically expressed N-terminal end of p55γ could bind Rb in cells and inhibit the binding of endogenous Rb to p55γ. Two GFP fusion proteins, one (N24-GFP) consisting of the 24-amino-acid N-terminal end of p55γ fused to the N-terminal end of GFP derived from pEGFP-N1 and the other (GFP-N24) consisting of the N-terminal end of p55γ fused to the C-terminal end of GFP derived from pEGFP-C1, were transiently expressed. GFP was also expressed as a control (GFP-N1). Expression of GFP and GFP fusion proteins was verified by Western blotting (data not shown). Cell lysates were then immunoprecipitated with an anti-GFP antibody. Data from Western blot analysis showed the association of N24-GFP and GFP-N24 with Rb in MCF-7 cells (Fig. 5A). We next immunoprecipitated these cell lysates with an antibody to p55γ to determine the effects of overexpression of the N-terminal 24 amino acids of p55 on binding of endogenous p55γ to Rb. Binding of endogenous p55γ with Rb was inhibited by expression of N24p55 but not by expression of the control GFP-N1 (Fig. 5B). Although hyper- and hypophosphorylated forms of Rb were observed in cell lysates by using the Pharminigen antibody, only the hypophosphorylated form of Rb bound p55γ (Fig. 5B), in accordance with previous data showing that the interaction of p55γ with Rb was mainly observed in quiescent cells when Rb was presumably hypophosphorylated.
FIG. 5.
The N-terminal end of p55γ associates with Rb and blocks the binding of p55γ with Rb in MCF-7 cells (A) Two vectors coding for different GFP fusion proteins were constructed. One, designated N24-GFP, consisted of the 24-amino-acid N-terminal end of p55 fused to the N terminus of GFP. The other, designated GFP-N24, consisted of the N-terminal end of p55 fused to the C terminus of GFP. These two constructs and the GFP-N1 plasmid expressing GFP as a control were transiently transfected into MCF-7 cells. Cell lysates were immunoprecipitated with an anti-GFP antibody. The blots were probed with an anti-Rb antibody. (B) Lysates of these transfected cells were immunoprecipitated with an anti-p55γ antibody, and the immunocomplexes were separated on SDS-polyacrylamide gels. Proteins were analyzed by Western blotting using an anti-Rb antibody (G3-245; PharMingen) capable of recognizing hyper- and hypophosphorylated Rb. Lysates from GFP-N1-transfected cells were also used to determine the position of hypophosphorylated or hyperphosphorylated (ppRb) Rb by Western blot analysis.
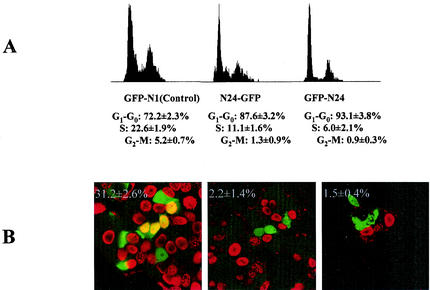
We predicted that an N24-GFP fusion protein might have a dominant-negative function in cell cycle progression by blocking Rb-p55γ interactions. We therefore examined the effects of ectopic expression of the Rb binding N-terminal end of p55γ on cell cycle progression by using MCF-7 cells, which have wild-type Rb. We were unable to establish cell lines stably expressing the N-terminal end of p55γ (N24-GFP and GFP-N24), consistent with its postulated inhibitory effects on the cell cycle. Therefore, transient transfection assays were used to examine the effects of N24-GFP and GFP-N24 on the cell cycle. Forty-eight hours after transfection, GFP-positive cells were selected by flow cytometry and cell cycle distribution was analyzed. There was an increase in the number of cells in the G0/G1 phase of the cell cycle in cells transfected with the N24-GFP fusion constructs, with a concomitant decrease in cells in S and G2/M compared to cells expressing GFP alone (Fig. 6A).
FIG. 6.
Expression of the N-terminal end of p55 blocks cell cycle progression in MCF-7 cells. (A) Plasmids expressing GFP alone (GFP-N1), N24-GFP, or GFP-N24 were transfected into MCF-7 cells. Cell cycle analysis of 10,000 GFP+ cells was performed 48 h after transfection. Data were averaged from three independent experiments (means ± SD), and histograms from one experiment using MCF-7 cells are shown. (B) Inhibition of DNA synthesis by expression of N24-GFP and GFP-N24 in MCF-7 cells. MCF-7 cells were transfected as described above. BrdU (10 μg/ml) was added 48 h after transfection, and cells were cultured for an additional 20 h. Cells were then immunostained with anti-GFP (green) and anti-BrdU (red) antibodies and the appropriate secondary antibodies. The images were taken on a laser-scanning confocal microscope. Two hundred GFP+ cells were counted on each of three slides for each experimental group. Means and SD were derived by pooling data from the three slides for each group. The number in each panel is the percentage of BrdU-positive transfected GFP+ cells (yellow staining) (mean ± SD) in each group.
To determine the numbers of cells in the proliferating population, 48 h posttransfection cells were incubated with BrdU overnight and the number of GFP+ cells incorporating BrdU was determined by immunofluorescence. Expression of the N24-GFP fusion protein decreased the number of cells synthesizing DNA by more than 90%, consistent with results indicating that N24p55 induced cell cycle arrest (Fig. 6B).
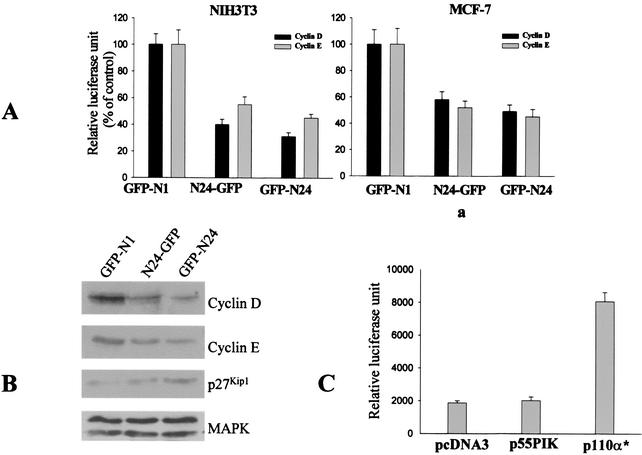
Cyclin D and cyclin E are two important CDK regulators that are involved in the phosphorylation of Rb during the cell cycle (18). Cyclins D and E are induced during mid- to late-G1, and this induction is required for cell cycle progression. As ectopic expression of the N-terminal 24-amino-acid domain of p55 inhibited cell cycle progression, we examined the activity of the cyclin D and E promoters in NIH 3T3 and MCF-7 cells transiently transfected with the N24-GFP fusion constructs. The activities of both the cyclin E and D promoters were significantly inhibited in cells transfected with either N24-GFP or GFP-N24 compared to those in cells transfected with GFP alone (Fig. 7A).
FIG. 7.
Effects of N24-GFP and GFP-N24 on the activities of the cyclin D and cyclin E promoters and expression of cell cycle-related proteins. (A) Cells were transfected with the GFP control expression vector (GFP-N1) or expression vectors for GFP-N24 fusion proteins with the N-terminal 24 amino acids of p55 cloned either N-terminal (N24-GFP) or C-terminal (GFP-N24) to GFP, cyclin D or E promoter luciferase reporter constructs, and β-Gal plasmids. Luciferase activity was assayed 48 h later as described in Materials and Methods. Aliquots of cell extracts were collected for β-Gal activity, and relative luciferase units were determined (means ± SD). (B) Expression of cell cycle-related proteins was examined in equal numbers of sorted cells expressing either GFP-N1, N24-GFP, or GFP-N24. Western blotting with an anti-MAPK antibody showed equal loading of proteins in each lane. (C) Effect of overexpression of p55PIK on the activity of the cyclin D promoter in MCF-7 cells. A cDNA construct encoding full-length p55PIKγ was cotransfected with a cyclin D promoter reporter construct and a β-Gal plasmid into MCF-7 cells. The cDNA plasmid encoding a constitutively activated p110α subunit of PI 3-kinase (p110α*) was used as a positive control. Forty-eight hours later, cell extracts were prepared, and luciferase and β-Gal activities were determined.
Next, the effects of N24p55 on the expression of proteins regulated during cell cycle progression were determined (Fig. 7B). NIH 3T3 cells were transfected with constructs expressing GFP or GFP fusion proteins as described above and were isolated by cell sorting 48 h later. Cell lysates were analyzed by Western blotting. Expression of cyclins D and E was significantly decreased in cells transfected with the fusion proteins, consistent with previous data indicating that the activities of the cyclin D and E promoters were inhibited by expression of N24p55. In addition, expression of p27Kip1, a CDK inhibitor (18), was increased in both N24-GFP transfectants.
To determine whether the effects of the N24-GFP constructs were due to the ability of these fusion proteins to mimic p55PIK functions, we examined the effects of overexpression of full-length p55γ on the activity of the cyclin D promoter. The ATG used for translation initiation of p50γ was replaced to eliminate the translation of p50 in cells (24). The cDNA construct was cotransfected into MCF-7 cells with the cyclin D promoter reporter construct and a β-Gal plasmid. No significant changes in cyclin D promoter activity were detected (Fig. 7C), in contrast to the effects of N24-GFP. On the other hand, expression of a constitutively activated p110α subunit of PI 3-kinase (p110α*) increased cyclin D promoter activity more than threefold, as previously reported (16).
The inhibitory effects of N24p55 on cell cycle progression depend on the presence of a functional Rb.
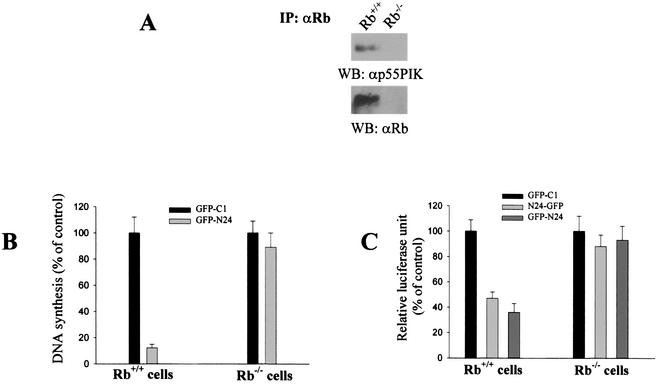
To determine if the effects of the expression of N24p55γ on cell cycle progression depended on the presence of a functional Rb, we used Rb-deficient MEFs (Rb−/− MEFs) (9). Rb−/− fibroblasts were transfected with the GFP-C1 control vector or GFP-N24. Rb was immunoprecipitated from Rb+/+ and Rb−/− fibroblasts and probed for p55γ. As expected, p55γ was found in Rb immunoprecipitates of Rb+/+ fibroblasts. Neither Rb nor p55γ was found in Rb immunoprecipitates of Rb−/− cells (Fig. 8A). DNA synthesis was determined in GFP+ cells by the BrdU incorporation assay. N24p55 did not inhibit DNA synthesis in Rb−/− cells (Fig. 8B). Similarly, expression of N24-GFP and GFP-N24 did not affect cyclin D promoter activity in Rb−/− cells (Fig. 8C). These results indicated that the effects of the N-terminal end of p55γ on the cell cycle depended on the presence of a functional Rb.
FIG. 8.
The inhibitory effects of N24p55 on cell cycle progression depend on the presence of a functional Rb. (A) Interaction of Rb with p55 in Rb+/+ MEFs. Extracts from Rb+/+ and Rb−/− MEFs were immunoprecipitated by an anti-Rb antibody. The presence of p55 and Rb in immunocomplexes was determined as described previously. (B) DNA synthesis is not affected by the expression of the N-terminal end of p55γ in Rb−/− cells. DNA synthesis in transfected Rb−/− and Rb+/+ MEFs was examined by BrdU staining as described for Fig. 6B. One hundred percent is equivalent to the number of BrdU-incorporating GFP+ cells transfected with the empty GFP-C1 plasmid. (C) Effects of expression of vector control GFP-C1, N24-GFP, and GFP-N24 on the activity of the cyclin D promoter in Rb−/− and Rb+/+ MEFs. Transfection of the cyclin D luciferase reporter plasmid and the N24-GFP constructs into Rb+/+ and Rb−/− MEFs was performed as described in the legend to Fig. 7A. Luciferase and β-Gal activities were determined as described previously.
DISCUSSION
We report here that Rb and the p55 regulatory subunits of PI 3-kinase associated in cultured cells. This association was regulated by serum and growth factors, such as heregulin and EGF. Moreover, interruption of Rb-p55 interactions by overexpression of the N-terminal 24-amino-acid Rb binding domain of p55 inhibited cell cycle progression. The inhibitory effects of the N-terminal 24 amino acids were observed only in cells with a functional Rb. We suggest that the signals mediated by the Rb-p55 interaction and downstream PI 3-kinase-mediated pathways might be important in cell cycle progression.
Other groups have shown that PI-3 kinase regulates cell cycle progression via indirect effects on Rb phosphorylation. For example, phosphorylation of Rb is inhibited by specific PI 3-kinase inhibitors and dominant-negative PI 3-kinase proteins in IL-2-stimulated T lymphocytes (5, 6). In these cases, PI 3-kinase effects were mediated by changes in AKT, ultimately resulting in decreases in cyclin levels (5, 6). Similarly, PI 3-kinase's effects on the cell cycle have been shown to be due to the effect of p110 on cyclin D induction (20). However, in this report we show a direct association of a PI 3-kinase regulatory subunit with Rb. The fact that Rb associated with p55γ in vivo and in vitro and that disruption of this complex by ectopic expression of the N-terminal 24-amino-acid Rb binding domain of p55 inhibited cell growth suggests that the Rb-p55 complex may play a role in cell cycle progression. The fact that the Rb-p55 interaction observed in quiescent cells appeared to be necessary for the initiation of cell cycle progression, but that p55 dissociated from Rb soon after addition of serum, appears at first to be paradoxical. However, it is possible that a threshold level of Rb-p55 complexes is needed before the initiation of the cell cycle.
In addition, the growth-inhibitory effects of N24 were observed only in cells with functional Rb. Overexpression of N24p55 had no effect on DNA synthesis or cyclin D promoter activity in Rb−/− MEFs compared with Rb +/+ MEFs. In addition, N24p55 did not affect growth of the Rb-negative MDA-MB-468 and SAOS-2 cells, while growth of MCF-7 cells was inhibited approximately 40% (unpublished data). However, these observations do not necessarily implicate the Rb-p55 complex as a prerequisite for cell cycle entry. For example, high levels of N24 may sequester Rb from its physiological interactions with other proteins. The physiological significance of the Rb-p55 interactions are still unclear and remain the focus of investigations in our laboratory.
It is now becoming apparent that the different isoforms of both the p110 catalytic subunit and the regulatory subunits of PI 3-kinase may perform unique functions. p55γ was first isolated by the ability of its SH2 domains to bind phosphorylated tyrosine in signaling proteins such as insulin receptor substrate 1 (IRS-1) (14). However, the function of the unique N-terminal end of p55 is largely unknown. Our data are the first to demonstrate that p55γ specifically associates with Rb via its 24-amino-acid N-terminal domain. The fact that the p85 and p50 regulatory subunits did not associate with Rb suggests that different regulatory subunits of PI 3-kinase have different functions and provides one mechanism of how PI 3-kinase is involved in diverse and often conflicting cellular processes, such as proliferation, differentiation, and cell survival.
We found that both p55α and p55γ associate with Rb, presumably due to the similarity in their N-terminal domains. The interaction of Rb with p55α in vivo suggests that p55α may have a function similar to that of p55γ in the regulation of cell cycle progression. It is interesting that the expression of p55γ and p55α varies significantly in different tissues and cells (11, 14), suggesting that these two proteins may have similar roles in different tissues. However, some tissues express neither isoform, suggesting that these proteins are not absolutely required for cell cycle progression.
The finding that the 24-amino-acid N-terminal end of p55γ was both necessary and sufficient to bind Rb excluded the possible involvement of SH2, SH3, bcr, and proline-rich domains in p85 in the interaction of Rb with PI 3-kinase. The fact that the N24 domain, which lacks the binding domain for the catalytic subunits of PI 3-kinase (p110), can bind Rb and have a biological function suggests that the p110 subunit does not contribute to the biological effects of the overexpressed N24 domain. There is no LXCXE consensus domain, present in many so-called pocket binding proteins such as CDK, RBP, and c-Abl, in the p55γ Rb-binding site, suggesting a new Rb binding domain. Conversely, although p55γ bound to GST-Rb379-928, it also bound Rb presumably in the presence of T antigen in COS7 cells, suggesting an alternate site of Rb-p55Rb interactions.
The mechanism that regulates the interaction of Rb with p55γ is not known. It is likely that the phosphorylation of Rb or p55γ might be a major factor in interaction regulation. Although there is a possible tyrosine phosphorylation site (Tyr-22) in the p55γ Rb-binding region, no significant tyrosine phosphorylation was detected in endogenous p55γ after serum treatment of AU565 cells (data not shown), making it unlikely that phosphorylation on this site disrupts the interaction. On the other hand, our data suggest that the phosphorylation of Rb may be a major factor in regulation of the interaction. Strong p55-Rbγ interactions were observed when cell growth was inhibited, such as when the cells were induced to differentiate by heregulin or were serum starved. During these times, Rb is hypophosphorylated. When the cells were induced to enter the cell cycle by the addition of mitogens, the dissociation of Rb from p55 was initiated as well, and binding was totally disrupted after 3 h, consistent with the time of Rb phosphorylation. There are more than 10 possible phosphorylation sites for serine/threonine kinases such as CDKs in Rb proteins. Phosphorylation on these sites disrupts the binding of Rb with other proteins (12), and it is possible that one of these sites may regulate the interaction of Rb with p55γ. Identification of the phosphorylation sites is under way in our current investigations.
Because of its important role in cell proliferation, PI 3-kinase is a potential target in cancer chemotherapy, and the therapeutic use of PI-3-kinase inhibitors is being extensively studied. The data reported here suggest that p55γ-mediated pathways may specifically mediate signals from growth factors. These pathways and the interaction of p55γ with Rb may provide molecular targets for the identification of new agents and drugs in cancer chemotherapy and gene therapy.
Acknowledgments
We thank S. Mittnacht, J. Downward, R. Livingston, T. Kouzarides, C. Kahn, and R. Pestell for various plasmid constructs used in this study; T. Jacks for the Rb−/− and Rb+/+ MEFs; N. Wehman for help in cell cycle analysis and cell sorting; and A. Passaniti, X. Wang, and H. Cheng for critical reading of the manuscript.
This work was supported in part by NIH grant R01 CA76047 and University of Maryland DRIF funds (to A.W.H.).
REFERENCES
- 1.Antonetti, D. A., P, Algenstaedt, and C. R. Kahn. 1996. Insulin receptor substrate 1 binds two novel splice variants of the regulatory subunit of phosphatidylinositol 3-kinase in muscle and brain. Mol. Cell. Biol. 16:2195-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacus, S. S., A. V. Gudkov, C. R. Zelnick, D. Chin, R. Stern, I. Stancovski, E. Peles, N. Ben-Baruch, H. Farbstein, R. Lupu, D. Wen, M. Sela, and Y. Yarden. 1993. Neu differentiation factor (heregulin) induces expression of intercellular adhesion molecule 1: implications for mammary tumors. Cancer Res. 53:5251-5261. [PubMed] [Google Scholar]
- 3.Bondeva, T., L. Pirola, G. Bulgarelli-Leva, I. Rubio, R. Wetzker, and M. P. Wymann. 1998. Bifurcation of lipid and protein kinase signals of PI-3K to the protein kinases PKB and MAPK. Science 282:293-296. [DOI] [PubMed] [Google Scholar]
- 4.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, P., J. W. Babbage, B. M. T. Burgering, B. Gronder, K. Reif, and D. A. Cantrell. 1997. Phosphatidylinositol 3-kinase couples the interleukin-2 receptor to the cell cycle regulator E2F. Immunity 7:679-689. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, P., J. W. Babbage, G. Thomas, and D. A. Cantrell. 1999. p70S6k integrates phosphatidylinositol 3-kinase and rapamycin-regulated signals for E2F regulation in T lymphocytes. Mol. Cell. Biol. 19:4729-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downward, J. 1998. Lipid-regulated kinases: some common theme at last. Science 279:673-674. [DOI] [PubMed] [Google Scholar]
- 8.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481-507. [DOI] [PubMed] [Google Scholar]
- 9.Herrera, R. E., V. P. Sah, B. O. Williams, T. P. Makela, R. A. Weinberg, and T. Jacks. 1996. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 16:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiebert, S. W., S. P. Chellappan, J. M. Horowitz, and J. R. Nevins. 1992. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 6:177-185. [DOI] [PubMed] [Google Scholar]
- 11.Inukai, K., M. E. Anai, T. Van Breda, H. Hosaka, M. Katagiri, Y. Funaki, T. Fukushima, Y. Ogihara, M. Yazaki, Y. Kikuchi, Y. Oka, and T. Asano. 1996. A novel 55KDa regulatory subunit for phosphatidyl inositol 3-kinase structurally similar to p55γ is generated by alternative splicing of the p85α gene. J. Biol. Chem. 271:5317-5320. [DOI] [PubMed] [Google Scholar]
- 12.Kaelin, W. G. 1999. Functions of the retinoblastoma proteins. Bioessays 21:950-958. [DOI] [PubMed] [Google Scholar]
- 13.Kraus, M. H., N. C. Popescu, S. C. Amsbaugh, and C. R. King. 1987. Overexpression of the EGF receptor-related proto-oncogene ErbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J. 6:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pons, S., T. Osano, E. Glasheen, M. Miralpeix, Y. Zhang, T. L. Fisher, M. G. Myers, X.-J. Sun, and M. White. 1995. The structure and function of p55γ reveals a new regulatory subunit for phosphatidylinositol 3-kinase. Mol. Cell. Biol. 15:4453-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsby, M. L., and G. S. Makowski. 1997. Differential detergent fractionation of eukaryotic cells. Methods Mol. Biol. 112: 53-67. [DOI] [PubMed] [Google Scholar]
- 16.Rodriquez-Viciana, P., P. H. Wanre, B. Vanhaesebroeck, and M. D. Waterfield. 1996. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutants. EMBO J. 15:2442-2451. [PMC free article] [PubMed] [Google Scholar]
- 17.Sellers, W. R., and W. Kaelin. 1996. Rb as a modulator of transcription. Biochim. Biophys. Acta 1288:M1-M5. [DOI] [PubMed] [Google Scholar]
- 18.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 19.Stephens, L. R., T. R. Jackson, and P. T. Hawkins. 1993. Agonist-stimulated synthesis of phosphatidylinositol (3,4,5)-triphosphate: a new intracellular signaling system? Biochim. Biophys. Acta 1179:27-75. [DOI] [PubMed] [Google Scholar]
- 20.Takuwa, N., Y. Fukui, and Y. Takuwa. 1999. Cyclin D1 expression is mediated by phosphatidylinositol 3-kinase through mTOR-p70S6K-independent signaling in growth factor-stimulated NIH 3T3 fibroblasts. Mol. Cell. Biol. 19:1346-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanhaesebroeck, B., S. J. Leevers, G. Panayotou, and M. D. Waterfield. 1997. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem. Sci. 22: 267-272. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe, G., C. Albanese, R. J. Lee, R. J. A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 24.Xia, X., and G. Serrero. 1999. Multiple forms of p55, a regulatory subunit of phosphoinositide 3-kinase, are generated by alternative initiation of translation. Biochem. J. 341:831-837. [PMC free article] [PubMed] [Google Scholar]
- 25.Xia, X., A. Cheng, T. Lessor, Y. Zhang, and A. W. Hamburger. 2001. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell. Physiol. 187:209-217. [DOI] [PubMed] [Google Scholar]
- 26.Zacksenhaus, E., Z. Jiang, D. Chung, J. D. Marth, R. A. Phillips, and B. L. Gallie. 1996. pRb controls proliferation, differentiation and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 10:3051-3064. [DOI] [PubMed] [Google Scholar]
- 27.Zarkowska, T., and S. Mittnacht. 1997. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J. Biol. Chem. 272:12738-12746. [DOI] [PubMed] [Google Scholar]