Abstract
The precellular Drosophila embryo contains ≈10 well characterized transcriptional repressors. At least half are short-range repressors that must bind within 100 bp of either upstream activators or the core transcription complex to inhibit (or quench) gene expression. The two long-range repressors can function over distances of 1 kilobase or more to silence transcription. Previous studies have shown that three of the five short-range repressors interact with a common corepressor protein, dCtBP. In contrast, the two long-range repressors, Hairy and Dorsal, recruit a different corepressor protein, Groucho. Hairy also was shown to interact with dCtBP, thereby raising the possibility that Groucho and dCtBP are components of a common corepressor complex. To investigate this issue, we have misexpressed wild-type and mutant forms of Hairy in transgenic embryos. Evidence is presented that Hairy-mediated repression depends on the Groucho interaction sequence (WRPW) but not the weak dCtBP motif (PLSLV) present in the native protein. Conversion of the PLSLV motif into an optimal dCtBP interaction sequence (PLDLS) disrupts the activity of an otherwise normal Hairy protein. These results suggest that dCtBP and Groucho mediate separate pathways of transcriptional repression and that the two proteins can inhibit one another when both bind the same repressor.
Localized patterns of gene expression are established by ≈10 different transcriptional repressors in the precellular Drosophila embryo (see refs. 1–4). These repressors represent a broad spectrum of DNA binding proteins, including members of the bHLH, rel, homeodomain, zinc finger, and nuclear receptor families. At least half of the repressors function over short distances, <100 bp, to inhibit (or quench) upstream activators or the core transcription complex (e.g., refs. 5 and 6). In contrast, two of the repressors can function over long distances, 1 kilobase or more, to silence gene expression (2, 7). The mechanisms underlying short-range and long-range repression remain uncertain, although recent studies suggest that unrelated repressors can function through common corepressor proteins. For example, three of the five short-range repressors, Snail, Kruppel, and Knirps, interact with the dCtBP corepressor (8–10). In contrast, both long-range repressors, Dorsal and Hairy, depend on a different corepressor protein, Groucho (1, 2, 11, 12). These observations raise the possibility that dCtBP and Groucho mediate separate pathways of transcriptional repression.
A potential complication of this simple view is that dCtBP originally was identified in yeast 2-hybrid screens using sequences from the Snail, Knirps, and Hairy repressors (8, 9). Thus, it has been suggested that Hairy interacts with both dCtBP and Groucho, thereby raising the possibility that the two proteins are components of a common corepressor complex (9). There are several potential arguments against this model. First, the dCtBP interaction sequence that was identified in Hairy, P-SLV-K (PLSLVK; see Fig. 1), is rather divergent from the conserved motif observed in Snail and Knirps and first was identified in the adenovirus E1A protein P-DLS-K (13, 14). Second, genetic interactions between hairy and dCtBP are enigmatic. hairy mutant embryos exhibit somewhat less severe segmentation defects when the maternal dose of dCtBP is lowered (9). A simple interpretation of this observation is that dCtBP somehow antagonizes hairy+ gene function. Another argument against a critical role for dCtBP in Hairy-mediated repression is the observation that Hairy continues to function as a repressor, at least in part, in mutant embryos containing reduced levels of maternal dCtBP products (10).
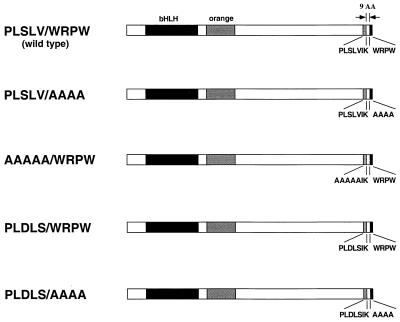
Figure 1.
Summary of wild-type and mutant Hairy proteins. Hairy is composed of 337 amino acid (AA) residues and contains four regions identified in previous studies (9, 11, 27–29). These include the bHLH domain that is required for dimerization and DNA binding and the orange domain, which is thought to mediate local repression of bHLH activators. The present study centers on two motifs located at the C terminus of the protein, PLSLVIK and WRPW. The latter sequence has been shown to mediate Hairy-Groucho interactions whereas the former sequence interacts with dCtBP. The two protein binding motifs are separated by nine AA residues. Four mutant forms of Hairy were examined, including one that lacks the Groucho motif (PLSLV/AAAA) and another that lacks the dCtBP motif (AAAAA/WRPW). Additional mutant proteins contain an optimal dCtBP motif (PLDLS) in place of the native weak interaction sequence (PLSLV). One of these lacks the Groucho motif (PLDLS/AAAA) whereas the other contains both motifs (PLDLS/WRPW).
In the present study, we examine the relative contributions of dCtBP and Groucho in Hairy-mediated repression. Wild-type and mutant Hairy proteins were expressed in transgenic embryos by using the maternal hsp83 promoter and the bicoid 3′ untranslated region (UTR). Misexpression of the wild-type Hairy protein leads to the repression of a number of potential target genes, including Sex lethal (Sxl; refs. 15 and 16), tailless (tll; ref. 17), forkhead (fkh; ref. 18), and huckebein (hkb; ref. 19) in anterior regions of transgenic embryos. Mutant forms of the protein lacking the Groucho interaction motif (WRPW) are significantly impaired whereas mutations in the dCtBP motif (P-SLV-K) have no obvious effect on the efficacy of Hairy-mediated repression. In fact, the removal of the dCtBP motif may augment repression activity. A mutant protein that lacks the WRPW motif but contains an optimal dCtBP motif (P-DLS-K) exhibits some repression activity whereas a modified Hairy protein containing both the WRPW motif and an optimal P-DLS-K motif is inactive. These results suggest that dCtBP and Groucho function antagonistically when both are bound to Hairy and that the two corepressors mediate separate pathways of transcriptional repression.
MATERIALS AND METHODS
Plasmid Construction.
A hairy-encoding plasmid for in vitro expression was made by inserting a BglII-EcoRI fragment containing the full-length hairy cDNA into the pBluescript II KS (−) vector (Stratagene) (S. Barolo, personal communication). Site-directed mutagenesis was done essentially as described by Kunkel (20). The pBluescript II KS (−)-hairy plasmid was used to transform the CJ236 strain of Escherichia coli to prepare uracil-containing single-stranded DNA. The following mutagenic oligonucleotides were used to mutagenize the Groucho and dCtBP interaction motifs in the Hairy protein, respectively: CATATGCAGACACCCTCTACGCGGCTGCCGCGGGCTGCTCCTCCTC, GATCTGCTTCTTGATCGCAGCTGCCGCGGCCTGCTGTTCCATGGG, and GATCTGCTTCTTGATCGACAGGTCCAGGGGCTGCTGTTCCATGGG. The underlined nucleotides indicate mutations in the normal Hairy coding sequence. Oligonucleotide 1 converts amino acid residues WRPW into AAAA, and oligonucleotides 2 and 3 convert PLSLV into AAAAA and PLDLS, respectively.
P-element transformation vectors were prepared by inserting wild-type or mutant forms of the hairy protein coding sequence into the KpnI site of pCaSpeR-hsp83-bcd3′ UTR, which contains the maternal hsp83 promoter sequence and the bicoid 3′ UTR (21, 22).
Glutathione S-Transferase (GST) Pull-Down Assays.
Wild-type and mutant forms of the Hairy protein were 35S-labeled with the TNT T7 Quick coupled transcription/translation system (Promega). GST-dCtBP (10) and GST-Groucho (2) were prepared as described (8), and binding assays were performed by incubating 5 μl of in vitro-translated protein with 5 μg of GST or GST fusion protein on glutathione-agarose beads at 4°C for 1 hr. Bound proteins were analyzed by SDS/PAGE and were visualized by autoradiography (8, 23).
P-Transformation and Whole-Mount in situ Hybridization.
P-element plasmids were introduced into the yw67c23 strain by using standard methods (e.g., ref. 24). At least three independent transgenic lines were examined for each construct. Whole-mount in situ hybridization was performed as described, using digoxigenin-labeled antisense riboprobes (25, 26).
RESULTS
The wild-type Hairy protein (Fig. 1) contains a bHLH DNA binding domain, the orange domain (which is thought to mediate short-range repression of bHLH activators), a weak dCtBP interaction motif (P-SLV-K), and the Groucho motif (WRPW; see refs. 9, 11, and 27–29). Four different hairy mutants were examined in this study, including PLSLV/AAAA and AAAAA/WRPW, which lack the Groucho and dCtBP interaction motifs, respectively (Fig. 1). Additional mutant forms of Hairy were prepared, including one that contains both the WRPW motif and an optimal dCtBP interaction motif, P-DLS-K, in place of the native P-SLV-K sequence (PLDLS/WRPW). A derivative of this protein lacks WRPW but contains the optimal dCtBP motif (PLDLS/AAAA).
GST Pull-Down Assays.
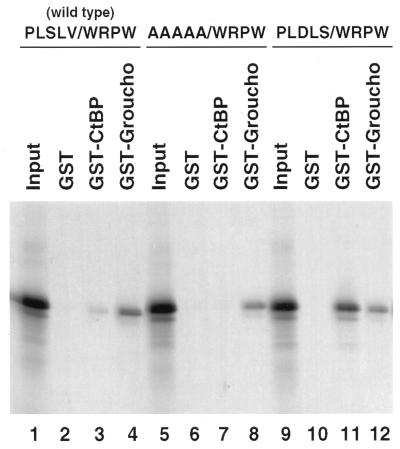
Protein binding assays were performed with GST-dCtBP and GST-Groucho fusion proteins (2, 10), and 35S-labeled hairy products were produced by in vitro translation (Fig. 2). The full length, wild-type Hairy protein binds quite well to the GST-Groucho fusion protein (Fig. 2, lane 4) as compared with a GST control protein (Fig. 2, lane 2). It exhibits only weak binding to the GST-dCtBP fusion protein (Fig. 2, lane 3). This weak interaction between Hairy and dCtBP is lost when mutations are introduced into the divergent dCtBP interaction motif contained in the normal Hairy protein (“AAAAA/WRPW,” Fig. 2, lane 7; compare with lane 3). In contrast, a modified Hairy protein that contains an optimal dCtBP interaction motif (“PLDLS”) in place of the native sequence (“PLSLV”) exhibits stronger binding to the GST-dCtBP fusion protein (Fig. 2, lane 11) than the native protein (Fig. 2, lane 3). Although these manipulations of the dCtBP interaction motif alter the strength of Hairy-dCtBP interactions (Fig. 2, lanes 3, 7, and 11), the different Hairy proteins bind about equally well to the GST-Groucho fusion protein (Fig. 2, lanes 4, 8, and 12). Thus, it is possible, at least in these in vitro assays, to uncouple the binding of Groucho and dCtBP to Hairy.
Figure 2.
GST pull-down assays. Different Hairy proteins were labeled with 35S via in vitro translation. Three different forms of Hairy were examined, including the wild-type protein (lanes 1–4), a mutant protein lacking the weak dCtBP motif (PLSLV was changed to AAAAA; lanes 5–8), and a modified protein that contains an optimal dCtBP interaction motif (PLDLS) in place of the native sequence (PLSLV; lanes 9–12). The wild-type protein binds to a GST-Groucho fusion protein (lane 4) but not to a GST nonfusion control protein (lane 2). It also binds weakly to a GST-dCtBP fusion protein (lane 3). The mutant protein lacking the PLSLV motif continues to bind the GST-Groucho fusion protein (lane 8) but no longer binds the GST-dCtBP fusion protein (lane 7). The modified protein containing the optimal dCtBP motif binds to the GST-Groucho fusion protein (lane 12) and exhibits enhanced binding to the GST-dCtBP protein (lane 11), as compared with the wild-type protein (lane 3). The lanes labeled “Input” (lanes 1, 5, and 9) contain 20% of the total amount of labeled protein used in the binding reactions.
Misexpression of hairy in Transgenic Embryos.
The five hairy protein coding sequences (summarized in Fig. 1) were inserted into the hsp83-bcd 3′ UTR P-element transformation vector (21, 22), and transgenic strains were established by using standard methods (e.g., ref. 24). The hsp83 promoter directs strong maternal expression, and the bcd UTR localizes the transcripts to the anterior pole of precellular embryos (30). Each of the five hairy transgenes exhibits intense expression in early embryos before the time when the endogenous gene is activated (Fig. 3). Multiple lines were established for each transgene, and although it is difficult to quantify expression, it appears that the different hairy transcripts are expressed at comparable levels (Fig. 3). For example, replacing the C-terminal WRPW motif with alanine residues does not appear to destabilize the hairy mRNA so that it is expressed at about the same levels as the wild-type transcript (Fig. 3, compare D with A). The ectopic hairy transcripts persist until the midpoint of cellularization at ≈3 hr after fertilization (Fig. 3B and data not shown). The misexpression of Hairy appears to cause a delay in the expression of the endogenous hairy stripes 1 and 2 (Fig. 3B; see ref. 31). By the onset of gastrulation, the ectopic hairy mRNAs at the anterior pole are lost (like the endogenous bcd mRNAs; ref. 32), and a normal endogenous hairy pattern is established (data not shown).
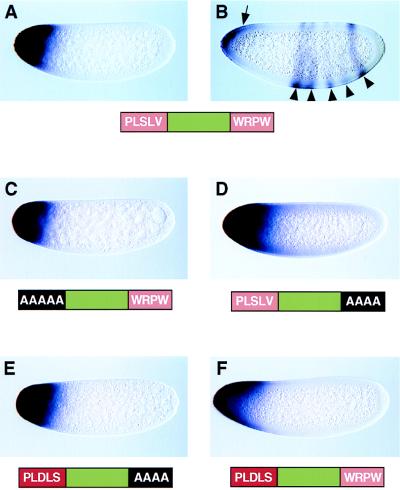
Figure 3.
Misexpression of hairy transcripts in precellular embryos. Embryos were collected from transgenic females that carry different hsp83-hairy-bcd 3′ UTR expression vectors. They are oriented with dorsal up and anterior to the left and were hybridized with a digoxigenin-labeled hairy antisense RNA probe. (A and B) Preblastoderm and midnuclear cleavage cycle-14 embryos, respectively, which express wild-type hairy transcripts. Transcripts are localized to the anterior pole before nuclear migration (A). During nuclear cleavage cycle 14 (B), these transcripts gradually fade (arrow), and the endogenous pattern begins to emerge (arrowheads). At this stage, hairy stripes 3–7 can be detected, but there is a delay in the appearance of stripes 1 and 2. (C) Preblastoderm embryo that expresses a mutant hairy product lacking the weak dCtBP interaction motif (PLSLV was converted to AAAAA). Strong expression is observed at the anterior pole. (D) Nuclear cleavage cycle-12/13 embryo that expresses a mutant hairy product lacking the Groucho interaction motif (WRPW was converted into AAAA). (E) Preblastoderm embryo that expresses a mutant hairy product that lacks the WRPW motif and contains an optimal dCtBP interaction sequence (PLDLS) in place of the native sequence (PLSLV). (F) Nuclear cleavage cycle-12/13 embryo that expresses a mutant hairy product that contains both WRPW and the optimal dCtBP motif (PLDLS). Note that this transcript is expressed at levels that are comparable to those observed for the other hairy products.
The consequence of misexpressing these different hairy products was investigated by analyzing the expression of the Sxl gene (33). Previous studies have shown that the ectopic expression of hairy by using the hunchback (hb) promoter results in female lethality caused by the repression of Sxl (16). As a result, adult males develop normally whereas transgenic females die during embryogenesis, presumably because of a breakdown in dosage compensation and the overexpression of X-linked genes (34). It has been proposed that ectopic hairy products mimic the activity of the Deadpan repressor, which is one of the “denominator” elements that normally keeps Sxl off in males (11, 35, 36).
A Sxl antisense RNA probe was used to monitor Sxl expression in wild-type and transgenic embryos (Fig. 4). During nuclear cleavage cycle 14, the early Sxl expression pattern is nonhomogenous and includes crude stripes and bands of staining (Fig. 4A; see ref. 37). Sxl expression was observed in about half of the control embryos, which presumably represent XX individuals. The other embryos, presumptive males, do not exhibit Sxl staining above background levels (data not shown).
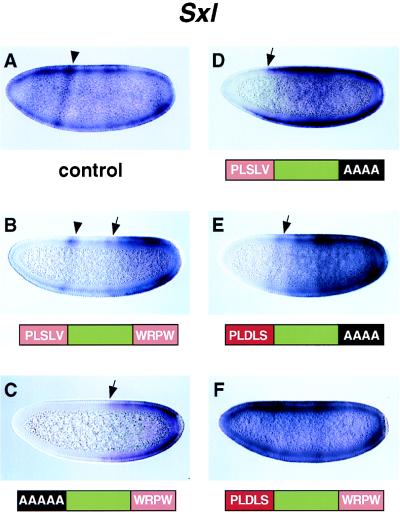
Figure 4.
Sxl expression is repressed by Hairy. Transgenic embryos that express different hairy products were hybridized with a digoxigenin-labeled Sxl antisense RNA probe and are oriented with dorsal up and anterior to the left. (A) Sxl expression in a wild-type, midnuclear cleavage cycle-14 embryo. Staining is somewhat nonhomogenous, and the pattern includes crude stripes and bands of expression (e.g., arrowhead). Sxl staining was observed in about half of the embryos; the others presumably correspond to males. (B) Sxl staining pattern in a cleavage cycle-14 embryo that expresses the wild-type hairy RNA. Sxl expression is repressed in the anterior half of the embryo (arrow), although a stripe persists in anterior regions (arrowhead). (C) Sxl staining in an early nuclear cleavage cycle-14 embryo that expresses a mutant form of Hairy that lacks the weak dCtBP interaction motif (PLSLV). Sxl is completely repressed in anterior regions (arrow), and the anterior stripe seen in B is absent. (D) Same as C except that the transgenic embryo expresses a mutant hairy product that lacks the Groucho interaction motif (WRPW) but retains the weak dCtBP interaction sequence (PLSLV). Sxl is repressed partially in the anterior-most regions of the embryo. More extensive repression was obtained with hairy products that retain the WRPW motif (see B and C). (E) Same as D except that the transgenic embryo expresses a mutant hairy product that contains an optimal dCtBP interaction sequence (PLDLS) in place of the native motif (PLSLV). Repression is observed in the anterior third of the embryo (arrow). Augmenting dCtBP binding in the absence of WRPW appears to increase the efficacy of repression (compare with D). (F) Late nuclear cleavage cylce-14 embryo that expresses a modified hairy product containing the optimal PLDLS motif in place of the weak PLSLV sequence. There is no repression of Sxl even in the anterior-most regions. This result suggests that the encoded protein, which contains optimal Groucho and dCtBP motifs, is inactive.
Transgenic embryos that express the wild-type hairy transgene exhibit an altered pattern of Sxl expression (Fig. 4B) whereby staining is reduced in anterior regions. It would appear that the localized hairy mRNA at the anterior pole (see Fig. 3A) serves as a source for an anteroposterior Hairy repressor gradient that inhibits Sxl expression. This repression might involve the binding of ectopic Hairy products to Deadpan binding sites in the Sxl Pe promoter (e.g., ref. 37; see Discussion). A similar pattern of Sxl repression was obtained with the AAAAA/WRPW protein, which lacks the dCtBP interaction motif (Fig. 4C). In fact, it appears that the modified protein, which retains the Groucho interaction motif (WRPW), might be a somewhat more potent repressor than native Hairy (Fig. 4, compare C with B). These results suggest that dCtBP is not essential for Sxl repression in this assay, and additional experiments were conducted to examine the role of the WRPW motif in Sxl regulation.
Embryos that express a mutant Hairy protein that lacks the Groucho interaction motif (PLSLV/AAAA) exhibit a slightly altered Sxl pattern whereby staining is lost in the anterior-most regions (Fig. 4, compare D with A). A modified version of this protein, containing an optimal dCtBP interaction motif (PLDLS/AAAA) mediates somewhat more efficient repression, so that staining is lost in the anterior third of the embryo (Fig. 4, compare E with D). However, the repression obtained with this protein is not as extensive as that obtained with Hairy proteins that contain the Groucho interactions motif (PLSLV/WRPW and AAAAA/WRPW; see Fig. 4 B and C). These results suggest that Groucho plays an important role in the Hairy-mediated repression of Sxl, as suggested in previous studies (11, 28).
A somewhat unexpected result was obtained with a modified Hairy protein that contains both the WRPW motif and the optimal dCtBP motif (PLDLS/WRPW). The modified mRNA is stably expressed at high levels in transgenic embryos (Fig. 3F) but completely lacks repression activity (Fig. 4F). There is no observable reduction in Sxl expression, even in the anterior-most regions in which there are high levels of the mutant RNA. In contrast, the PLSLV/AAAA protein, which lacks the WRPW motif, is able to mediate at least some repression of Sxl at the anterior pole (Fig. 4, compare D with F). This result is consistent with the possibility that dCtBP and Groucho function in an antagonistic manner to mediate transcriptional repression (see Discussion).
Hairy Represses Terminal Patterning Genes.
Transgenic embryos that misexpress mutant forms of Hairy lacking the WRPW motif (both the PLSLV/AAAA and the PLDLS/AAAA proteins) do not exhibit defective cuticles (data not shown). However, the PLDLS/AAAA protein, which contains an optimal dCtBP interaction motif, results in female lethality (data not shown) whereby only transgenic males are viable. This lethality correlates with the repression of Sxl in the anterior third of presumptive XX embryos (Fig. 4E). There is only a mild distortion in the sex ratio observed for the PLSLV/AAAA mutant protein, and in situ hybridization assays indicate that Sxl is repressed only at the anterior pole (Fig. 4D; data not shown). Similarly, the PLDLS/WRPW protein, which contains an optimal dCtBP motif, does not produce distortions in the sex ratio and does not repress Sxl expression (Fig. 4F).
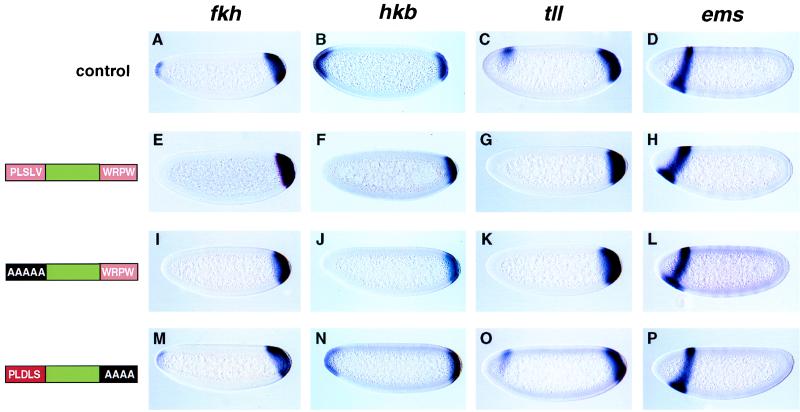
Transgenic embryos that express either wild-type Hairy or the AAAA/WRPW mutant protein (which lacks the weak dCtBP motif) are lethal and exhibit severe head defects (data not shown). To determine the basis for this lethality, we analyzed the expression of a number of patterning genes that are active in anterior regions, including fkh (18), hkb (19), tll (17), and empty spiracles (ems; refs. 38 and 39). The normal staining patterns are presented in Fig. 5 A–D. The fkh, hkb, and tll genes normally are expressed in both anterior and posterior regions. Transgenic embryos that express either the wild-type (Fig. 5 E–G) or AAAAA/WRPW protein (Fig. 5 I–K) exhibit abnormal fkh, hkb, and tll patterns whereby staining is selectively lost in anterior regions. In all cases, the posterior patterns are unaffected. Neither the PLSLV/WRPW (wild-type) nor the AAAAA/WRPW transgene inhibits ems expression (Fig. 5 H and L), although in both cases there is an anterior shift in the staining pattern as compared with control embryos (Fig. 5D).
Figure 5.
Hairy represses terminal patterning genes. Nuclear cleavage cycle-14 embryos were hybridized with the indicated digoxigenin-labeled antisense RNA probes. The wild-type staining patterns (“control”) are shown in A–D. fkh, hkb, and tll are expressed in both the anterior and posterior poles whereas ems is expressed in a single stripe near the head. (E–H) Expression of the terminal patterning genes in a transgenic embryo that expresses the wild-type form of hairy. The anterior, but not posterior, patterns of fkh, hkb, and tll are repressed. In addition, there is an anterior shift in the ems pattern (compare H with D). (I–L) Same as E–H except that the transgenic embryos express a mutant form of hairy that lacks the PLSLV motif. Efficient repression of fkh, hkb, and tll is observed. (M–P) Same as I–L except that the embryos express a mutant form of hairy that lacks the WRPW motif and contains an optimal dCtBP interaction sequence (PLSLV was changed to PLDLS). There may be a slight attenuation in the anterior staining of fkh and hkb, but otherwise the expression patterns appear normal (see A–C). In addition, there is no anterior shift in the ems pattern. Although this hairy product is essentially inactive in these assays, it is able to mediate substantial repression of Sxl (see Fig. 4E).
The PLDLS/AAAA protein contains an optimal dCtBP motif but does not repress the anterior expression of fkh, hkb, and tll or cause an obvious shift in the ems pattern (Fig. 5 M–P), even though it is reasonably effective in repressing Sxl expression (Fig. 4E). These results provide evidence that dCtBP-mediated repression is somewhat more selective than Groucho-mediated repression (see Discussion). The modified PLDLS/WRPW transgene, which contains both WRPW and the optimal dCtBP motif, fails to repress fkh, hkb, and tll (data not shown) and, as shown earlier, also fails to repress Sxl (see Fig. 4F). These results suggest that dCtBP and Groucho somehow interfere with one another when both interact with Hairy (see below).
DISCUSSION
We have presented evidence that dCtBP is not essential for Hairy-mediated repression in the early embryo. Elimination of the weak dCtBP interaction motif, PLSLV, does not disrupt Hairy activity when the Groucho interaction motif, WRPW, is intact. These observations suggest that dCtBP and Groucho mediate separate pathways of transcriptional repression. In fact, several lines of evidence suggest that dCtBP and Groucho may function antagonistically to mediate repression. For example, replacing the weak dCtBP motif (PLSLV) with an optimal sequence (PLDLS) inactivates an otherwise normal Hairy protein.
Repression of Terminal Patterning Genes.
The repression of Sxl expression by ectopic Hairy products is consistent with previous studies that used the zygotic hb promoter to misexpress Hairy in anterior regions of transgenic embryos (16). Both the hb promoter and the maternal hsp83-bcd UTR expression vector used in this study result in the misexpression of Hairy in anterior regions and the concomitant repression of Sxl. However, hb-hairy fusion genes did not cause patterning defects (31) whereas the maternal expression vector results in the repression of both Sxl and terminal genes (see Figs. 4 and 5). There are several possible explanations for the different findings. Perhaps higher concentrations of the Hairy repressor are expressed at the anterior pole by using the bcd 3′ UTR as compared with the hb promoter. Alternatively, it is possible that the repression of fkh, hkb, and tll depends on the early expression of Hairy, which is obtained with the maternal hsp83 promoter. It is unclear whether ectopic Hairy products work directly or indirectly to repress the terminal patterning genes, although there are potential Hairy binding sites in the promoter regions of tll and fkh (H.Z., unpublished observations).
Repression of fkh, hkb, and tll depends on the presence of an intact Groucho interaction motif (WRPW). A mutant form of Hairy (PLDLS/AAAA) that lacks this motif but contains an optimal dCtBP sequence does not cause head patterning defects, even though it is reasonably effective in repressing Sxl (see Fig. 4E). There are several possible explanations for this regulatory specificity. Previous studies suggest that the repression of the Sxl Pe promoter can be obtained with a variety of disparate repressors, including those that function over short or long distances (1). In vitro binding assays suggest that the Deadpan repressor sites map quite close to the Sis-a and Sis-b activator sites within the Sxl promoter (36, 37). Consequently, short-range repressors should be able to quench the adjacent activators and work just as effectively as long-range repressors in blocking Sxl expression. In contrast, repression of fkh, hkb, and tll might depend on long-range repressors.
dCtBP-Groucho Interactions.
The removal of the weak dCtBP interaction motif (PLSLV) does not impair Hairy-mediated repression of Sxl, fkh, hkb, and tll (see Figs. 4 and 5). If anything, removal of this motif augments Hairy function (e.g., Fig. 4 B and C). This observation suggests that the binding of dCtBP somehow interferes with Groucho-mediated repression. Additional support for this view stems from the observation that the PLDLS/WRPW protein, which contains an optimal dCtBP motif, is inactive and fails to repress any of the target genes that were examined. Moreover, there were no distortions in the sex ratio observed among different transgenic strains that express this form of Hairy (data not shown). The simplest interpretation of these results is that the dCtBP and Groucho corepressors interfere with one another when both are bound to Hairy. Such antagonistic interactions are supported by previous genetic studies, which suggest that lowering the dose of maternal dCtBP products can partially suppress the embryonic phenotypes of hairy mutants (9).
The P-SLV-K and WRPW motifs are separated by just nine amino acid residues within the C terminus of the Hairy protein (Fig. 1). When dCtBP and Groucho both bind, they might be unable to interact with additional corepressors or with their target proteins in the core transcription complex. In the course of normal development, post-translational modification might determine which corepressor can interact with Hairy and whether Hairy functions as a short-range or long-range repressor. A similar situation might apply to the adenovirus E1A protein, which interacts with two different corepressors, the retinoblastoma protein (40) and CtBP (13, 14). The retinoblastoma protein, like Groucho, appears to mediate long-range repression, suggesting that E1A might function as both a short-range and long-range repressor.
Acknowledgments
We thank Jumin Zhou, Al Courey, Lisa Komenda, and Jessica Dines for plasmids and Tina Liang and Pearline Chong for technical assistance. This work was supported by Grants GM46638 and 34431 from the National Institutes of Health.
ABBREVIATIONS
- UTR
untranslated region
- GST
glutathione S-transferase
- Sxl
Sex lethal gene
- hb
hunchback gene
- fkh
forkhead gene
- hkb
huckebein gene
- tll
tailless gene
- ems
empty spiracles gene
References
- 1.Jimenez G, Paroush Z, Ish-Horowicz D. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubnicoff T, Valentine S A, Chen G, Shi T, Lengyel J A, Paroush Z, Courey A J. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkhurst S M. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 4.Gray S, Levine M. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 5.Gray S, Levine M. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 6.Arnosti D N, Gray S, Barolo S, Zhou J, Levine M. EMBO J. 1996;15:3659–3666. [PMC free article] [PubMed] [Google Scholar]
- 7.Cai H N, Arnosti D N, Levine M. Proc Natl Acad Sci USA. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nibu Y, Zhang H, Levine M. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 9.Poortinga G, Watanabe M, Parkhurst S M. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paroush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 12.Fisher A L, Caudy M. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 13.Schaeper U, Boyd J M, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeper U, Subramanian T, Lim L, Boyd J M, Chinnadurai G. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 15.Cline T W, Meyer B J. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 16.Parkhurst S M, Bopp D, Ish-Horowicz D. Cell. 1990;63:1179–1191. doi: 10.1016/0092-8674(90)90414-a. [DOI] [PubMed] [Google Scholar]
- 17.Pignoni F, Baldarelli R M, Steingrimsson E, Diaz R J, Patapoutian A, Merriam J R, Lengyel J A. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- 18.Weigel D, Seifert E, Reuter D, Jäckle H. EMBO J. 1990;9:1199–1207. doi: 10.1002/j.1460-2075.1990.tb08227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigel D, Jurgens G, Klingler M, Jäckle H. Science. 1990;248:495–498. doi: 10.1126/science.2158673. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang A M, Rusch J, Levine M. Genes Dev. 1997;11:1963–1973. doi: 10.1101/gad.11.15.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusch J, Levine M. Development (Cambridge, UK) 1997;124:303–311. doi: 10.1242/dev.124.2.303. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Catron K M, Abate-Shen C. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small S, Blair A, Levine M. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Kosman D, Ip Y T, Levine M. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 27.Wainwright S M, Ish-Horowicz D. Mol Cell Biol. 1992;12:2475–2483. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson S R, Turner D L, Weintraub H, Parkhurst S M. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher A L, Ohsako S, Caudy M. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macdonald P M, Struhl G. Nature (London) 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 31.Parkhurst S M, Ish-Horowicz D. Development (Cambridge, UK) 1991;111:1121–1135. doi: 10.1242/dev.111.4.1121. [DOI] [PubMed] [Google Scholar]
- 32.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bopp D, Bell L R, Cline T W, Schedl P. Genes Dev. 1991;5:403–415. doi: 10.1101/gad.5.3.403. [DOI] [PubMed] [Google Scholar]
- 34.Kelley R L, Wang J, Bell L, Kuroda M I. Nature (London) 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 35.Barbash D A, Cline T W. Genetics. 1995;141:1451–1471. doi: 10.1093/genetics/141.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoshijima K, Kohyama A, Watakabe I, Inoue K, Sakamoto H, Shimura Y. Nucleic Acids Res. 1995;23:3441–3448. doi: 10.1093/nar/23.17.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estes P A, Keyes L N, Schedl P. Mol Cell Biol. 1995;15:904–917. doi: 10.1128/mcb.15.2.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walldorf U, Gehring W J. EMBO J. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wimmer E A, Jäckle H, Pfeifle C, Cohen S M. Nature (London) 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 40.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]