Abstract
Gene activation in eukaryotes requires chromatin remodeling, in part via histone modifications. To study the events at the promoter of a mitogen-inducible gene, we examined the induction of expression of the collagenase gene. It has been established that the collagenase gene can be activated by c-Jun and c-Fos and that the transcriptional coactivator p300 is involved in the activation. As expected, we found histone acetyltransferase activity at the collagenase promoter during activation. Interestingly, we also found histone methyltransferase and kinase activity. Strikingly, the first modification observed is methylation of histone H3 lysine 4, which correlates with the binding of the SET9 methyltransferase and the assembly of a complex consisting of c-Jun, c-Fos, TATA binding protein, and RNA polymerase II. The assembly of the preinitiation complex also shows an ordered binding of the acetyltransferase p300, the RSK2 kinase, and the SWI/SNF component Brg-1. Our results suggest that collagenase gene activation involves a dynamic recruitment of different factors and that in addition to acetylation, histone H3 lysine 4 di- and trimethylation and histone H3 serine 10 phosphorylation are important steps in the activation of this gene.
In eukaryotic cells, genomic DNA is packaged into nucleosomes, the basic unit of chromatin structure. The packaging of DNA into chromatin has a repressive effect on gene expression, and therefore, reconfiguration of chromatin has been postulated as mandatory for transcriptional initiation (24, 34, 45). One of the central questions in this process is how RNA polymerase and associated proteins gain access to chromatin templates. Increasing evidence indicates that gene expression is regulated by means of facilitating access of DNA-binding factors to DNA via chromatin-remodeling factors and covalent histone modifications (14).
The major chromatin-remodeling factors are the SWI/SNF ATP-dependent remodeling complexes. Upon recruitment, these complexes are thought to alter the structure of the promoter-bound nucleosome in an ATP-dependent manner (34).
The covalent-histone modifications are brought about by proteins that, upon recruitment, can phosphorylate, acetylate, and methylate histone tails (5, 41). The best-studied histone modification involved in transcriptional activation is acetylation. Levels of acetylated histones H3 and H4 have been correlated with the transcription status of many genes: transcriptionally active euchromatin regions of the genome are often associated with hyperacetylated histones, whereas transcriptionally silent regions are associated with hypoacetylated histones (17). In vivo, the steady-state levels of histone acetylation are maintained by the balance of the opposing histone acetyltransferase (HAT) and histone deacetylase activities, and these enzymes are associated with gene activation and repression, respectively (21, 44). Besides acetylation, phosphorylation of histone H3 on serine 10 is also associated with gene activation. Phosphorylation, mediated via ERK or p38 MAP kinase cascades, has thus far been associated only with rapidly inducible genes like c-fos and c-jun (7, 50). Histone methylation occurs at several lysine and arginine residues, and only recently have the roles of some of these modifications been established (3, 15, 42, 43). For example, methylation of histone H3 lysine 9 has been implicated in gene repression (3, 22, 29) whereas methylation of lysine 4 of histone H3 plays an important role in gene activation (31, 43). The mechanism by which methylation at lysine 4 of histone H3 results in transcription activation is still unclear. Methylation at histone H3 lysine 4 could mark a gene for the recruitment of complexes involved in transcription activation and/or might lead to the displacement of complexes involved in transcriptional repression such as histone deacetylases (30).
Histone-modifying complexes are thought to cooperate with histone-remodeling complexes to reconfigure chromatin, thereby establishing a local chromatin structure that is permissive for the subsequent assembly of an active preinitiation complex (PIC) at the promoter (24, 45). The in vivo sequence of nucleosomal modifying and remodeling events relative to this PIC formation and transcriptional activation has been studied for only four types of promoters. First, in the cell cycle-regulated Saccharomyces cerevisiae HO promoter, the Swi5p activator recruits the SWI/SNF remodeling complex, which then recruits the SAGA HAT complex. These two factors then facilitate the binding of a second activator (SBF) and the recruitment of the SRB/mediator complex. This is followed by the Cdk1-dependent association of RNA polymerase II to the promoter followed by transcription (6, 8, 9). Second, in the case of the virus-induced beta interferon promoter, transient H4 hyperacetylation is required for remodeling of the neighboring nucleosome, which is essential for recruitment of the general transcription factor TFIID and transcription initiation (1). Third, in order to induce expression of the differentiation marker α1-antitrypsin, a complete PIC is assembled at the promoter before recruitment of the histone acetyltransferases CBP and p/CAF. Following recruitment of the human Brahma homologue and nucleosome remodeling, transcription initiation starts (40). Finally, for cathepsin D it was shown that the hormone-activated estrogen receptor and a number of coactivators, including histone acetyltransferases CBP, p300, and the SWI/SNF component Brg-1, rapidly associate with the promoter followed by binding of RNA polymerase II and transcription (11, 38).
In this study we investigated the ordered recruitment of transcription regulatory proteins in relation to the presence of histone modifications. We have focused on the collagenase type I gene, an early gene known to be activated by c-Jun, c-Fos, and the transcriptional coactivator p300 (18, 23, 39, 52). We show that during activation of the collagenase gene multiple histone modifications occur specifically at the collagenase promoter. These modifications correlate with the assembly of a PIC consisting of c-Jun, c-Fos, RNA polymerase II, SET9, p300, RSK2, and Brg-1. The results show that activation of the collagenase gene is a coordinated event involving an ordered recruitment of DNA binding factors, remodeling enzymes, and multiple histone-modifying enzymes.
MATERIALS AND METHODS
Cells, stimulation, reverse transcription-PCR (RT-PCR), and transfection.
Human glioblastoma T98G cells were maintained on Dulbecco's modified Eagle's medium with 8% fetal calf serum (FCS). Subconfluent cells were arrested for 24 h in 0.1% FCS before stimulation for the indicated times with tetradecanoyl phorbol acetate (TPA) (Sigma) and 20% serum or, for the assay depicted in Fig. 1B, either TPA or serum. In experiments with U0126 (Promega), SB203580 (Promega), trichostatin A (TSA) (Sigma), and 5′-deoxy-(5′-methylthio)adenosine (MTA) (Sigma), the inhibitor was added as indicated in the text. RNA isolation and cDNA preparation were as described previously (26). The primers used were as follows: collagenase, GGCATGGTCCACATCTGCTC and CTCACGGACTACACCGAGTC; GAPDH, AATCCCATCACCATCTTCC and ATGAGTCCTTCCACGATACC; c-Jun, GAGCTAGCGCCTGTGGCTCC and CTCTGCCACTTGTCTCCGGTC; c-Fos, CTGGCCGTCTCCAGTGCCAAC and CATGGTCTTCACAACGCCAGC; p300, CGGGATCCGCTGCATCCAGTCTCTG and GCTCTAGATCAAGGGAGGCCCTGTTGCTG.
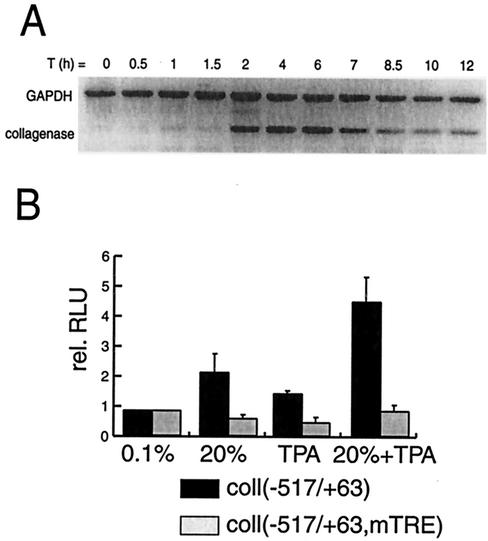
FIG. 1.
Induction of collagenase expression in T98G cells by TPA and/or serum. (A) Time course (in hours) of endogenous collagenase mRNA induction by TPA and 20% serum. Collagenase and GAPDH mRNA levels were assessed by RT-PCR. (B) Activation of transfected collagenase (−517 to +63)-Luc or collagenase (−517 to +63, mTRE)-Luc reporter in response to treatment with TPA and/or 20% serum.
Transient transfections and immunoprecipitation assays.
Transient transfections using the calcium phosphate method (16) were as described previously (19). After transfection, cells were kept on Dulbecco's modified Eagle's medium with 8% FCS for 16 h, after which they were arrested for 24 h in 0.1% FCS before subsequent stimulation. Plasmid collagenase promoter (coll −517/+63) pGL3 was described by Vries et al. (52); plasmid collagenase promoter mutant TPA response element (TRE) was constructed by cloning the promoter fragment sequence (coll −517/+63, mTRE) (32) in front of the luciferase reporter gene of pGL3. Immunoprecipitations were performed, as described before, in an assay buffer (0.1% NP-40, 250 mM NaCl, 50 mM Tris-HCl [pH 7.5]) containing a mixture of protease inhibitors (26).
ChIPs.
Chromatin immunoprecipitation assays (ChIPs) were as described previously (26). For PCR analysis, either 1/100 (input) or 1/20 (immunoprecipitates) of the DNA was amplified by using 50 pmol of the indicated primers in 40-μl reaction mixtures containing 200 μM deoxynucleoside triphosphates, 2.7 mM MgCl2, and 0.25 U of AmpliTaq (Perkin-Elmer). After 4 min at 94°C, 28 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C were performed. PCR products were electrophoresed on 2.5% agarose gels containing ethidium bromide, analyzed under UV light, and presented in inversed intensity. For the assay depicted in Fig. 3C, bands were quantified and corrected for the slight variation in input and presented in arbitrary units. All ChIP experiments presented were performed at least twice. Primers used were as follows: collagenase promoter (coll1), GTGTGTCTCCTTCGCACACATCTTG and GAGTCCTTGCCCTTCCAGAAAGCC; collagenase 3′ untranslated region (coll2), CCCAGAGAGCAGCTTCAGTGACAAAC and CCAGGGTGACACCAGTGACTGCAC.
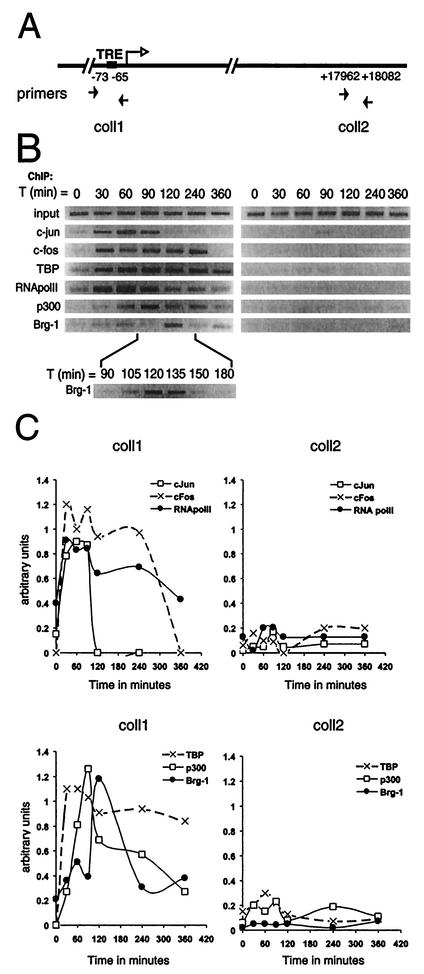
FIG. 3.
The dynamics of transcription complex assembly at the endogenous collagenase promoter. (A) Schematic representation of the collagenase promoter indicating the primers used in ChIP assays. (B) Occupancy of the collagenase promoter by c-Jun, c-Fos, TBP, RNA polymerase II, p300, and Brg-1 at different times (in minutes) after induction of T98G cells with TPA and 20% serum as assessed by ChIP. The final DNA extractions were amplified with pairs of primers that cover the regions of the collagenase gene as indicated. (C) Bands from Fig. 3B were quantified, and data were corrected for the slight variation in input and presented in arbitrary units.
Antibodies.
Antibody p300 (3) was described previously (12, 26). p300 (N15), CBP (C20), c-Jun (H79), and ATF-2 (C19) antibodies were obtained from Santa Cruz; Ac-H3 (catalog no. 06-599), Ac-H4 (catalog no. 06-866) and c-fos (catalog no. 06-341) antibodies were obtained from Upstate Biotech; P-H3 antibodies were from Cell Signaling, and Me2(K4)-H3 antibodies (ab7766) and Me3(K4)-H3 antibodies (ab8580) were from Abcam. Anti-Brg-1 (54) was a kind gift of Weidong Wang, anti-RNAPolII 8WG16 (48) was a kind gift from Marc Timmers, anti-TATA binding protein (TBP) (36) was a kind gift of Henk Stunnenberg, and anti-SET9 (53) was a kind gift from Yi Zhang. The antihistone polyclonal antiserum raised against purified Drosophila melanogaster core histones (20) was a kind gift from G. E. Chalkley and C. P. Verrijzer.
Histone extraction and Western blotting.
Acid extraction of histones was performed according to the manufacturer's protocol for acetylated histone H3 extraction (Upstate Biotech). Proteins, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were transferred onto Immobilon membranes (Millipore). Blots were blocked in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.2% Tween 20) containing 20% nonfat dried milk powder and subsequently incubated with the appropriate antibodies. After being washed, blots were incubated with peroxidase-conjugated antibodies (1:10,000; Jackson ImmunoResearch Laboratories). Blots were washed again, and immunoreactive bands were visualized by enhanced chemoluminescence.
RESULTS
Induction of collagenase expression in T98G cells.
To investigate the sequence of events occurring at the human collagenase I promoter upon induction of the gene, we used the glioblastoma cell line T98G, a cell line which can be effectively arrested by serum deprivation and subsequently stimulated by addition of serum (35, 47). Levels of endogenous collagenase mRNA were determined by RT-PCR analysis at various time points after induction with TPA and 20% serum. Collagenase mRNA was detected at 2 h after stimulation with a further increase up to 6 h, while at later time points collagenase expression was clearly reduced (Fig. 1A). Next, transcription activation of the collagenase gene was monitored by using a luciferase reporter construct containing the collagenase promoter DNA sequence spanning nucleotides −517 to +63. This reporter was transiently transfected into T98G cells, and luciferase activity was assayed 6 h after treatment with 20% serum and/or TPA. As shown in Fig. 1B, treatment with either 20% serum or TPA alone resulted in a twofold increase in luciferase expression while the combination of 20% serum and TPA resulted in a fivefold increase in expression. Mutation of the TRE abolished the stimulation by TPA and/or serum, indicating that factors binding to this DNA element play a pivotal role in the induction of this gene. These results clearly show that the collagenase gene can be readily activated in serum-arrested T98G cells after treatment with TPA and serum and that the maximal activation occurs 4 to 6 h after induction.
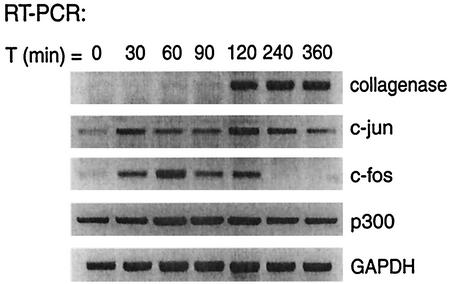
Collagenase activation has been shown to require binding of the AP-1 transcription factors c-Fos and c-Jun to the TRE. We monitored the expression of these transcription factors by RT-PCR during the first 6 h following stimulation with TPA and serum. As shown in Fig. 2, c-Fos was expressed 30 min after stimulation, reaching its highest level at 60 min. c-Jun expression was observed prior to treatment with TPA and 20% serum but was readily enhanced by stimulation with TPA and serum. The transcriptional coactivator p300, which has also been implicated in collagenase activation, is constitutively expressed during the stimulation of T98G cells. These results show that the AP-1 factors c-Fos and c-Jun and the transcriptional coactivator p300 are expressed in T98G cells after stimulation with TPA and serum.
FIG. 2.
Induction of c-Fos and c-Jun expression in T98G cells by TPA and 20% serum. Shown is the time course (in minutes) of c-Fos, c-Jun, p300, collagenase, and GAPDH mRNA expression as assessed by RT-PCR after treatment of cells with TPA and 20% serum.
p300 is targeted to the collagenase promoter during activation.
To examine the recruitment of factors like c-Jun and c-Fos to the endogenous collagenase promoter, we performed ChIP assays at different time points after induction with 20% serum and TPA. Both the collagenase promoter (coll1) and a downstream fragment encoding the 3′ untranslated region of collagenase (coll2) were examined (Fig. 3A). We first studied AP-1 factors involved in activating the collagenase promoter and found c-Fos and c-Jun to be rapidly and transiently associated with the collagenase promoter within 30 min after stimulation (Fig. 3B), indicating their involvement in the onset of collagenase transcription. Moreover, TBP and RNA polymerase II were also found to be present at the collagenase promoter within 30 min of stimulation. While RNA polymerase II and TBP were detected at 30 min following stimulation, collagenase mRNA expression was detected only after 2 h (Fig. 1A), indicating that TFIID recruitment alone is not sufficient for activation of the collagenase gene. Consistent with its presumed role in collagenase activation, the transcriptional coactivator p300 was also found at the collagenase promoter ∼60 min after stimulation. In addition, Brg-1, a component of the ATP-dependent remodeling complexes, was recruited to the collagenase promoter during a very restricted time window at ∼120 min after induction (Fig. 3B). A quantitative representation of these data clearly shows that binding of c-Jun, c-Fos, RNA polymerase II, and TBP to the collagenase promoter is followed by binding of p300 and finally Brg-1 (Fig. 3C), illustrating that an ordered recruitment of regulatory proteins is required for the onset of transcription of this gene.
Transcriptional induction of collagenase is associated with methylation, phosphorylation, and acetylation of histones at the collagenase promoter.
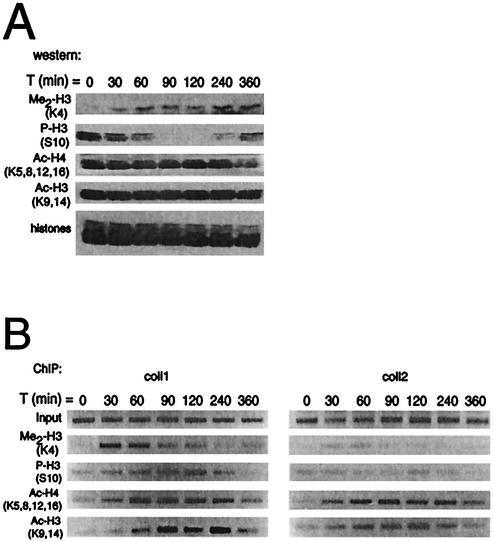
To examine if recruitment of p300 (Fig. 3B) coincides with histone acetylation, we performed additional ChIP assays. In addition, we tested whether other histone modifications like serine phosphorylation and lysine methylation might be involved in collagenase activation. First, global levels of histone modifications upon stimulation with TPA and serum were examined by Western blotting with antibodies specific for dimethylated histone H3 (Lys 4), phosphorylated histone H3 (Ser 10), acetylated histone H4 (Lys 5, 8, 12, 16), and acetylated histone H3 (Lys 9, 14). As shown in Fig. 4A, global levels of histone H3 methylated at lysine 4 increased during the time course. Surprisingly, global levels of histone H3 phosphorylation decreased dramatically within 1.5 h after stimulation but increased again at 4 h after stimulation. In agreement with other studies (4, 50), we found acetylated histone levels to be high in quiescent cells, and serum and TPA stimulation did not significantly change this (Fig. 4A).
FIG. 4.
The dynamics of histone modifications after collagenase induction. (A) Western blot analysis of total levels of dimethylated histone H3 (Me2-H3), phosphorylated histone H3 (P-H3), acetylated histones H4 (Ac-H4) and H3 (Ac-H3) in histone preparations of T98G cells following stimulation with TPA and 20% serum for the indicated times. (B) Occupancy of the collagenase gene by dimethylated histone H3, phosphorylated histone H3, acetylated histones H4 and H3 in T98G cells at different times following TPA and serum treatment as assessed by ChIP. The final DNA extractions were amplified with pairs of primers that cover regions of the collagenase gene as indicated in Fig. 3A.
Next, we performed ChIP assays with the same antibodies to examine histone modifications that are specifically associated with induction of the collagenase gene. Strikingly, the earliest modification that we observed at the collagenase promoter was histone H3 lysine 4 dimethylation (Fig. 4B; coll1). This modification was predominantly promoter specific, as only very low levels of methylation were detected in the region of the collagenase gene encoding the 3′ untranslated region (coll2). ChIP assays with antibodies against phosphorylated histone H3 showed a considerable increase of phosphorylated histone H3 at the collagenase promoter over time. This increase is in sharp contrast with the overall levels of phosphorylated histone H3, which were dramatically decreased at time points when specific occupancy is highest at the collagenase promoter (90 to 120 min after induction). Together with the absence of histone H3 phosphorylation downstream of the collagenase gene, these findings suggest that serine phosphorylation at the collagenase promoter is a targeted event. Acetylated histones H3 and H4 could also be detected at the collagenase promoter, as would be expected from the recruitment of a HAT enzyme to the collagenase promoter (Fig. 3B). The time points at which acetylation of histones H3 and H4 were observed (from 30 to 90 min) correlate with the appearance of p300 (Fig. 3B). Methylated and phosphorylated histones were not observed downstream of the collagenase gene, whereas acetylation, possibly due to HAT activities accompanying elongating polymerase II (33), was observed. Taken together, these results suggest that nucleosomal changes mediated by histone modifications play an important role in mediating the transcriptional activation of the collagenase gene. These effects are promoter specific, since the levels of global modifications do not correspond to the observed site-specific alterations. Moreover, lysine 4 dimethylation of histone H3 precedes histone acetylation and phosphorylation, suggesting that for collagenase expression histone methylation is the first step in gene induction. The cascade of histone modifications at the promoter suggests that gene activation is an ordered event in which different factors are recruited sequentially, resulting in transcription.
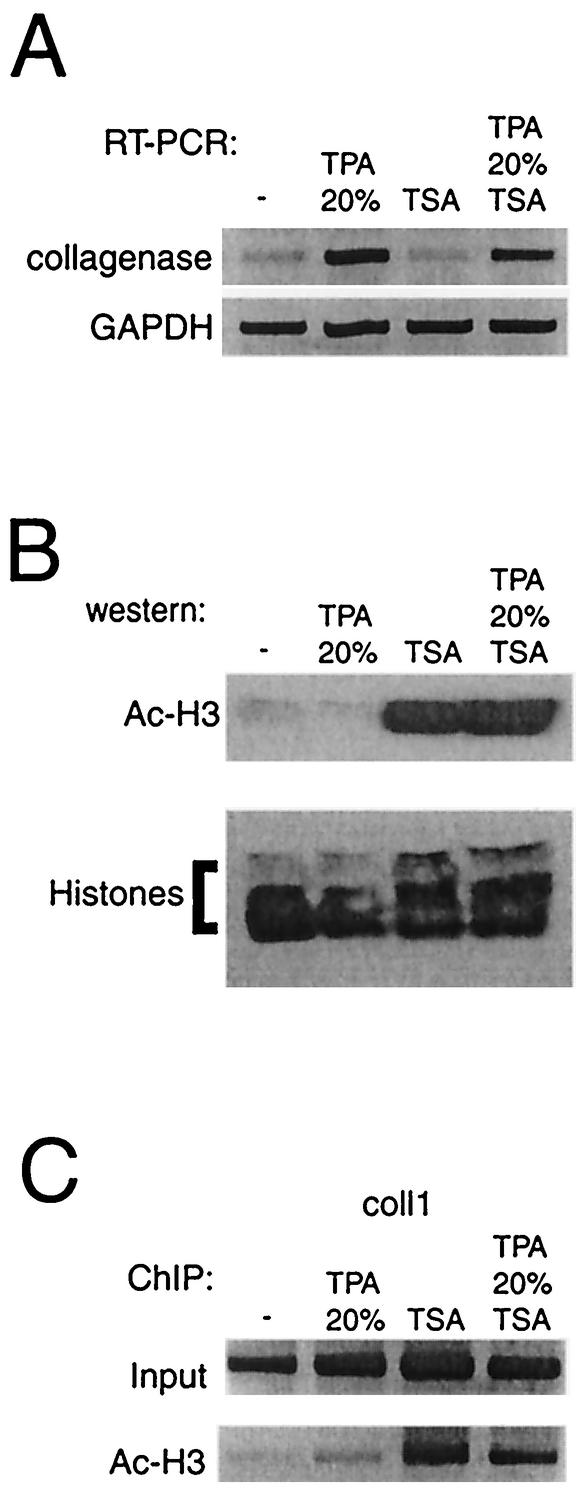
Histone deacetylase inhibitor TSA does not augment collagenase activation.
As shown in Fig. 3B, the HAT enzyme p300 is recruited to the collagenase promoter. To assess the effect of histone acetylation alone on collagenase expression, we inhibited deacetylation of the collagenase promoter by using the inhibitor TSA. RT-PCR shows that TSA alone did not enhance basal collagenase mRNA levels (Fig. 5A) while treatment of cells with TSA did augment both global histone H3 acetylation (Fig. 5B) and local histone H3 acetylation levels at the collagenase promoter (Fig. 5C). These findings suggest that histone acetyltransferase recruitment is not sufficient for collagenase gene expression, suggesting that other histone-configuring activities like methylation, phosphorylation, and remodeling are also necessary.
FIG. 5.
TSA does not augment collagenase activation in T98G cells. (A) Collagenase mRNA expression after treatment of cells for 6 h with TSA, TPA and 20% serum, or both. (B) Western blot analysis of acetylated histone H3 present in total histone preparations of cells after treatment with TSA. (C) ChIP analysis of occupancy of the collagenase gene by acetylated histone H3. The final DNA extracts were amplified with a pair of primers that cover the region of the collagenase promoter as indicated in Fig. 3A.
p300 and RSK2 are simultaneously recruited to the collagenase promoter.
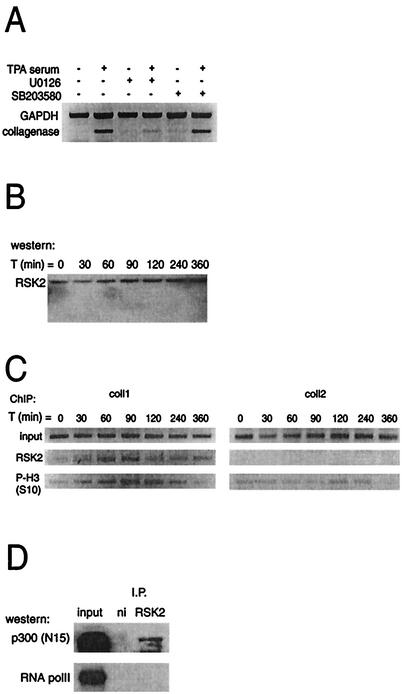
The phosphorylation of histone H3 at the collagenase promoter indicates that a kinase is recruited to this site. Two candidate histone kinases have been identified: MSK1/2, which is phosphorylated and activated by ERK or p38 (10, 49), and RSK2, which is activated only by ERKs (37, 51). Using the mitogen-activated kinase/ERK kinase 1/2 (MEK1/2) inhibitor U0126, we found that inhibition of MEK1/2 blocked collagenase activation whereas blocking p38 with the inhibitor SB203580 had no effect (Fig. 6A). Since the latter compound readily inhibited MAPKAP-2 phosphorylation (data not shown), these results suggest that activation of RSK2 via ERK is involved in activation of the collagenase gene. U0126 has also been shown to inhibit expression of c-Fos (13), but this is not the primary mechanism of inhibition by U0126 in this case, since addition of U0126 following c-Fos expression (e.g., 1.5 h after induction [Fig. 2]) also prevented collagenase expression (data not shown). Furthermore, by using ChIPs we could indeed show that RSK2, which is constitutively expressed in T98G cells (Fig. 6B), is bound to the collagenase promoter, a binding that correlated with the appearance of histone H3 phosphorylation (Fig. 6C). Since the p300 homologue CBP has been shown to bind RSK2 (27), it is conceivable that RSK2 is corecruited with p300 to the collagenase promoter. Figure 6D shows that p300 and RSK2 indeed could be coimmunoprecipitated from lysates prepared from cells after stimulation (Fig. 6D). As a negative control, the same blot was probed with antibodies against RNA polymerase II, showing no interaction between RNA polymerase II and RSK2. Taken together, these results strongly suggest that the kinase involved in activation of the collagenase gene is RSK2, which might be recruited by p300 to the collagenase promoter.
FIG. 6.
RSK2 is recruited to the endogenous collagenase promoter. (A) Effects of inhibitors U0126 (10 μM) and SB203580 (10 μM) on collagenase mRNA induction. The expressions of collagenase and GAPDH mRNA were assessed by RT-PCR. The inhibitors were added with TPA and serum treatment. (B) Western blot analysis of RSK2 expression at different times after treatment. (C) ChIP analysis of occupancy of the collagenase promoter by RSK2 and phosphorylated histone H3 (P-H3). The final DNA extracts were amplified with pairs of primers that cover regions of the collagenase gene as indicated in Fig. 3A. (D) Endogenous p300 coimmunoprecipitates with RSK2. T98G whole-cell extracts stimulated for 1.5 h with TPA and 20% serum were immunoprecipitated with antibodies against RSK2 and a nonimmune control (ni) and tested by Western blotting for coimmunoprecipitated p300 and RNA polymerase II.
The lysine 4 histone H3-specific methyltransferase, SET9, is recruited to the collagenase promoter.
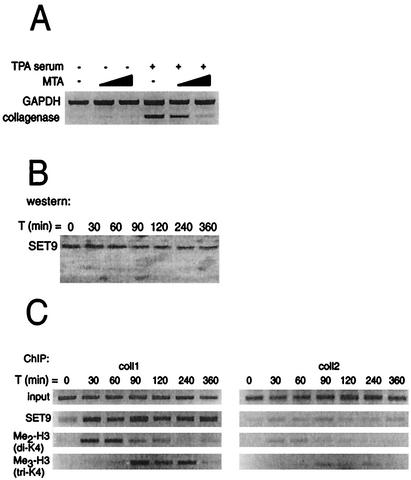
We have established that histone kinase and acetyltransferase activity are recruited to and modulate the collagenase promoter. We next assessed histone methylation activity at the promoter. First, we examined the importance of methyltransferase activity in collagenase activation by using the specific protein methyltransferase inhibitor, MTA (25, 28, 55). As shown in Fig. 7A, pretreatment of T98G cells with increasing amounts of MTA was able to efficiently block induction of collagenase expression, suggesting that methyltransferase activity is indeed involved in collagenase activation. Only one enzyme possessing specific histone H3 lysine 4 methyltransferase activity has been described thus far, SET9 (30, 53), which is present in T98G cells (Fig. 7B). By using ChIP assays, SET9 binding was detected early during collagenase activation, concomitant with the appearance of methylated histone H3 at the promoter site (Fig. 7C). Strikingly, SET9 appears to be present at the collagenase promoter when dimethylation of histone H3 has decreased. To examine whether this is due to subsequent trimethylation at the same lysine residue, we performed additional ChIP assays using an antibody specifically recognizing trimethylated lysine 4 of histone H3. As shown in Fig. 7C, we observed trimethylated histone H3 at the collagenase promoter while only very low levels of methylation were detected in the region of the collagenase gene encoding the 3′ untranslated region (coll2). Note that the appearance of trimethylated histone H3 was observed at time points when the signal with the dimethylated histone H3 antibody decreased. These results therefore suggest that the histone H3 lysine 4 methyltransferase involved in activation of the collagenase gene is SET9, which is recruited early after stimulation to the collagenase promoter and that, in addition to dimethylation, trimethylation of lysine 4 of histone H3 is also involved in collagenase activation.
FIG. 7.
SET9 recruitment to the endogenous collagenase promoter correlates with the presence of methylated histone H3. (A) Effect of the methyltransferase inhibitor MTA on collagenase induction. Collagenase and GAPDH mRNA were assessed by RT-PCR. MTA (1 and 2 mM) was added 1.5 h before TPA and serum treatment. (B) Western blot analysis of SET9 expression at different times after treatment. (C) ChIP analysis of occupancy of the collagenase promoter by SET9 and di- and trimethylated histone H3 (Me2-H3; Me3-H3). The final DNA extracts were amplified with pairs of primers that cover regions of the collagenase gene as indicated in Fig. 3A.
DISCUSSION
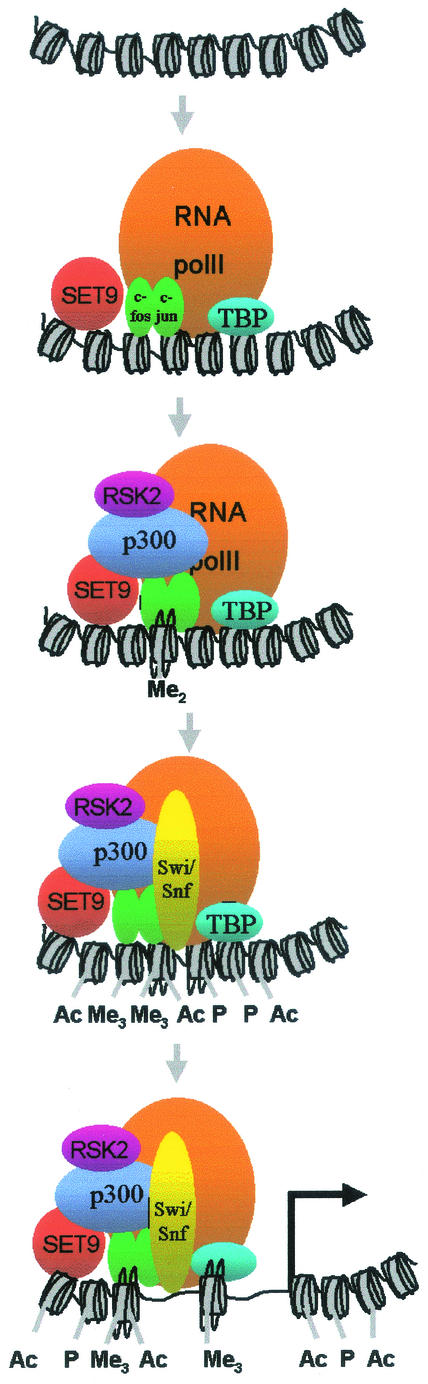
In order to analyze the events occurring at the promoter of a gene upon mitogenic stimulation, we investigated the mitogen-inducible collagenase promoter. Using ChIPs, we established that first the transcription factors c-Jun and c-Fos, the basal transcription factor TBP, RNA polymerase II, and the methyltransferase SET9 are recruited to the collagenase promoter (Fig. 8). Simultaneous with the appearance of SET9 at the collagenase promoter, first dimethylation and later trimethylation of lysine 4 of histone H3 are observed. Next, the HAT p300 and the kinase RSK2 are recruited. The recruitment of p300 might occur via its interaction with c-Jun (23), while RSK2 might be recruited via its interaction with p300 (Fig. 6D). The presence of p300 and RSK2 at the collagenase promoter correlates with the appearance of acetylated and phosphorylated histones. Finally, the cascade of recruitment events culminates with the association of the remodeling complex component Brg-1, which coincides with initiation of transcription. The timing of recruitment suggests that recruitment of the nucleosome remodeling machinery requires prior histone methylation, acetylation, and phosphorylation of the nucleosome.
FIG.8.
Model depicting the ordered recruitment of chromatin-modifying and basal factors to the collagenase promoter. Shortly after TPA and 20% serum treatment, c-Jun, c-Fos, TBP, RNA polymerase II, and SET9 assemble on the collagenase promoter and histones are first dimethylated and later trimethylated. Next, p300 and RSK2 are recruited, and this event correlates with the appearance of acetylated and phosphorylated nucleosomes. Finally, SWI/SNF associates with the promoter, thereby completing PIC formation, resulting in initiation of transcription.
Thus far, the in vivo sequence of nucleosomal modifying and remodeling events leading to transcriptional activation has been studied for four types of promoters: the cell cycle-regulated yeast HO promoter, the virus-induced beta interferon promoter, the differentiation-induced α1-antitrypsin promoter, and the hormone-induced cathepsin D promoter (see the introduction). The recruitment results described here predominantly resemble the situation described for the α1-antitrypsin gene (40), in which TBP and RNA polymerase II appear at the promoter before transcriptional activation. Transcription is started only after the recruitment of HAT and remodeling complexes, suggesting that at these promoters chromatin reconfiguration is the defining step of the initiation process. Interestingly, although the recruitment of factors is an ordered event for all promoters examined so far, the order and timing of recruitment are different in all cases (HO, beta interferon, α1-antitrypsin, cathepsin D, and collagenase), suggesting that each gene has its own order and timing of events leading to transcription.
The chromatin reconfigurations that are required for collagenase expression to be induced are most likely brought about by nucleosomal modifications. Two observations are relevant in this respect. First, the modifications at the collagenase gene are controlled at the local level and do not reflect the average level of these modifications in the cell (for example, compare in Fig. 5 the total level of phosphorylated histone H3 with phosphorylated histone H3 levels at the collagenase promoter). Second, it seems that different histone modifications follow each other during gene activation and there is no single modification event associated with gene activation. While acetylation and phosphorylation represent late modifications, methylation is an early event. As methylation of lysine 4 has been suggested not only to preclude repression complex formation but also to be involved in the recruitment of other chromatin-modifying enzymes (30), it might be the earliest step in collagenase gene activation. This is supported by the observation that lysine-4-methylated histone H3 is a preferred target for subsequent acetylation over unmodified histone H3 (53). SET9 recruitment prior to association of p300 as observed at the collagenase promoter (Fig. 3 and 7) supports this model.
The nucleosomal changes observed at the collagenase promoter are brought about by specific histone-modifying factors. Apparently, in each case recruitment is sufficient to get enzymatic activity, since the presence of the kinase, acetyltransferase, and methyltransferase activity coincided with the appearance of the associated histone modification. However, RSK2 and SET9 appear to be still present at the collagenase promoter when the modification of histones by these enzymes is already decreasing. This might suggest the involvement of specific demethylase and phosphatase activities, but it could also be due to the fact that more modifications occur at the single histone tail (2, 50, 53). Hence, the presence of RSK2 and SET9 at the promoter could still reflect enzymatically active molecules but proof of functionality would require the use of antibodies that are not affected by additional modifications. Our results also indicate that p300 and RSK2 are both enzymatically active at the collagenase promoter, possibly as a complex. This is in apparent contrast to a study by Merienne et al. (27) that showed the preferential interaction of the p300 homologue CBP with unphosphorylated RSK2, forming a complex in which both kinase activity and acetyltransferase activity are inhibited. This apparent contradiction might be explained by the differences between p300 and CBP as well as by cell type-specific effects, affecting both binding and activity. For example, under conditions where we demonstrate binding of p300 to RSK2 (Fig. 6D) we could not detect an interaction between CBP and RSK2 (data not shown).
One of the questions remaining is how SET9 is recruited to the collagenase promoter. As our ChIP results indicate that SET9 is not present before stimulation (T = 0), it should be targeted to the collagenase promoter. One possibility is that SET9 associates with a putative repression complex present at the collagenase promoter before stimulation. In this complex it could methylate lysine 4 of histone H3 and thereby prevent the repression complex from binding to the collagenase promoter. Alternatively, SET9 could be recruited by another complex containing specific DNA binding factors. Finally, SET9 might also be a component of the PIC, thereby precluding formation of a repression complex and stimulating PIC formation at the collagenase promoter. As our ChIP assays indicate that SET9 is present at the collagenase promoter during the entire PIC formation, we favor the last explanation, although our attempts to identify the specific recruitment factor have thus far failed. Consistent with the idea of SET9 being a component of the PIC is the observation that SET9 can methylate only core histones and not nucleosomes (30, 53), implying that SET9 functions in concert with other chromatin-reconfiguring enzymes, as was recently shown for the yeast homologue SET1 (46). Together with histone acetylation and phosphorylation, SET9-mediated histone methylation would then create the conditions which preclude formation of repression complexes and stimulate stable PIC assembly and subsequent transcription activation.
In conclusion, we show for the first time that in addition to the well-established requirement of acetylation, induction of collagenase expression also involves di- and trimethylation of lysine 4 of histone H3 and phosphorylation of serine 10 of histone H3. These modifications are brought about by different histone-modifying enzymes like SET9, p300, and RSK2, which are recruited sequentially to the collagenase promoter culminating in the initiation of transcription.
Acknowledgments
We thank M. Timmers, H. Stunnenberg, G. E. Chalkley, W. Wang, and Y. Zhang for gifts of antibodies and D. Baker, A. J. van der Eb, and C. P. Verrijzer for helpful discussions.
This work was supported by the Counsel Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW).
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Barratt, M. J., C. A. Hazzalin, E. Cano, and L. C. Mahadevan. 1994. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc. Natl. Acad. Sci. USA 91:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton, A. L., S. Rose, M. J. Barratt, and L. C. Mahadevan. 2000. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 9.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 10.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiRenzo, J., Y. Shang, M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20:7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duyndam, M. C., H. van Dam, P. H. Smits, M. Verlaan, A. J. van der Eb, and A. Zantema. 1999. The N-terminal transactivation domain of ATF2 is a target for the co-operative activation of the c-jun promoter by p300 and 12S E1A. Oncogene 18:2311-2321. [DOI] [PubMed] [Google Scholar]
- 13.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 14.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 15.Gary, J. D., and S. Clarke. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61:65-131. [DOI] [PubMed] [Google Scholar]
- 16.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 17.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 18.Hagmeyer, B. M., H. Konig, I. Herr, R. Offringa, A. Zantema, A. van der Eb, P. Herrlich, and P. Angel. 1993. Adenovirus E1A negatively and positively modulates transcription of AP-1 dependent genes by dimer-specific regulation of the DNA binding and transactivation activities of Jun. EMBO J. 12:3559-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalkhoven, E., H. Teunissen, A. Houweling, C. P. Verrijzer, and A. Zantema. 2002. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol. Cell. Biol. 22:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsani, K. R., J. J. Arredondo, A. J. Kal, and C. P. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornberg, R. D., and Y. Lorch. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9:148-151. [DOI] [PubMed] [Google Scholar]
- 22.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J.-S., R. H. See, T. Deng, and Y. Shi. 1996. Adenovirus E1A downregulates cJun- and JunB-mediated transcription by targeting their coactivator p300. Mol. Cell. Biol. 16:4312-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 25.Maher, P. A. 1993. Inhibition of the tyrosine kinase activity of the fibroblast growth factor receptor by the methyltransferase inhibitor 5′-methylthioadenosine. J. Biol. Chem. 268:4244-4249. [PubMed] [Google Scholar]
- 26.Martens, J. H., M. Verlaan, E. Kalkhoven, J. C. Dorsman, and A. Zantema. 2002. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol. Cell. Biol. 22:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merienne, K., S. Pannetier, A. Harel-Bellan, and P. Sassone-Corsi. 2001. Mitogen-regulated RSK2-CBP interaction controls their kinase and acetylase activities. Mol. Cell. Biol. 21:7089-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mowen, K. A., J. Tang, W. Zhu, B. T. Schurter, K. Shuai, H. R. Herschman, and M. David. 2001. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 104:731-741. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 30.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 32.Offringa, R., S. Gebel, H. van Dam, M. Timmers, A. Smits, R. Zwart, B. Stein, J. L. Bos, A. van der Eb, and P. Herrlich. 1990. A novel function of the transforming domain of E1a: repression of AP-1 activity. Cell 62:527-538. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 34.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 35.Potapova, O., S. Basu, D. Mercola, and N. J. Holbrook. 2001. Protective role for c-Jun in the cellular response to DNA damage. J. Biol. Chem. 276:28546-28553. [DOI] [PubMed] [Google Scholar]
- 36.Ruppert, S. M., V. McCulloch, M. Meyer, C. Bautista, M. Falkowski, H. G. Stunnenberg, and N. Hernandez. 1996. Monoclonal antibodies directed against the amino-terminal domain of human TBP cross-react with TBP from other species. Hybridoma 15:55-68. [DOI] [PubMed] [Google Scholar]
- 37.Sassone-Corsi, P., C. A. Mizzen, P. Cheung, C. Crosio, L. Monaco, S. Jacquot, A. Hanauer, and C. D. Allis. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285:886-891. [DOI] [PubMed] [Google Scholar]
- 38.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 39.Smits, P. H., L. de Wit, A. J. van der Eb, and A. Zantema. 1996. The adenovirus E1A-associated 300 kDa adaptor protein counteracts the inhibition of the collagenase promoter by E1A and represses transformation. Oncogene 12:1529-1535. [PubMed] [Google Scholar]
- 40.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 41.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 42.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 43.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 45.Struhl, K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98:1-4. [DOI] [PubMed] [Google Scholar]
- 46.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, N. E., T. H. Steinberg, D. B. Aronson, and R. R. Burgess. 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 264:11511-11520. [PubMed] [Google Scholar]
- 49.Thomson, S., A. L. Clayton, C. A. Hazzalin, S. Rose, M. J. Barratt, and L. C. Mahadevan. 1999. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 18:4779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomson, S., A. L. Clayton, and L. C. Mahadevan. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 8:1231-1241. [DOI] [PubMed] [Google Scholar]
- 51.Trivier, E., D. De Cesare, S. Jacquot, S. Pannetier, E. Zackai, I. Young, J. L. Mandel, P. Sassone-Corsi, and A. Hanauer. 1996. Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature 384:567-570. [DOI] [PubMed] [Google Scholar]
- 52.Vries, R. G., M. Prudenziati, C. Zwartjes, M. Verlaan, E. Kalkhoven, and A. Zantema. 2001. A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J. 20:6095-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 54.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 55.Williams-Ashman, H. G., J. Seidenfeld, and P. Galletti. 1982. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem. Pharmacol. 31:277-288. [DOI] [PubMed] [Google Scholar]