Abstract
The Saccharomyces cerevisiae ATP-binding cassette (ABC) transporter Pdr12p effluxes weak acids such as sorbate and benzoate, thus mediating stress adaptation. In this study, we identify a novel transcription factor, War1p, as the regulator of this stress adaptation through transcriptional induction of PDR12. Cells lacking War1p are weak acid hypersensitive, since they fail to induce Pdr12p. The nuclear Zn2Cys6 transcriptional regulator War1p forms homodimers and is rapidly phosphorylated upon sorbate stress. The appearance of phosphorylated War1p isoforms coincides with transcriptional activation of PDR12. Promoter deletion analysis identified a novel cis-acting weak acid response element (WARE) in the PDR12 promoter required for PDR12 induction. War1p recognizes and decorates the WARE both in vitro and in vivo, as demonstrated by band shift assays and in vivo footprinting. Importantly, War1p occupies the WARE in the presence and absence of stress, demonstrating constitutive DNA binding in vivo. Our results suggest that weak acid stress triggers phosphorylation and perhaps activation of War1p. In turn, War1p activation is necessary for the induction of PDR12 through a novel signal transduction event that elicits weak organic acid stress adaptation.
Weak organic acids are naturally occurring compounds that prevent microbial growth (40). For instance, sorbate, benzoate and propionate are commonly used as preservatives in the food and beverage industries. At low pH, weak acids exist mainly in their undissociated RCOOH state, in which they enter the cell by passive diffusion. The higher intracellular pH dissociates weak acids, generating protons and RCOO− anions that accumulate intracellularly. The antimicrobial effects of these acids are generally attributed to the release of protons and subsequent cytoplasmic acidification, which inhibit essential metabolic functions (24). In addition, the intracellular anion accumulation exerts inhibitory effects on glycolysis, as benzoate-treated cells suffer from severe energy depletion, partly caused by inhibition of phosphofructokinase (29). More-lipophilic weak acids such as sorbate and benzoate may cause membrane-disruptive effects, which, in the presence of oxygen, lead to endogenous production of superoxide free radicals (31, 33).
Saccharomyces cerevisiae can be induced to adapt to weak acid stress by exogenous sorbate or benzoate. S. cerevisiae cells usually exit the cell cycle and enter a long period of stasis. Eventually, after several hours, they become adapted, allowing cells to resume growth (18, 32). Stress adaptation sometimes allows yeasts such as various Saccharomyces and Zygosaccharomyces spp. to grow in the presence of the highest concentrations of weak acids allowed in food preservation, leading to food spoilage and enormous economic losses (6).
In S. cerevisiae sorbate adaptation utilizes induction of two plasma membrane proteins, the heat shock protein Hsp30p (34) and the ATP-binding cassette (ABC) anion efflux pump Pdr12p (32). It also involves activation of the Pma1p plasma membrane H+-transporting ATPase, since cells with reduced Pma1p levels display weak acid hypersensitivity (19). While Pdr12p is present at very low levels in unstressed cells, it becomes one of the most abundant plasma membrane proteins upon sorbate stress (31, 32). Pdr12p is essential for growth in the presence of weak organic acids, as cells lacking this efflux pump are hypersensitive to water-soluble monocarboxylic acids with chain lengths from C1 to C7 (18). Notably, the apparent substrate spectrum of Pdr12p shows no overlap with those of other well-characterized yeast ABC drug efflux pumps such as Pdr5p (2, 3), Snq2p (38), and Yor1p (22). These pumps are the main mediators of pleiotropic drug resistance in yeast (47), displaying a broad substrate specificity with hundreds of different hydrophobic drug substrates (23). In contrast, the Pdr12p substrate spectrum appears distinct and rather narrow, since it effluxes mainly weak organic acid anions such as benzoate, sorbate, and fluorescein (18).
The family of Zn2Cys6 zinc finger regulators constitutes a major class of fungal transcriptional regulators, with 54 putative members in S. cerevisiae. These regulators control a wide variety of cellular processes, including galactose metabolism (25), pleiotropic drug resistance (47), amino acid metabolism (13), and respiration (30). Notably, certain regulators exhibit both activating and repressing effects on their target genes (41). In Gal4p, the best-studied family member, the DNA-binding zinc finger is close to the N terminus, immediately followed by a coiled-coil motif that mediates homodimerization (27). The C terminus usually contains an activation domain (37), and the large, less-conserved middle homology region may regulate transcriptional activity through an interaction with the C terminus.
The induction of PDR12 following weak acid stress is extremely rapid, causing, within a few minutes, Pdr12p levels comparable to those of the most abundant plasma membrane protein, Pma1p (31, 32). However, the signal transduction mechanisms and the transcription factors mediating Pdr12p induction have remained obscure. In this study, we identify a novel nuclear Zn2Cys6 zinc finger transcription factor, War1p, as the only mediator of Pdr12p stress induction. War1p is required for Pdr12p induction in response to weak organic acid stress, and its absence causes hypersensitivity to a range of acids. Activated War1p acts through a novel cis-acting weak acid response element (WARE) in the PDR12 promoter, which is necessary and sufficient for PDR12 induction. This is the first report of a eukaryotic transcription factor dedicated to the adaptation to weak organic acid stress. Its posttranslational activation is also specifically triggered by exposure to the same stress conditions.
MATERIALS AND METHODS
Growth conditions and cytotoxicity assays.
Rich medium (yeast extract-peptone-dextrose [YPD]) and synthetic complete medium (SC) were prepared as described elsewhere (21). Unless otherwise indicated, all yeast strains were grown routinely at 30°C. Tests for weak acid resistance phenotypes were performed with cells grown to the exponential growth phase and diluted to an optical density at 600 nm (OD600) of 0.2. Identical volumes of cultures, as well as 1:10, 1:100, and 1:1,000 serial dilutions, were spotted onto agar plates containing various concentrations of weak acids. Colony growth was inspected after a 48-h incubation at 30°C.
Yeast strains and plasmid constructions.
All yeast strains used in this study are listed in Table 1. Gene deletions were carried out by a PCR-based method using disruption cassettes derived from plasmid pFA6a-HIS3Mx6 (42). Epitope tagging of YML076c (WAR1) at the C terminus with a triple-hemagglutinin (HA) or green fluorescent protein (GFP) tag or 13 copies of a c-myc tag used plasmids pFA6a-3HA-KanMx6, pFA6a-GFP-His3Mx6, and pFA6a-13-c-myc-TRP1, respectively, and routine PCR methods with appropriate oligonucleotide primers (26). YAK3 was obtained by genomic integration into URA3 of a pRS316-based reporter plasmid driving lacZ expression from a 1,000-bp PDR12 promoter. Transformation of yeast strains was as described elsewhere (21). Correct genomic integration was verified by PCR, and functionality of fusion proteins was tested by determining their ability to restore sorbate tolerance. Screening of transcription factor deletion strains for their Pdr12p expression levels employed the EUROSCARF (Frankfurt, Germany) collection.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| FY1679-28c | MATaura3-52 his3-Δ200 leu2-Δ1 trp1-Δ63 | 32 |
| YAK100 | Isogenic to FY1679-28c; Δwar1::His3Mx6 | This study |
| YAK101 | Isogenic to FY1679-28c; WAR1-3HA-Kan-Mx4 | This study |
| YCG1 | Isogenic to FY1679-28c; WAR1-13c-myc-TRP1 | This study |
| W303-1A | MATaura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 | 32 |
| YBB14 | Isogenic to W303-1A; Δpdr12::hisG-URA3-hisG | This study |
| YBB15 | Isogenic to W303-1A; Δpdr12::hisG-URA3-hisG Δwar1::HIS3-Mx6 | This study |
| YAK120 | Isogenic to W303-1A; Δwar1::HIS3-Mx6 | This study |
| YAK3 | Isogenic to W303-1A; ura3-52::PDR12-lacZ-URA3 | This study |
| YAK122 | Isogenic to W303-1A; WAR1-GFP-HIS3-Mx6 | This study |
| YAK3 | Isogenic to W303-1A; ura3-52::PDR12-lacZ-URA3 | This study |
| BY4741 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 | EUROSCARF collection |
| Y06734 | Isogenic to BY-4741; Δwar1::Kan-Mx4 | EUROSCARF collection |
A 4.32-kb PCR fragment containing the entire WAR1 open reading frame (ORF), as well as 1 kb of the 5′ region and 0.45 kb of the 3′ region was isolated by PCR using genomic DNA from W303-1A. The PCR fragment was cloned after a partial digestion with EcoRI into the corresponding sites of pRS313 (39) and YEp352 (17), resulting in the CEN plasmid pYA313WAR1 and the multicopy plasmid pYA352WAR1, respectively. The WAR1 plasmids were sequenced to exclude PCR errors. To construct plasmid pYA22 carrying a glutathione S-transferase (GST)-War1p fusion protein, the complete WAR1 ORF from start to stop was amplified by PCR from pYA313WAR1, introducing EcoRI sites via the primers. The PCR fragment was cloned into the EcoRI site of pGEX-3X.
For the deletion analysis of the PDR12 promoter, PDR12 upstream regions were amplified by PCR using genomic DNA isolated from strain FY1679-28c and oligonucleotide primers containing EcoRI or BamHI cleavage sites. PCR products were digested and ligated into plasmid pUP41a (44), thereby replacing the HSP12 promoter of the YCp50-based plasmid by various upstream regions of PDR12. The plasmid containing the longest PDR12 promoter fragment (−772 to −1) was named pPWP1. For the construction of plasmids containing the weak acid-responsive region of PDR12, we used appropriate oligonucleotides spanning the PDR12 promoter region from −646 to −603 and introduced XhoI/BglII sites for simplified cloning. The annealed double-stranded oligonucleotides were cloned into the corresponding sites of the disabled CYC1 promoter of the CEN-based TRP1 vector pRW95-W1 (45). The resulting plasmid, pRW95-3, contained the wild-type WARE sequence, which was verified by DNA sequencing.
β-Galactosidase measurements and RNA methods.
Cells carrying a PDR12-lacZ reporter gene were grown in YPD medium to the mid-exponential growth phase. Cells were treated with various compounds or left untreated. β-Galactosidase assays were carried out in triplicate either with total-cell extracts (21) or with permeabilized cells (1).
Preparation of RNA for Northern analysis and fractionation by agarose gel electrophoresis were done by routine methods previously described (46). Nylon membranes were prehybridized in 10 ml containing 10× Denhardt's buffer (1 g of Ficoll 400, 1 g of polyvinylpyrrolidone, 1% [wt/vol] bovine serum albumin fraction V), 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% sodium dodecyl sulfate (SDS), and 20 μg of salmon sperm DNA/ml for 3 h at 65°C. Appropriate DNA fragments from PDR12, WAR1, and ACT1 were radiolabeled and directly added to the prehybridization solution after purification on NICK columns (Pharmacia) and subsequent heat denaturation. After an overnight incubation at 65°C, membranes were washed three times with 2× SSC-1% SDS and three times with 1× SSC-1% SDS at 65°C, followed by detection with the STORM phosphorimager (Molecular Dynamics).
Preparation of cell extracts for immunoblotting and phosphatase treatment.
The preparation of cell extracts for immunoblotting was performed exactly as described elsewhere (10). Cell lysates from solutions with an OD600 of 0.5 were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on a 7.5% gel and transferred to nitrocellulose membranes. War1p and Pdr12p were visualized on immunoblots with the ECL chemiluminescence detection system (Amersham) and appropriate antibodies, such as anti-HA antibody 12CA5, or polyclonal anti-Pdr12p antisera (32).
For phosphatase treatment, 5 to 10 OD600 equivalents of cells were harvested, washed, and resuspended in 150 μl of chilled breaking buffer I (50 mM Tris-HCl [pH 7.6], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride), followed by addition of an equal volume of prechilled glass beads. Cell lysis was achieved by vigorous vortex mixing four times for 1 min each, with 1 min on ice in between. After the addition of 150 μl of breaking buffer II (breaking buffer I containing 2% SDS), cell lysis was completed by vortex mixing for another 2 min. Cell extracts were cleared by a centrifugation step at 13,000 × g for 5 min. For calf intestinal phosphatase (CIP) treatment, 30 μl of lysate was treated with 2 U of alkaline phosphatase in a total volume of 500 μl (50 mM Tris-HCl [pH 7.5], 1 mM MgCl2). Mock phosphatase treatment was performed in the absence of the CIP enzyme. Phosphatase treatment was also done in the presence of 10 mM sodium orthophosphate as an inhibitor. After 1 h of incubation at 37°C, proteins were precipitated by addition of 500 μl of cold 50% (wt/vol) trichloroacetic acid, heated in SDS-PAGE sample buffer (40 mM Tris-HCl [pH 6.8], 8 M urea, 5% SDS, 0.1 mM EDTA, 2% mercaptoethanol, 0.01% bromphenol blue), resolved by SDS-PAGE, and subjected to immunoblotting.
In vitro GST pull-down assays.
For in vitro binding assays, FY1679-28c (wild-type) cells and YCG1 cells expressing War1p-c-Myc were grown in 100 ml of YPD to an OD600 of 1.0, harvested, and washed. Cells were lysed with glass beads in 5 ml of buffer G (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1 mM ZnSO4, 1 mM phenylmethylsulfonyl fluoride, 0.5% NP-40) as described above. To assay the interaction of yeast War1p-c-Myc with bacterially derived GST-War1p, bacterial lysates containing either GST alone or GST-War1p were attached to glutathione-Sepharose beads as previously described (7). The beads were extensively washed, and 80-μl resin aliquots corresponding to 10 μg of the GST-War1p fusion protein were incubated for 2 h at 4°C with total yeast extract corresponding to an OD600 of 5. After the beads were washed four times with 2 ml of buffer G, bound proteins were resolved in 40 μl of 2× Laemmli buffer, separated by 10% SDS-PAGE, and further analyzed by immunoblotting using monoclonal 9E10 anti-c-Myc antibodies.
Fluorescence microscopy.
Fluorescence experiments were performed as described previously (14). Briefly, YAK122 cells were grown in YPD medium and stressed with 8 mM sorbate for 2 h or left untreated and War1p-GFP was visualized in living cells without prior fixation. Nuclear DNA was stained for 10 min by addition of 2 μg of 4,6-diamidino-2-phenylindole (DAPI)/ml prior to microscope inspection. Cells were viewed with a Zeiss Axiovert microscope with DAPI and fluorescein isothiocyanate filter sets. Images were recorded with a Quantix charge-coupled device camera (Roper Scientific) with the IP LAB software (Spectra Services) and further processed in Adobe Photoshop, version 6.0.
Purification of bacterial GST-War1p.
The expression and purification of GST-War1p fusion proteins in Escherichia coli were done essentially as previously described (46) with minor modifications. Bacteria transformed with plasmid pYA22 or control vector pGEX-3X (Pharmacia) were grown at 37°C to an OD600 of 0.4 in Luria broth plus ampicillin to maintain plasmids. Cells were shifted to room temperature, and addition of 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induced production of GST-War1p for 40 min. Cells were harvested, washed once in water, and resuspended in 2 ml of buffer TpG (0.2 M Tris-Cl [pH 8], 0.1 M NaCl, 1 mM EDTA, 1 mM DTT, 0.1 mM ZnCl2, protease inhibitor cocktail). After sonication to lyse bacteria, extracts were cleared by centrifugation, diluted to 10 ml in TpG, and incubated with 500 μl of a 50% slurry of glutathione beads (Pharmacia) overnight at 4°C. Beads were washed twice with TpG buffer, and the fusion protein was eluted twice in 250 μl of Tris-HCl, pH 8, containing 10 mM reduced glutathione.
Gel mobility shift assays.
The oligonucleotide harboring the entire WARE (WAREs; −655 to −604 of the PDR12 promoter; 5′-AATTCTCGCCGCTCACGGTTCCTAACCTGGTCCCTTACCAACCGGTCCCTTGGTG-3′) was end labeled with polynucleotide kinase and [γ-32P]ATP, annealed to the complementary antisense strand (WAREas; 5′-AATTCACCAAGGGACCGGTTGGTAAGGGACCAGGTTAGGAACCGTGAGCGGCGAG-3′), and purified via MicroSpin G-25 columns (Pharmacia). Various amounts of purified GST-War1p fusion protein and GST as a control were incubated for 30 min at 25°C with 1 ng of end-labeled probe in assay buffer (100 mM KCl, 20 mM Tris-Cl [pH 8], 5 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA, 5% glycerol) in the presence of 25 ng of poly(dI-dC). To verify the specificity of WARE binding, an excess of unlabeled WARE oligonucleotide or an unspecific competitor fragment (US) was added to the incubation mixture. After a 20-min incubation on ice, the mixture was loaded onto a 6% native polyacrylamide gel. Electrophoresis proceeded at 180 V in 0.5× Tris-borate-EDTA; the gel was dried, and signals were detected with the STORM phosphorimager (Molecular Dynamics).
In vivo footprint experiments.
In vivo footprint analysis was carried out by a method described earlier (14). Briefly, cultures were grown in the exponential growth phase in YPD until an OD600 of 1.0 to 1.8 was reached. Samples were either treated with 8 mM sorbate for 10 min or left untreated. Cells were harvested, resuspended in 5 ml of culture medium, and treated with 5 μl of dimethyl sulfoxide (DMS) (in the presence of sorbate for stressed cells) for 5 min to methylate DNA. The reaction was stopped by rinsing cells with ice-cold TNEβ (10 mM Tris-HCl [pH 7.5], 40 mM NaCl, 1 mM EDTA, 100 mM β-mercaptoethanol). DNA was isolated after the spheroblasting of cells in SCEβ (1 M sorbitol, 100 mM trisodium citrate, 10 mM EDTA, 100 mM β-mercaptoethanol [pH 7.0 to 7.5]) and subsequent lysis (2% SDS, 100 mM Tris-Cl [pH 9.0], 50 mM EDTA) at 65°C for 5 min. Proteins were precipitated in 0.63 M potassium acetate at 4°C overnight. DNA from the supernatant was purified by several cycles of ethanol precipitation and RNase digestion, followed by digestion with EcoRI. Primer extension was carried out with 32P-end-labeled oligonucleotides for 40 cycles (1 min at 94°C, 2 min at 58°C, 3 min at 72°C). Oligonucleotides used for this reaction were Pdr12-497as and Pdr12-770s, which bind to the PDR12 promoter. The products were precipitated twice with ethanol for purification, dissolved in denaturing loading buffer (50 mM NaOH, 0.5 mM EDTA, 4 M urea, 0.1% bromphenol blue, 0.1% xylene cyanol), and resolved in 8% sequencing gels. Dried gels were scanned in a STORM phosphorimager (Molecular Dynamics) and quantified with standard software (ImageQuant, version 5.0).
RESULTS
Weak acid induction of Pdr12p is mediated by the novel transcription factor War1p.
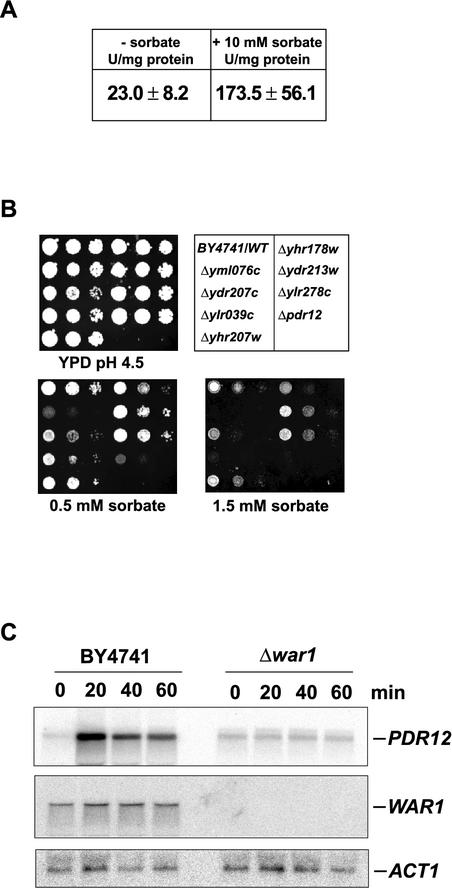
We have previously shown that levels of the Pdr12p ABC pump and its mRNA are dramatically increased following weak organic acid stress (32). To verify that these high Pdr12p levels are due to transcriptional activation, we performed β-galactosidase reporter assays with YAK3 cells, harboring genomically integrated lacZ driven by the PDR12 promoter. Cells were grown in YPD, pH 6.8, and treated with 10 mM sorbate for 30 min. β-Galactosidase activity was around 23 U/mg in uninduced control cells. However, PDR12 promoter activity sharply increased about eightfold after sorbate addition (Fig. 1A), demonstrating that weak acid stress triggers transcriptional activation of the PDR12 promoter.
FIG. 1.
Deletion of WAR1 abolishes stress-induced Pdr12p expression. (A) YAK3 cells were grown on YPD and stressed with 10 mM sorbate or left untreated. β-Galactosidase activity in three independent samples was determined. (B) BY4741 and the single-deletion strains were grown in YPD to an OD600 of 1.0 and spotted onto pH 4.5 YPD plates containing various concentrations of sorbate. Cell growth was inspected after a 48-h incubation. WT, wild type. (C) BY4741 and Δyml076c (Δwar1) cells were grown on YPD and treated with 8 mM sorbate for the indicated times. RNA was isolated and blotted and PDR12, WAR1, and ACT1 mRNAs were detected by Northern analysis.
Known regulators such as Msn2p and Msn4p (14), Yap1p to Yap8p (12), Pdr1p and Pdr3p (47), and Yrr1p (5) fail to control Pdr12p expression (data not shown). This suggests that PDR12 is controlled by another yet-unknown transcription factor. We thus tested a total of 261 putative or confirmed nonessential transcription factors, as listed in the yeast protein database (http://www.incyte.com/sequence/proteome/), for a sorbate sensitivity phenotype. Haploid single-deletion strains of the EUROSCARF knockout collection (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/) were grown in microtiter plates to an OD600 of 1.0, diluted in water to an OD600 of 0.1, and spotted onto YPD control plates, as well as onto pH 4.5 YPD plates containing 1.0 or 1.5 mM sorbate. After 48 h of incubation, seven strains displayed reduced growth in the presence of sorbate and were thus further investigated. These seven strains were again cultured to the exponential growth phase and spotted along with wild-type (BY4741) and Δpdr12 control cells onto sorbate-containing pH 4.5 YPD agar plates (Fig. 1B). At the lowest concentration of 0.5 mM sorbate, growth of the wild type and most deletion strains was unaffected, while Δpdr12 control cells failed to grow. However, only one deletion strain, the Δyml076c strain, showed severely impaired growth comparable to that of Δpdr12 cells in the presence of low concentrations of sorbate (Fig. 1B).
To verify that sorbate sensitivity was due to reduced Pdr12p levels, we performed immunoblotting on extracts prepared from deletion strains, demonstrating a lack of Pdr12p induction in Δyml076c cells (data not shown). Furthermore, we analyzed PDR12 mRNA levels in wild-type and Δyml076c cells treated with 8 mM sorbate. Samples were taken at the indicated time points, and isolated RNA was subjected to Northern analysis (Fig. 1C). PDR12 mRNA levels were strongly elevated during sorbate stress in wild-type cells. By contrast, in the absence of YML076c, no increase of PDR12 mRNA was detected (Fig. 1C), confirming the results of the cytotoxicity assays and the immunoblot analysis.
Notably, we have also identified YML076c in a conventional mutagenesis screen employing the strain YAK3, with a genomically integrated PDR12 promoter-lacZ reporter construct (data not shown). This screen resulted in the isolation of two yeast mutants carrying distinct loss-of-function alleles of YML076c (B. E. Bauer et al., unpublished data). Together with the data from the functional screening (Fig. 1A), this mutagenesis study revealed that YML076c encodes a novel yeast regulator mediating weak acid resistance through the induction of Pdr12p. We thus named the gene WAR1, for weak acid resistance.
War1p mediates resistance to several weak acids through Pdr12p.
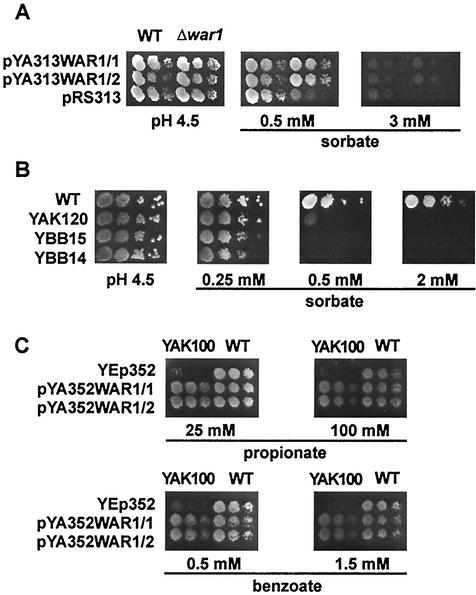
WAR1 potentially encodes the 944-residue Zn2Cys6 zinc finger transcription factor War1p. To further study the role of War1p in stress response, we cloned the WAR1 gene into appropriate multicopy and single-copy plasmids. Wild-type BY-4741 and Δyml076c (Δwar1) cells were transformed with either the CEN-based plasmid pYA313WAR1 or the empty control vector pRS313. Serial dilutions of independent transformants were spotted onto pH 4.5 SC agar plates containing various sorbate concentrations (Fig. 2A). While Δwar1 cells carrying the control vector displayed debilitated growth in the presence of 0.5 mM sorbate, wild-type cells, as well as Δwar1 cells harboring pYA313WAR1, grew at up to 3 mM sorbate. No difference between wild-type and Δwar1 cells expressing WAR1 from the plasmid was observed (Fig. 2A). The plasmid pYA313WAR1 also restored Pdr12p induction in Δwar1 cells after sorbate stress to levels observed in wild-type cells (data not shown). Notably, overexpression of War1p failed to enhance either sorbate resistance or Pdr12p levels (data not shown).
FIG. 2.
War1p mediates resistance to several weak organic acids. (A) BY4741 (wild type [WT]) and Δyml076c (Δwar1) cells transformed with pYA313WAR1 and the empty vector control pRS313 were grown in SC to an OD600 of 1.0 and spotted onto pH 4.5 SC plates containing sorbate. Cell growth was inspected after a 48-h incubation. (B) W303-1A (WT), YAK120 (Δwar1), YBB14 (Δpdr12), and YBB15 (Δwar1 Δpdr12) cells growing exponentially in YPD were spotted onto pH 4.5 YPD plates containing the indicated amounts of sorbate. Colonies were inspected after a 48-h incubation. (C) FY1679-28c (WT) and YAK100 (Δwar1) transformed with YEp352 or pYA352WAR1 were grown to an OD600 of 1.0 and spotted on pH 4.5 YPD plates containing weak acids as indicated.
To test whether the weak acid hypersensitivity of Δwar1 cells results only from low Pdr12p levels or whether War1p controls other genes mediating weak acid resistance, we constructed a set of isogenic deletion strains carrying the Δwar1 (YAK120) or Δpdr12 (YBB14) single deletion or the Δwar1 Δpdr12 (YBB15) double deletion. Immunoblot analysis showed that loss of War1p almost abolished Pdr12p expression, although basal Pdr12p levels remained detectable (data not shown). Growing these cells on pH 4.5 YPD plates in the presence of sorbate, we observed that all strains grew at 0.25 mM sorbate. Increasing the sorbate concentration to 0.5 mM abolished growth of the Δpdr12 single-deletion strain and the Δwar1 Δpdr12 double-deletion strain, while a Δwar1 single-deletion strain exhibited residual growth at this concentration. This is consistent with low Pdr12p levels in these cells (Fig. 2B). At 2.0 mM sorbate, only the W303-1A wild-type control showed normal cell growth. Since deletion of WAR1 does not further exacerbate the hypersensitivity of Δpdr12 cells, Pdr12p is perhaps the major weak acid resistance determinant under the control of War1p (Fig. 2B). The extremely low Pdr12p levels present in Δwar1 cells indicate a low constitutive PDR12 promoter activity that causes a detectable weak acid tolerance.
Since sorbate is not the only Pdr12p substrate (33), we tested whether War1p also confers resistance to other weak organic acids. Wild-type FY1679-28c cells and YAK100 (Δwar1) cells carrying multicopy pYA352WAR or the empty vector control were spotted onto pH 4.5 SC plates containing the indicated concentrations of benzoate and propionate (Fig. 2C). As with sorbate, War1p mediated pronounced resistance to both substances. Δwar1 cells with the empty vector were unable to grow on 0.5 mM benzoate and 25 mM propionate. Wild-type cells and Δwar1 cells transformed with pYA352WAR1 still grew on 1.5 mM benzoate and 100 mM propionate. Furthermore, β-galactosidase measurements on cells harboring a PDR12 promoter-lacZ fusion plasmid (pPWP1) demonstrated that several other stresses, including organic alcohols (ethanol, propanol, and butanol), high osmolarity (1 M NaCl), oxidative stress (menadione and H2O2), and heat shock, fail to drive detectable PDR12 promoter activity (K. Hatzixanthis et al., unpublished data). The regulation of PDR12 by War1p is therefore highly specific for weak acid stress, and Pdr12p appears to be the major anion efflux pump.
War1p is constitutively localized in the nucleus.
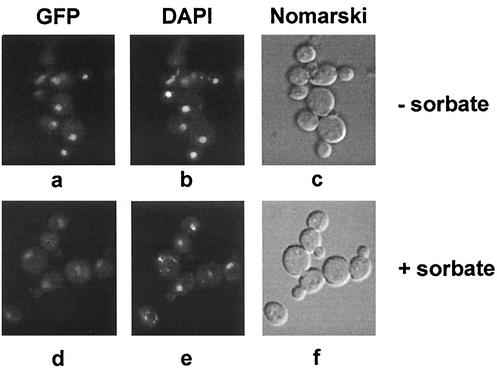
To investigate the subcellular localization of War1p, we introduced a C-terminal in-frame fusion of GFP with chromosomal WAR1, resulting in strain YAK122. The functionality of this War1p-GFP fusion protein was verified under sorbate stress conditions (data not shown). For fluorescence experiments, cells were grown in YPD and either treated with 8 mM sorbate for 2 h or left untreated. Under both growth conditions, the signal of War1p-GFP localized in distinct spots (Fig. 3a and d). For visualization of the nuclear DNA, cells were stained for 10 min with DAPI. Microscopic inspection of living cells revealed that the War1p-GFP signal entirely colocalized with the nuclear DAPI staining, both in the absence (Fig. 3a and b) and presence (Fig. 3d and e) of sorbate stress. GFP staining was observed only in YAK122 cells, not in wild-type W303-1A cells lacking War1p-GFP (data not shown). Taken together, these data demonstrate that War1p is constitutively localized in the nucleus under both normal and adverse conditions.
FIG. 3.
War1p localizes to the nucleus and is able to form homodimers. (A) YAK122 cells expressing a War1p-GFP fusion were grown in YPD to the exponential growth phase and stressed with 8 mM sorbate for 2 h (d to f) or left untreated (a to c). Living cells were treated with DAPI for 15 min to stain nuclear DNA and inspected in a Zeiss Axiovert 10 fluorescence microscope. (a and d) Fluorescein isothiocyanate filter, War1-GFP; (b and e) DAPI staining; (c and f) Nomarski image.
Many zinc finger transcription factors, including Gal4p (27) and Pip2p/Oaf1p (35) form homo- or heterodimers to bind to their palindromic target DNA. To test whether War1p has the ability to interact with itself, we performed pull-down experiments using bacterially expressed GST-War1p. Indeed, a GST-War1p fusion protein purified from E. coli was able to specifically bind a War1p-c-Myc present in crude cell extracts (data not shown). Immunoblot detection of War1p-c-Myc with a c-Myc antibody revealed a single band, which was detectable only in the presence of GST-War1p (data not shown). These results indicate that War1p might act at least as a homodimer, similar to several other fungal binuclear zinc cluster regulators.
Identification of the promoter element mediating PDR12 induction.
Since War1p appears to be a transcriptional activator, it may directly recognize a distinct cis-acting element in the PDR12 promoter. To identify the WARE that mediates sorbate induction of Pdr12p, we determined the transcriptional activity of PDR12 promoter deletion constructs. These reporter constructs were based on the CEN-based plasmid pPWP1, harboring lacZ under the control of several PDR12 promoters with deletions (Fig. 4A). Expression levels were measured in cells cultured at pH 4.5, both in the absence and presence of sorbic acid concentrations yielding maximal transcriptional activity of the full-length promoter (1 mM sorbate). Cells containing the PDR12 reporter with a length of 772 bp (pPWP1) displayed a β-galactosidase activity of about 28 Miller units upon stress under both experimental conditions, with undetectable activity in the absence of stress (Fig. 4A). While a deletion to bp −681 had no effect on induction, deletion to bp −606 caused severe reductions of sorbate-dependent transcription to 3 Miller units. Promoter truncations to bp −527 did not further decrease β-galactosidase activity. A PDR12 promoter deletion to bp −445, as well as all greater deletions (producing shorter constructs), resulted in a complete loss of promoter activity and hence also weak acid regulation (Fig. 4A), suggesting the existence of a second WARE in the PDR12 promoter. Taken together, these data indicate that a major cis-acting element responsible for sorbate induction localizes to a 74-bp region (−681 to −607) in the PDR12 promoter, as well as to a second stretch from bp −527 and −445 relative to the translational start site. Deletion of both putative WARE sequences resulted in complete loss of sorbate induction. To confirm that the major WARE drives PDR12 induction, we inserted the −646 to −603 sequence into the disabled CYC1 promoter of the lacZ reporter plasmid pRW95-3 (45) to yield plasmid pRW95-W1. This insert conferred strong sorbate inducibility on the otherwise-dormant CYC1 promoter (Fig. 4B). The −646 to −603 promoter region hence contains all sequence elements necessary and sufficient to drive induction of a reporter gene upon stress challenge and must therefore contain a functional WARE.
FIG. 4.
War1p recognizes a cis-acting WARE in the PDR12 promoter. (A) FY1679-28c transformed with plasmids carrying the indicated fragments of the PDR12 promoter fused to lacZ were grown to the logarithmic growth phase, shifted to pH 4.5 YPD, and treated with 1 mM sorbate. β-Galactosidase assays were performed in triplicate in the absence (−) and presence (+) of sorbate, and activity is given in Miller units. (B) FY1679-28c (wild type [WT]) and YAK100 (Δwar1) cells transformed with pRW95-3 and pRW95-W1 were grown in SC medium to the logarithmic growth phase, shifted to pH 6.8 YPD, and stressed with 8 mM sorbate. β-Galactosidase activity was assayed, and activities are given in Miller units. Three independent experiments were performed with each construct, and standard deviations are given in parentheses.
War1p binds to the cis-acting WARE in vitro and in vivo.
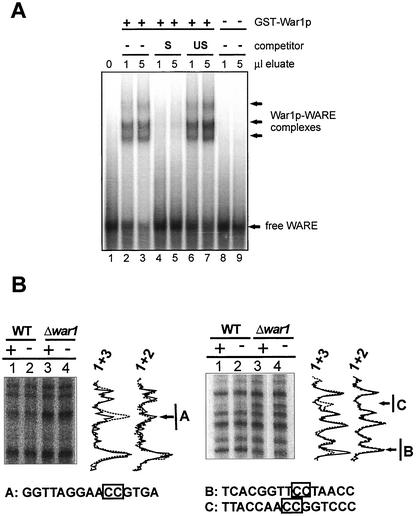
To verify that War1p recognizes the WARE sequence, we performed both in vitro band shift experiments using a GST-War1p fusion protein and in vivo footprinting. GST-War1p was purified from bacteria and analyzed by immunoblotting using polyclonal GST antibodies (11). The fusion protein underwent proteolysis, as 50-kDa degradation products containing the N-terminal zinc finger domain were seen in addition to the predicted 150-kDa full-length protein (data not shown). For band shift experiments, a double-stranded 32P-end-labeled probe containing the WARE region was incubated with or without purified GST-War1p, as well as with purified GST as a control. Acrylamide gel electrophoresis showed that complexes of slower mobility appeared with labeled DNA only in the presence of GST-War1p, not with the bacterial control lysate (Fig. 5A) or purified GST (data not shown). The different complexes probably reflected the full-length War1p protein, as well as different truncations of GST-War1p. To verify the specificity of the reaction, we performed competition experiments using a 50-fold excess of the unlabeled WARE probe (S; Fig. 5A, lanes 4 and 5), which almost abolished WARE binding. By contrast, a similar excess of unspecific competitor DNA (US; lanes 6 and 7) failed to impair WARE binding. Thus, War1p specifically binds to the WARE sequence in the PDR12 promoter.
FIG. 5.
War1p binds to the WARE in the PDR12 promoter. (A) For mobility shift experiments, the 32P-end-labeled WARE probe (bp −655 to −604 of the PDR12 promoter) was incubated with GST-War1p alone (lanes 2 and 3) or in the presence of a specific (S; lanes 5 and 6) or unspecific (US; lanes 6 and 7) competitor probe for 30 min. WARE alone (lane 1) or extracts from uninduced E. coli (lanes 8 and 9) served as controls. Complexes were separated by native gel electrophoresis and detected by autoradiography. (B) For in vivo footprint experiments, FY1679-28c (lanes 1 and 2) or YAK100 (Δwar1; lanes 3 and 4) cells were grown to the logarithmic growth phase and stressed with 8 mM sorbate for 10 min (+; lanes 1 and 3) or left unstressed (−; lanes 2 and 4). Cells were treated with DMS to methylate DNA. Primer extension was carried out on both strands (left, antisense; right, sense) to detect DNA methylation. Products were separated by native acrylamide gel electrophoresis and autoradiographed. Graphs reflect intensities of the bands (lane 1, solid line; lanes 2 and 3, dashed line). A, B, and C, sequences in the WARE. Boxed nucleotides show changes in methylation intensity corresponding to protection or deprotection.
To investigate whether the PDR12 promoter is also occupied in living cells, we used in vivo footprint analysis. Experiments were carried out with FY1679-28c wild-type and YAK100 (Δwar1) cells. Both strains were grown to the exponential growth phase and either treated with 8 mM sorbate for 10 min to induce the PDR12 promoter or left unstressed. Cells were then treated with DMS, which methylates guanine and to a lesser extent adenine in the chromosomal DNA. After DNA purification, the methylation status was determined by primer extension analysis (Fig. 5B). Comparing the methylation patterns of wild-type and Δwar1 cells treated with sorbate (Fig. 5B, lanes 1 and 3), we observed a strong War1p-dependent protection at two positions. These corresponded to nucleotides −617 and −618 and nucleotides −642 and −643. By contrast, nucleotides −638 and −639 were strongly deprotected in the wild-type strain, hinting at changes in the DNA structure. Strikingly, the protected guanines identified by the in vivo footprint matched precisely to a 5′-CCG-Nx-CGG-3′ repeat (where subscript x indicates a spacing of 23 bp), a palindromic repeat, which is typically recognized by zinc finger transcription factors (Fig. 5). Surprisingly, the comparison of sorbate-treated and untreated wild-type cells revealed that the WARE was occupied even in the absence of stress (Fig. 5B, lanes 1 and 2). These results confirm that the WARE is decorated by War1p and demonstrate that War1p constitutively binds to the WARE in vivo.
Phosphorylation of War1p coincides with PDR12 induction.
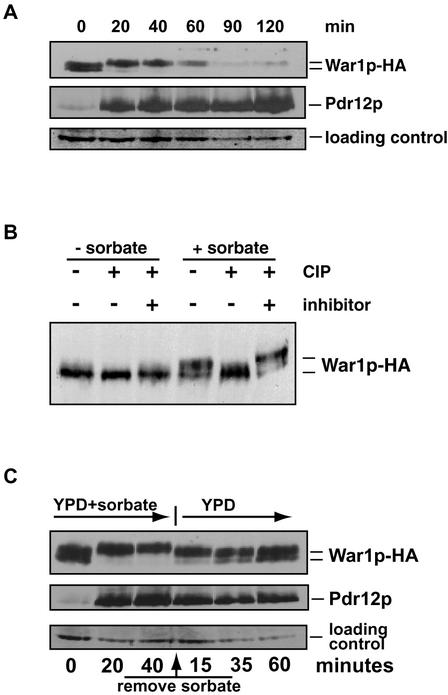
To study War1p function, we tagged chromosomal WAR1 with an epitope by fusing a triple-HA at the C terminus of this regulator to generate strain YAK101. The War1p-HA variant was fully functional, since sorbate tolerance and Pdr12p induction were not compromised (data not shown). YAK101 cells were grown in YPD to the exponential growth phase and exposed to 8 mM sorbate for 2 h. Samples were taken at the indicated time points and used for immunodetection of both War1p and Pdr12p (Fig. 6A). In unstressed cells, War1p migrated as a double band with an apparent molecular mass of about 130 kDa, indicating two distinct isoforms of the protein. Within 20 min of sorbate treatment, the higher-mobility War1p form disappeared, while slower-migrating bands emerged, suggesting posttranslational modifications of War1p in response to stress (Fig. 6A). During 120 min of stress, War1p gradually disappeared, suggesting that War1p levels decrease during stress adaptation, with the fastest-migrating form of War1p disappearing after 40 min. Cycloheximide chase experiments of stressed and unstressed cells indicated similar stabilities of War1p and Pdr12p under both conditions (data not shown). Furthermore, WAR1 mRNA levels, both in stressed and unstressed cells, were unaffected during the first 60 min (Fig. 1C), suggesting that the War1p decay was not caused by reduced transcription or increased degradation. As expected, Pdr12p levels sharply increased after 20 min of weak acid stress and then followed a slight but steady increase over the entire period.
FIG. 6.
War1p is reversibly phosphorylated during weak acid stress. (A) Exponentially growing YAK101 (WAR1-HA) cells were treated with 8 mM sorbate for the time periods indicated. War1p-HA and Pdr12p were immunodetected in cell extracts with monoclonal anti-HA antibody 12CA5 and polyclonal anti-Pdr12p antibodies, respectively. (B) YAK101 cells were stressed with 8 mM sorbate for 1 h in YPD or left unstressed. Protein extracts were incubated with phosphatase (CIP) for 1 h at 37°C, mock treated, or treated with CIP in the presence of orthophosphate as an inhibitor. (C) YAK101 cells were grown to an OD600 of 1.0; then, 8 mM sorbate was added for the time periods indicated (sorbate). Sorbate was removed by spinning out cells and resuspending them in fresh prewarmed YPD medium (YPD). Samples were taken, and protein extracts were analyzed by immunoblotting with anti-HA antibody 12CA5 and anti-Pdr12p antibodies.
To identify the possible modifications of War1p isoforms, we performed phosphatase treatment of extracts prepared from strain YAK101 as described in Materials and Methods. In unstressed cells, we observed two distinct War1p isoforms (Fig. 6B, left). Phosphatase treatment caused the slower-migrating form to disappear, demonstrating that War1p is phosphorylated even in the absence of stress. In sorbate-stressed cells, War1p appeared as a fast-migrating form, as well as several slower-migrating bands (Fig. 6B, right). Phosphatase treatment removed all slower-migrating bands, yielding a single War1p immunoreactive band. Therefore, War1p is rapidly and extensively phosphorylated in response to weak acid stress.
To analyze a possible correlation between War1p phosphorylation and Pdr12p induction, we performed stress release experiments. YAK101 cells were treated with 8 mM sorbate for 40 min. After this period, the stress agent was removed by changing the growth medium to fresh YPD. Cell extracts were taken before, during, and after the stress period, followed by immunodetection of War1p and Pdr12p (Fig. 6C). Consistent with earlier results, sorbate stress caused a shift of the slower-migrating War1p isoform. While the mobility of the fastest War1p band remained unchanged, slower-migrating bands appeared within 20 min. Interestingly, only 15 min after removal of sorbate, War1p immunoreactive bands showed again a higher mobility, similar to those observed in unstressed conditions. Hence, War1p phosphorylation strictly requires stress conditions and is rapidly lost after stress relief. Taken together, our data suggest that War1p phosphorylation must be tightly linked to the activation of War1p, which subsequently triggers Pdr12p induction and stress adaptation.
DISCUSSION
The War1p regulator is essential for weak acid resistance and Pdr12p induction.
We originally identified the Pdr12p ABC transporter as the major determinant of weak organic acid tolerance in S. cerevisiae. Pdr12p is strongly induced by certain weak organic acids and actively pumps acid anions to the extracellular space (32). In this study, we identify a novel zinc finger regulator, War1p, required for the transcriptional induction of PDR12 in response to weak acid stress. The loss of War1p abolishes PDR12 induction (Fig. 1C) and results in hypersensitivity to various weak organic acids (Fig. 1B and 2C). Thus, our data also identify a new stress response in yeast, which is essential for adaptation to growth in the presence of weak organic acids and which is regulated by a novel stress-specific transcription factor.
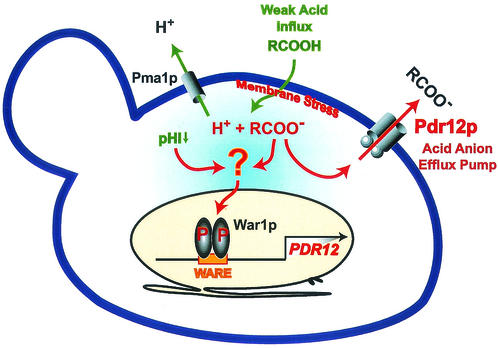
War1p is constitutively localized in the nucleus (Fig. 3), and it is able to form homodimers in vitro (data not shown). Cytotoxicity assays of cells lacking Pdr12p and/or War1p establish PDR12 as the major target of War1p (Fig. 2B), which binds to a 48-bp WARE in the PDR12 promoter (Fig. 4B and 5A). Moreover, we show that War1p decorates the WARE both in the presence and absence of weak acid stress (Fig. 5B). Because phosphorylation of War1p is tightly linked to its transactivation activity, we propose that War1p receives and transmits a signal through a novel signaling pathway dedicated to stress adaptation (Fig. 7).
FIG. 7.
Model for War1p-mediated weak acid stress adaptation. Uncharged weak acids enter the cell and dissociate in the cytoplasm, generating a lower intracellular pH. The resulting protons are extruded by Pma1p, and acid anions are effluxed by induced Pdr12p in cells adapting to weak acid stress. We propose that acid anions directly or indirectly activate War1p, which is constitutively bound to the WARE in the PDR12 promoter. War1p is rapidly phosphorylated upon weak acid stress and thus activated to induce PDR12, leading to a massive upregulation of Pdr12p in the plasma membrane.
Although we analyzed 261 putative transcription factor deletion strains for sorbate sensitivity, only the Δyml076c (Δwar1) strain failed to induce Pdr12p (Fig. 1B). The importance of War1p in weak acid resistance development was also demonstrated by a conventional UV mutagenesis screen, which yielded two defective war1 mutant alleles unable to induce PDR12 (B. E. Bauer et al., unpublished data). While loss of War1p abolishes PDR12 induction, it does not interfere with the basal expression of PDR12 (Fig. 1C), consistent with a low but detectable sorbate resistance in the Δwar1 strain (Fig. 2B). Moreover, a Δwar1 Δpdr12 strain does not display increased sorbate sensitivity compared to a Δpdr12 single-deletion strain, suggesting that Pdr12p is the most important War1p target.
Indeed, expression of a GAL1pro-driven Pdr12p in Δwar1 cells suppresses sorbate hypersensitivity. Moreover, global transcriptome analysis of wild-type and Δwar1 cells under normal and stress conditions demonstrates that the WAR1 regulon comprises a rather small number of genes (C. Schüller et al., unpublished data). Interestingly, War1p confers resistance to sorbate, benzoate, and propionate (Fig. 2C). In addition, only a limited spectrum of carboxylic acids with C3 to C8 chains strongly activate War1p-dependent PDR12 induction (data not shown). Furthermore, Pdr12p levels are not influenced by other adverse conditions such as oxidative stress, high osmolarity, and heat shock (data not shown), demonstrating that Pdr12p induction by War1p is a highly specific process, and thus distinct from the well-described general stress response (28).
War1p binds the WARE in the PDR12 promoter.
We demonstrate that War1p controls stress adaptation through the WAREs in the PDR12 promoter, as depicted in the model (Fig. 7). The WARE is directly decorated by War1p in vitro, as shown by band shift experiments using bacterially expressed GST-War1p (Fig. 5A). Interestingly, War1p binding does not require any additional factors, posttranslational modifications, or sorbate stress in vitro.
Most importantly, in vivo footprint analysis demonstrates that the occupancy of WARE is constitutive and strictly requires the presence of War1p in living cells (Fig. 5B). The in vivo results confirm the in vitro data regarding the War1p binding region, which coincides with the cis-acting WAREs required for weak acid stress adaptation. For the PDR12 WARE, the in vitro and in vivo experiments suggest that the cis-acting element 5′-CGG-Nx-CCG-3′ (where subscript x indicates a spacing of 23 bp) is essential for PDR12 transactivation. However, additional experiments are clearly required to delineate the sequence constraints within this motif. This is consistent with the nature of DNA-binding sites of other zinc finger transcription factors, all of which are direct, inverted, or everted palindromic repeats containing a 5′-CGG-3′ motif, with various numbers of nucleotides in the intervening spacer region. For example, Hap1p recognizes the direct repeat of 5′-CGGNNNTANCGG-3′ (48), while the heterodimer Pip2p/Oaf1p binds to inverted repeats of the consensus 5′-CGG-N15-18-CCG-3′ (35). Finally, Pdr1p and Pdr3p regulate their targets via the everted sequence motif 5′-TCCG/aC/tGG/cA/g-3′ (variable bases in a consensus motif are in lowercase) (8). Interestingly, many zinc finger regulators recognizing inverted or everted palindromic repeats, including Gal4p and Pip2p/Oaf1p, bind the DNA as dimers. Likewise, we show a specific interaction of War1p-c-Myc with GST-War1p, demonstrating the ability of War1p to homodimerize (Fig. 3). This further supports the idea of the palindromic 5′-CCG-Nx-CGG-3′ repeat as the War1p binding site.
The mechanism of War1p activation.
War1p is 1 out of 54 putative Zn2Cys6 regulators present in the yeast genome (http://genome-www.stanford.edu/Saccharomyces/). Like most family members, War1p contains a zinc finger at the N terminus, ranging from residues 75 to 111, and a predicted C-terminal activation domain stretching from residues 911 to 937. The means to activate zinc finger regulators include mechanisms such as nuclear-cytoplasmic shuttling, DNA binding, phosphorylation, and, most importantly, the unmasking of the activation domain. For War1p, we show that induction of its transactivation activity does not involve increases in WAR1 mRNA (Fig. 1C) or protein (Fig. 6A). Likewise, War1p overexpression fails to increase either weak acid tolerance or Pdr12p levels (data not shown). Therefore, the mechanisms that regulate War1p activity are most likely posttranslational and may involve phosphorylation or perhaps other posttranslational modifications (see also below). A War1p-GFP fusion localizes exclusively to the nucleus (Fig. 3), excluding the possibility of nuclear-cytoplasmic shuttling, as observed for other stress factors such as Msn2p (14). Interestingly, some zinc finger regulators bind constitutively to their cis-acting elements. For instance, the Leu3p regulator of amino acid biosynthesis is always bound to the LEU2 promoter. Leu3p transcriptional activity is regulated by alpha-isopropylmalate (α-IPM), an intermediate of leucine biosynthesis. Depending on the presence or absence of α-IPM, Leu3p acts as an activator or inhibitor of transcription (41). Similarly, we demonstrate direct and constitutive binding of War1p to the PDR12 promoter (Fig. 5) both in the presence and absence of a stress signal. However, since PDR12 mRNA levels are not increased in unstressed Δwar1 cells (Fig. 1C), War1p is highly unlikely to exert any inhibitory effect on PDR12 expression in the absence of stress.
Phosphatase treatment shows that War1p exists in several phosphorylated isoforms in unstressed cells (Fig. 6A and B). We propose that War1p transactivation capacity is tightly linked to its phosphorylation state. This idea is supported by several observations. First, upon addition of sorbate, the slower-migrating War1p forms are replaced by differently phosphorylated War1p isoforms (Fig. 6A). Second, phosphorylated War1p appears only after the imposition of stress, whereas removal of sorbate results in an immediate loss of phosphorylation (Fig. 6C). Third, War1p phosphorylation occurs in response to sorbate, benzoate, and propionate but not acetate, and acetate also fails to induce Pdr12p (data not shown). Our data resemble those obtained for other regulators such as Put3p. This regulator of the proline utilization pathway is constitutively bound to the promoter DNA and differentially phosphorylated in response to the quality of nitrogen sources (20). Likewise, the transcriptional activation state of the Cat8p regulator appears to change during carbon repression and derepression (16). Hence, stress-induced modifications such as phosphorylation might induce conformational changes that activate War1p on the PDR12 promoter, similar to the situation for Put3p (9). Alternatively, phosphorylation could influence protein-protein interactions that modulate War1p activity. Similar observations were made for Gal4p, where phosphorylation of a single residue (Ser699) destabilizes the binding of Gal80p. The latter then dissociates, unmasking the activation domain of the Gal4p regulator (36).
While our studies demonstrate that weak acids elicit a novel stress response, the upstream sensor and the components transducing the signal to War1p remain elusive. Since only a few monocarboxylic acids strongly activate War1p, it seems reasonable to speculate that the sensor of this pathway, whether intracellular or membrane bound, may respond to a limited substrate spectrum, with special requirements as to size and charge. Otherwise War1p activity could be directly modulated by organic monocarboxylate anions. One could envision that their binding to War1p induces conformational changes or modifies interactions with other proteins, including the kinase that phosphorylates War1p. Experiments with Leu3p in mammalian and yeast cells show that the effector α-IPM directly influences the conformation to unmask the C-terminal Leu3p activation domain (15, 43).
Weak acid stress elicits a novel response signaling event in yeast.
As summarized in Fig. 7, the War1p response system appears to be the major, if not the only, stress regulator of Pdr12p. In vitro pull-down experiments using GST-War1p suggest that War1p interacts with itself (data not shown) and may therefore form homodimers like several other fungal zinc finger regulators. Since WARE occupancy is constitutive, stress-induced phosphorylation perhaps functions as the signal for War1p activation. However, the actual signal transduction mechanism triggering War1p activation remains unknown. Cytoplasmic weak acids cause a variety of effects, including a lowering of intracellular pH (pHi). We can exclude the possibility that low pHi triggers Pdr12p induction, since acetate effectively lowers pHi (31) but fails to induce Pdr12p. By contrast, sorbate, which gives no pHi reduction (4), induces Pdr12p. We cannot entirely rule out the possibility that a large cytoplasmic anion pool acts as the inducer. Data from global transcriptome analysis (C. Schüller et al., unpublished data) and our results presented in this study suggest that PDR12 is the only major target gene of War1p mediating ATP-dependent efflux of weak acids. Hence, S. cerevisiae counteracts weak acid stress by utilizing a strongly inducible ABC transporter that effluxes the stress agent. In turn, this anion efflux pump is tightly controlled by a highly specialized stress response pathway with the downstream effector War1p. Constitutive high expression of Pdr12p is thus prevented through the tight regulation of War1p activity. In fact, high Pdr12p levels under normal-growth conditions might even be disadvantageous, since futile ATP consumption or the efflux of physiological weak acid metabolites otherwise required for normal cell growth or intermediary metabolism might be detrimental to cells.
Acknowledgments
We thank Chantal Schwartz and Gerd Krapf for expert technical assistance. Michael Schuster is acknowledged for help with computers and software.
This work was supported by grants from the Fonds zur Förderung der wissenschaftlichen Forschung (P-12661-BIO and P-15934-B08) and in part by funds from DSM Life Sciences and the Herzfelder'sche Familienstiftung. P.P. was supported by BBSRC grants 31/D10371 and 31/D13918.
The first two authors equally contributed to this work.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Balzi, E., M. Wang, S. Leterme, L. Van Dyck, and A. Goffeau. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269:2206-2214. [PubMed] [Google Scholar]
- 3.Bissinger, P. H., and K. Kuchler. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J. Biol. Chem. 269:4180-4186. [PubMed] [Google Scholar]
- 4.Bracey, D., C. D. Holyoak, and P. J. Coote. 1998. Comparison of the inhibitory effect of sorbic acid and amphotericin B on Saccharomyces cerevisiae: is growth inhibition dependent on reduced intracellular pH? J. Appl. Microbiol. 85:1056-1066. [DOI] [PubMed] [Google Scholar]
- 5.Cui, Z., T. Shiraki, D. Hirata, and T. Miyakawa. 1998. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol. 29:1307-1315. [DOI] [PubMed] [Google Scholar]
- 6.Deak, T. 1991. Food-borne yeasts. Adv. Appl. Microbiol. 36:179-278. [DOI] [PubMed] [Google Scholar]
- 7.Delahodde, A., T. Delaveau, and C. Jacq. 1995. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 15:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 9.Des Etages, S. A., D. Saxena, H. L. Huang, D. A. Falvey, D. Barber, and M. C. Brandriss. 2001. Conformational changes play a role in regulating the activity of the proline utilization pathway-specific regulator in Saccharomyces cerevisiae. Mol. Microbiol. 40:890-899. [DOI] [PubMed] [Google Scholar]
- 10.Egner, R., and K. Kuchler. 1996. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 378:177-181. [DOI] [PubMed] [Google Scholar]
- 11.Egner, R., Y. Mahé, R. Pandjaitan, and K. Kuchler. 1995. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5879-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes, L., C. Rodrigues-Pousada, and K. Struhl. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17:6982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friden, P., and P. Schimmel. 1987. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol. Cell. Biol. 7:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, H., and G. B. Kohlhaw. 1996. Regulation of transcription in mammalian cells by yeast Leu3p and externally supplied inducer. FEBS Lett. 390:191-195. [DOI] [PubMed] [Google Scholar]
- 16.Hedges, D., M. Proft, and K. D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 18.Holyoak, C. D., D. Bracey, P. W. Piper, K. Kuchler, and P. J. Coote. 1999. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J. Bacteriol. 181:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holyoak, C. D., M. Stratford, Z. McMullin, M. B. Cole, K. Crimmins, A. J. Brown, and P. J. Coote. 1996. Activity of the plasma membrane H+-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak acid preservative sorbic acid. Appl. Environ. Microbiol. 62:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, H. L., and M. C. Brandriss. 2000. The regulator of the yeast proline utilization pathway is differentially phosphorylated in response to the quality of the nitrogen source. Mol. Cell. Biol. 20:892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Katzmann, D. J., T. C. Hallström, M. Voet, W. Wysock, J. Golin, G. Volckaert, and W. S. Moye-Rowley. 1995. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6875-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolaczkowski, M., A. Kolaczowska, J. Luczynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4:143-158. [DOI] [PubMed] [Google Scholar]
- 24.Krebs, H. A., D. Wiggins, M. Stubbs, A. Sols, and F. Bedoya. 1983. Studies on the mechanism of the antifungal action of benzoate. Biochem. J. 214:657-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohr, D., P. Venkov, and J. Zlatanova. 1995. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9:777-787. [DOI] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 27.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Pastor, M. T., G. Marchler, C. Schüller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce, A. K., I. R. Booth, and A. J. Brown. 2001. Genetic manipulation of 6-phosphofructo-1-kinase and fructose 2,6-bisphosphate levels affects the extent to which benzoic acid inhibits the growth of Saccharomyces cerevisiae. Microbiology 147:403-410. [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer, K., K. S. Kim, S. Kogan, and L. Guarente. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56:291-301. [DOI] [PubMed] [Google Scholar]
- 31.Piper, P., C. O. Calderon, K. Hatzixanthis, and M. Mollapour. 2001. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology 147:2635-2642. [DOI] [PubMed] [Google Scholar]
- 32.Piper, P., Y. Mahé, S. Thompson, R. Pandjaitan, C. Holyoak, R. Egner, M. Mühlbauer, P. Coote, and K. Kuchler. 1998. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piper, P. W. 1999. Yeast superoxide dismutase mutants reveal a pro-oxidant action of weak organic acid food preservatives. Free Radic. Biol. Med. 27:1219-1227. [DOI] [PubMed] [Google Scholar]
- 34.Piper, P. W., C. Ortiz-Calderon, C. Holyoak, P. Coote, and M. Cole. 1997. Hsp30, the integral plasma membrane heat shock protein of Saccharomyces cerevisiae, is a stress-inducible regulator of plasma membrane H+-ATPase. Cell Stress Chaperones 2:12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rottensteiner, H., A. J. Kal, B. Hamilton, H. Ruis, and H. F. Tabak. 1997. A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur. J. Biochem. 247:776-783. [DOI] [PubMed] [Google Scholar]
- 36.Sadowski, I., C. Costa, and R. Dhanawansa. 1996. Phosphorylation of Ga14p at a single C-terminal residue is necessary for galactose-inducible transcription. Mol. Cell. Biol. 16:4879-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Servos, J., E. Haase, and M. Brendel. 1993. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol. Gen. Genet. 236:214-218. [DOI] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stratford, M., and P. A. Anslow. 1996. Comparison of the inhibitory action on Saccharomyces cerevisiae of weak-acid preservatives, uncouplers, and medium-chain fatty acids. FEMS Microbiol. Lett. 142:53-58. [DOI] [PubMed] [Google Scholar]
- 41.Sze, J. Y., M. Woontner, J. A. Jaehning, and G. B. Kohlhaw. 1992. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258:1143-1145. [DOI] [PubMed] [Google Scholar]
- 42.Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065-1075. [DOI] [PubMed] [Google Scholar]
- 43.Wang, D., Y. Hu, F. Zheng, K. Zhou, and G. B. Kohlhaw. 1997. Evidence that intramolecular interactions are involved in masking the activation domain of transcriptional activator Leu3p. J. Biol. Chem. 272:19383-19392. [DOI] [PubMed] [Google Scholar]
- 44.Watt, R., and P. W. Piper. 1997. UBI4, the polyubiquitin gene of Saccharomyces cerevisiae, is a heat shock gene that is also subject to catabolite derepression control. Mol. Gen. Genet. 253:439-447. [DOI] [PubMed] [Google Scholar]
- 45.Wolf, S. S., K. Roder, and M. Schweizer. 1996. Construction of a reporter plasmid that allows expression libraries to be exploited for the one-hybrid system. BioTechniques 20:568-574. [DOI] [PubMed] [Google Scholar]
- 46.Wolfger, H., Y. Mahé, A. Parle-McDermott, A. Delahodde, and K. Kuchler. 1997. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 418:269-274. [DOI] [PubMed] [Google Scholar]
- 47.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2001. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 152:375-389. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, L., and L. Guarente. 1994. The yeast activator HAP1—a GAL4 family member—binds DNA in a directly repeated orientation. Genes Dev. 8:2110-2119. [DOI] [PubMed] [Google Scholar]