Abstract
Cyclin T1, together with the kinase CDK9, is a component of the transcription elongation factor P-TEFb which binds the human immunodeficiency virus type 1 (HIV-1) transactivator Tat. P-TEFb facilitates transcription by phosphorylating the carboxy-terminal domain (CTD) of RNA polymerase II. Cyclin T1 is an exceptionally large cyclin and is therefore a candidate for interactions with regulatory proteins. We identified granulin as a cyclin T1-interacting protein that represses expression from the HIV-1 promoter in transfected cells. The granulins, mitogenic growth factors containing repeats of a cysteine-rich motif, were reported previously to interact with Tat. We show that granulin formed stable complexes in vivo and in vitro with cyclin T1 and Tat. Granulin bound to the histidine-rich domain of cyclin T1, which was recently found to bind to the CTD, but not to cyclin T2. Binding of granulin to P-TEFb inhibited the phosphorylation of a CTD peptide. Granulin expression inhibited Tat transactivation, and tethering experiments showed that this effect was due, at least in part, to a direct action on cyclin T1 in the absence of Tat. In addition, granulin was a substrate for CDK9 but not for the other transcription-related kinases CDK7 and CDK8. Thus, granulin is a cellular protein that interacts with cyclin T1 to inhibit transcription.
Human cyclin T1 is a component of positive transcription elongation factor b (P-TEFb) and plays a key role in the activation of human immunodeficiency virus type 1 (HIV-1) transcription by the viral protein Tat (trans-activator of transcription). Cyclin T1 was first isolated as a Tat-binding protein (61) and an orthologue of Drosophila cyclin T (39, 46, 47). P-TEFb contains cyclin T1 and the cyclin-dependent kinase CDK9. This kinase phosphorylates the carboxy-terminal domain (CTD) of the large subunit of RNA polymerase II, thereby facilitating the transition of polymerase II into a productive elongation mode (22, 43, 44, 48-50, 55, 70). The stimulation by Tat of HIV-1 transcriptional elongation and replication is dependent on P-TEFb that contains functional CDK9 and cyclin T1 (9, 11).
CDK9 also associates with two additional related cyclins, T2a and T2b, which share their first 642 amino acids. Cyclin T2-CDK9 complexes phosphorylate polymerase II but do not participate in HIV transactivation. The cyclin boxes in the N-terminal regions of cyclins T1 and T2 are 81% identical, while their C-terminal regions are less conserved (47). In spite of this high degree of identity, Tat fails to bind to the T2 cyclins because they lack a crucial cysteine residue at position 261 (14, 62). This cysteine is in the Tat-TAR recognition motif of human cyclin T1 that is necessary for its interactions with Tat and TAR, the transactivation response element in the 5′ untranslated region of all HIV-1 mRNAs (for a review, see reference 26).
The activity of the ternary complex P-TEFb-Tat-TAR is modulated by multiple interactions among its components (10, 13, 22, 68). Furthermore, cyclin T1 and CDK9 are present in large complexes (50), suggestive of additional regulatory functions. Besides CTD phosphorylation and CDK9 autophosphorylation, the cyclin T1-CDK9 complex can also phosphorylate cyclin T1, Tat-SF1, and human SPT5 in vitro (13, 28). SPT5 is a component of human DSIF (composed of human SPT4 and human SPT5) which, together with the negative elongation factor NELF, inhibits elongation by polymerase II. This inhibition is relieved by phosphorylation of the polymerase II CTD by P-TEFb (58, 59, 65). SPT5 and SPT6 are also associated with the elongating polymerase, and SPT5 has a positive role in Tat transactivation in vitro (16, 25). Thus, P-TEFb is a pivotal regulator of transcription elongation, which is reflected in its structure.
The cyclin T family contains the longest cyclins known to date, about twice the size of cyclins C and H that are also involved in transcription. Most of the expansion appears to be in the proteins' C-terminal region. This region harbors a few consensus sequences and structural motifs (Fig. 1A) but for the most part is devoid of recognizable domains identified with distinct functions. On the premise that the C-terminal region is likely to interact with cellular regulatory proteins, possibly including some that participate in Tat transactivation, we carried out a yeast two-hybrid screen with cyclin T1 as the bait (T. M. Young, T. Pe'ery, and M. B. Mathews, submitted for publication). One clone isolated from this screen was a cDNA corresponding to part of a growth factor known as granulin. Remarkably, Trinh et al. recently found that a portion of granulin is able to bind Tat in Saccharomyces cerevisiae and in vitro (57).
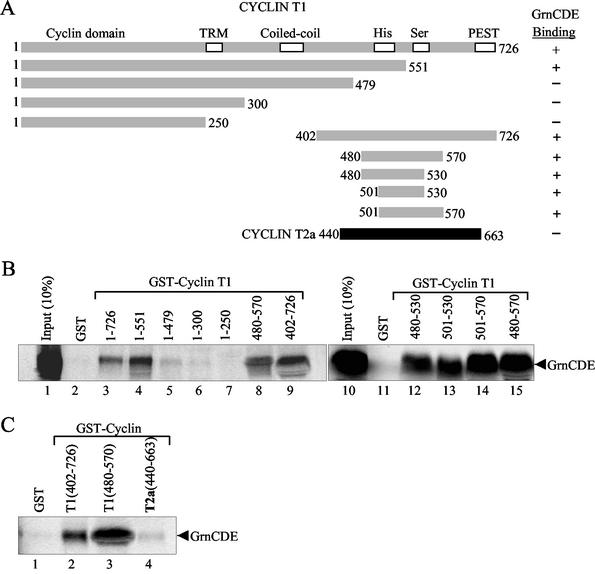
FIG. 1.
Granulin interacts with cyclin T1. (A) Schematic representation of granulins. (B) GrnCDE and GEP binding to GST-cyclin T1. 35S-labeled GrnCDE and GEP (lanes 1 and 5) were synthesized in vitro and pulled down by GST-cyclin T1 (lanes 3 and 7) but not by GST or GST-CDK9 (lanes 2 and 6 and 4 and 8, respectively). GST-CDK9 pulled down 35S-labeled, in vitro-synthesized cyclin T1 (lane 10). Bound proteins were separated in SDS-polyacrylamide gels, and the gels were subjected to autoradiography. Lanes 1, 5, and 9 contained 10% of the input labeled proteins. (C) Mapping the domain of cyclin T1 that interacts with granulin by yeast two-hybrid (Y2H) analysis with deleted forms of cyclin T1. Beneath the schematic representation of cyclin T1, the horizontal lines indicate the regions tested for interaction with GrnCDE. Interactions are indicated by + in the column on the right; * signifies that the result was confirmed with GEP. TRM. Tat-TAR recognition motif.
Granulin was originally purified as a growth factor from conditioned tissue culture medium (64, 69). The protein contains seven and a half repeats of a cysteine-rich motif in the order P-G-F-B-A-C-D-E (where P is the half motif). Several of these repeat units have been isolated from tissue culture medium, blood, and urine as approximately 6-kDa peptides known as granulins or epithelins, and some of them are individually biologically active (for a review, see reference 4). Most of its biological activities are attributed to full-length granulin (also termed granulin/epithelin precursor [GEP], progranulin, acrogranin, and PC cell-derived growth factor [PCDGF]). For example, GEP secreted by murine teratoma-derived PC cells acts as an autocrine growth factor for the producing cells (67, 69). GEP produced by murine BRL-3A cells is sufficient to support the growth of 3T3 cells null for the type 1 insulin-like growth factor receptor in serum-free medium, and no other growth factor by itself can stimulate the growth of these cells (66).
High levels of granulin expression are found in several cancers (7, 32, 33). Overexpression of granulin has been linked to the growth and tumorigenicity of human breast carcinomas (34, 36) and to the acquisition of estrogen independence by estrogen receptor-positive breast cancer cells (35). Granulin was also isolated from sperm acrosomes (1, 2) and regulates blastocyst formation in preimplantation mouse embryos (6). Despite these strong connections with cancer and growth control, granulin's mode of action is not well understood.
We show here that granulin is present intracellularly and interacts specifically with cyclin T1. These interactions are detected in S. cerevisiae, in vitro, and in cell culture. The histidine-rich domain of cyclin T1, which is conserved among the human, mouse, and equine species and has recently been shown to interact with polymerase II (56), is necessary and sufficient for granulin binding. Granulin binds to intact P-TEFb from human cell extracts and serves as a substrate for the CDK9-cyclin T1 complex, but prevents P-TEFb from phosphorylating the CTD. Accordingly, granulin inhibits Tat transactivation in human T cells and 3T3 cells supplemented with human cyclin T1. Granulin also binds to the activation domain of HIV-1 Tat in S. cerevisiae, in vitro, and in human cells, but this interaction is dispensable for granulin's effect on transactivation of the viral promoter. Thus, granulin is a unique cellular protein that interacts with two transcription elongation regulators, P-TEFb and HIV-1 Tat, and modulates transcription elongation by binding to cyclin T1. These data suggest that it plays a role in regulating transcription elongation in both uninfected and infected cells.
MATERIALS AND METHODS
Cell lines and culture.
CEM and U937 cells were obtained from the American Type Culture Collection (Manassas, Va.), while 293T, 3T3, MCF-7, Daudi, and COS7 cell lines were from our laboratory stocks. Suspension cultures (U937, CEM, and Daudi cells) were maintained in RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, Calif.) supplemented with 2 mM l-glutamine and 10% fetal bovine serum (Sigma-Aldrich, St. Louis, Mo.). Adherent cell lines (293T, 3T3, COS7, and MCF-7) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Plasmids and plasmid construction.
The plasmids pGBT9, pGAD GH, and pACT2 and the S. cerevisiae strain CG1945 for yeast two-hybrid system experiments were from Clontech Laboratories Inc. Truncated versions of cyclin T1 were cloned into the S. cerevisiae vector pGBT9, and cyclin T1 amino acids 1 to 708 [cyclin T1(1-708)] was also cloned into pGAD GH to confirm the interactions. Full-length CDK9 was cloned into pACT2. GrnCDE and GEP were also cloned into pGBT9. Plasmids for the expression of glutathione S-transferase (GST) fusion proteins were the full-length cyclin T1 [T1(1-726)] from K. A. Jones (61) and the C-terminal truncations T1(1-250), T1(1-300), T1(1-479), and T1(1-555) from B. M. Peterlin (12). Plasmids for expression in mammalian cells were pcDNA3-HATatWT from B. M. Peterlin (12), pLTR-luciferase (firefly), GST-cyclin T1(402-726) and GST-cyclin T2a(440-633) from D. H. Price (47), and GST-CDK9 from A. Giordano (15).
The GST-cyclin T1 deletion mutants T1(480-570), T1(480-530), T1(501-530), and T1(501-570) were constructed by PCR amplification of the corresponding cyclin T1 sequences followed by ligation into pGEX-4T-3 (Promega). Plasmids containing GST-Tat72, GST-Tat48, GST-Tat86, and GST-Tat86C22G were obtained from the AIDS Research and Reference Reagent Program, National Institutes of Health. The plasmid encoding GST-CTD (containing the 52 repeats from murine RNA polymerase II) was from J. L. Manley (19). The GrnCDE, GrnPGFBA, and GrnC sequences were cloned into the bacterial expression vector pGEX-4T-3, into the mammalian expression vector pFLAG-CMV-2 (Sigma), and into the S. cerevisiae vector pGBT9. The plasmid pcDNA3-PCDGF (encoding full-length human granulin) for expression in mammalian cells was described before (34). GEP was cloned into the S. cerevisiae vector pGAD GH and the mammalian expression vector pFLAG-CMV-2.
To construct the different fused enhanced green fluorescent protein (EGFP)-granulin vectors, EGFP sequence (from pBI-EGFP; Clontech) was first cloned into pcDNA3.1(+). GrnCDE and GrnDE were subcloned C-terminally to the EGFP, and GEP was subcloned N-terminally to the EGFP. pRFP-cyclinT1(1-708) was generated by PCR amplification and cloning into the pDSRed1-C1 vector (Clontech). Tat72 under the control of the Rous sarcoma virus (RSV) promoter and pRSV-luciferase (Renilla) were constructed by S. M. Reza (50a). The PCNA-luciferase (firefly) plasmid contains the promoter (−560 to +60) of the proliferating cell nuclear antigen gene (41) and was provided by T. W. Reichman in our laboratory. Gal4-cyclin T1, Gal4-CDK9, and G5-HIV-luciferase were from L. Lania (37). For in vitro transcription and translation, GrnCDE was subcloned into the expression vector pRSETA. All constructs used in this study were confirmed by sequencing.
Yeast two-hybrid interactions.
Yeast two-hybrid screens were performed according to the Match Maker Gal4 two-hybrid user manual (Clontech). Briefly, cyclin T1 (amino acids 1 to 708) was cloned into the Gal4 binding domain (BD)-containing vector pGBT9 and used as the bait to screen the HeLa cDNA library (Clontech) contained in the Gal4 activation domain (AD) vector pGAD-GH. Interactions of the bait with prey proteins resulted in growth on selective medium and β-galactosidase expression. β-Galactosidase assays were performed according to the manufacturer's instructions (Clontech). Positive clones were identified by sequencing.
Expression and purification of GST fusion proteins.
Escherichia coli strain BL21(DE3) was transformed with recombinant GST fusion protein-expressing plasmids. Overnight cultures (grown at 37°C) were diluted 100-fold in Luria-Bertani (LB) medium containing 50 μg of ampicillin per ml and grown for 3 to 4 h until the A600 reached 0.6. Cells were induced with 0.1 mM isopropylthiogalactopyranoside (IPTG) and grown for an additional 4 h at 30°C for GST and GST-GrnCDE and 15 h at 15°C for GST-CTD. Bacterial pellets were washed twice with phosphate-buffered saline, and the cells were lysed in 5 volumes of EBCD buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40, 5 mM dithiothreitol [DTT]) containing 2 mg of lysozyme per ml, 0.1 mM phenylmethylsulfonyl fluoride, and 1 μg each of leupeptin, pepstatin A, and antipain per ml. After sonication and 20 min of centrifugation (13,000 rpm, 4°C), the clear supernatants were collected and stored at −80°C.
Bacterial extracts containing GST or GST-GrnCDE were mixed with glutathione-Sepharose beads (Amersham Pharmacia Biotech AB) and equilibrated in EBCD buffer. The bound proteins were washed five times with EBCD containing 0.03% sodium dodecyl sulfate (SDS), followed by two washes with 50 mM Tris (pH 7.6) and 5 mM DTT, and eluted with 10 mM glutathione diluted in the same buffer. Eluted proteins were dialyzed against buffer containing 50 mM Tris (pH 7.6) and 1 mM DTT. After dialysis, MgCl2, MnCl2, and DTT were added to final concentrations of 4 mM, 5 mM, and 5 mM, respectively. Protein concentrations were determined by comparison with known amounts of bovine serum albumin (fraction V; Sigma) after Coomassie blue staining in denaturing 15% polyacrylamide gels (30%:0.15% acrylamide/bisacrylamide).
In vitro binding assay.
GrnCDE and GEP were synthesized with the in vitro TNT quick coupled transcription and translation systems (Promega). We mixed 10 μl of in vitro-synthesized [35S]methionine-labeled GrnCDE or [35S]methionine-labeled GEP with GST or GST fusion proteins bound to glutathione-Sepharose beads and incubated them for 2 h at 4°C with gentle rocking. The beads were washed extensively in EBCD plus 0.03% SDS, denatured, and resolved in denaturing 15% polyacrylamide gels. 35S-labeled proteins were detected by autoradiography. The amounts of GST and GST fusion proteins used in the GST pulldown assays were approximately the same.
Preparation of WCE.
For preparation of whole-cell extracts (WCE), cells were grown to 100% confluency in 100-mm tissue culture dishes or in suspension to a density of 106 cells/ml and washed twice with 10 ml of phosphate-buffered saline (minus calcium and magnesium). Cells were then lysed in 1 ml of EBC buffer containing 1 mM DTT, 1 mM sodium vanadate, 0.1 M phenylmethylsulfonyl fluoride, and 1 μg of each of leupeptin, pepstatin A, and antipain per ml. After 30 min of incubation on ice, the cell lysate was sonicated for 10 s (medium frequency) to homogenize the suspension. Clear supernatants were collected after centrifugation in an Eppendorf centrifuge (15 min, 13,000 rpm at 4°C) and stored at −80°C.
Kinase assays.
Tat-associated kinase (TAK) assays were described earlier (70). The substrates used in the TAK assays were CTD4 (four repeats of the heptamer YSPTSPS), purified GST-GrnCDE, or purified GST. To study GrnCDE-associated kinase activity, cell extract was mixed with GST-GrnCDE bound to glutathione-Sepharose beads. After 2 h of incubation at 4°C with rocking, the beads were washed extensively with EBCD buffer containing 0.03% SDS, followed by washing with TKB buffer (50 mM Tris-HCl [pH 7.6], 5 mM DTT, 4 mM MgCl2, 5 mM MnCl2). The kinase reaction was performed in the presence of 2 μM ATP and 10 μCi of [γ-32P]ATP at 25°C for 40 min. The proteins were denatured and resolved in denaturing 15% polyacrylamide gels. 32P-labeled proteins were detected by autoradiography.
Immunoprecipitation and kinase assays.
Protein A- or protein G-Sepharose beads (10 μl) equilibrated in EBCD buffer were mixed with 1 μg of anti-CDK9, 1 μg of anti-CDK8, or 1 μg of anti-CDK7 (anti C-terminal peptide polyclonal antibodies; Santa Cruz Biotechnology). After 1 h of incubation, beads were washed once in EBCD, mixed with 100 μg of cell extract, and incubated for 2 h at 4°C with rocking. The unbound proteins were washed extensively in EBCD containing 0.03% SDS, followed by equilibration in TKB. Kinase reaction mixes were incubated with 1 μg of purified GST, GST-GrnCDE, or GST-CTD at 25°C for 40 min as described before (49). We used 10 μM DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole; Sigma) as the kinase inhibitor (70).
Immunodepletion.
Immunodepletion was performed with CDK9 antibody (Santa Cruz Biotechnology) as described previously (70). The proteins were depleted from 293T cell extract, and EBCD buffer was used for binding and washing.
Coimmunoprecipitation.
293T cells (8 × 106) were transfected with 5 μg of each plasmid using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Cells were harvested at 24 h posttransfection and lysed in 1 ml of EBCD containing 0.1 mM phenylmethylsulfonyl fluoride, plus 1 μg each of pepstatin, leupeptin, and antipain per ml. Extracts were cleared and supernatants were collected after 15 min of centrifugation at 4°C (Eppendorf, 13,000 rpm). Antibody (1 μg) was bound to protein A-Sepharose or protein G-Sepharose by incubation at 4°C for 2 h with rocking. The conjugates were mixed with cell extract (100 μg of protein) and incubated overnight at 4°C with rocking. Unbound proteins were removed by extensive washing with EBCD buffer containing 0.03% SDS. Protein complexes bound to the conjugated antibody were separated by denaturing polyacrylamide gel electrophoresis and subjected to immunoblotting. Daudi cells were grown to 106 cells/ml, and WCE were prepared as above. Immunoprecipitates from 1 mg of WCE were washed exhaustively with EBCD to remove unbound proteins.
Western blot analysis.
Immunoblotting was performed as described previously (50). Anti-FLAG M2 monoclonal antibody was obtained from Sigma, anti-HA rabbit polyclonal antibody was from Santa Cruz Biotechnology, and anti-GEP rabbit polyclonal antibody was from G. Serrero (34).
Localization of granulin and cyclin T1.
Intracellular localization of cyclin T1 and GrnCDE, GEP, and GrnDE was carried out in COS7 cells. Cells were grown on collagen-coated coverslips in tissue culture plates to a confluency of 30 to 40% and transfected with the indicated plasmids with Cytofectene (Bio-Rad) according to the manufacturer's instructions. The expression and localization or colocalization of the fluorescent label-tagged proteins were directly analyzed under a Nikon PCM 2000 confocal microscope. Digital images were processed with Adobe Photoshop.
Transfection and luciferase assays.
CEM and U937 cells (107 cells in 250 μl of RPMI without serum) were transfected by electroporation with a Bio-Rad Gene Pulser at 960 μF and 250 V. The DNA amounts of all the plasmids were equalized with pUC19 DNA. After electroporation, cells were transferred into plates containing 10 ml of complete RPMI. Cells were harvested after 20 h of transfection and lysed in 1 ml of 1× Promega lysis buffer. MCF-7 and 3T3 cells (3 × 105 cells) were transfected with Lipofectamine. After 20 h, the cells were harvested and lysed as above. Luciferase assays were performed with the Promega dual luciferase reporter system according to the manufacturer's instructions.
RESULTS
Human cyclin T1 interacts with granulin.
As an approach to the identification of cellular components that might regulate cyclin T1, we used the yeast two-hybrid system to screen for cyclin T1-interacting proteins. Cyclin T1 lacking its C-terminal PEST sequence (amino acids 1 to 708; Fig. 1C), which is required for CDK9 degradation (27), was cloned into a vector containing the GAL4 binding domain (BD) to form the fusion protein GAL4BD-T1. The PEST sequence was deleted to avoid the possibility that it would destabilize the cyclin T1 fusion protein in S. cerevisiae.
The ability of the fusion protein to interact with CDK9 fused to the GAL4 activation domain (AD) in S. cerevisiae cells was confirmed and served as a positive control in all yeast two-hybrid experiments (Table 1). Only S. cerevisiae cells that carry vectors expressing interacting proteins grow on medium lacking leucine, tryptophan, and histidine and produce the enzyme β-galactosidase. A standard positive control was provided by the interaction between p53 and simian virus 40 T antigen. Negative controls were vectors that lacked a GAL4-fused gene or encoded the irrelevant “sticky” protein lamin C.
TABLE 1.
Interaction of human cyclin T1 and HIV-1 Tat with granulin
| Gal4 BD fusion | Gal4 AD fusion | Growth | β-Galactosidase activity |
|---|---|---|---|
| None | None | − | − |
| Cyclin T1 | CDK9 | + | + |
| p53 | Simian virus 40 T antigen | + | + |
| Lamin C | GrnCDE | − | − |
| Cyclin T1 | GrnCDE | + | + |
| GrnCDE | Cyclin T1 | + | + |
| Cyclin T1 | GEP | + | + |
| Cyclin T1 | GrnPGFBA | + | + |
| Cyclin T1 | GrnC | − | − |
| GrnCDE | CDK9 | − | − |
| Wild-type Tat72 | GrnCDE | + | + |
| Wild-type Tat72 | GEP | + | + |
| Wild-type Tat72 | GrnPGFBA | + | + |
| Wild-type Tat72 | GrnC | − | − |
| Wild-type Tat72 | CDK9 | − | − |
The cyclin T1 construct was used as a bait to screen a HeLa cDNA library linked to the GAL4 AD. Among the clones isolated, a polypeptide that interacts strongly with cyclin T1 (as determined by β-galactosidase activity levels) was found to be expressed from a partial cDNA sequence derived from the growth factor granulin. The 251 granulin residues encoded in the clone encompassed the protein's C-terminal portion (amino acids 342 to 593) and contained the C, D, and E granulin repeats. This segment is referred to as GrnCDE, and full-length granulin is referred to as GEP (Fig. 1A). Interaction was maintained when the fused proteins and GAL4 domains were exchanged (i.e., GAL4BD-GrnCDE and GAL4AD-cyclin T1). GEP, 593 amino acids long and composed of seven and one-half repeats of the cysteine-rich granulin motif (Fig. 1A), interacted with cyclin T1 in the yeast two-hybrid system (Table 1). The N-terminal portion of GEP, GrnPGFBA, also interacted, but the isolated C repeat (GrnC) did not (Table 1, Fig. 1A).
Since CDK9 is the kinase partner of cyclin T1, we tested whether GrnCDE could interact with CDK9. No such interaction was detected in the yeast two-hybrid system (Table 1). On the other hand, the HIV-1 protein Tat, which binds cyclin T1, also interacted with GEP, GrnCDE, and GrnPGFBA in the yeast two-hybrid system but not with GrnC. As expected, Tat did not interact with CDK9 (Table 1).
To verify the interactions between full-length cyclin T1 and granulin in vitro, we performed GST pulldown assays (Fig. 1B). 35S-labeled GrnCDE (lane 1) and GEP (lane 5) were both pulled down by GST-cyclin T1 (lanes 3 and 7) but not by GST alone (lanes 2 and 6) or GST-CDK9 (lanes 4 and 8). In this assay, GST-CDK9 pulled down 35S-labeled cyclin T1 (lane 10), as expected. These results confirm that granulin interacts specifically with cyclin T1 but not with CDK9.
Mapping of granulin-binding domain within cyclin T1.
The human cyclin T1 cDNA used as the bait in the yeast two-hybrid screen contains several defined domains and sequences that could participate in granulin binding (Fig. 1C). These include a cyclin domain (amino acids 1 to 254) homologous to that of other cyclins, the adjacent Tat-TAR recognition motif (amino acids 254 to 272), which also includes a potential nuclear localization signal, a potential coiled-coil region (amino acids 384 to 425), a histidine-rich domain (His; amino acids 506 to 530), and a serine-rich sequence (Ser; amino acids 560 to 570).
To identify regions of cyclin T1 that interact with granulin, we constructed a series of C-terminal and N-terminal truncations of cyclin T1 that could be expressed in S. cerevisiae and tested them in the yeast two-hybrid system. The results, summarized in Fig. 1C, showed that C-terminal truncations containing amino acids 1 to 288 and 1 to 418 did not bind to GrnCDE, indicating that the cyclin domain, Tat-TAR recognition motif, and part of the coiled-coil region are not sufficient for binding. Correspondingly, the N-terminal truncations 229 to 708 and 364 to 708 retained GrnCDE-binding activity, confirming that the cyclin domain and the Tat-TAR recognition motif are not important for binding. On the other hand, the C-terminal truncation 1 to 573 interacted with GrnCDE, while the complementary N-terminal truncation, amino acids 573 to 708, did not bind. These data indicate that the region of cyclin T1 (amino acids 364 to 573) that contains the coiled-coil and His-rich domains is important for binding.
To determine whether both of these elements are necessary for the interaction, we constructed a deletion mutant, amino acids 415 to 573, which lacked most of the coiled-coil region but contained the intact His-rich domain and surrounding sequences. This region was sufficient for GrnCDE binding. Further deletion constructs (amino acids 480 to 570 and 480 to 530) fully excluded the involvement of the coiled-coil region and localized the binding activity to residues 480 to 530. Similar data were obtained with GEP, indicating that the region of cyclin T1 that interacts with granulin includes its His-rich domain (amino acids 506 to 530).
Cyclin T1's histidine-rich domain is necessary and sufficient for granulin binding.
Similar cyclin T1 truncations were produced as GST fusion proteins (Fig. 2A). In keeping with the observations made in S. cerevisiae, 35S-labeled GrnCDE was pulled down only when the His-rich domain and its surroundings were included in the GST-cyclin T1 deletion construct (Fig. 2B, lanes 3 to 9). To further narrow down the region responsible, additional cyclin T1 deletions were made around the His-rich domain. Pulldown experiments with labeled GrnCDE (lanes 12 to 15) refined the binding site on cyclin T1 to between amino acids 501 and 530 (lane 13), which contain the His-rich stretch (amino acids 517 to 526). The sequence THPSNH5NHHSH is absolutely conserved among the mouse, horse, and human cyclin T1 proteins. Human cyclin T2, however, contains the sequence HPSSRHHTSSHKHSHSHS, which has fewer histidine residues overall and no uninterrupted run of more than two histidines. When tested in a pulldown assay with 35S-labeled GrnCDE, GST-cyclin T2a (440-663) interacted only weakly relative to the C-terminal domain of cyclin T1 (Fig. 2C, compare lane 4 with lanes 2 and 3). Since the two cyclins are largely homologous, this observation supports the inference that the His-rich domain of cyclin T1 is required for binding to granulin.
FIG. 2.
Granulin binds to the His region of cyclin T1. (A) Schematic representation of GST-cyclin T1 constructs (shaded bars) and GST-cyclin T2a construct (solid bar) used in pulldown assays with GrnCDE. Results (illustrated in parts B and C) are summarized in the column on the right. (B) In vitro-synthesized 35S-labeled GrnCDE was pulled down by GST fusions with full-length or truncated cyclin T1 as indicated. (C) Comparison of GrnCDE pulldowns by GST fusions with the C-terminal regions of cyclin T1 and cyclin T2a. Bound proteins were separated and detected as in Fig. 1.
For direct evidence, we synthesized a peptide of 20 residues corresponding to amino acids 511 to 530 of cyclin T1 (Fig. 3A). When added together with 35S-labeled GrnCDE, this peptide efficiently competed for binding to GST-cyclin T1 (Fig. 3B, left panel). Increasing amounts of the peptide progressively decreased the amount of GrnCDE that was pulled down by the fusion proteins GST-cyclin T1(480-570) and GST-cyclin T1(501-530). It did not compete effectively with GrnCDE binding to the HIV-1 Tat activation domain (Fig. 3B, right panel). The mutant peptide 7H-7A, in which seven of the histidine residues at the core of the His peptide were changed to alanines, failed to compete (Fig. 3C). A truncated version of the wild-type peptide (amino acids 516 to 525) that contained the seven histidine residues but lacked flanking sequences, competed very inefficiently (Fig. 3C). We conclude that granulin binds specifically to the 20 amino acids comprising the His-rich domain of cyclin T1 and that the series of seven histidines within it, as well as flanking residues, participate in the interaction.
FIG. 3.
Effect of His region peptides on granulin-cyclin T1 interactions. (A) Sequences of the 20-amino-acid His peptide that corresponds to residues 511 to 530 of human cyclin T1 and contains the His-rich sequence; the 7H-7A peptide, in which seven histidine residues were changed to alanines; and the 10-amino-acid peptide containing residues 516 to 525. The peptides were chemically synthesized. (B) Increasing amounts of His peptide 511 to 530 inhibited the binding of 35S-labeled, in vitro-synthesized GrnCDE to the His-rich domain in GST-cyclin T1(480-570) or GST-cyclin T1(501-530) (left panel) but did not inhibit the binding of GrnCDE to GST-Tat48Δ (right panel). (C) Increasing amounts of His peptide 511 to 530 or the peptides 7H-7A or 516 to 525 were added, together with 35S-labeled, in vitro-synthesized GrnCDE, to GST-cyclin T1(501-530) beads.
P-TEFb interacts with granulin in vivo.
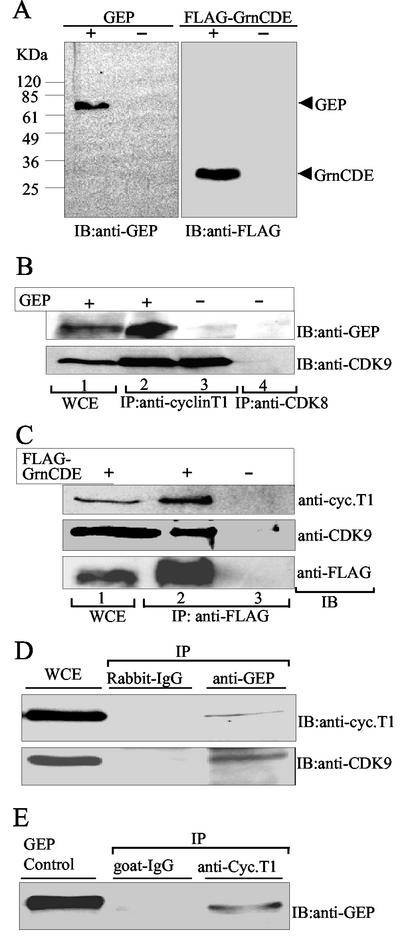
Most of the cyclin T1 in HeLa cell nuclear extracts is bound to CDK9 in the P-TEFb complex (47). To determine whether GEP interacts with P-TEFb in human cells, we expressed GEP or FLAG-tagged GrnCDE proteins in 293T cells and conducted sequential immunoprecipitation and immunoblotting analyses. WCE prepared from transfected and mock-transfected cells were probed with antibody against GEP, which detected an intact ≈70-kDa protein, or with anti-FLAG antibody, which detected GrnCDE migrating at ≈30 kDa (Fig. 4A). GEP was detected in transfected cells at least until 72 h posttransfection, but no endogenous GEP was detected in untransfected cells, suggesting that the level of this protein in these cells is low (data not shown). Extracts prepared from mock-transfected cells or cells overexpressing GEP were immunoprecipitated with anti-cyclin T1 antibody, and the resulting complexes were probed with anti-GEP and anti-CDK9 antibodies (Fig. 4B). Complexes from overexpressing cells gave a strong GEP signal (lane 2), and a weak signal was visible in the immunoprecipitate from mock-transfected cells (lane 3). CDK9 was present in both cases, as expected. Neither protein was detectable in control immunoprecipitates with anti-CDK8 (lane 4). Thus, granulin is present in cells in complexes with cyclin T1.
FIG. 4.
Granulin interacts with P-TEFb in vivo. (A) Immunoblot (IB) analysis of WCE from 293T cells overexpressing GEP or GrnCDE. GEP and FLAG-tagged GrnCDE were detected in extracts of transfected cells but not mock-transfected cells. (B) GEP coimmunoprecipitates with cyclin T1. Cyclin T1 complexes were immunoprecipitated (IP) from WCE prepared from 293T cells overexpressing GEP or mock transfected, as indicated (lanes 2 and 3). The immunoprecipitated complexes were separated in SDS-polyacrylamide gels, transferred to a membrane, and probed with anti-GEP or anti-CDK9 antibodies by immunoblotting (IB). Controls included unfractionated WCE (lane 1) and immunoprecipitation with anti-CDK8 (lane 4). (C) GrnCDE coimmunoprecipitates with CDK9 and cyclin T1. The immunoprecipitated complexes were probed with anti-FLAG, anti-CDK9. and anti-cyclin T1 antibodies, as indicated. Complexes containing GrnCDE, CDK9, and cyclin T1 were detected in extracts from 293T cells overexpressing FLAG-GrnCDE (lane 2) but not from cells transfected with empty vector (lane 3). (D) Endogenous cyclin T1 and GEP coimmunoprecipitate. Cyclin T1 complexes immunoprecipitated from WCE prepared from Daudi cells were probed with anti-GEP as described in for B. Controls included goat immunoglobulin G and purified GEP. (E) Endogenous GrnCDE coimmunoprecipitates with CDK9 and cyclin T1. Complexes were immunoprecipitated with anti-GEP and probed with anti-cyclin T1 and anti-CDK9 antibodies, as indicated. Rabbit immunoglobulin G was used as the control.
To determine whether granulin forms a complex with intact P-TEFb containing CDK9 as well as cyclin T1, cells were transfected with FLAG-tagged GrnCDE. Immunoblotting confirmed the presence of all three proteins, FLAG-GrnCDE, endogenous cyclin T1, and CDK9, in the extracts (Fig. 4C, lane 1). Anti-FLAG antibody coimmunoprecipitated cyclin T1 and CDK9 from transfected cell extracts (lane 2) but not from mock-transfected cell extracts (lane 3). Therefore, cyclin T1 can simultaneously bind both GrnCDE and CDK9, implying that granulin interacts with P-TEFb in vivo. In case these data resulted from the expression of exogenous tagged proteins, we repeated the coimmunoprecipitation experiments with untransfected Daudi cells. These cells express higher levels of GEP than HeLa, 293T, 3T3, or U937 cells (data not shown). Anti-cyclin T1 precipitated GEP from Daudi cell extracts (Fig. 4D), and reciprocally, anti-GEP antibody precipitated cyclin T1 (Fig. 4E). Complexes precipitated with anti-GEP also contained CDK9 (Fig. 4E), confirming that granulin binds to P-TEFb in vivo.
Colocalization of granulin and cyclin T1.
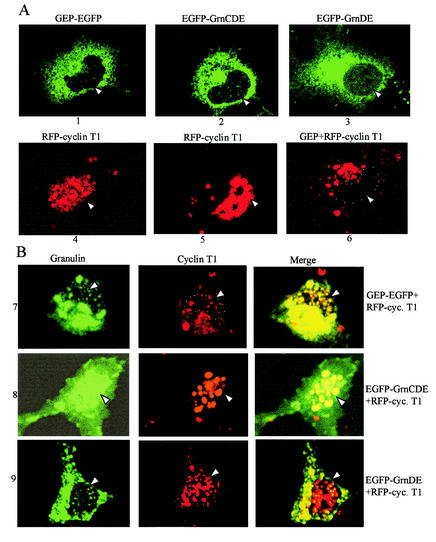
Granulin is made with a signal sequence for export and is known to be a secreted protein (69), so it was surprising to find it coprecipitating with cellular proteins from cell extracts. However, secreted proteins can be reimported to the cytosol from the endoplasmic reticulum (51) or by receptor-ligand internalization, and there are precedents for intracellular proteins doubling as cytokines (60). We therefore examined the distribution of GEP, GrnCDE, GrnDE, and cyclin T1 in living cells by confocal microscopy with fluorescently tagged proteins. Cyclin T1 tagged with red fluorescent protein (RFP) supported Tat transactivation in 3T3 cells about half as well as untagged cyclin T1 (data not shown). All three granulins were seen predominantly in the cytoplasm (Fig. 5A, panels 1 to 3), whereas RFP-cyclin T1 was predominantly nuclear (panels 4 and 5). In agreement with previous observations made with either endogenous or fluorescently tagged cyclin T1 (17, 38, 42, 45), RFP-cyclin T1 was present in the nonnucleolar nucleoplasm, giving a dotted pattern. Coexpression of untagged GEP led to a dramatic shift in the distribution of RFP-cyclin T1 into the cytoplasm (panel 6), consistent with a biochemical interaction in this compartment.
FIG. 5.
Intracellular localization of granulins and cyclin T1. (A) EGFP-fused granulins and RFP-fused cyclin T1 were expressed individually in COS7 cells (panels 1 to 5). The dotted nuclear distribution of cyclin T1 is seen in panel 4, whereas the overexposed image from another cell shows the presence of cyclin T1 in the nucleoplasm but not in the nucleoli (panel 5). In panel 6, untagged GEP was coexpressed with RFP-fused cyclin T1. Monochromatic images are shown in each case. (B) Distribution of cyclin T1 and the granulins in cells coexpressing RFP-fused cyclin T1 with EGFP-tagged GEP, GrnCDE, or GrnDE (panels 7 to 9, respectively). Monochromatic images for EGFP and for RFP and the merged images are displayed from left to right. Arrows and white dots show the position of the nucleus in individual cells.
To determine whether granulin and cyclin T1 colocalize within the cell, the two proteins were coexpressed in tagged form (Fig. 5B). In the presence of GEP, much of the cyclin T1 moved into the cytoplasm and colocalized there with the GEP (panels 7). Conversely, when GrnCDE was coexpressed with cyclin T1, most of the GrnCDE moved into the nucleus and colocalized with the cyclin T1 (panels 8). Thus, the proteins colocalized to a considerable extent, and some redistribution took place within the cell. As a control, we used GrnDE, which does not interact with cyclin T1 in the yeast two-hybrid assay (unpublished data). Its distribution was largely cytoplasmic and was not greatly perturbed by coexpression with cyclin T1, although some overlap was seen with the minority of the cyclin T1 resident in the cytoplasm (panels 9). These data provide confirmation of granulin-cyclin T1 interactions in living cells.
Granulin blocks CTD phosphorylation but is a P-TEFb substrate.
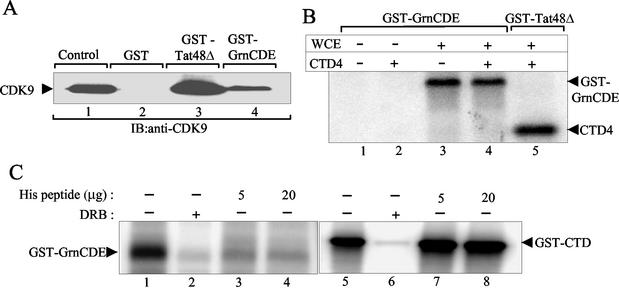
P-TEFb was initially identified as a Tat-associated kinase that binds tightly to GST-Tat fusion protein and phosphorylates the CTD of polymerase II (18, 49). Since granulin interacts with P-TEFb in cell extracts, it seemed likely that GST-GrnCDE would also be able to bind P-TEFb from human cell extracts. Depending on the nature of the interaction, the resultant complex might have altered ability to phosphorylate the CTD. Immunoblotting showed that CDK9 was pulled down by GST-GrnCDE (Fig. 6A, lane 4), albeit less efficiently than by GST-Tat 48Δ (amino acids 1 to 48), which contains the activation domain of HIV-1 Tat (lane 3). Since granulin interacts directly with cyclin T1 but not with CDK9 (Fig. 1B and Table 1), this result implies that the GST-GrnCDE complex contains P-TEFb.
FIG. 6.
Effect of granulin binding on kinase activity of P-TEFb. (A) GST-GrnCDE pulls down P-TEFb from 293T cell WCE. Immunoblot (IB) analysis was performed with anti-CDK9 antibody. CDK9 was detected in complexes bound to GST-GrnCDE (lane 4) and GST-Tat48Δ (lane 3) but not GST alone (lane 2). WCE was used as control for the presence of CDK9 (lane 1). (B) Complexes bound to GST-GrnCDE and GST-Tat48Δ were tested in kinase assays in the absence and presence of CTD4 peptide. CTD4 was phosphorylated by GST-Tat48Δ-bound complexes (lane 5) but not by GST-GrnCDE-bound complexes (lane 4). GST-GrnCDE was phosphorylated in the absence or presence of CTD4 (lanes 3 and 4). Neither GST-GrnCDE nor CTD4 was phosphorylated in the absence of WCE (lanes 1 and 2). (C) CDK9 immunocomplexes from 293T WCE phosphorylated purified GST-GrnCDE (lane 1) or GST-CTD (lane 5). The phosphorylation was inhibited by 10 μM DRB (lanes 2 and 6). Increasing concentration of His peptide 511 to 530 inhibited the phosphorylation of GST-GrnCDE but not of GST-CTD.
We then examined the kinase activity of the P-TEFb complexes bound to GST-GrnCDE. Surprisingly, these complexes failed to phosphorylate CTD4 (Fig. 6B, lane 4), a peptide containing four repeats of the CTD heptad that was phosphorylated efficiently by P-TEFb complexes bound to GST-Tat 48Δ (lane 5). Similar results were obtained with different amounts of the P-TEFb complexes (data not shown). However, a phosphorylated band with the mobility of GST-GrnCDE itself was labeled in reactions containing GST-GrnCDE-bound P-TEFb complexes irrespective of the presence of the CTD peptide (lanes 3 and 4). Immunoblotting the gel in Fig. 6B with antibody against GST confirmed that the labeled band contained the GST fusion protein (data not shown). These data indicate that granulin binds P-TEFb and serves as a substrate for the bound kinase but inhibits its ability to phosphorylate the CTD4 peptide.
To confirm the phosphorylation of Grn-CDE by CDK9, P-TEFb was immunoprecipitated with antibody to CDK9 and used in kinase assays to which purified GST-GrnCDE was added as a substrate. GST-GrnCDE, like GST-CTD, served as a substrate for P-TEFb (Fig. 6C, lanes 1 and 5). The His peptide (511 to 530), which blocked the interaction of cyclin T1 with granulin (Fig. 3B and 3C), also blocked the phosphorylation of GST-GrnCDE by P-TEFb (Fig. 6C, lanes 3 and 4), as would be expected. On the other hand, the same peptide did not affect the phosphorylation of GST-CTD (lanes 7 and 8). Taube et al. (56) recently reported that the CTD of polymerase II interacts with the His-rich region of cyclin T1, specifically amino acids 481 to 551. Our data imply that residues 511 to 530, comprising the core of the His-rich region, are not sufficient for the cyclin T1-CTD interaction, at least in our in vitro assay.
Specificity of granulin phosphorylation.
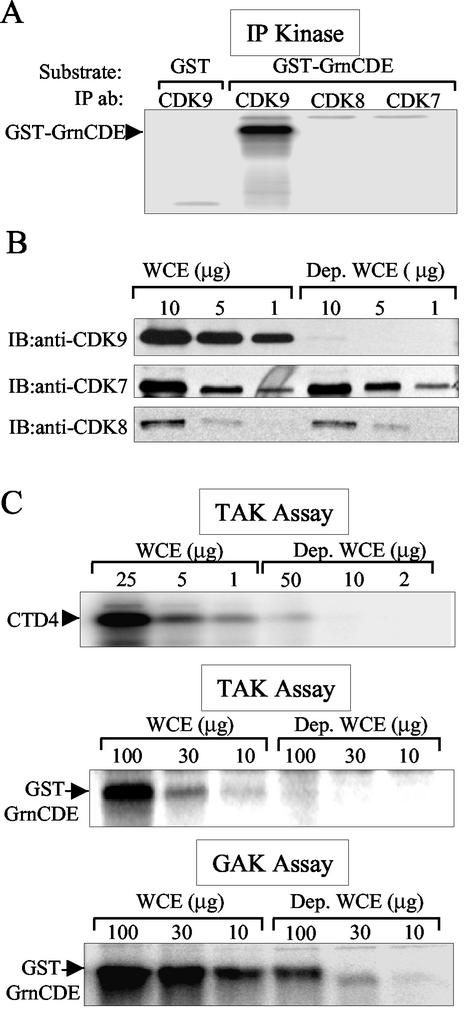
Like the CTD, phosphorylation of granulin by P-TEFb was sensitive to the adenosine analogue DRB, a strong inhibitor of this kinase (Fig. 6C, lanes 2 and 6). To determine whether other kinases are capable of phosphorylating granulin, cellular complexes containing CDK7, CDK8, or CDK9 were immunoprecipitated with appropriate antibodies (49). The results depicted in Fig. 7A show that CDK9 complexes phosphorylated GST-GrnCDE, whereas the CDK7 and CDK8 complexes did not.
FIG. 7.
GST-GrnCDE is phosphorylated by P-TEFb and TAK. (A) GST-GrnCDE is a substrate for CDK9 but not CDK8 or CDK7. CDK9, CDK8, and CDK7 immunocomplexes from 293T WCE were used in kinase assays with purified GST or GST-GrnCDE as the substrate. (B) Depletion of CDK9 from 293T cell WCE. Immunoblotting (IB) detected only trace amounts of CDK9 in depleted (Dep.) WCE (upper panel), whereas the levels of CDK7 and CDK 8 remained the same (middle and lower panels). (C) GST-GrnCDE phosphorylation is greatly reduced after P-TEFb depletion. After binding to GST-Tat48Δ, kinase assays (TAK assays) were conducted with CTD4 peptide (upper panel) or GST-GrnCDE (middle panel) as the substrate. GST-GrnCDE phosphorylation was also examined after binding to GST-GrnCDE (GAK assay; bottom panel).
We next conducted depletion experiments to determine whether this substrate can be phosphorylated by other cellular kinases. If GST-GrnCDE is phosphorylated only by CDK9, removal of CDK9 complexes from cell extracts should eliminate GST-GrnCDE phosphorylation. Depletion of CDK9 from 293 cell extracts by anti-CDK9 antibody removed nearly all of this protein, while the amounts of CDK7 and CDK8 were unchanged (Fig. 7B). Quantitation of the immunoblots revealed that the depleted extracts contained almost 100-fold less CDK9 than the undepleted extracts (data not shown). Like the CTD, GST-GrnCDE is phosphorylated by CDK9 bound to Tat48Δ (i.e., in TAK assays; Fig. 7C, top and middle panels, respectively). A 50- to 100-fold decrease in the phosphorylation of both CTD4 and GST-GrnCDE substrates was observed when the CDK9-depleted extracts were bound to GST-Tat 48Δ (Fig. 7C). These data confirm that GST-GrnCDE is phosphorylated by P-TEFb, specifically by CDK9-cyclin T1, the only P-TEFb complex that binds to Tat. They also show that Tat binding does not inhibit the phosphorylation of granulin by P-TEFb.
We also examined the possibility that kinases other than P-TEFb can bind to granulin and phosphorylate it. The depleted and control extracts were compared in a GST-GrnCDE pulldown kinase assay (GrnCDE-associated kinase assay; Fig. 7C, lower panel). Depletion of CDK9 complexes reduced the level of GST-GrnCDE phosphorylation by at least 10-fold, but the remaining amount of GST-GrnCDE phosphorylation indicates that an additional kinase(s) may bind to it and recognize it as a substrate.
Interactions of granulin with Tat.
It has been reported that a truncated form of granulin lacking the first 84 amino acids and GrnBA (containing repeats B and A) interact with exon 1 of HIV-2 Tat in the yeast two-hybrid system. The cysteine-rich domains of both HIV-1 and HIV-2 Tat were found to be essential for this interaction in pulldown assays with GST-GrnBA (57). In accord with the yeast two-hybrid data shown in Table 1, GEP and GrnCDE interacted in GST pulldown assays with full-length HIV-1 Tat72 and Tat86 (Fig. 8A). The Tat activation domain (GST-Tat48Δ), containing the Cys-rich domain (amino acids 22 to 37), was sufficient for granulin binding (Fig. 8A, lane 4, and Fig. 3B). To determine whether such interactions occur in human cells, we cotransfected 293T cells with FLAG-GrnCDE and hemagglutinin (HA)-Tat72. Complexes immunoprecipitated with antibody to GrnCDE contained Tat (Fig. 8B, lane 4), showing that the proteins can interact in vivo.
FIG. 8.
Granulin interacts with Tat in vitro and in vivo. (A) The Tat activation domain is sufficient for binding to GEP and GrnCDE. 35S-labeled, in vitro-synthesized GrnCDE and GEP (upper and lower panels, respectively) were pulled down by GST-Tat fusion proteins as indicated. Bound proteins were separated in a denaturing polyacrylamide gel and autoradiographed. (B) Tat coimmunoprecipitates with GrnCDE from 293T cell extracts. GrnCDE complexes were immunoprecipitated from cells overexpressing FLAG-GrnCDE, FLAG-GrnCDE together with HA-Tat, or the corresponding empty vectors. Immunoblot analysis of these complexes with anti-HA antibody detected the presence of Tat in GrnCDE complexes when Tat was coexpressed with GrnCDE (lane 4) and not when it was expressed with the empty vector (lane 3).
Granulin inhibits Tat transactivation.
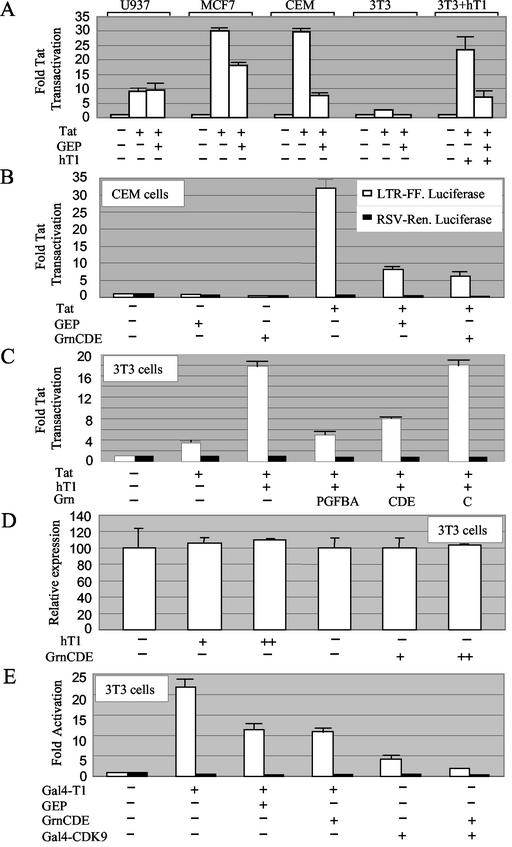
The observations that granulin binds the His-rich domain of cyclin T1, blocks the phosphorylation of CTD4 by P-TEFb, and interacts with the Tat activation domain suggested that it might inhibit Tat transactivation. Anticipating that any response would be more evident in cells that express less endogenous granulin, we surveyed a series of cell lines for the influence of GEP overexpression on Tat transactivation (Fig. 9A). Transactivation was monitored by assaying the expression of luciferase under the control of the HIV-1 long terminal repeat. Tat transactivation was observed in three human cell lines, U937 (promonocytic), MCF-7 (breast carcinoma), and CEM (T cell). Cotransfecting a GEP expression vector reduced transactivation in both MCF-7 and CEM cells, which express intermediate and low levels of granulin mRNA, respectively, but had little or no effect on transactivation in U937 cells, which express relatively high levels of granulin mRNA (5).
FIG. 9.
Inhibition of gene expression by granulin. (A) Effect of GEP on Tat transactivation in different cell types. CEM and U937 cells were transfected with 4 μg of HIV long terminal repeat-firefly luciferase reporter plasmid, 0.1 μg of RSV-Tat, and 10 μg of CMV-GEP plasmid as indicated. MCF-7 and 3T3 cells were transfected with 0.1 μg, 0.02 μg, and 2 μg of these plasmids, respectively. Where indicated, 3T3 cell transfections were supplemented with 0.1 μg of human cyclin T1 (hT1). Each cell type was tested in two or three experiments. Results are the average of duplicates, with standard errors, expressed relative to the respective controls without Tat. (B) Both GEP and GrnCDE inhibit Tat transactivation in CEM cells. Transfections were performed as in A except that 1 μg of RSV-Renilla luciferase plasmid was included in all cases (black bars). (C) Inhibition of Tat transactivation correlates with granulin-cyclin T1 interactions. Mouse 3T3 cells were transfected as in A except that 0.1 μg of RSV-Renilla luciferase plasmid was included. (D) GrnCDE has no effect on the expression of firefly luciferase from the human PCNA promoter. 3T3 cells were transfected with 0.1 μg of PCNA-firefly luciferase, 0.1 μg or 0.5 μg of human cyclin T1, and 1 or 2 μg of GrnCDE as indicated. Results are expressed as the ratio of firefly to Renilla luciferase activity, with standard errors. The value for controls lacking human cyclin T1 and GrnCDE was set at 100. (E) Inhibition of cyclin T1 function by granulin. NIH 3T3 cells were transfected with 0.1 μg of G5-HIV luciferase, 0.1 μg of RSV-Renilla, 0.25 μg of Gal4-cyclin T1 or 0.25 μg of Gal4-CDK9, and 1.6 μg of GEP or equimolar amounts of GrnCDE or empty vector, as indicated.
The actions of GEP and GrnCDE on basal and Tat-transactivated gene expression in CEM cells were compared in the experiment in Fig. 9B. Tat stimulated transcription from the HIV-1 promoter by ≈30-fold, as measured by luciferase expression. Cotransfection with GEP or GrnCDE reduced the Tat stimulation by ≈75%. In contrast, GEP and GrnCDE had only a small inhibitory effect on the basal expression of firefly luciferase from the HIV-1 promoter and on Renilla luciferase expression from the RSV promoter.
Specificity of granulin's transcriptional effect.
Mouse NIH 3T3 cells elicited only a weak response to Tat, as expected (61), but coexpression of human cyclin T1 allowed a robust transactivation which was repressed by GEP (Fig. 9A). These data are in keeping with the low level of endogenous granulin mRNA in NIH 3T3 cells (5) and with the involvement of cyclin T1 in the granulin response. Results of yeast two-hybrid assays showed that either the N- or C-terminal portion of GEP, GrnPGFBA and GrnCDE, respectively, can interact with cyclin T1, whereas GrnC does not (Table 1). Correspondingly, GrnPGFBA and GrnCDE both inhibited transactivation mediated by cyclin T1 in 3T3 cells were transfected as in Fig. 9A except that 0.1 μg of RSV-Renilla was included in all cases (Fig. 9C). There was no effect of GrnCDE on the expression of firefly luciferase from the human PCNA promoter, which is not responsive to the exogenous expression of cyclin T1 (Fig. 9D). We conclude that granulin can selectively repress Tat transactivation in human and mouse cells.
Since granulin also interacts with Tat, we wanted to determine whether the transcriptional inhibition by granulin required the presence of Tat. For this purpose, we conducted transactivation assays in the absence of Tat using a Gal4BD-cyclin T1 fusion protein to tether cyclin T1 to the HIV-1 promoter via Gal4 binding sites inserted in the reporter construct (37). As shown in Fig. 9D, cotransfection of GEP or GrnCDE reduced luciferase expression in the absence of Tat. Similarly, transcriptional activation by tethered CDK9 was also inhibited by GrnCDE. These findings are consistent with a direct and specific action of granulin on transcription via its interaction with cyclin T1.
DISCUSSION
Cyclin T1 is a subunit of the essential transcription factor P-TEFb as well as the longest cyclin known to date, implying that it is likely to engage in interactions with numerous regulatory proteins. Granulin was identified in a screen for cyclin T1-interacting proteins. Remarkably, this protein also interacts with HIV-1 Tat, an established viral cofactor of cyclin T1. We found that granulin inhibits CTD phosphorylation in vitro as well as the human cyclin T1-dependent activation of the HIV-1 promoter in vivo, suggesting that it can act as a cellular repressor of transcription elongation. Although it was originally purified as a growth factor from conditioned medium and its biological effects were assessed after its addition to tissue culture medium, granulin also works as a growth factor when expressed inside cells (35, 66). Our data demonstrate that granulin is present within cells, whether expressed endogenously or from plasmid vectors. These findings suggest a model in which granulin's growth-promoting effects are elicited by repressing the transcriptional elongation of genes involved in limiting cell growth, such as tumor suppressor genes.
Cyclin T1 interaction with granulin.
Since the isolation of cyclin T1 as a CDK9 partner and Tat-interacting protein, three other cellular proteins have been identified that bind directly to the cyclin box (amino acids 1 to 254) of cyclin T1, the major histocompatibility complex class II transactivator CIITA, the NF-κB RelA/p65 subunit, and the glucocorticoid receptor interacting polypeptide 1 GRIP-1 (3, 24, 30). Tat binding also requires the cyclin box in addition to the adjacent Tat-TAR recognition motif (amino acids 250 to 262) (14). Like the CTD of polymerase II (56), granulin binds outside this N-terminal region in the His-rich region. The interaction sites for these two proteins on cyclin T1 may not be congruent, however, since the phosphorylation by P-TEFb of granulin is inhibited by the His-rich peptide (amino acids 511 to 530), whereas CTD phosphorylation is resistant (Fig. 6C). Granulin interacts directly with the histidine-rich domain of cyclin T1 (amino acids 501 to 530), but not with the corresponding region of cyclin T2a, which is less rich in histidine residues (Fig. 2); this suggests that P-TEFb complexes containing cyclin T2 may not recognize granulin as a substrate. These findings suggest that the His-rich region may be a regulatory domain and substrate-binding site for cyclin T1. Competition experiments indicate that the highly histidine-rich core of the His-rich peptide (amino acids 516 to 525) is necessary but not sufficient for the interaction.
The mapping of the granulin interaction site has structural and functional implications. Granulin is a cysteine-rich protein, containing seven and a half repeats of the consensus sequence CX5-6CX5CCX8CCX6CCX5CCX5CX5-6C (with the exception of the G repeat, which has 10 cysteines instead of 12). The three-dimensional structure of carp granulin 1 (one repeat) displays a novel superhelix consisting of four stacked β-hairpins linked by an axial rod of six disulfide bonds. In its N-terminal subdomain, granulin shows structural homology with the N-terminal part of epidermal growth factor and transforming growth factor alpha, which are involved in receptor binding. The C-terminal subdomain contains the conserved sequence CCXDX2HCCP, and it has been suggested to have a metal binding site and to be involved in regulatory function (20).
Cysteines and histidines coordinate zinc ions to form structural elements such as zinc fingers and RING fingers, which participate in protein-protein interactions that are bridged by binding to zinc ions (23, 31, 52). In the cyclin T1-granulin interaction, the His-rich domain of cyclin T1 interacts with cysteine-rich granulin motifs. We speculate that granulin-cyclin T1 binding is mediated by zinc ions bridging between cysteines present in one or more of granulin's repeats and the polyhistidine stretch of cyclin T1. Such zinc-mediated association between one protein containing a cysteine-rich region and another containing a histidine-rich sequence could be a novel general type of protein-protein interaction.
Granulin interaction with Tat.
Granulin has also been reported to interact with the mammalian type III hexokinase, possibly as part of a large multiprotein complex (54), but the significance of this interaction is unknown. It also binds to Tat, as shown here (Table 1 and Fig. 8) and previously (57). As with cyclin T1, the granulin interaction is with the activation domain of Tat (Fig. 8). Although Tat is not required for granulin's inhibitory effect on transcription (Fig. 9E), the possibility that Tat modulates the transcriptional action of granulin remains to be explored.
The requirements for granulin binding to Tat and cyclin T1 are similar but not identical. In contrast to the three subunits in GrnCDE, one unit of granulin, GrnC, is not sufficient to bind to cyclin T1 or Tat in S. cerevisiae (Table 1), and this subunit does not inhibit Tat transactivation (Fig. 9C). On the other hand, although GrnDE fails to interact with cyclin T1, it does interact with Tat in the yeast two-hybrid system (Fig. 5B and data not shown). As with cyclin T1, Tat-granulin interactions may be dependant on metal ions. The interaction between Tat and cyclin T1 in vitro is dependent on zinc, and it has been suggested that Cys-261 in cyclin T1, which is crucial for Tat binding to cyclin T1, is involved in Tat binding via a zinc ion bridge (14, 50). Examination of the binding of zinc to a Tat-derived peptide (amino acids 21 to 38) led to a structural model in which five cysteines and one histidine were postulated to bind to two zinc ions (21). Similarly, the binding of Tat to granulin might be dependent on the coordinated binding of its cysteines to zinc ions bridging between cysteines and histidines in the interacting molecules.
How does granulin inhibit transcription?
Granulin's effect on transcription correlates with cyclin T1 binding and is at least partly independent of Tat binding (Table 1 and Fig. 9). Since granulin binds to intact P-TEFb (Fig. 4), one possible explanation for the transcriptional inhibition is that granulin reduces the availability of P-TEFb for Tat binding or transcription. This could be accomplished by sequestering P-TEFb or altering its subcellular distribution, a notion that is supported by confocal microscopy data (Fig. 5). A second possibility is that granulin alters the activity of P-TEFb. In contrast to CIITA, which binds to the same region of P-TEFb as Tat and therefore competes with Tat for P-TEFb binding, granulin binds to the histidine-rich domain of cyclin T1 and presumably allows Tat binding to another surface of P-TEFb at the same time. Consistent with this view, P-TEFb bound to the Tat activation domain can phosphorylate GST-GrnCDE, indicating that a ternary complex can form, at least transiently. When P-TEFb is bound to GST-GrnCDE, however, there is no detectable phosphorylation of CTD4 (Fig. 6), possibly because of interference with the binding of the CTD to cyclin T1 (56). Thus, the interaction of granulin with P-TEFb may block the activity of CDK9 towards the polymerase II CTD and thereby inhibit transcription elongation. These findings suggest that granulin could be a paradigm for natural regulators and pharmaceuticals that control transcription via P-TEFb.
Granulin phosphorylation.
Granulin is phosphorylated by immunocomplexes containing P-TEFb but not by two other CTD kinases that are involved in transcription (Fig. 7). This suggests that granulin may be involved in the transcription elongation step where P-TEFb is known to function. P-TEFb also phosphorylates human SPT5, an elongation factor which is necessary for in vitro Tat transactivation and can enhance Tat transactivation in vivo (28, 29, 63). In competition experiments, human SPT5 is phosphorylated more effectively than the GST-CTD. Similarly, our experiments suggest that granulin may be a preferred substrate for P-TEFb (Fig. 6), in keeping with the observation that tightly bound substrates are more readily phosphorylated by CDK2 (53). The well-studied CDK2-cyclin A complex interacts stably with its substrates E2F-1 and pRb. These substrates bind directly to cyclin A, and their binding is necessary for their recognition as substrates by CDK2 (8, 53).
P-TEFb may not be the only kinase that phosphorylates granulin, since cell extract depleted of P-TEFb still contained activity that could associate with GST-GrnCDE and phosphorylate it (Fig. 7C). Hence, granulin may be regulated by multiple kinases via its phosphorylation on multiple sites. By the same token, it is conceivable that cyclin T1 binds many more cellular and viral proteins which can serve as substrates and regulators of CDK9, as found for other cyclins (40).
Acknowledgments
We thank B. Matija Peterlin, David H. Price, Katherine A. Jones, James L. Manley, Antonio Giordano, and Luigi Lania for generously providing plasmids and Lawrence D. Gaspers and Andrew P. Thomas for advice and help with confocal microscopy.
This work was supported by grant AI31802 from the National Institutes of Health.
REFERENCES
- 1.Anakwe, O. O., and G. L. Gerton. 1990. Acrosome biogenesis begins during meiosis: evidence from the synthesis and distribution of an acrosomal glycoprotein, acrogranin, during guinea pig spermatogenesis. Biol. Reprod. 42:317-328. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., H. B. Hoff 3rd, H. Nemoto, H. Lee, J. Orth, Y. Arai, and G. L. Gerton. 1993. Acrogranin, an acrosomal cysteine-rich glycoprotein, is the precursor of the growth-modulating peptides, granulins, and epithelins, and is expressed in somatic as well as male germ cells. Mol. Reprod. Dev. 34:233-243. [DOI] [PubMed] [Google Scholar]
- 3.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., and H. P. Bennett. 1998. Granulins: the structure and function of an emerging family of growth factors. J. Endocrinol. 158:145-151. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, R., Z. He, K. P. Carmichael, J. Halper, and A. Bateman. 2000. Cellular localization of gene expression for progranulin. J. Histochem. Cytochem. 48:999-1009. [DOI] [PubMed] [Google Scholar]
- 6.Diaz-Cueto, L., P. Stein, A. Jacobs, R. M. Schultz, and G. L. Gerton. 2000. Modulation of mouse preimplantation embryo development by acrogranin (epithelin/granulin precursor). Dev. Biol. 217:406-418. [DOI] [PubMed] [Google Scholar]
- 7.Donald, C. D., A. Laddu, P. Chandham, S. D. Lim, C. Cohen, M. Amin, G. L. Gerton, F. F. Marshall, and J. A. Petros. 2001. Expression of progranulin and the epithelin/granulin precursor acrogranin correlates with neoplastic state in renal epithelium. Anticancer Res. 21:3739-3742. [PubMed] [Google Scholar]
- 8.Endicott, J. A., M. E. Noble, and J. A. Tucker. 1999. Cyclin-dependent kinases: inhibition and substrate recognition. Curr. Opin. Struct. Biol. 9:738-744. [DOI] [PubMed] [Google Scholar]
- 9.Flores, O., G. Lee, J. Kessler, M. Miller, W. Schlief, J. Tomassini, and D. Hazuda. 1999. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc. Natl. Acad. Sci. USA 96:7208-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-In autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor b To transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graña, X., A. DeLuca, N. Sang, Y. Fu, P. P. Claudio, J. Rosenblatt, D. O. Morgan, and A. Giordano. 1994. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc. Natl. Acad. Sci. USA 91:3834-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann, C. H., and M. A. Mancini. 2001. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci 114:1491-1503. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature (London) 395:93-96. [DOI] [PubMed] [Google Scholar]
- 20.Hrabal, R., Z. Chen, S. James, H. P. Bennett, and F. Ni. 1996. The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat. Struct. Biol. 3:747-752. [DOI] [PubMed] [Google Scholar]
- 21.Huang, H. W., and K. T. Wang. 1996. Structural characterization of the metal binding site in the cysteine-rich region of HIV-1 Tat protein. Biochem. Biophys. Res. Commun. 227:615-621. [DOI] [PubMed] [Google Scholar]
- 22.Isel, C., and J. Karn. 1999. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 290:929-941. [DOI] [PubMed] [Google Scholar]
- 23.Iuchi, S. 2001. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 27.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbe, T. Lorca, K. Nakayama Ki, and M. Benkirane. 2001. Interaction between cyclin T1 and SCF.(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 21:7956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor B phosphorylates human SPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 29.Kim, J. B., Y. Yamaguchi, T. Wada, H. Handa, and P. A. Sharp. 1999. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19:5960-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kino, T., O. Slobodskaya, G. N. Pavlakis, and G. P. Chrousos. 2002. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J. Biol. Chem. 277:2396-2405. [DOI] [PubMed] [Google Scholar]
- 31.Laity, J. H., B. M. Lee, and P. E. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39-46. [DOI] [PubMed] [Google Scholar]
- 32.Liau, L. M., R. L. Lallone, R. S. Seitz, A. Buznikov, J. P. Gregg, H. I. Kornblum, S. F. Nelson, and J. M. Bronstein. 2000. Identification of a human glioma-associated growth factor gene, granulin, with differential immuno-absorption. Cancer Res. 60:1353-1360. [PubMed] [Google Scholar]
- 33.Line, A., A. Stengrevics, Z. Slucka, G. Li, E. Jankevics, and R. C. Rees. 2002. Serological identification and expression analysis of gastric cancer-associated genes. Br. J. Cancer 86:1824-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, R., and G. Serrero. 2000. Inhibition of PC cell-derived growth factor (PCDGF, epithelin/granulin precursor) expression by antisense PCDGF cDNA transfection inhibits tumorigenicity of the human breast carcinoma cell line MDA-MB-468. Proc. Natl. Acad. Sci. USA 97:3993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, R., and G. Serrero. 2001. Mediation of estrogen mitogenic effect in human breast cancer MCF-7 cells by PC-cell-derived growth factor (PCDGF/granulin precursor). Proc. Natl. Acad. Sci. USA 98:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, R., and G. Serrero. 1999. Stimulation of PC cell-derived growth factor (epithelin/granulin precursor) expression by estradiol in human breast cancer cells. Biochem. Biophys. Res. Commun. 256:204-207. [DOI] [PubMed] [Google Scholar]
- 37.Majello, B., G. Napolitano, A. Giordano, and L. Lania. 1999. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene 18:4598-4605. [DOI] [PubMed] [Google Scholar]
- 38.Marcello, A., R. A. Cinelli, A. Ferrari, A. Signorelli, M. Tyagi, V. Pellegrini, F. Beltram, and M. Giacca. 2001. Visualization of in vivo direct interaction between HIV-1 TAT and human cyclin T1 in specific subcellular compartments by fluorescence resonance energy transfer. J. Biol. Chem. 276:39220-39225. [DOI] [PubMed] [Google Scholar]
- 39.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 40.Miller, M. E., and F. R. Cross. 2001. Cyclin specificity: how many wheels do you need on a unicycle? J. Cell Sci. 114:1811-1820. [DOI] [PubMed] [Google Scholar]
- 41.Morris, G. F., and M. B. Mathews. 1990. Analysis of the proliferating cell nuclear antigen promoter and its response to adenovirus early region 1. J. Biol. Chem. 265:16116-16125. [PubMed] [Google Scholar]
- 42.Napolitano, G., P. Licciardo, R. Carbone, B. Majello, and L. Lania. 2002. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J. Cell. Physiol. 192:209-215. [DOI] [PubMed] [Google Scholar]
- 43.Napolitano, G., B. Majello, P. Licciardo, A. Giordano, and L. Lania. 2000. Transcriptional activity of positive transcription elongation factor b kinase in vivo requires the C-terminal domain of RNA polymerase II. Gene 254:139-145. [DOI] [PubMed] [Google Scholar]
- 44.Parada, C. A., and R. G. Roeder. 1999. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18:3688-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pendergrast, P. S., C. Wang, N. Hernandez, and S. Huang. 2002. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol. Biol. Cell 13:915-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273:13855-13860. [DOI] [PubMed] [Google Scholar]
- 47.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 50.Ramanathan, Y., S. M. Reza, T. M. Young, M. B. Mathews, and T. Pe'ery. 1999. Human and rodent transcription elongation factor P-TEFb: interactions with human immunodeficiency virus type 1 tat and carboxy-terminal domain substrate. J. Virol. 73:5448-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Reza, S. M., M. Rosetti, M. B. Mathews, and T. Pe'ery. Differential activation of Tat variants in mitogen-stimulated cells: implications for HIV-1 postintegration latency. Virology, in press. [DOI] [PubMed]
- 51.Romisch, K. 1999. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 112:4185-4191. [DOI] [PubMed] [Google Scholar]
- 52.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 53.Schulman, B. A., D. L. Lindstrom, and E. Harlow. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95:10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sui, D., and J. E. Wilson. 2000. Interaction of insulin-like growth factor binding protein-4, Miz-1, leptin, lipocalin-type prostaglandin D synthase, and granulin precursor with the N-terminal half of type III hexokinase. Arch. Biochem. Biophys. 382:262-274. [DOI] [PubMed] [Google Scholar]
- 55.Sune, C., A. C. Goldstrohm, J. Peng, D. H. Price, and M. A. Garcia-Blanco. 2000. An in vitro transcription system that recapitulates equine infectious anemia virus tat-mediated inhibition of human immunodeficiency virus type 1 Tat activity demonstrates a role for positive transcription elongation factor b and associated proteins in the mechanism of Tat activation. Virology 274:356-366. [DOI] [PubMed] [Google Scholar]
- 56.Taube, R., X. Lin, D. Irwin, K. Fujinaga, and B. M. Peterlin. 2002. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 22:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trinh, D. P., K. M. Brown, and K. T. Jeang. 1999. Epithelin/granulin growth factors: extracellular cofactors for HIV-1 and HIV-2 Tat proteins. Biochem. Biophys. Res. Commun. 256:299-306. [DOI] [PubMed] [Google Scholar]
- 58.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wakasugi, K., and P. Schimmel. 1999. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284:147-151. [DOI] [PubMed] [Google Scholar]
- 61.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 62.Wimmer, J., K. Fujinaga, R. Taube, T. P. Cujec, Y. Zhu, J. Peng, D. H. Price, and B. M. Peterlin. 1999. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology 255:182-189. [DOI] [PubMed] [Google Scholar]
- 63.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 277:179-197. [DOI] [PubMed] [Google Scholar]
- 64.Xu, S. Q., D. Tang, S. Chamberlain, G. Pronk, F. R. Masiarz, S. Kaur, M. Prisco, T. ZanoccoMarani, and R. Baserga. 1998. The granulin/epithelin precursor abrogates the requirement for the insulin-like growth factor 1 receptor for growth in vitro. J. Biol. Chem. 273:20078-20083. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 66.Zanocco-Marani, T., A. Bateman, G. Romano, B. Valentinis, Z. H. He, and R. Baserga. 1999. Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res.. 59:5331-5340. [PubMed] [Google Scholar]
- 67.Zhang, H., and G. Serrero. 1998. Inhibition of tumorigenicity of the teratoma PC cell line by transfection with antisense cDNA for PC cell-derived growth factor (PCDGF, epithelin/granulin precursor). Proc. Natl. Acad. Sci. USA 95:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, J., N. Tamilarasu, S. Hwang, M. E. Garber, I. Huq, K. A. Jones, and T. M. Rana. 2000. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J. Biol. Chem. 275:34314-34319. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, J., G. Gao, J. W. Crabb, and G. Serrero. 1993. Purification of an autocrine growth factor homologous with mouse epithelin precursor from a highly tumorigenic cell line. J. Biol. Chem. 268:10863-10869. [PubMed] [Google Scholar]
- 70.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]