Abstract
In budding yeast, diffusible mating pheromones initiate a signaling pathway that culminates in several responses, including cell cycle arrest. Only a handful of genes required for the interface between pheromone response and the cell cycle have been identified, among them FAR1 and FAR3; of these, only FAR1 has been extensively characterized. In an effort to learn about the mechanism by which Far3 acts, we used the two-hybrid method to identify interacting proteins. We identified five previously uncharacterized open reading frames, dubbed FAR7, FAR8, FAR9, FAR10, and FAR11, that cause a far3-like pheromone arrest defect when disrupted. Using two-hybrid and coimmunoprecipitation analysis, we found that all six Far proteins interact with each other. Moreover, velocity sedimentation experiments suggest that Far3 and Far7 to Far11 form a complex. The phenotype of a sextuple far3far7-far11 mutant is no more severe than any single mutant. Thus, FAR3 and FAR7 to FAR11 all participate in the same pathway leading to G1 arrest. These mutants initially arrest in response to pheromone but resume budding after 10 h. Under these conditions, wild-type cells fail to resume budding even after several days whereas far1 mutant cells resume budding within 1 h. We conclude that the FAR3-dependent arrest pathway is functionally distinct from that which employs FAR1.
Signal transduction pathways are ubiquitous mechanisms that allow cells to respond appropriately to intracellular and environmental cues. The pheromone response pathway in the budding yeast Saccharomyces cerevisiae permits haploid cells to sense a potential mating partner and initiate key cellular changes that facilitate mating. These changes include transcriptional induction of genes whose products promote mating, inhibition of cell cycle progression, and reorientation of cell polarity (8, 16, 43).
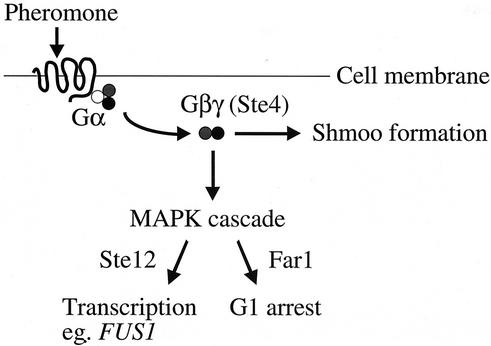
Pheromone response (summarized in Fig. 1) begins at the cell surface, with binding of peptide pheromone a-factor or α-factor to its cognate G protein-coupled serpentine receptor. Binding of the ligand causes Gβγ to be released from the inhibitory Gα subunit (47, 65). Gβγ activates the pheromone response mitogen-activated protein kinase cascade (17, 36), which controls two important physiological responses: activation of the transcription factor Ste12 to allow induction of mating-specific genes, such as FUS1; and cell cycle arrest (18, 42). The activated G protein also controls a second pathway branch that serves to repolarize growth in the direction of the mating partner, ultimately leading to shmoo formation (8). Several decades of thorough study has permitted the identification and characterization of most components of pheromone response. Nonetheless, important questions remain, particularly regarding the ways in which pheromone response interfaces with vegetative cellular processes (24, 43, 68). The goal of this work is to better understand the connection between pheromone response and the cell cycle.
FIG. 1.
Pheromone response pathway. Aspects of pheromone signaling relevant to this work are depicted. Although not indicated in this diagram, the α and γ subunits of the heterotrimeric G protein are tethered to the plasma membrane. MAPK, mitogen-activated protein kinase.
The cell cycle in yeast is controlled by a single cyclin-dependent kinase, Cdc28, whose functional specificity is determined by its association with various cyclin partners (43). Passage through Start requires that Cdc28 associate with the G1 cyclins Cln1, Cln2, and Cln3. Transcription of the structural genes and proteolysis of the innately unstable protein products are regulated such that these cyclins accumulate to significant levels only in G1 (56, 61, 68). Pheromone response has several consequences for the cell cycle machinery, including inhibition of Cln-Cdc28 activity, repression of CLN1 and CLN2 transcription, and turnover of the Cln1 and Cln2 proteins, resulting in G1 arrest (44, 61, 68). Exactly how these cell cycle components are regulated by pheromone is unclear.
To address this question, several groups have isolated far mutants, which are defective in pheromone-mediated cell cycle arrest but still competent to transduce signal, as revealed by transcriptional induction of FUS1, suggesting that pheromone response has been uncoupled from the cell cycle. FAR1, the first gene identified by this strategy (7), is itself transcriptionally induced by pheromone. Moreover, during the pheromone response, Far1 protein is phosphorylated by Fus3 and can then bind to Cln-Cdc28 complexes (49, 50, 60). This binding was reported to inhibit the Cln-Cdc28 protein kinase activity, leading to the view that Far1 is a pheromone-dedicated cyclin-dependent kinase inhibitor for Cdc28 (50). Recently, this satisfying picture has been challenged by biochemical evidence suggesting that Far1 does not directly inactivate Cln-Cdc28 in vivo (20, 43). The understanding of FAR1 has been further complicated by the realization that it also functions in cell polarity, during both mating and vegetative growth (45, 48, 57). Moreover, it is thought that pheromone-mediated G1 arrest does not rely solely on FAR1 because this gene is not required for downregulation of the G1 cyclins, at either the RNA or protein level (49, 63).
Several other genes have been identified as potential players in pheromone arrest by virtue of their Far− mutant phenotypes (10, 30). The work presented here focuses on FAR3. Like far1, the far3 null mutant is not compromised for pheromone signal transduction, yet it continues to grow in the presence of pheromone. FAR3 does not appear to regulate G1 cyclin levels, nor does it participate in a FAR1-dependent pathway; therefore, it must affect the cell cycle by using a novel mechanism (30). To identify proteins that physically interact with Far3, and might therefore shed light on its role in pheromone arrest, we carried out a two-hybrid screen. Our studies reveal that five previously uncharacterized genes have a null-mutant phenotype indistinguishable from far3, encode proteins that physically interact with Far3 and each other, and participate in a FAR3-dependent pathway which prevents premature recovery from pheromone arrest.
MATERIALS AND METHODS
Strains, media, and microbiological techniques.
The yeast strains used are listed in Table 1. The identification and characterization of FAR3 was done in SY2227, a derivative of the Jim Hopper laboratory strain, Sc252 (30). Characterization of FAR7, FAR8, FAR9, FAR10, and FAR11 was carried out in the same background. Yeast two-hybrid was performed in the strains indicated below. Most genomic alterations, such as gene deletions and epitope tagging, were engineered by PCR-based mutagenesis, using published cassettes (23, 35, 39). The far3::LEU2 null allele was introduced using NheI-SstI-digested pSL2310, as described previously (30). The ste12::LEU2 null allele was introduced by transformation of SY2227 with SacI-SphI-digested pSUL-16 (19). Most multiply mutant haploids were obtained by crossing single or multiple mutants of opposite mating type, sporulating the diploid, and dissecting tetrads. Strain genotypes were confirmed by PCR Southern blot analysis (55) and, where possible, on the basis of phenotype. The successful construction of genes encoding epitope-tagged proteins was further confirmed by Western blot analysis.
TABLE 1.
Yeast strains used in this study
| Straina | Genotype | Source or reference |
|---|---|---|
| PJ69-4A | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 33 |
| SY2227 | MATaade1-1 leu2-2,113 trp1 ura3-52 bar1 HIS3::pFUS1::HIS3 mfa2-Δ1::FUS1-lacZ rad16::pGAL1::STE4 | 30 |
| SY2673 | far1Δhb | 30 |
| SY4059 | far7::URA3 | This study |
| SY4060 | far9::URA3 | This study |
| SY4061 | far8::URA3 | This study |
| SY4062 | far3::LEU2 | This study |
| SY4063 | far10::KAN | This study |
| SY4064 | far11::KAN | This study |
| SY4065 | far3::LEU2 FAR7-MYC13-KAN 〈YEp352〉 | This study |
| SY4066 | far3::LEU2 FAR7-MYC13-KAN 〈pSL2784〉 | This study |
| SY4067 | far3::LEU2 FAR8-MYC13-KAN 〈YEp352〉 | This study |
| SY4068 | far3::LEU2 FAR8-MYC13-KAN 〈pSL2784〉 | This study |
| SY4069 | far3::LEU2 FAR9-MYC13-KAN 〈YEp352〉 | This study |
| SY4070 | far3::LEU2 FAR9-MYC13-KAN 〈pSL2784〉 | This study |
| SY4071 | far3::LEU2 FAR10-MYC13-KAN 〈YEp352〉 | This study |
| SY4072 | far3::LEU2 FAR10-MYC13-KAN 〈pSL2784〉 | This study |
| SY4073 | far3::LEU2 FAR11-MYC13-KAN 〈YEp352〉 | This study |
| SY4074 | far3::LEU2 FAR11-MYC13-KAN 〈pSL2784〉 | This study |
| SY4075 | far3::LEU2 far7::CgTRP1 far8::URA3 far9::HYGB far10::KAN far11::NAT | This study |
| SY4076 | far3::LEU2 far9::HYGB FAR10-MYC13-KAN 〈YEp352〉 | This study |
| SY4077 | far3::LEU2 far9::HYGB FAR10-MYC13-KAN 〈pSL2784〉 | This study |
| SY4078 | FAR7-MYC13-KAN 〈pSL2771〉 | This study |
| SY4079 | FAR8-MYC13-KAN 〈pSL2771〉 | This study |
| SY4080 | FAR9-MYC13-KAN 〈pSL2771〉 | This study |
| SY4081 | FAR10-MYC13-KAN 〈pSL2771〉 | This study |
| SY4082 | FAR11-MYC13-KAN 〈pSL2771〉 | This study |
| SY4083 | far3::LEU2 far9::HYGB FAR11-MYC13-KAN 〈YEp352〉 | This study |
| SY4084 | far3::LEU2 far9::HYGB FAR11-MYC13-KAN 〈pSL2784〉 | This study |
| SY4085 | ste12::LEU2 | This study |
All SY strains listed here are MATa derived from SY2227, previously published as YDH121 (30).
Yeast and bacterial strains were propagated using standard media and methods (5, 52, 54). Yeast strains were grown on yeast extract-peptone-dextrose (YEPD) unless selection was required. Plasmid-bearing strains were maintained on the appropriate SD omission medium. Integration of drug resistance cassettes was selected using YEPD supplemented with 200 μg of Geneticin per ml, 250 μg of hygromycin B per ml, or 100 μg of clonNAT per ml. YEPGal and SGal contained 2% galactose. Yeast transformations were performed using modifications of the lithium acetate method (9, 22). 5-Fluoroorotic acid (5-FOA) synthetic complete and omission plates were used in some selections and during two-hybrid screening. Bacterial transformations, DNA preparations, and plasmid constructions were performed by standard methods (54).
Plasmids.
The plasmids used in this work are listed in Table 2. pSL2784 is a high-copy-number YEp352-derived vector, in which Far3 is expressed from its own promoter, and has a C-terminal triple HA epitope tag. pSL2784 was constructed by ligating a 1.8-kb SacI-SalI fragment of pSL2783, which contains the entire FAR3 coding sequence, a C-terminal triple HA epitope tag, plus 300 bp of upstream genomic sequence and 870 bp of downstream sequence, into SacI-SalI-digested YEp352. The expression of the Far3-HA fusion protein was confirmed by Western blotting as described below. To confirm that Far3-HA was functional, we transformed pSL2784 into a far3 null mutant and found that it could rescue pheromone-induced G1 arrest, as assessed by halo formation. pSL2783 is a single-copy plasmid obtained by ligating the 100-bp fragment of HpaI-digested pKEB35 (3), containing a triple-HA epitope tag flanked by HpaI sites, into HpaI-digested pSL2367. pSL2367 was made by introducing a HpaI site into pSL2252, immediately prior to the FAR3 stop codon (J. Horecka, unpublished data). pSL2252 has been described previously (30) and consists of a 1.8-kb SalI-BglII genomic fragment which contains the FAR3 open reading frame (ORF) plus 300 bp of upstream sequence and 870 bp of downstream sequence ligated into SalI-BamHI-digested pRS316.
TABLE 2.
Plasmids used in this study
| Name | Description | Source or reference |
|---|---|---|
| pGBDU-C1 | Two-hybrid Gal4BD plasmid | 33 |
| pGAD-C1 | Two-hybrid Gal4AD plasmid | 33 |
| pGAD-C3 | Two-hybrid Gal4AD plasmid | 33 |
| pKEB35 | HA epitope with HpaI sites | 3 |
| pOBDCYH | Two-hybrid Gal4BD plasmid | 6 |
| pSL2252 | Single-copy Far3 | 30 |
| pSL2310 | far3::LEU2 replacement plasmid | 30 |
| pSL2367 | Single-copy Far3 with C-terminal HpaI sites | Horecka, un- published |
| pSL2771 | HA epitope expression plasmid | 34 |
| pSL2783 | Single-copy Far3-HA | This study |
| pSL2784 | High-copy-number Far3-HA | This study |
| pSL2785 | Two-hybrid Gal4BD-Far3 fusion | This study |
| pSL2786 | Two-hybrid Gal4BD-Far7 fusion | This study |
| pSL2787 | Two-hybrid Gal4BD-Far9 fusion | This study |
| pSL2788 | Two-hybrid Gal4BD-Far3 fusion | This study |
| pSL2789 | Two-hybrid Gal4AD-Far7A fusion | This study |
| pSL2790 | Two-hybrid Gal4BD-Far8 fusion | This study |
| pSL2791 | Two-hybrid Gal4BD-Far9A fusion (larger) | This study |
| pSL2792 | Two-hybrid Gal4BD-Far9B fusion (smaller) | This study |
| pSUL-16 | ste12::LEU2 replacement plasmid | 19 |
pSL2771 was a kind gift from Megan Keniry and contains a triple-HA epitope tag driven by 446 bp of STE20 upstream sequence subcloned into the ApaI-SacII restriction sites of pRS315 (34).
Two-hybrid bait constructs pSL2785, pSL2786, and pSL2787 were made by gap repair (41) of pOBDCYH, essentially as described previously (6). The constructs express a fusion protein consisting of full-length Far3, Far7, or Far9, respectively, fused at its N terminus to the Gal4 binding domain (Gal4BD).
A second Far3 two-hybrid bait, pSL2788, was constructed by ligating a 1.8-kb fragment of EcoRI-SalI-digested pSL2785 into EcoRI-SalI-digested pGBDU-C1. pGBDU-C1 has been described previously (33). The expression of all two-hybrid bait constructs was confirmed by Western blotting as described below. We also confirmed that the bait constructs rescued the corresponding null mutant for pheromone-induced G1 arrest, as assessed by halo formation.
Plasmids pSL2789 to pSL2792, expressing Far3 interacting proteins fused to the Gal4 activation domain (Gal4AD), consist of genomic DNA ligated into pGAD-C1 or pGAD-C3 and were generated during construction of the Phil James two-hybrid libraries, as described previously (33). pSL2789 expresses the C-terminal 157 amino acids of Far7/Yfr008w (codons 65 to 221) in pGAD-C1. pSL2790 expresses a full-length version of Far8/Ymr029c (codons 1 to 523) plus three N-terminal amino acids encoded by 9 bp of FAR8 upstream genomic sequence (Glu Lys Asn) in pGAD-C3. pSL2791 encodes the C-terminal 528 amino acids of Far9/Ydr200c (codons 75 to 604) in pGAD-C1. pSL2792 encodes an internal fragment of Far9/Ydr200c (codons 138 to 572) in pGAD-C3. pGAD-C1 and pGAD-C3 have been described previously (33).
The far3::LEU2 replacement plasmid, pSL2310, has been described previously (30). The ste12::LEU2 replacement plasmid, pSUL-16, has been described previously (19).
All plasmids generated during the course of this work were confirmed by both restriction analysis and sequencing. Where indicated, protein expression was also confirmed by Western blot analysis and protein function was confirmed by complementation of null mutants.
Pheromone response assays.
Three outputs of pheromone response were assayed: pheromone-mediated G1 arrest, transcriptional induction of FUS1, and shmoo formation.
The loss of G1 arrest, or the Far− phenotype, was assayed by growth on pheromone or under conditions that simulate pheromone exposure. Standard plate halo assays were performed as previously described (30, 58). Briefly, 2 μg of α-factor was spotted onto a lawn of cells on a YEPD plate. To examine several strains in parallel, spot assays were done as follows. Tenfold serial dilutions of fresh overnight cultures were made, with optical densities at 600 nm (OD600) ranging from 0.1 to 0.0001. Then aliquots (2 μl) of each dilution were spotted onto a YEPD plate containing 0.1 μg of α-factor per ml. Strains containing plasmids were assayed on the appropriate SD omission plate. Pheromone exposure could be simulated in SY2227-derived strains, which contain the rad16::pGAL1::STE4 allele, by spotting serial dilutions onto YEPGal or SGal omission plates.
Levels of basal and induced flux through the pheromone response pathway were determined by β-galactosidase assays of SY2227-derived strains, which contain a genomic copy of the pFUS1::lacZ reporter. Saturated cultures were diluted into YEPD (OD600, approximately 0.1), grown to log phase (OD600, approximately 0.4), and then split in two. One sample (“basal”) was assayed immediately. The other sample (“induced”) was induced by adding α-factor to a final concentration of 0.6 μg/ml and incubating for 2 h at 30°C. For both sets of samples, cell density was determined as absorbance at 650 nm. β-Galactosidase assays were performed using chlorophenolred-β-d-galactopyranoside (CPRG) substrate in a 96-well microtiter plate format as described previously (31). For each strain tested, four independent isolates were assayed per experiment, and the experiment was repeated no fewer than three times. β-Galactosidase activity is given in arbitrary units similar in principle to Miller units (31). The cultures were also examined by microscopy to determine the extent of shmoo formation.
Time course of recovery from cell cycle arrest during pheromone treatment.
To analyze the timing of recovery from pheromone treatment, exponentially growing MATa cells (OD600, approximately 0.6) were spread onto YEPD or YEPD containing 0.1 μg of α-factor per ml. For each strain, a minimum of 30 unbudded cells were isolated using a tetrad dissection needle. The plates were then incubated at 30°C. At the indicated time points, the cells were examined under a light microscope to determine the timing of appearance of initial buds and the formation of microcolonies. In every trial, the wild-type cells never resumed budding, even after several extra days of incubation. For the mutants, the percent budding was determined at each time point by dividing the number of budded cells by the total number of cells that eventually formed a visible colony. Budded cells scored at 25 h consisted of microcolonies containing anywhere from two cells to several thousand, depending on the strain and the timing of rebudding. Cells which had not resumed budding by the 25-h time point never did, suggesting that they were irreversibly arrested or damaged by the isolation protocol. Visible colony formation was confirmed no later than 32 h from the zero time point.
Two-hybrid screens.
A two-hybrid screen was initiated using full-length Far3 fused to the Gal4BD as bait. The pSL2788 bait plasmid was introduced into the PJ69-4a strain to screen a set of three libraries, C1, C2, and C3, essentially as described previously (33). A total of 107, 2 × 106, and 3 × 106 transformants were screened for the C1, C2, and C3 libraries respectively. A minimum of 2 × 106 transformants were needed to ensure complete coverage of each library. A summary of the results appears in Table 3. Two-hybrid interactions, defined as the ability of a pair of constructs to interact and thereby activate transcription from pGAL4-driven reporters, were first detected by growth of the PJ69-4a strain on SD lacking histidine and supplemented with 3 mM 3-amino-1,2,4-triazole (3-AT). Positives were rescreened on SD lacking adenine. Bait dependence of the His+ Ade+ phenotype was confirmed as follows. The URA3-marked bait plasmid was lost by growth on 5-FOA medium lacking leucine (to maintain the library plasmid), and the strain was tested for restoration of His− Ade− auxotrophy. The LEU2-marked library plasmid was rescued from the positive strain. After amplification in bacteria, library plasmids were digested with a variety of enzymes and classed according to restriction pattern. Representatives from each class were retransformed into PJ69-4a along with the pSL2788 bait plasmid. In every case, retransformation was accompanied by complementation of the strain to His+ Ade+ prototrophy. Library plasmids were then sequenced to identify the Far3-interacting protein. In some cases, different fragments of the same gene were identified. In total, eight unique ORFs were represented. Directed two-hybrid assays were likewise done in the PJ69-4a strain background, using many of the “target” plasmids identified in the two-hybrid screen; in this case, bait plasmids were marked with TRP1. Interactions were scored by patching independent isolates in duplicate to SD-Trp Leu, then replica plating to SD-Trp Leu His and SD-Trp Leu Ade. Growth on both tester media indicated a positive result. Patch tests were repeated at least four times, using different isolates each time.
TABLE 3.
Two-hybrid screen results
| Library (no. of transformants) | No. that were:
|
Genes identified | ||
|---|---|---|---|---|
| His+ | His+ Ade+ | Unique classes | ||
| C1 (107) | 73 | 27 | 5 | YFR008W, YDR200C |
| C2 (2 × 106) | 59 | 0 | 0 | None |
| C3 (3 × 106) | 66 | 13 | 10 | YMR029C, YDR200C, YPR049C, YDR295C, YPR083W, FIR1, TID3 |
Western blotting.
Western blotting was done essentially as previously described (54). Protein samples were resuspended in Thorner buffer (8 M urea, 5% sodium dodecyl sulfate SDS, 40 mM Tris [pH 6.8], 0.1 mM EDTA, 04 mg of bromophenol blue per ml, β-mercaptoethanol added fresh to 1%), heated at 95°C for 3 min, and analyzed on sodium dodecyl sulfate-6 or 10% polyacrylamide gel electrophoresis (SDS-PAGE) gels, using the Protean II minigel system as recommended by the manufacturer (Bio-Rad). Gels were electroblotted to a 0.45-μm-pore-size nitrocellulose membrane (Intermountain Scientific Corp.). The blots were incubated for 30 min at room temperature in blocking buffer (5% nonfat dried milk resuspended in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20) and then incubated with the indicated antibodies diluted in blocking buffer. Primary antibody incubation was done at 4°C overnight and was followed by several washes in TBST; incubation with secondary antibody was done at room temperature for 1 h, and was followed by several washes in TBST. Secondary antibody was detected using the Supersignal chemiluminescent substrate (Pierce, Rockford, Ill.). Primary antibodies were all mouse monoclonal antibodies. HA-tagged proteins were detected using 12CA5 anti-HA monoclonal antibody. Myc-tagged proteins were detected with 9E10 anti-myc monoclonal antibody. Both HA and myc monoclonal antibodies were culture supernatants prepared from hybridomas obtained from the American Type Culture Collection. Two-hybrid baits fused to Gal4BD were detected using anti-GAL4BD antibody (51). Antibodies against the 30- and 39-kDa subunits of bovine mitochondrial respiratory complex I (encoded by bovine NDUFJ3 and NDUFA9, respectively) were obtained from Mike Marusich (Monoclonal Antibody Facility, University of Oregon). The Dpm1 loading control was detected using monoclonal anti-Dpm1 (Molecular Probes, Eugene, Oreg.). In all cases, the secondary antibody was goat anti-mouse horseradish peroxidase conjugate (Bio-Rad).
Coimmunoprecipitation.
Strains were grown to saturation in the appropriate SD omission medium, diluted to an OD600 of 0.25 in YEPD, and grown at 30°C to an OD600 of 0.6. Then 100 ml of exponentially growing cells (OD600, 0.6) was harvested and spheroplasted as follows. Cells were resuspended in 4 ml of dithiothreitol buffer (50 mM Tris [pH 9.5], 10 mM dithiothreitol) and incubated for 15 min at room temperature. The cells were pelleted, resuspended in 3.75 ml of spheroplast buffer (1.2 M sorbitol, 50 mM potassium phosphate [pH 7.4], 1 mM MgCl, 250 μg of zymolyase per ml), and incubated for 1 h at 30°C on a roller drum. Spheroplasts were harvested by centrifugation for 2 min at 5,000 rpm in a tabletop microcentrifuge (IEC Micromax rotor), washed in 1 ml of 1.2 M sorbitol, and then pelleted at low speed as before. All subsequent steps were performed at 4°C, unless otherwise indicated. Spheroplasts were lysed for 15 min on ice in 1 ml of cold IP buffer (50 mM Tris [pH 8], 1 mM EDTA, 50 mM NaCl, 1% NP-40) to which a final concentration of 1× protease inhibitor cocktail and 1 mM phenylmethylsulfonyl fluoride (PMSF) was added immediately before use. Lysates were centrifuged in a tabletop microcentrifuge at 13,000 rpm for 5 min, and the pellet was discarded. Supernatants were precleared by incubation for 30 min with 40 μl of protein A-Sepharose beads prepared as a 50% slurry in bead blocking buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 0.1% sodium azide, 0.1% bovine serum albumin [BSA] fraction V). Beads were removed by centrifugation for 1 min at 13,000 rpm. The bead pellet was kept as the no-antibody control and treated as described below for the immunoprecipitate (IP) pellet. Far3-HA was immunoprecipitated from supernatant by incubation with 3 μl of rabbit anti-HA polyclonal serum (3) for 30 min on ice, after which 40 μl of protein A-Sepharose beads was added for another 30 min. All bead incubations were done on a roller drum to prevent beads from settling. The IP was pelleted by centrifugation for 1 min at 13,000 rpm in a tabletop microcentrifuge. Both no-antibody and IP pellets were washed twice in 0.5 ml of IP buffer and then resuspended in 40 μl of Thorner buffer. Protein was released from beads by a 3-min boiling step, and the beads were removed by centrifugation for 1 min 13,000 rpm, after which the pellet was discarded. Supernatant was either used immediately or frozen at −20°C. Protein samples were analyzed by Western blotting as described above.
Sucrose velocity gradients.
Exponentially growing cells (OD600, 0.6; 100 ml) were spheroplasted essentially as described above. All subsequent steps were performed at 4°C, and both lysis buffer and solubilization buffer contained a final concentration of 1× protease inhibitor cocktail and 1 mM PMSF, added immediately before use. Spheroplasts were resuspended in 1.8 ml of lysis buffer (0.2 M sorbitol, 50 mM Tris [pH 7.5], 1 mM EDTA), left on ice for 10 min, and centrifuged in a tabletop microcentrifuge at 13,000 rpm for 10 min to collect the membranes and organelles. The pellet was resuspended in 0.65 ml of solubilization buffer (100 mM Tris [pH 7.5], 1 mM EDTA, 1% NP-40) and left on ice for 5 min; unsolubilized protein complexes were removed by centrifugation at 13,000 rpm for 5 min. The clarified lysate was analyzed for protein concentration by using a protein assay dye as specified by the manufacturer (Bio-Rad). A 50-μl volume of lysate was added to the same volume of Thorner buffer for later Western analysis. Lysate (0.5 ml, containing approximately 1 mg of total protein) was gently layered onto a 5-ml 15 to 40% continuous sucrose gradient. Sucrose solutions contained a final concentration of 1× protease inhibitor cocktail. The gradients were centrifuged at 55,000 rpm in an SW55Ti swinging-bucket rotor (Beckman) at 4°C for 4 h. Then 10 0.5-ml fractions were collected from the top and saved at −20°C for later IP. Samples were taken from each fraction, added to an equal volume of Thorner buffer, heated to 65°C for 3 min, and analyzed by Western blotting. Fractions were prepared for IP by adding 0.25 ml of 2× IP buffer (100 mM Tris [pH 8.0], 2 mM EDTA, 50 mM NaCl, 2% NP-40 detergent); IP and subsequent Western analysis were performed as described above. To determine the size of the complex, a mix of molecular weight markers (BSA, catalase, and thyroglobulin) was loaded on a duplicate gradient; fractions were analyzed by SDS-PAGE and stained with Coomassie blue. Bovine heart mitochondria were also lysed, treated as described above, and loaded onto a third gradient to separate mitochondrial complexes of known size (14, 59). Sedimentation of mitochondrial complex I was determined by Western blotting using monoclonal antibodies to the 30- and 39-kDa subunits of the complex (59). Bovine heart mitochondria were prepared by differential centrifugation as described previously (25). Marker sizes are as follows: BSA, 67 kDa; catalase, 232 kDa; thyroglobulin, 669 kDa; mitochondrial complex I, 900 kDa (59, 64).
Sequence analysis.
Predicted protein domains in Far3 and Far7 to Far11 were previously annotated in the literature or were generated by using online protein prediction programs as follows. FHA (forkhead-associated) domains in Far9 and Far10 have been described previously (28). The CC (coiled-coil) domains in Far8 to Far10 were annotated previously (46) and subsequently confirmed using COILS (http://www.ch.embnet.org/software/COILS_form.html) (40). A CC domain in Far3 is also predicted by the COILS program. All predicted transmembrane domains were obtained using TMpred: http://www.ch.embnet.org/software/TMPRED_form.html (29). Homology searches were done using iterative BLAST at NCBI Blast (http://www.ncbi.nlm.nih.gov/BLAST). Protein sequence alignments were generated by Multalin (http://prodes.toulouse.inra.fr/multalin/multalin.html) (12).
Reagents.
The following chemicals and reagents were used in this study: 3-AT (Sigma); BSA (electrophoresis grade) (Sigma); ClonNat (Werner Bioagents); CPRG (Boehringer Mannheim); α-factor (synthesized as the following peptide: H-Trp-His-Trp-Leu-Gln-Leu-Lys-Pro-Gly-Gln-Pro-Met-Tyr-OH) (Genemed Synthesis); 5-FOA (BioVectra DCL); Geneticin (BioVectra DCL); hygromycin B (Sigma); lauryl maltoside (n-docecyl-β-d-maltoside, ULTROL grade) (Calbiochem); NP-40 (Igepal CA-630) (Sigma); PMSF (Sigma); protease inhibitor cocktail tablets (Beohringer Mannheim); protein A-Sepharose CL-4B beads (Amersham Pharmacia Biotech); trichloroacetic acid, SigmaUltra grade (Sigma); Tween 20 (polyoxyethylenesorbitan monolaurate) (Sigma); and Zymolyase 100T (Seikagaku America, Inc.). Catalase and thyroglobulin were obtained as part of a molecular weight marker kit (Sigma). Purified beef heart mitochondria were a generous gift from James Murray and Roderick Capaldi.
RESULTS
Identification of Far3-interacting proteins by two-hybrid analysis.
Using full-length Far3 as bait, we initiated a two-hybrid screen. A Gal4BD-Far3 fusion construct, pSL2788, was screened against a library consisting of yeast genomic DNA fragments ligated into a vector expressing the Gal4AD (33). This screen was performed as described in Materials and Methods and yielded eight candidates (Table 3). Two of these, FIR1 and TID3, have previously been characterized, the former with respect to its role in mRNA processing (53) and the latter as a spindle pole-associated protein that plays a role in chromosome segregation (26, 66, 67). The six remaining ORFs are predicted to encode proteins with no known function and no extensive homology to proteins with known function.
A similar screen using the Gal4BD-Far3 fusion construct, pSL2785, against an ordered array of all full-length yeast ORFs fused to the Gal4AD (6) yielded three candidates. Of these, two (YDR200C and YFR008W) had previously been isolated as Far3 interactors in our screen; the third ORF was YJL184W, another protein of unknown function and no known homologs (B. Drees and S. Fields, personal communication).
Far3 interactors function like Far3 and define three new FAR genes.
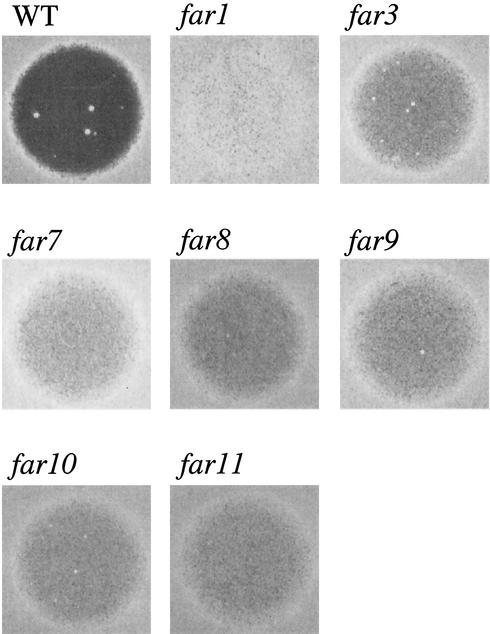
Null mutants of each Far3-interacting protein were constructed and analyzed for pheromone arrest defects by using a standard halo assay. YFR008W, YMR029C, and YDR200C each had a far3-like null mutant phenotype (Fig. 2); consequently we named these genes FAR7, FAR8, and FAR9, respectively. (FAR4 to FAR6 had been identified in a screen by another group, although, unlike FAR1 and FAR3, disruption of these genes simultaneously perturbed pheromone signaling and arrest [10].) TID3 could not be assayed in this manner because the null mutation is lethal. Null mutants for the remaining five candidates, FIR1, YDR295C, YJL184W, YPR049C, and YPR083W, were phenotypically indistinguishable from wild type with respect to halo formation.
FIG. 2.
Characterization of far7 to far11 null mutants by halo assay. Halo assays were performed as described in Materials and Methods, using 2 μg of α-factor applied to a lawn of MATa cells. Strains were as follows: wild type (WT), SY2227; far1, SY2673; far3, SY4062; far7, SY4059; far8, SY4061; far9, SY4060; far10, SY4063; and far11, SY4064.
Far9 interactors are also required for pheromone arrest.
While this work was under way, several other groups completed large-scale genome-wide two-hybrid screens (32, 62). These studies identified two of the same proteins as our work did, but they also identified potential Far3 interaction partners that our work did not detect. In addition, these studies identified potential Far7, Far8, and Far9 interactors. To test whether any of the Far3, Far7, Far8, or Far9 interactors identified in these other studies also participate in pheromone-mediated cell cycle arrest, we constructed null alleles and assessed pheromone arrest by halo assay. By this means, we were able to identify two more previously uncharacterized FAR genes, FAR10/YLR238W and FAR11/YNL127W, each of which encodes a Far9-interacting protein (62). Neither Far10 nor Far11 has previously had a function ascribed to it, but loss of either protein confers a Far− phenotype (Fig. 2).
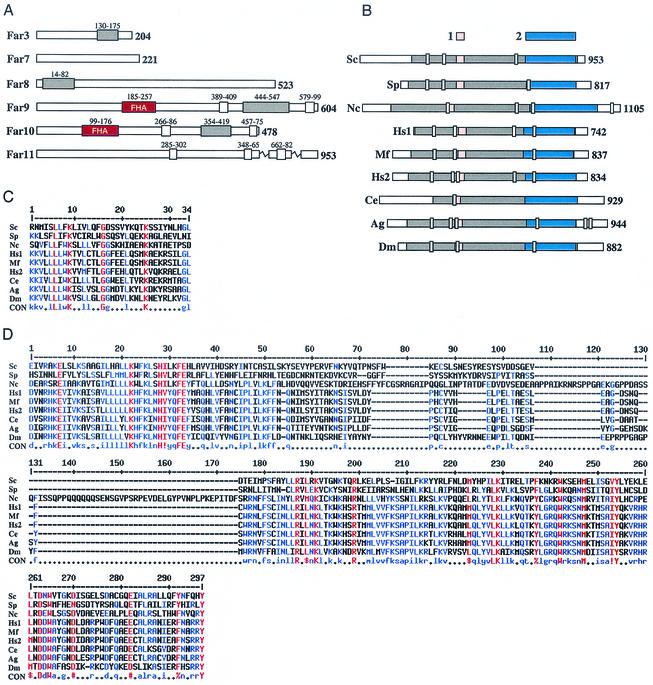
FAR11/YNL127W has recently been identified as a member of a highly conserved family of eukaryotic genes which encode transmembrane proteins (69). Our BLAST searches revealed a region of ∼700 amino acids in Far11 which is between 15 and 27% identical to a similar stretch of amino acids in a number of putative proteins in other systems (Fig. 3B). Furthermore, multiple sequence alignment revealed two highly conserved regions (Fig. 3C and D). While FAR11 homologs exist in many eukaryotic systems, including Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens, only the founding member, ham-2, has been previously characterized. ham-2 is required for hyphal fusion in Neurospora crassa (69). It has been suggested that FAR11 might be required for cell fusion during mating (69).
FIG. 3.
Far3 and Far7 to Far11 protein structure and homology. (A) Far3 and Far7 to Far11 protein domains. Regions predicted to encode CC, FHA, and TM domains are indicated by grey, red, and white boxes, respectively. Residues spanning each domain are indicated above the box in smaller type. (B) Far11 homologs in eukaryotes. A ∼700-amino-acid region of similarity was identified in Sc/Far11 to contain between 15 and 27% identity to the indicated homologous sequences from other organisms; in Sc/Far11, this region is indicated by grey shading. In protein sequences from other organisms, the region of homology, identified by iterative BLAST, is also indicated by grey shading. Domains 1 and 2, each containing absolutely conserved amino acid residues, are indicated by pink and blue shading, respectively. Predicted TM domains are indicated by white boxes. For panels A and B, the number of residues in the protein product is indicated in bold type. (C) Alignment of domain 1, containing four absolutely conserved amino acid residues over 33-amino-acid stretch. (D) Alignment of domain 2, containing 14 absolutely conserved residues over ∼250 amino acids. For panels C and D, red capital letters in the consensus indicate absolutely conserved residues and blue indicates weakly conserved amino acids. ! is either I or V; # represents any of NDQE; $ is L or M; % is either F or Y. For panels B to D, proteins of known function are as follows: Sc, S. cerevisiae Far11; Nc, N. crassa HAM-2. Hypothetical gene products in other eukaryotes are as follows: Sp, S. pombe SPBC27B12.04c; Hs1, H. sapiens FLJ14743; Hs2, H. sapiens KIAA1170; Mf, M. fascicularis hypothetical product of the AB050515 locus; Ag, A. gambiae agCP10939; Dm, D. melanogaster CG11526; Ce, C. elegans F10E7.8. For all panels, sequence analysis was performed as described in Materials and Methods.
FAR10, like FAR7-FAR9, has no extensive homology to genes of known function. However, it is highly homologous to FAR9, and both genes are predicted to encode an FHA domain (28), which recent structural studies suggest is a phosphoprotein interaction domain (14, 37, 38). Moreover, FAR3 and FAR8 to FAR10 are each predicted to encode a CC domain, also thought to be involved in protein-protein interaction. The predicted presence of either an FHA or CC domain does not suggest an obvious function for the encoded proteins, although some of these domains may be involved in the Far-Far interactions. Structural features predicted to be encoded by FAR3 and FAR7 to FAR11 are diagrammed in Fig. 3A.
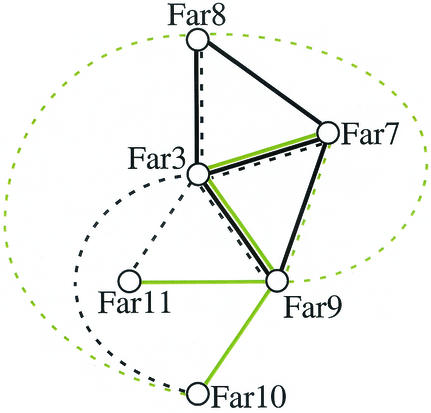
A network of Far proteins is revealed by two-hybrid analysis and coprecipitation.
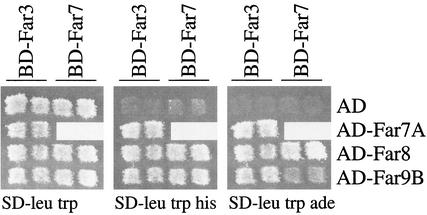
Because Far3 exhibits two-hybrid interaction with several proteins that also function in pheromone-mediated cell cycle arrest, we wondered whether these proteins interact with each other. We tested for two-hybrid interactions among Far7, Far8, and Far9. The FAR7 coding region was subcloned into the Gal4BD vector and tested against Far7, Far8, and Far9 Gal4AD fusion constructs previously obtained as targets in our two-hybrid screen (see Materials and Methods). Full-length Far7 interacted with both full-length Far8 and truncated Far9 (Far9B) (Fig. 4), but not with Far7A, a truncated version of itself (data not shown). Interestingly, Far7 did not interact with Far9A, a truncation which encompasses the C-terminal Far9B region and contains all but the first 74 amino acids of the N terminus (data not shown). We also tested a full-length version of Far9 against Far7, Far8, and Far9 but could detect no interactions (data not shown). These results suggest that the N terminus of Far9 inhibits interactions. It is also possible that other factors or modifications required to facilitate interaction with other Far proteins are absent when full-length Far9 is expressed as a fusion with Gal4BD. In either case, the two-hybrid interaction data support the notion that Far3, Far7, Far8, and Far9 form a complex.
FIG. 4.
Two-hybrid interactions among Far3 and Far7 to Far9. Two-hybrid tests were performed as described in Materials and Methods. Fusion proteins were tested in pairwise combinations consisting of a bait (Gal4BD fused to the N terminus of full-length Far3 or Far7) and a target (Gal4AD alone or Gal4AD fused to the N terminus of a Far protein or protein fragment). BD-Far3 and BD-Far7 baits were expressed from pSL2785 and pSL2786, respectively. Target proteins were expressed from plasmids as follows: AD alone from pGAD-C1; AD-Far7A (AD plus the C terminus of Far7, amino acids 65 to 222) from pSL2789; AD-Far8 (AD fused to full-length Far8) from pSL2790; AD-Far9B (AD plus the internal fragment of Far9, amino acids 138 to 572) from pSL2792. Interactions were assayed by patching independent isolates in duplicate to SD-Trp Leu and then replica plating to SD-Trp Leu His and SD-Trp Leu Ade. Growth on both tester media indicated a positive result. Patch tests were repeated at least four times using different isolates each time; the results of a typical patch test are shown.
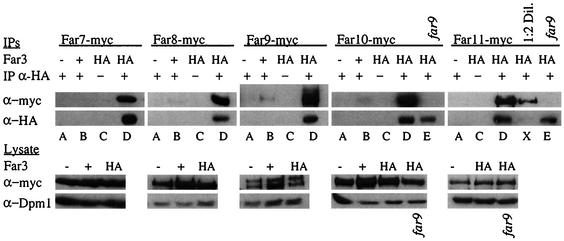
To validate and expand the two-hybrid analysis results, we expressed epitope-tagged versions of the Far proteins in vivo and examined the ability of Far3 to interact with Far7 to Far11 by coimmunoprecipitation (coIP). We constructed five strains that express a myc-tagged Far protein—Far7, Far8, Far9, Far10, or Far11—from its native locus. Western analysis revealed that all five proteins were expressed. Because Far3 protein levels are too low to detect when the gene is expressed in single copy, we introduced HA-tagged Far3, under the control of its own promoter, on a high-copy-number plasmid. All strains carried a deletion of the FAR3 locus. Halo formation was normal in all these strains, confirming that the epitope-tagged Far proteins are functional, including the plasmid-borne Far3. Far3-HA was immunoprecipitated from extracts (see Materials and Methods), and then the IP was analyzed by Western blotting to determine if any of Far7 to Far11 were present. We found that Far7 to Far11 all coprecipitated with Far3 (Fig. 5, lanes D). If the rabbit anti-HA IP antibody was not used (lanes C) or if the Far3-HA plasmid was replaced with an empty vector (lanes A), the IP no longer contained myc-tagged Far7 to Far11. Furthermore, in isogenic strains with an untagged copy of FAR3 at its native locus, the expression of the triple-HA epitope from a plasmid was not sufficient to coprecipitate myc-tagged proteins with the HA antibody (lanes B). Western analysis of the lysate revealed that HA- and myc-tagged proteins were expressed at similar levels in all strains, as was the Dpm1 loading control. Therefore, the presence of myc-tagged Far7 to Far11 in the HA IP is entirely dependent on Far3-HA.
FIG. 5.
Coprecipitation of Far3 with Far7 to Far11. Strains containing myc-tagged Far7 to Far11 were subjected to IP with a polyclonal rabbit HA antibody. Proteins contained in the IP were separated on duplicate SDS-PAGE gels and analyzed by Western blotting using monoclonal mouse antibodies to detect Far3-HA and myc-tagged Far7 to Far11. Pre-IP lysates were also analyzed by Western blotting to detect myc-tagged Far proteins or the loading control, Dpm1. Each myc-tagged Far protein was tested in isogenic strains with the following alterations: far3 mutant plus empty vector (YEp352), far3 mutant plus high-copy-number Far3-HA (pSL2784), and wild-type plus plasmid-borne HA tag (pSL2771). Far10-myc and Far11-myc were also tested in a far3 far9 background that contained the high-copy-number Far3-HA plasmid (pSL2784). Lane letters indicate strain details and experimental conditions, as follows: A, far3 〈YEp352〉 + IP α-HA (polyclonal rabbit anti-HA antibody); B, FAR3 〈pSL2771〉 + IP α-HA; C, far3 〈pSL2784〉 no IP α-HA; D, far3 〈pSL2784〉 + IP α-HA; E, far3 far9 〈pSL2784〉 + IP α-HA; X, 1:2 dilution of the concentrations in panel D. Strains used in each lane were as follows: Far7-myc (A, SY4065; B, SY4078; C and D, SY4066); Far8-myc (A, SY4067; B, SY4079; C and D, SY4068); Far9-myc (A, SY4069; B, SY4080; C and D, SY4070); Far10-myc (A, SY4071; B, SY4081; C and D, SY4072; E, SY4077); Far11-myc (A, SY4073; C, D, and X, SY4074; E, SY4084).
Our results were further supported by two recent high-throughput mass spectrometry studies using a subset of all yeast proteins as bait (21, 27). The large number of physical interactions among Far3 and Far7 to Far11, corroborated by two different in vivo assays, two-hybrid and coIP assays, strongly suggest that these proteins form a complex (Fig. 6). However, these data do not rule out the possibility that Far3 and Far7 to Far11 participate in a complicated series of pairwise interactions. Therefore, we sought to establish whether any of the Far-Far interactions might be indirect. Because Far3 interacted with Far10 and Far11 in the coIP experiment but not in the two-hybrid screens, while Far9 was shown to interact with Far10 and Far11 by two-hybrid analysis (62), we considered the possibility that Far9 might be required to facilitate the Far3-Far10 and Far3-Far11 interactions. We therefore examined the ability of Far3 to coprecipitate Far10 and Far11 in a far9 background. As shown in Fig. 5, the Far3 IP contained no detectable Far10 or Far11 when Far9 was not present (lanes E). Each far9 strain had similar levels of Far10 or Far11 compared to the wild type; however, the Far3 levels were reduced in the lysate (data not shown) and there was a slight reduction in the amount of Far3 in the IP. This suggests that the stability of Far3 may be dependent on Far9. Nonetheless, the loss of a small amount of Far3 cannot account for the complete loss of the Far3-Far10 and Far3-Far11 interactions. This conclusion was confirmed by diluting the Far3-Far11 IP by half (lane X). The amount of Far3 was nearly below the level of detection, yet the Far11 band was still clearly visible. In the far9 strain, Far3 was clearly present, yet no Far11 was found in the precipitate. This result supports our hypothesis that Far9 bridges the interactions between Far3 and each of Far10 and Far11. We infer that at least a subset of the Far proteins associate in higher-order structures.
FIG. 6.
Physical interactions among Far3 and Far7 to Far11. Solid lines represent two-hybrid interactions. Dashed lines indicate interactions found by coIP. Black lines denote interactions discovered in the course of this study. Green lines denote interactions, either two-hybrid (32, 62) or coIP (21, 27), found by other groups.
Far3 is part of a high-molecular-weight complex that also contains other Far proteins.
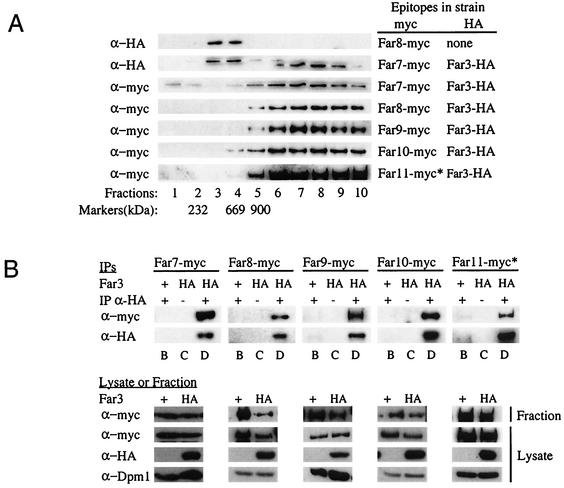
The data presented thus far are consistent with the possibility that the Far proteins are present in a large complex. To test this idea directly, we performed velocity sedimentation experiments. From subcellular fractionation experiments, we knew that Far3 and the majority of its interactors are found mainly in the subcellular fraction consisting of membranes and organelles (data not shown). As described in Materials and Methods, cell lysates were prepared and the membrane and organelle fraction was separated from the cytosol by centrifugation. The membrane and organelle fraction was solubilized with a mild detergent to release protein complexes from membranes and organelles, centrifuged to remove unsolubilized complexes, and loaded onto a 15 to 40% continuous sucrose velocity gradient. A far3 strain containing high-copy-number Far3-HA yielded a Far3 peak in fraction 8 (Fig. 7A). (A band of a slightly higher molecular weight than Far3 was present in fractions 3 and 4; since this band was also seen in control strains expressing untagged Far3, it is assumed to be nonspecific background.) Given that all size markers peaked no later than fraction 5, Far3 appeared to be associated with a complex considerably larger than 900 kDa (Fig. 7A). A complex consisting exclusively of Far3 and Far7 to Far11 at a single copy apiece would be approximately 600 kDa. Thus, Far3 is present in a complex much larger than would be expected if the complex contained a single molecule of each Far protein. Perhaps other proteins are also present, or perhaps more than one copy of each Far protein is represented in the complex.
FIG. 7.
Sucrose velocity gradients of Far3 and Far7 to Far11. (A) Cosedimentation of the Far proteins on sucrose velocity gradients. Lysates from strains containing epitope-tagged Far3 and Far7 to Far11 were separated on 15 to 40% continuous sucrose velocity gradients. Fractions were collected from the top and analyzed by Western blotting to detect each of the tagged Far proteins. Markers of known size were also separated on sucrose gradients. The fraction in which a particular size marker peaked is indicated below the corresponding lane. (B) Coprecipitation of Far3 with Far7 to Far11 in IPs from the sucrose gradient fraction peak. Far3-HA was immunoprecipitated from sucrose gradient fractions in which it was the most abundant; the presence of Far proteins in the IP was detected by Western blotting as described in Materials and Methods. Pregradient lysates and the gradient fractions used for IP were also analyzed by Western blotting to detect epitope-tagged Far proteins or the loading control, Dpm1. Lane letters indicate strain details and experimental conditions, as follows: B, FAR3 〈pSL2771〉 + IP α-HA; C, far3 〈pSL2784〉 no IP α-HA; D, far3 〈pSL2784〉 + IP α-HA. For both panels A and B, antibodies and strains are the same as those used in the original coIP experiments (Fig. 5). The asterisk (∗) next to the Far11-myc lanes in both panels A and B indicates that the 130-kDa cleavage product is shown, as opposed to the 160-kDa full-length protein.
To determine whether the high-molecular-weight Far3 complex contains the other Far proteins, we first determined where they fractionated in the sucrose gradient. Far7 to Far10 were all found in the same high-molecular-weight fractions as Far3 (Fig. 7A). Full-length Far11 (160 kDa) was concentrated in the same fractions as Far3, although this band was extremely faint (data not shown); in addition, a 130-kDa Far11 cleavage product was clearly visible in these same fractions (Fig. 7A). We then immunoprecipitated Far3 from the sucrose gradient fraction in which it was the most abundant and asked whether Far7-myc to Far11-myc were all present in the Far3 IP. We observed that the high-molecular-weight Far3 complex contained Far7 to Far10 (Fig. 7B). Full-length Far11 was not detected in these IPs, possibly due to its low initial concentration in the fraction. However, we were able to clearly see the 130-kDa cleavage product of Far11 in the Far3 IP (Fig. 7B). None of these interactions were detected in the isogenic Far-myc strain expressing untagged Far3 from its native locus; these control strains also contain a plasmid expressing the HA epitope alone. Western analysis of the lysates from all strains revealed that Far3-HA, Far7-myc, and Far9-myc to Far11-myc proteins were expressed at equivalent levels in both the Far3-HA and Far3-untagged strains. We routinely observed higher levels of Far8 in the Far8-myc Far3-untagged control strain, than in the double-tagged strain used for the coIP experiment. Nonetheless, there was no Far8 background in the control IP. Similarly, the control strains did not exhibit an altered gradient distribution of myc-tagged protein (data not shown); in both the Far3-HA and Far3-untagged strains, each of Far7 to Far10 and the truncated Far11 were present in the fraction where Far3 peaked, yet these myc-tagged proteins were detected in the IP only when Far3 was tagged. Dpm1 was used as a loading control, and its levels did not vary among strains. Therefore, the coprecipitation is specific, and all of Far7 to Far10, and at least a truncated version of Far11, are associated with Far3 in a high-molecular-weight complex.
Pheromone response in the single and sextuple mutants of far3 and far7 to far11.
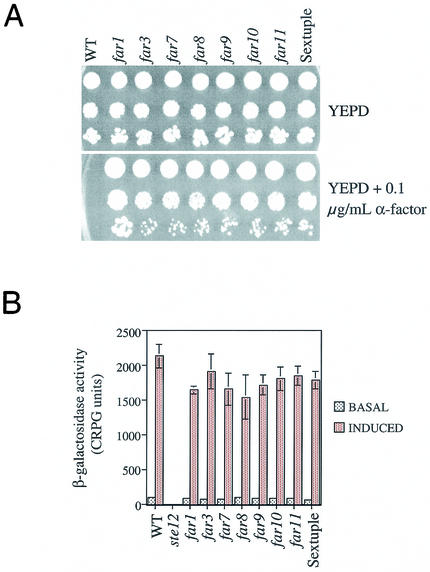
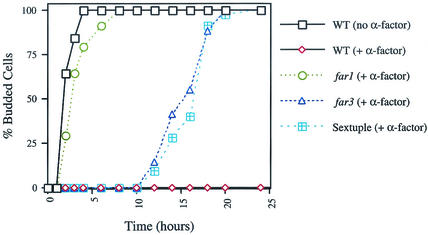
The five new FAR genes identified in this study exhibited the same null mutant phenotype as far3. We sought to determine whether they participate in the same pathway as FAR3. If this were the case, a strain lacking all six genes would have essentially the same phenotype as the single mutants. To test this hypothesis a sextuple far3 far7 to far11 mutant was constructed. When grown on YEPD plates containing levels of pheromone that inhibit the wild type, the sextuple mutant exhibited essentially the same level of recovery from G1 arrest as did the single mutants (Fig. 8A). In comparison, the isogenic far1 mutant displayed a more severe defect, as previously observed (30).
FIG. 8.
Pheromone response in single and multiple mutants of far3 and far7 to far11. (A) Growth on pheromone. Serial dilutions of wild-type (WT) and mutant strains were spotted onto YEPD alone and YEPD + 0.1 μg of α-factor per ml. Growth was assessed after 3 days at 30°C. The experiment was repeated more than 20 times with the same results. The results of a typical experiment are shown. (B) Signal transduction. Wild-type and mutant strains containing a genomically integrated FUS1::lacZ reporter were tested for basal (no α-factor) and induced (plus α-factor) levels of β-galactosidase, as described in Materials and Methods. Each time the assay was performed, four independent isolates of each strain were grown to log phase and then either assayed directly or induced with α-factor before the β-galactosidase levels were measured. The assay was repeated three times, with similar results. The results of a typical experiment is shown. For all experiments, strains were as follows: WT, SY2227; ste12, SY4085; far1, SY2673; far3, SY4062; far7, SY4059; far8, SY4061; far9, SY4060; far10, SY4063; far11, SY4064; sextuple far3 far7 to far11, SY4075.
We also tested the single and multiple mutants for their ability to respond to pheromone as assessed by FUS1-lacZ expression and by shmoo formation. As previously reported for far3 (30), all single mutants and the sextuple mutant had wild-type levels of FUS1-lacZ, both before (basal) and after (induced) pheromone exposure (Fig. 8B). Similarly, the far1 mutant was not significantly impaired for signal transduction. In contrast, ste12, which is defective for signal transduction, had essentially no β-galactosidase activity (Fig. 8B). Likewise, shmoo formation was normal in the far3 far7 to far11 mutant but absent in ste12 (data not shown). All of this supports the initial finding that FAR7 to FAR11 function like FAR3 with respect to pheromone response and are specifically defective for arrest but not for other outputs of the pathway. By all these assays, the newly discovered FAR genes are shown to function exactly like FAR3. The phenotypes observed in the sextuple mutant argue that all six genes are in the same FAR3-dependent pathway leading to G1 arrest.
Far3 and Far1 pathways are functionally distinct.
We next investigated the nature of the G1 arrest defect in far1 and far3 mutants. Previous work had shown that a far3 far1 double mutant has a more severe phenotype than either single mutant and that the FAR1 and FAR3 pathways must therefore be independent. We considered two alternate possibilities: either the far mutants might exhibit an accelerated recovery from G1 arrest or they might exhibit a defect in initial arrest. Therefore we examined the timing of recovery from arrest more closely (see Materials and Methods). Briefly, a minimum of 30 unbudded cells were isolated on YEPD alone or YEPD containing 0.1 μg of α-factor per ml and incubated at 30°C. At the indicated time points, the cells were examined under a light microscope to determine the timing of the appearance of initial buds and the formation of microcolonies. We found that far3 and the sextuple mutant initially underwent G1 arrest when treated with pheromone (Fig. 9). If continually exposed to pheromone, far3 and the sextuple mutant resumed budding approximately 10 h after the initial arrest whereas the wild-type cells failed to recover. In contrast, the far1 mutant exhibited little or no lag in cell cycle reentry when continuously exposed to mating pheromone. To control for other factors which might delay budding, we also examined the signaling defective mutant, ste12, and found that in the presence of pheromone it began rebudding with the same kinetics as the wild-type control without pheromone (data not shown). The timing of their respective G1 arrest defects underscores that the FAR1 and FAR3 pathways are distinct and suggests a functional basis for the observed phenotypic difference between far3 and far1 mutants. It is reasonable to suppose that FAR1 participates in initial G1 arrest whereas FAR3 and FAR7 to FAR11 are required later to prevent premature recovery from G1 arrest.
FIG. 9.
Kinetics of far3 and far1 recovery from cell cycle arrest during pheromone exposure. Strains were grown on YEPD with or without α-factor, and the percentage of budded cells was determined over time as described in Materials and Methods. Briefly, 30 or more unbudded cells from each strain were dissected onto the appropriate agar plate and budding was determined by microscopic examination. Budded cells scored at 25 h consisted of microcolonies containing two or more cells. Cells which had not resumed budding by the 25-h time point never did, suggesting that they were irreversibly arrested or damaged by the isolation protocol. Visible colony formation was confirmed no later than 32 h from the zero time point. The experiment was repeated three times with similar results; the results of a typical trial are shown. Strains were as follows: wild type (WT), SY2227; far1, SY2673; far3, SY4062; sextuple far3 far7 to far11, SY4075.
DISCUSSION
To investigate the role of Far3 in pheromone-mediated cell cycle arrest, we carried out a two-hybrid screen to identify interacting proteins. This effort revealed five new proteins that, like Far3, participate in pheromone arrest. We have named the genes encoding these proteins FAR7, FAR8, FAR9, FAR10, and FAR11. As discussed below, our studies indicate that these proteins may form a complex that controls reentry into the cell cycle following pheromone treatment.
We characterized the physical and genetic relationships among these proteins to learn more about the basis of Far protein action. Physical interaction data suggest that Far3 and Far7 to Far11 form a protein complex. Our results are further supported by two recent high-throughput mass spectrometry studies using a subset of all yeast protein baits (21, 27). While neither study expressed Far3 as a bait in their screen, Far3 was found in association with the Far11/Ynl127w bait (21). Consistent with our directed two-hybrid results, Far7/Yfr008c was found to associate with Far9/Ydr200c (27). In addition, Far8/Ymr029c was detected after IP of either Far9/Ydr200c or Far10/Ylr238w (27).
We have also shown that the interactions between Far3 and Far10 and between Far3 and Far11 require Far9, providing evidence that Far3, Far9, Far10, and Far11 do not just participate in pairwise interactions but are part of a larger complex. In sucrose velocity gradients, Far3 and Far7 to Far11 cosedimented and were subsequently detected in fractions which would be expected to contain protein complexes at least large enough to house all six proteins. Furthermore, when Far3 was immunoprecipitated from the gradient fraction in which it was most abundant, Far7 to Far10 and a 130-kDa cleavage product of Far11 all coprecipitated with Far3, strongly suggesting that all six proteins are present in the same high-molecular-weight complex. Given that Far9, Far10, and Far11 are all predicted transmembrane proteins (Fig. 3), it is possible that the Far complex assembles at the plasma membrane or is attached to a membrane-bound organelle. Either possibility is consistent with the observation that these proteins are found in a subcellular fraction consisting of plasma membrane and organelles (data not shown).
Genetic studies support the notion that the Far proteins form a complex with a common function. Comparison of the single-mutant phenotypes with the sextuple-mutant phenotype argues that FAR7 to FAR11 are all in the same pathway with FAR3. The fact that the sextuple-mutant phenotype is no more severe than that of any single-mutant suggests that removal of a single component is sufficient to cripple the putative complex. This is especially interesting in light of the fact that Far9 and Far10 are 41% identical and, in particular, are predicted to possess a similar arrangement of FHA (14, 28, 37, 38), CC (40, 46), and TM (29) domains (Fig. 3A). Despite this similarity, they are not functionally redundant. Furthermore, we have observed that far3 and far7 to far9 single mutants cannot be rescued by overexpressing Far3, Far7, or Far8 (data not shown), suggesting that FAR3 and FAR7 to FAR9 do not operate in a hierarchical, linear pathway but instead are required simultaneously.
Genetic studies and phenotypic analysis also strongly suggest that the five new FAR genes discussed here are not allelic to the recently discovered, although not yet cloned, FAR4 to FAR6 genes (10). Each of the far4, far5, and far6 alleles perturb pheromone response signaling in some way, which functionally distinguishes them from conventional far mutants. In particular, far4 mutants exhibit constitutive signaling, so that basal FUS1 levels are equivalent to what is normally seen during pheromone exposure, whereas far5 and far6 mutants exhibit a twofold reduction in basal signaling (10). In contrast, all the single and multiple far mutants studied in this report exhibit wild-type levels of pheromone signaling under both basal and induced conditions.
Although both FAR1 and FAR3 are required for pheromone arrest, the phenotypes of the mutants are not identical; the far3 mutant is considerably less resistant to pheromone than is the far1 mutant (30). Analysis of the far1 far3 double mutant has already shown that the FAR3 pathway operates independently of FAR1 (30). We extended this observation by comparing the kinetics of arrest and reentry into the cell cycle. Both far3 and the sextuple far3 far7 to far11 mutant initially arrested in G1 when treated with pheromone but then recovered approximately 10 h after the initial arrest. In contrast, far1 cells exhibited little or no lag in cell cycle progression and wild-type cells arrested and failed to reenter the cell cycle during the time course examined. Therefore, the FAR3-dependent pathway functions at a distinctly different time from FAR1. Since FAR1 is responsible for inactivating the G1 cyclin-dependent kinase, Cln-Cdc28, it is reasonable to propose that it is part of the initial G1 arrest program. FAR3, on the other hand, may be required to prevent premature recovery from pheromone arrest. The underlying mechanism may therefore be expected to be quite different from that employed by FAR1. Interestingly, pheromone arrest can be restored to far3 mutants by the additional loss of cln3 or cln1 and cln2 together (30). Thus, it seems likely that at the time far3 and far7 to far11 mutants reenter the cell cycle, Cln-Cdc28 activity returns, perhaps as a secondary consequence of the misregulation of some FAR3 or FAR7 to FAR11 target.
The observation that far3 and far7 to far11 mutants do not exhibit a pheromone arrest defect until 10 h raises a question regarding epistasis. Our assertion that the Far complex operates as an intermediary between pheromone response and cell cycle components is based in part on an experiment at 2 h, in which FUS1 levels are indistinguishable from wild-type levels. Solely on the basis of this experiment, one cannot determine whether the Far3 complex is specifically dedicated to preventing recovery at 10 h or whether it is involved in preventing general pathway attenuation. Unfortunately, it is impossible (for technical reasons) for us to examine the levels of FUS1 at 10 h. Nonetheless, the initial characterization of FAR3 revealed that FUS1 levels do not significantly decrease even when the cells begin to rebud (30). Specifically, far3 mutants were assayed in a His− background containing pGAL-STE4 construct where HIS3 is controlled by the FUS1 promoter. When grown on galactose, these cells constitutively express STE4 and activate the pheromone response pathway, initiating FUS1 transcription and G1 arrest. This is lethal for a wild-type cell, which undergoes terminal G1 arrest. far3 mutants, on the other hand, continue to grow (30). This is not merely a case of pathway attenuation, because the cells are grown on galactose medium lacking histidine, where survival requires that the FUS1 promoter drive HIS3 expression. far3 mutants continue to grow even after several days, suggesting that the transcriptional arm of the pheromone response pathway remains active during recovery from G1 arrest (30). We have verified that this holds true for each of the far7 to far11 single mutants, as well as the sextuple mutant (data not shown).
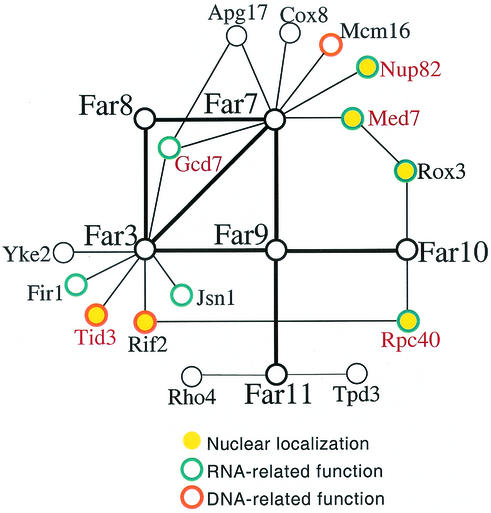
The two-hybrid interactions in which the Far proteins are known to participate involve many nuclear proteins and/or RNA-regulatory proteins (Fig. 10). Because the complex may be membrane associated, we have considered that the Far proteins may be localized to the nuclear envelope or the endoplasmic reticular membrane contiguous with the nuclear envelope. Hence, although it is still not possible to build a detailed model for the FAR3, FAR7 to FAR11 mechanism, one possibility is that the Far3 and Far7 to Far11 proteins are also localized at or near the nucleus and play a general role in RNA metabolism. In light of this possibility, it is noteworthy that genes whose overexpression allow cells to bypass pheromone arrest include several that encode transcription and translation factors, as well as various RNA-binding proteins (15). It is not without precedent for perturbation of a general process to have very specific physiological consequences. SRM1 and CDC33 have both been characterized with respect to the pheromone arrest defects of particular alleles (2, 11). In fact, the Srm1 protein is an integral part of the nuclear RNA transport machinery (1), whereas CDC33 encodes the translation initiation factor eIF4E (4).
FIG. 10.
Far3 and Far7 to Far11 with all two-hybrid interactors of known function. Two-hybrid interactions were identified in one or more published large-scale screens (32, 62). Protein function and localization (13) are indicated as follows. Filled yellow circles indicate nuclearly localized proteins. Proteins with an RNA-related function (transcription, RNA processing, nuclear export, or translation) are indicated by blue borders. Proteins with a DNA-related function (chromosome segregation; silencing; telomere length) are indicated by orange borders. Essential proteins (13) are denoted by red type.
Another intriguing possibility is that Far11, and perhaps the other members of the Far3 complex, may be required for cell fusion during mating. This suggestion stems from the observation that FAR11 is the budding-yeast homolog of a family of conserved eukaryotic genes, whose founding member, ham-2, is required for hyphal fusion in N. crassa (69). Currently, FAR11 and ham-2 represent the only characterized members of this gene family, but homologs are found in D. melanogaster, H. sapiens, S. pombe, and C. elegans as well as Anopheles gambiae (the malaria mosquito) and Macaca fascicularis (a species of macaque monkey). If FAR11 were determined to function during cell fusion in budding yeast, this would strengthen the proposal that the ham-2 gene family is required for cell fusion in general (69). Such a finding could also explain the timing of the arrest defect in far3 and far7 to far11 mutants. The Far3 and Far7 to Far11 complex could operate as part of a checkpoint that moniters fusion and prevents premature reentry into mitosis, thereby ensuring that mating cells have sufficient time to fuse.
Acknowledgments
We gratefully acknowledge Becky Drees and Stan Fields for performing the comprehensive two-hybrid screen. We thank Kathy Chicas-Cruz for making purified bovine heart mitochondria, and we thank Douglass Forbes for performing preliminary sequence analysis of the Far protein sequences. We are grateful to members of the laboratories of Tom Stevens and Rod Capaldi—particularly Kate Bowers, Liz Connibear, Laurie Graham, and James Murray—for sharing antibodies and protocols. Paul Cullen, April Goehring, Joe Horecka, Megan Keniry, Dave Mitchell, David Rivers, and Greg Smith all provided invaluable discussion, reagents, and technical support.
This work was supported by a research grant (GM-30027) from the U.S. Public Health Service.
REFERENCES
- 1.Amberg, D. C., M. Fleischmann, I. Stagljar, C. N. Cole, and M. Aebi. 1993. Nuclear PRP20 protein is required for mRNA export. EMBO J. 12:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, C., Q. Zong, and A. De Benedetti. 2001. Overexpression of eIF4E in Saccharomyces cerevisiae causes slow growth and decreased alpha-factor response through alterations in CLN3 expression. J. Biol. Chem. 276:39645-39652. [DOI] [PubMed] [Google Scholar]
- 3.Bowers, K., B. P. Levi, F. I. Patel, and T. H. Stevens. 2000. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 11:4277-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner, C., N. Nakayama, M. Goebl, K. Tanaka, A. Toh-e, and K. Matsumoto. 1988. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:3556-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Habor Laboratory Press, Plainview, N.Y.
- 6.Cagney, G., P. Uetz, and S. Fields. 2000. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 328:3-14. [DOI] [PubMed] [Google Scholar]
- 7.Chang, F., and I. Herskowitz. 1990. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell 63:999-1011. [DOI] [PubMed] [Google Scholar]
- 8.Chant, J. 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15:365-391. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 10.Cherkasova, V., and E. A. Elion. 2001. far4, far5, and far6 define three genes required for efficient activation of MAPKs Fus3 and Kss1 and accumulation of glycogen. Curr. Genet. 40:13-26. [DOI] [PubMed] [Google Scholar]
- 11.Clark, K. L., and G. F. Sprague, Jr. 1989. Yeast pheromone response pathway: characterization of a suppressor that restores mating to receptorless mutants. Mol. Cell. Biol. 9:2682-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo, M. C., M. E. Crawford, J. E. Hirschman, J. E. Kranz, P. Olsen, L. S. Robertson, M. S. Skrzypek, B. R. Braun, K. L. Hopkins, P. Kondu, C. Lengieza, J. E. Lew-Smith, M. Tillberg, and J. I. Garrels. 2001. YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29:75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, M. C., N. Liegeois, J. Horecka, R. A. DePinho, G. F. Sprague, Jr., M. Tyers, and S. J. Elledge. 1997. Human CPR (cell cycle progression restoration) genes impart a Far− phenotype on yeast cells. Genetics 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 17.Elion, E. A. 2001. The Ste5p scaffold. J. Cell Sci. 114:3967-3978. [DOI] [PubMed] [Google Scholar]
- 18.Elion, E. A., B. Satterberg, and J. E. Kranz. 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell 4:495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields, S., and I. Herskowitz. 1987. Regulation by the yeast mating-type locus of STE12, a gene required for cell-type-specific expression. Mol. Cell. Biol. 7:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gartner, A., A. Jovanovic, D. I. Jeoung, S. Bourlat, F. R. Cross, and G. Ammerer. 1998. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol. Cell. Biol. 18:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 22.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 24.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. Mitogen-activated protein kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson, B. J., B. Schulenberg, W. F. Patton, and R. A. Capaldi. 2001. A novel subfractionation approach for mitochondrial proteins: a three-dimensional mitochondrial proteome map. Electrophoresis 22:950-959. [DOI] [PubMed] [Google Scholar]
- 26.He, X., D. R. Rines, C. W. Espelin, and P. K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106:195-206. [DOI] [PubMed] [Google Scholar]
- 27.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann, K., and P. Bucher. 1995. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 20:347-349. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. 374:166.
- 30.Horecka, J., and G. F. Sprague, Jr. 1996. Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics 144:905-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iniguez-Lluhi, J. A., D. Y. Lou, and K. R. Yamamoto. 1997. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J. Biol. Chem. 272:4149-4156. [DOI] [PubMed] [Google Scholar]
- 32.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keniry, M., and G. F. Sprague. 2003. Identification of p-21 activated kinase specificity determinants in budding yeast: A single amino acid substitution imparts Ste20 specificity to Cla4. Mol. Cell. Biol. 23:1569-1580. [DOI] [PMC free article] [PubMed]
- 35.Kitada, K., E. Yamaguchi, and M. Arisawa. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165:203-206. [DOI] [PubMed] [Google Scholar]
- 36.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, J., G. I. Lee, S. R. Van Doren, and J. C. Walker. 2000. The FHA domain mediates phosphoprotein interactions. J. Cell Sci. 113:4143-4149. [DOI] [PubMed] [Google Scholar]
- 38.Liao, H., I. J. Byeon, and M. D. Tsai. 1999. Structure and function of a new phosphopeptide-binding domain containing the FHA2 of Rad53. J. Mol. Biol. 294:1041-1049. [DOI] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 40.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 41.Ma, H., S. Kunes, P. J. Schatz, and D. Botstein. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201-216. [DOI] [PubMed] [Google Scholar]
- 42.McCaffrey, G., F. J. Clay, K. Kelsay, and G. F. Sprague, Jr. 1987. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2680-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendenhall, M. D., C. A. Jones, and S. I. Reed. 1987. Dual regulation of the yeast CDC28-p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell 50:927-935. [DOI] [PubMed] [Google Scholar]
- 45.Nern, A., and R. A. Arkowitz. 2000. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J. Cell Biol. 148:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman, J. R., E. Wolf, and P. S. Kim. 2000. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:13203-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nomoto, S., N. Nakayama, K. Arai, and K. Matsumoto. 1990. Regulation of the yeast pheromone response pathway by G protein subunits. EMBO J. 9:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Shea, E. K., and I. Herskowitz. 2000. The ins and outs of cell-polarity decisions. Nat. Cell Biol. 2:E39-E41. [DOI] [PubMed] [Google Scholar]
- 49.Peter, M., A. Gartner, J. Horecka, G. Ammerer, and I. Herskowitz. 1993. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73:747-760. [DOI] [PubMed] [Google Scholar]
- 50.Peter, M., and I. Herskowitz. 1994. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science 265:1228-1231. [DOI] [PubMed] [Google Scholar]
- 51.Printen, J. A., and G. F. Sprague, Jr. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 53.Russnak, R., S. Pereira, and T. Platt. 1996. RNA binding analysis of yeast REF2 and its two-hybrid interaction with a new gene product, FIR1. Gene Expr. 6:241-58. [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsh, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 55.Sathe, G. M., S. O'Brien, M. M. McLaughlin, F. Watson, and G. P. Livi. 1991. Use of polymerase chain reaction for rapid detection of gene insertions in whole yeast cells. Nucleic Acids Res. 19:4775.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider, B. L., E. E. Patton, S. Lanker, M. D. Mendenhall, C. Wittenberg, B. Futcher, and M. Tyers. 1998. Yeast G1 cyclins are unstable in G1 phase. Nature 395:86-89. [DOI] [PubMed] [Google Scholar]
- 57.Shimada, Y., M. P. Gulli, and M. Peter. 2000. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat. Cell Biol. 2:117-124. [DOI] [PubMed] [Google Scholar]
- 58.Sprague, G. F., Jr. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 59.Triepels, R. H., B. J. Hanson, L. P. van den Heuvel, L. Sundell, M. F. Marusich, J. A. Smeitink, and R. A. Capaldi. 2001. Human complex I defects can be resolved by monoclonal antibody analysis into distinct subunit assembly patterns. J. Biol. Chem. 276:8892-8897. [DOI] [PubMed] [Google Scholar]
- 60.Tyers, M., and B. Futcher. 1993. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol. Cell. Biol. 13:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12:1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 63.Valdivieso, M. H., K. Sugimoto, K. Y. Jahng, P. M. Fernandes, and C. Wittenberg. 1993. FAR1 is required for posttranscriptional regulation of CLN2 gene expression in response to mating pheromone. Mol. Cell. Biol. 13:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker, J. E. 1992. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25:253-324. [DOI] [PubMed] [Google Scholar]
- 65.Whiteway, M., L. Hougan, D. Dignard, D. Y. Thomas, L. Bell, G. C. Saari, F. J. Grant, P. O'Hara, and V. L. MacKay. 1989. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell 56:467-477. [DOI] [PubMed] [Google Scholar]
- 66.Wigge, P. A., O. N. Jensen, S. Holmes, S. Soues, M. Mann, and J. V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wittenberg, C., K. Sugimoto, and S. I. Reed. 1990. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell 62:225-237. [DOI] [PubMed] [Google Scholar]
- 69.Xiang, Q., C. Rasmussen, and N. L. Glass. 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160:169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]