Abstract
The replication-dependent histone mRNAs, the only eukaryotic mRNAs that do not have poly(A) tails, are present only in S-phase cells. Coordinate posttranscriptional regulation of histone mRNAs is mediated by the stem-loop at the 3′ end of histone mRNAs. The protein that binds the 3′ end of histone mRNA, stem-loop binding protein (SLBP), is required for histone pre-mRNA processing and is involved in multiple aspects of histone mRNA metabolism. SLBP is also regulated during the cell cycle, accumulating as cells enter S phase and being rapidly degraded as cells exit S phase. Mutation of any residues in a TTP sequence (amino acids 60 to 62) or mutation of a consensus cyclin binding site (amino acids 99 to 104) stabilizes SLBP in G2 and mitosis. These two threonines are phosphorylated in late S phase, as determined by mass spectrometry (MS) of purified SLBP from late S-phase cells, triggering SLBP degradation. Cells that express a stable SLBP still degrade histone mRNA at the end of S phase, demonstrating that degradation of SLBP is not required for histone mRNA degradation. Nuclear extracts from G1 and G2 cells are deficient in histone pre-mRNA processing, which is restored by addition of recombinant SLBP, indicating that SLBP is the only cell cycle-regulated factor required for histone pre-mRNA processing.
Histone mRNAs are tightly regulated during the cell cycle, allowing the synthesis of histone protein to occur coordinately with the replication of DNA. The metazoan replication-dependent histone mRNAs are unique in that they are the only eukaryotic mRNAs that do not end in poly(A) tails. Instead they end in a highly conserved 26-nucleotide sequence that contains a stem-loop (25). The mRNAs for all five histone proteins are coordinately regulated, and the majority of the regulation is posttranscriptional (17). There is only a three- to fivefold change in the rate of transcription of the histone genes during the cell cycle (7, 18) but a 35- to 50-fold increase in the steady-state levels of histone mRNA. The 3′ end of histone mRNAs is the cis-acting element that coordinates the posttranscriptional regulation of the histone mRNAs (3). The efficiency of histone pre-mRNA processing is regulated as cells progress from G1 to S phase (17, 23, 40), accounting for the additional 10-fold increase in histone mRNA levels during S phase. At the end of S phase, the half-life of histone mRNA becomes very short, resulting in a rapid decrease in histone mRNA levels (17, 44).
The only processing event required for formation of mature histone mRNA is an endonucleolytic cleavage after the stem-loop (13). Cleavage of the pre-mRNA requires two cis elements: the stem-loop and a purine-rich region 3′ of the stem-loop, termed the histone downstream element, that binds U7 snRNP (6, 31, 32, 39). The only protein that binds the 3′ end of histone mRNA is the stem-loop binding protein (SLBP) (24, 43). SLBP is required for histone pre-mRNA processing (10) and remains with the mature mRNA, stimulating histone mRNA translation (37) as part of the histone messenger RNP (mRNP) (16), making it an ideal factor to participate in coordinate regulation of histone mRNAs. We have recently shown that SLBP is a cell cycle-regulated protein, accumulating just prior to entry into S phase and being rapidly degraded by the proteasome at the end of S phase, similar to the timing of degradation of histone mRNAs (44). The translation of the SLBP mRNA is activated as cells approach S phase, and SLBP is rapidly degraded as cells exit S phase (44).
Here we demonstrate that a sequence in the amino-terminal domain of SLBP is necessary for the rapid degradation of SLBP at the end of S phase. This region contains a consensus cyclin phosphorylation site and a consensus cyclin binding site. Mutation of either of these sequences stabilizes SLBP. Despite the fact that SLBP is stabilized, histone mRNA is degraded at the appropriate time. We also show that SLBP is the only factor necessary for histone pre-mRNA processing that is missing from either G1 or G2 cells that are continuously cycling. Thus, SLBP is the critical factor responsible for the posttranscriptional component of the cell cycle regulation of histone mRNA.
MATERIALS AND METHODS
Cell culture and synchronization.
HeLa cells were grown in Dulbecco modified Eagle medium plus 10% fetal calf serum and penicillin-streptomycin on 10-cm-diameter plates. The cells were plated at a low density 48 h before the double-thymidine block was initiated and were synchronized exactly as previously described (44). After release from the block, cells were harvested at 2-h intervals, and protein and/or RNA was prepared. An aliquot of the cells from each time point was taken for flow cytometry which was performed after the cells were fixed and stained with propidium iodide as described previously (44).
Cloning of SLBP mutants.
To construct pcDNA3-HA-SLBP, the NcoI fragment of SLBP cDNA was ligated to pcDNA3-HA digested with HpaI. To generate pcDNA-His-SLBP or pcDNA-Myc-SLBP, the PCR fragment of the SLBP coding region was ligated either to pcDNA-Myc (3) digested with EcoRV/XbaI or pcDNA-His vector digested with HpaI/XbaI. To construct the expression plasmid for the hemagglutinin (HA)-tagged SLBP deletion mutants, Δ54N, Δ124N, Δ13C, Δ52C, Δ72C, Δ65N/Δ13C, the PCR fragments of the coding region for these deletions were ligated to pcDNA-HA digested with HpaI and XbaI. All of these expression plasmids were checked by restriction digestion with appropriate enzymes and verified by DNA sequencing.
All site-directed mutations were made starting with pcDNA-His-SLBP according to the QuickChange Site Directed Mutagenesis protocols (Stratagene) using appropriate complementary oligonucleotides ranging from 30 to 50 nucleotides in length. Dimethyl sulfoxide (2%) was added to all the PCR mixtures. All mutations were verified by DNA sequencing.
Transfection and selection of stable transfectants.
Transfections were done with Lipofectamine (GibcoBRL) according to the manufacturer's protocol. Ten micrograms of each plasmid was used with each 10-cm-diameter plate. For selection of stably transfected cells, 24 h after transfection, cells were replated onto fresh plates and allowed to grow for another 24 h. Then 500 μg of G418 per ml was added to the medium. The medium containing G418 was changed every 2 days to remove dead cells. Cells were kept under selection until separate colonies could be observed by eye on the plates, and these stably transfected cells (at least 20 colonies per plate) were pooled together. To maintain the stably transfected cells, 200 μg of G418 per ml was kept in the medium and was removed just prior to synchronization.
Analysis of SLBP protein and histone mRNA.
Stably transfected HeLa cells were synchronized by a double-thymidine block as previously described (44). After release from the thymidine block, cells were collected at 2-h intervals, and the levels of SLBP were determined by Western blotting with anti-SLBP antibody or anti-HA antibody for the HA-tagged deletions. Detection with the SLBP antibody allowed the direct comparison of the levels of transfected SLBP mutants and endogenous SLBP.
For analysis of histone mRNA after the cells were released from the thymidine block, cells were collected at 2-h intervals, and total RNA was isolated using TRIzol (GibcoBRL). An aliquot of the same cell suspension was used for analysis of SLBP. Histone mRNA levels were analyzed by Northern hybridization using randomly labeled mouse H3.2-614 DNA as a probe.
Nuclear extract preparation and in vitro processing.
An 86-nucleotide histone pre-mRNA substrate was synthesized and labeled at the 5′ end as previously described (10). To prepare nuclear extracts from synchronized HeLa cells, HeLa cells were split at a low density 2 days before synchronization. Cells were synchronized by double-thymidine block. By the end of the synchronization process, cells reached 80 to 90% confluence. Cells were collected 2 to 3 h after release from thymidine block for S-phase cells, 8 to 8.5 h after release for G2-phase cells, and 12 to 14 h after release for G1-phase cells. Twenty to forty p150 plates were used for each cell cycle point. The nuclear extracts were prepared as previously described (27), and assayed for SLBP by Western blotting and mobility shift assay.
Each processing reaction was performed in a total volume of 10 μl containing 5 μl of the nuclear extract, 5 ng of the pre-mRNA substrate labeled at the 5′ end with [γ-32P]ATP, and 20 mM EDTA. Samples were incubated at 32°C for 60 min, and the RNA was isolated by phenol extraction and analyzed as previously described (10). A complete set of experiments was performed with extracts prepared in parallel from cells in S, G2, or G1 phase synchronized starting from the same initial cell culture.
Expression and purification of recombinant SLBP proteins.
Recombinant His-tagged SLBP was prepared as previously described and purified by affinity chromatography on Ni-agarose (10).
Cell fractionation.
For fractionation of cells into nuclear and cytoplasmic fractions, nuclear extracts were prepared as described above. For fractionation of cells into nuclear, polyribosomal, and cytosol fractions, the cells were lysed with 0.1% Nonidet P-40 (NP-40) in 0.32 M sucrose-1 mM spermidine, the nuclei were removed by centrifugation at 1,000 × g for 10 min, and polyribosomes and cytosol were prepared as described previously (16).
Analysis of phosphorylated proteins.
His-tagged SLBP was purified from synchronous cells that had been treated with MG132 for 4 h, 4 h after release from the double-thymidine block. Total cell extracts in 20 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 50 mM NaF, 0.5% NP-40) were passed over a Ni-agarose column, the column was washed with 10 volumes of buffer containing 20 mM Tris-HCl (pH 8.5), 500 mM KCl, 5 mM imidazole, 10 mM 2-mercaptoethanol, and 10% glycerol, followed by 2 volumes of buffer containing 20 mM Tris-HCl (pH 8.5), 1 M KCl, 10 mM 2-mercaptoethanol, and 10% glycerol. The bound proteins were eluted with a solution containing 100 mM imidazole, 20 mM Tris-HCl (pH 8.5), 100 mM KCl, 10 mM 2-mercaptoethanol, and 10% glycerol. The peak fractions were combined, EDTA was added to a concentration of 20 mM, and then 5 μg of biotinylated stem-loop RNA (synthesized by Dharmacon, Boulder, Colo.) was added and incubated on ice for 2 h. Streptavidin beads were added to bind the biotinylated RNA, washed extensively with buffer containing 20 mM HEPES (pH 7.9), 20% glycerol, 100 mM KCl, 0.2 mM EDTA, and 0.5 mM dithiothreitol, followed by several washes with phosphate-buffered saline to remove all traces of glycerol. An aliquot of the beads was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The remainder was digested with trypsin and then analyzed by mass spectrometry (MS).
The tandem mass spectrometric analyses were performed on an ABI QSTAR-Pulsar (QSTAR) (Applied Biosystems Division, Perkin-Elmer Corp., Foster City, Calif.), with nitrogen as the collision gas. The QSTAR was coupled on line to an Agilent 1100 capillary liquid chromatography (LC) system (Agilent, Palo Alto, Calif.) equipped with an external LC Packings (San Francisco, Calif.) splitter to produce a flow rate of 200 nl/min. The chromatography was performed using a 75-μm-diameter capillary column (PepMap C18; 3-μm particle size) from LC Packings and a water-acetonitrile gradient with both mobile phases containing 0.1% formic acid. The mobile-phase composition was held at 5% acetonitrile for 5 min. The concentration of acetonitrile was then increased at a rate of 1% acetonitrile per min, until 75% acetonitrile was reached. The acetonitrile concentration was then increased by 3% per min for 5 min to 90% acetonitrile, held at 90% acetonitrile for 30 min, and then reduced to 5% to recondition the column. For the diphosphorylated T13 peptide, the gradient used was different, with the composition being held at 5% acetonitrile for the first 30 min. The concentration of acetonitrile was then increased to 70% over 65 min (1% per min). The acetonitrile concentration was then increased to 90% over 20 min (1% per min) and then dropped to 5% to allow the column to reequilibrate to the initial conditions. Liquid chromatography coupled on line to tandem MS (LC-MS/MS) was performed by selecting a precursor ion, combined with the Pulsar feature of the Q-STAR, to maximize both the duty cycle of the mass spectrometer for the selected precursor ion and the sensitivity for any product ions formed which are within the selected mass range, thus optimizing the mass spectrometric sensitivity for these selected product ions. In these experiments, only the mass spectrometric cleavage products of the m/z 560.27 ion (the m/z of the triple-charged monophosphorylated T13 peptide) and the m/z 586.9 ion (the m/z of the triple-charged diphosphorylated T13 peptide) were monitored. As standards, an unphosphorylated tryptic peptide (RPESFTTPEGPKPR) corresponding to amino acids 55 to 68 from SLBP, as well the mono- (T61) and diphosphorylated (T60T61) peptides on the equivalent of T60 and T61 were synthesized at the University of North Carolina Peptide Synthesis Facility.
RESULTS
A region of the amino-terminal domain of SLBP is required for SLBP degradation at the end of S phase.
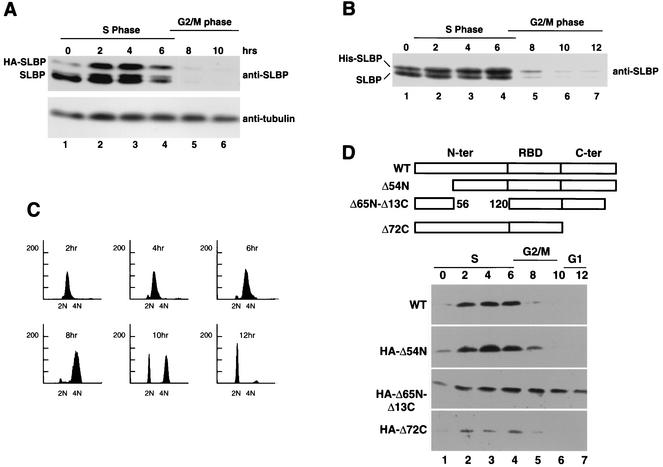
To determine the requirements for degradation of SLBP and the relationship of SLBP degradation to histone mRNA degradation, we constructed several epitope-tagged mutants of SLBP and expressed them stably in HeLa cells. These cells were then synchronized using a double-thymidine block, and the levels of the mutant SLBPs were determined, using the anti-SLBP antibody that detects both the endogenous and exogenous SLBP proteins (Fig. 1). In S-phase cells, both HA-tagged and His-tagged SLBPs were expressed at levels similar to the level of endogenous SLBP during S phase (Fig. 1A and B, lanes 2 to 4). The levels of both the endogenous and tagged SLBPs rapidly declined as cells exited S phase. In cells blocked at the G1/S border, the tagged proteins accumulated to a lesser extent than the endogenous SLBPs. There was very little HA-tagged SLBP present in the cells blocked at the G1/S border, although levels of HA-tagged SLBP were rapidly increased as cells were released and entered S phase (Fig. 1A, lanes 1 and 2). The low level of HA-tagged SLBP in blocked cells is a result of the instability of the tagged protein, which is rapidly degraded in cells not synthesizing DNA (unpublished results with M. L. Whitfield). In contrast, the His-tagged protein accumulated to significant levels in the cells blocked at the G1/S border (Fig. 1B, lane 1), as did the endogenous SLBP (44).
FIG. 1.
Cell cycle regulation of HA-tagged and His-tagged SLBP. SLBP was cloned into the pcDNA3-HA (A) or pcDNA3-his (B) vector and transfected into HeLa cells, and stably transfected cells were selected. The cells were synchronized by a double-thymidine block, arresting the cells at the G1/S border (time zero [lane 1]). Cells were released from the block, and total proteins were prepared at 2-h intervals after release (times shown above the lanes). The proteins were resolved by gel electrophoresis on an SDS-12.5% polyacrylamide gel, and the proteins were transferred to nitrocellulose filters and assayed by Western blotting using an SLBP antibody. The positions of the HA-tagged SLBP (HA-SLBP) or His-tagged SLBP (His-SLBP) and endogenous SLBP are indicated to the left of the gels. (C) Flow cytometry analysis of the synchronized cells in panel B. Chromosome content is shown on the x axis, and cell number is shown on the y axis. (D) Three deletion mutants of SLBP tagged with HA at the amino terminus and missing the first 54 amino acids (HA-Δ54N), the last 72 amino acids (HA-Δ72C), or amino acids 57 to 121 and the last 13 amino acids (HA-Δ65N-Δ13C) were transfected into HeLa cells, and stably transfected cells were selected. The cells were synchronized by a double-thymidine block, arresting the cells at the G1/S border (time zero [lane 1]). Cells were released from the block, and total proteins were prepared at 2-h intervals after release (times shown above the lanes). The proteins were resolved by gel electrophoresis on an SDS-12.5% polyacrylamide gel, transferred to nitrocellulose filters, and assayed by Western blotting using an anti-HA antibody. WT, wild type; N-ter and C-ter, N- and C-terminal regions, respectively; RBD, RNA binding domain.
The expression of the exogenous SLBP resulted in at most a two- to fourfold increase in SLBP levels, depending on the construct, suggesting that this level of overexpression should not overwhelm the normal cellular regulatory systems. There was no effect on cell growth or progression through the cell cycle in cells transfected with the full-length SLBPs. Progression through the cell cycle was monitored by flow cytometry in all experiments, and a typical profile is shown in Fig. 1C for cells transfected with the His-tagged SLBP.
A series of deletion mutants of SLBP with an HA tag on the amino terminus were constructed and stably introduced into HeLa cells. Since many of these constructs lacked the C terminus of SLBP that contains the peptide antigen used for preparation of the SLBP antibody, we used an antibody directed against the HA tag to detect these proteins. Deletion of amino acids 58 to 123 resulted in a protein that was not rapidly degraded at the end of S phase (Fig. 1D), while deletions of amino acids 1 to 54 (Fig. 1D) or deletion of the last 72 amino acids had no effect on SLBP metabolism (Fig. 1D). We analyzed the region from amino acids 58 to 123 to identify the amino acids required for SLBP degradation.
Identification of a potential phosphorylation site responsible for SLBP degradation.
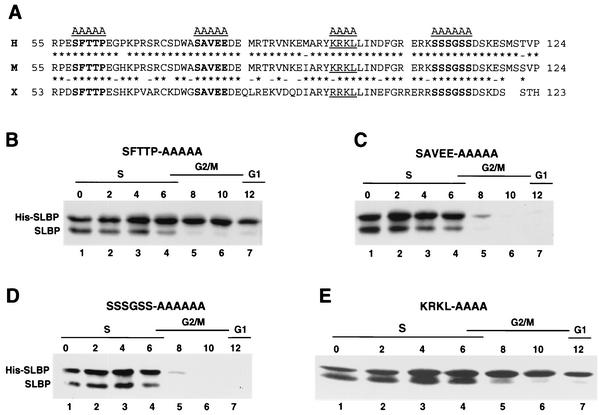
We previously showed that inhibition of the proteasome in late S-phase cells resulted in the accumulation of a phosphorylated form of SLBP, suggesting that phosphorylation of SLBP may be required for its degradation at the end of S phase (44) (see Fig. 4). We compared the sequences of the mammalian and Xenopus SLBP1 (42) between amino acids 57 to 122 (deleted in the Δ65N mutation) to identify possible conserved phosphorylation sites (Fig. 2A). This region is not required for histone pre-mRNA processing either in vivo (T. C. Ingledue and W. F. Marzluff, unpublished results) or in vitro (10). Three conserved regions containing serine or threonine were identified, and the amino acids in these regions were mutated to alanine (Fig. 2A). Each of these mutant SLBPs was transfected into HeLa cells, and stable cell cultures expressing the His-tagged SLBP were generated. These cultures were synchronized, and the levels of His-tagged and endogenous SLBP during the cell cycle were analyzed by Western blotting using the anti-SLBP antibody to simultaneously detect the endogenous and exogenous SLBP. Mutation of either the SAVEE region (amino acids 76 to 81) or the patch of five serines at amino acids 109 to 114 had no effect on the cell cycle regulation of SLBP (Fig. 2C and D). However, mutation of the conserved SFTTP sequence at amino acids 58 to 62 resulted in an SLBP which was not degraded at the end of S phase (Fig. 2B), suggesting that phosphorylation of one or more amino acids in this region was essential for SLBP degradation.
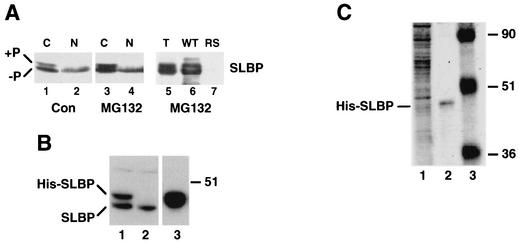
FIG. 4.
Purification of His-tagged SLBP. (A) HeLa cells were synchronized by a double-thymidine block and released for 4 h, and then MG132 was added for an additional 4 h. Cells were fractionated into nuclear and cytoplasmic fractions, and samples from equal amounts of cells were resolved by gel electrophoresis, and SLBP was detected by Western blotting. Lanes 1 and 2 contain cells harvested 4 h after release from the thymidine block (control). Lanes 3 and 4 contain cells harvested after treatment with MG132 for 6 h, starting 4 h after release. The cytoplasmic lysate from cells treated with MG132 for 6 h (lane 5) was incubated with a biotinylated wild-type (WT) stem-loop RNA (lane 6) or a biotinylated mutant stem-loop RNA with the stem sequence reversed (RS) (lane 7) and then the RNA bound to streptavidin beads. An equivalent amount of total lysate and protein bound to the beads was analyzed by gel electrophoresis, and the SLBP was detected by Western blotting. (B) Cells expressing the His-tagged SLBP (His-SLBP) were synchronized by a double-thymidine block. Four hours after release from the block, MG132 was added for 4 h. Total cell extract was fractionated on Ni-agarose, and the bound proteins were eluted with imidazole. Aliquots of the total lysate (0.1% [lane 1]), unbound protein (0.1% [lane 2]), and bound protein (1% [lane 3]) were analyzed by gel electrophoresis, and the SLBP was detected by Western blotting. The position of the 51-kDa molecular mass marker is indicated to the right of the blot. (C) A biotinylated stem-loop RNA was incubated with the proteins eluted from Ni-agarose, and then streptavidin agarose was added. The resin was washed extensively, and an aliquot of the beads analyzed by SDS-polyacrylamide gel electrophoresis (5% of the total [lane 2]) together with an aliquot of the proteins bound to Ni-agarose (1% of the total [lane 1]). The proteins were detected by staining with Coomassie blue. Note that the His-tagged SLBP (His-SLBP) is not detectable by staining of the total proteins bound to Ni-agarose. The positions of molecular mass markers (in kilodaltons) (lane 3) are shown to the left of the gel.
FIG. 2.
Amino acids 58 to 62 and 95 to 98 are required for SLBP degradation. (A) The sequences of the N-terminal regions of human (H), mouse (M), and Xenopus (X) SLBPs are shown (amino acids 55 to 124 in the human and mouse protein). The three conserved possible phosphorylation sites that were mutated are shown in bold type. The putative cyclin binding site (KRKL) is underlined. Conserved (asterisks) and similar (dashes) amino acids between two sequences are indicated. (B to E) Plasmids encoding the mutant His-tagged SLBPs (His-SLBPs) indicated above each panel were stably transfected into HeLa cells, and the cells were synchronized by a double-thymidine block. Protein extracts were made from the blocked cells (lanes 1) and from cells every 2 h after release from the block (lanes 2 to 7). Equal amounts of protein were resolved by SDS-polyacrylamide gel electrophoresis, proteins were transferred to nitrocellulose filters, and the endogenous and His-tagged SLBPs were detected by Western blotting with the anti-SLBP antibody. The time after release and the cell cycle stage is indicated above each lane.
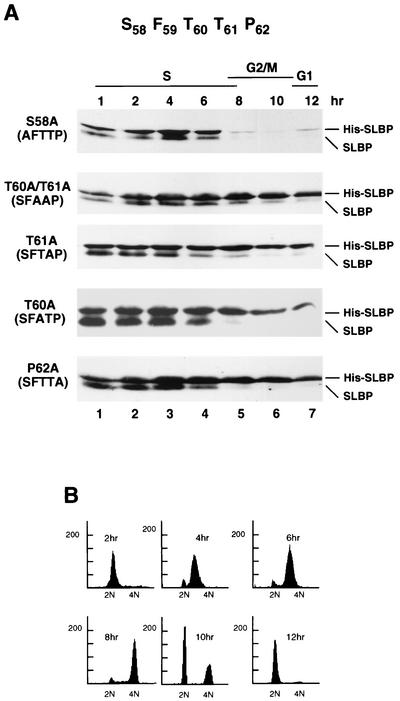
To further delineate the requirements for SLBP degradation, we mutated either one or two amino acids in the SFTTP sequence (Fig. 3A). In addition to mutation of the serines or threonines, we also mutated proline 62, since many protein kinases, including the cyclin-dependent kinases, phosphorylate a TP dipeptide. Each of these mutant proteins was stably transfected into HeLa cells, the cells were synchronized by a double-thymidine block, and the SLBP levels were analyzed by Western blotting. The results for all the mutants are summarized in Table 1.
FIG. 3.
Threonines 60 and 61 and proline 62 are required for SLBP degradation. (A) Mutations in the SFTTP sequence are shown at the left of each blot. The mutant SLBP constructs shown in panel A were stably transfected into HeLa cells. The cells were synchronized by a double-thymidine block, and protein extracts were prepared at 2-h intervals after release from the double-thymidine block. The levels of SLBP were analyzed by Western blotting using the anti-SLBP antibody that detects both the endogenous and His-tagged SLBP (His-SLBP). Lane 1 contains cells blocked at the G1/S border, and lanes 2 to 7 contain cells after release from the block. The time after release is indicated above each lane. (B) Flow cytometry analysis of the cells transfected with the T60/T61 mutant of SLBP. Chromosome content is shown on the x axis, and cell number is shown on the y axis.
TABLE 1.
Cell cycle expression of mutant SLBPsa
| Mutant | Cell cycle expression |
|---|---|
| Δ54N | Regulated |
| Δ13C | Regulated |
| Δ52C | Regulated |
| Δ65N Δ13C | Stable |
| Δ72C | Regulated |
| SFTTP (58-62) | Stable |
| SAVEE | Regulated |
| KRKL | Stable |
| SSSGSS | Regulated |
| S58A | Regulated |
| T60A | Stable |
| T61A | Stable |
| P62A | Stable |
| T60A/T61A | Stable |
| T60E/T61E | Stable |
Pools of stable cells that express the indicated SLBPs tagged with either HA (the deletion mutants) or His (the site-specific mutations) were synchronized by a double-thymidine block. The levels of the mutant and endogenous SLBP levels were measured by Western blotting using the antibody against SLBP. A regulated mutant is degraded in parallel with the endogenous SLBP, and a stable mutant is degraded slowly only at the end of S phase.
Mutation of serine 58 to alanine did not affect SLBP stability (Fig. 3A). However, mutation of both threonine 60 and 61 to alanine, mutation of either threonine 60 or 61 to alanine, or mutation of proline 62 to alanine resulted in an SLBP that was stable at the end of S phase. The amount of these mutant SLBPs only slowly decreased as cells progressed through G2 and mitosis, while the endogenous SLBP was rapidly degraded (Fig. 3A). There was no change in the time these cells went through mitosis as assayed by flow cytometry (Fig. 3B). We also changed both threonine 60 and 61 to glutamic acid, which might mimic the phosphorylated protein. This mutant SLBP was expressed identically to the SFAAP mutant; the protein was not degraded at the end of S phase. Thus, the substitution of two glutamic acids for threonines was not able to mimic the phosphorylation of the threonines (Table 1; data not shown).
These results strongly implicate phosphorylation of threonines 60 and/or 61 in SLBP degradation. Since mutation of either of these residues to alanine had an identical effect on SLBP stability, it is possible that phosphorylation of both of these residues is required for the rapid degradation of SLBP at the end of S phase.
A cyclin recognition motif is required for SLBP degradation.
The requirement for a TP dipeptide and the fact that the degradation of SLBP is a cell cycle-regulated event suggested the possible involvement of a cyclin-dependent kinase in SLBP degradation. Recently, a cyclin binding motif, ZRXL, (where Z is a Lys or Arg) has been identified in many cyclin binding proteins (36, 38), including several cyclin/cdk substrates as well as the p21 class of cdk inhibitors (Table 2). The substrates include the pRb family, the E2F family, poly(A) polymerase (4), and a novel cell cycle regulatory protein, HIRA (15). Inspection of the SLBP sequence reveals a putative cyclin binding site, KRKL (amino acids 95 to 98 [Fig. 2A]), present in the region deleted in the Δ65N mutant (Table 2). We mutated these amino acids to alanine and stably expressed the mutant protein in HeLa cells. Mutation of these four amino acids resulted in an SLBP that was stable as cells exited S phase (Fig. 2E), i.e., that behaved identically to the mutants in the SFTTP sequence. This result suggests that phosphorylation of SLBP by a member of the cyclin/cdk family is required for degradation of SLBP at the end of S phase.
TABLE 2.
Cyclin binding sites in cyclin/cdk substrates and inhibitorsa
| Cyclin/cdk substrateor inhibitor | Sequence |
|---|---|
| p21 (N)b | KACRRLFG |
| p21 (C)b | HSKRRLIF |
| p27 | SACRNLFG |
| p57 | SACRSLFG |
| pRB | KPLKKLRF |
| p107 | SAKRRLFG |
| p130 | TTRRRLFV |
| E2F1 | PVKRRLDL |
| E2F2 | PAKRKLDL |
| E2F3 | PAKRRLEL |
| p45 (SKP2) | PTLKTLQV |
| Cdc25A | PAPRRLLF |
| PAP | SKIRILVG |
| HIRA | LSKRKLEL |
| E1 (HPV)c | KAKRRLFT |
| Consensus | ZRXL |
| SLBP | RYKRKLLI |
The cyclin binding sites from cdk inhibitors (p21, p27, and p57) and cdk substrates are shown and compared with the sequence in SLBP (amino acids 95 to 100).
The N-terminal (N) and C-terminal (C) regions of p21 are shown.
HPV, human papillomavirus.
Phosphorylation of SLBP at the end of S phase.
A phosphorylated form of SLBP accumulates in cells treated with MG132 near the end of S phase (44). In control cells, there is a small amount of phosphorylated SLBP, identified by its reduced electrophoretic mobility (44), and the phosphorylated SLBP is present exclusively in the cytoplasm (Fig. 4A, lanes 1 and 2). In the late S-phase cells treated with MG132, there is an increase in the amount of the phosphorylated form of SLBP, and all of the phosphorylated SLBP is present in the cytoplasm (Fig. 4A, lanes 3 and 4). To determine whether the phosphorylated SLBP that accumulates in these cells is capable of binding to the stem-loop, we incubated the extract with a biotinylated RNA, isolated the RNA with streptavidin agarose, and then analyzed the beads for the presence of SLBP (Fig. 4A, lanes 5 and 6). The phosphorylated form of SLBP was selected as efficiently as the unphosphorylated form of SLBP (more than 85% of each was bound to the beads), and neither form bound to a biotinylated RNA with the stem sequence reversed (Fig. 4A, lane 7).
To explicitly determine whether SLBP is phosphorylated on the SFTTP sequence at the end of S phase, we used MS to detect the phosphorylated peptide. Since the phosphorylated protein accumulates only in cells treated with MG132 near the end of S phase (unpublished results), we synchronized large amounts of cells, treated them with MG132 for 4 h to accumulate phosphorylated SLBP, and then purified the His-tagged phosphorylated protein for analysis by MS. We could not routinely detect phosphorylated forms of the His-tagged SLBPs by changes in electrophoretic mobility, presumably an effect of the epitope tag on electrophoretic behavior of the protein. We analyzed two samples, one expressing the wild-type protein and the other expressing the SLBPKRKL-AAAA, which has the cyclin binding motif mutated. Each of these proteins contains the same sequence in the tryptic peptide that contains the SFTTP sequence. For standards, we synthesized peptides that contained the SFTTP sequence with both threonines or one threonine (T61) phosphorylated. The His-tagged SLBP was purified in two steps. The whole-cell extract was passed over Ni-agarose to enrich for the His-tagged SLBP and remove the endogenous SLBP (Fig. 4B, lanes 2 and 3). The bound fraction which contained the His-tagged SLBP plus other proteins (Fig. 4C, lane 1) was incubated with a biotinylated stem-loop to isolate the proteins that bind specifically to this RNA. An aliquot of the proteins was analyzed by gel electrophoresis. After the two fractionation steps, the major protein remaining is the His-tagged SLBP as detected by staining with Coomassie blue (Fig. 4C, lane 2).
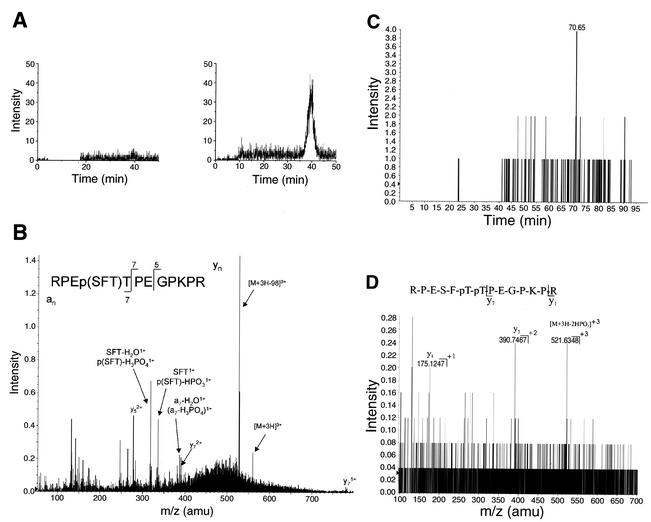
The remainder of the sample was then digested with trypsin and analyzed by capillary LC-MS/MS. This technique was used to determine the relative amounts of the mono- and diphosphorylated peptide found in wild-type versus mutant His-tagged SLBP. The same amount of wild-type and mutant protein was injected. The extracted ion chromatograms show that there was only a trace of the monophosphorylated peptide in the wild-type sample (Fig. 5A, left panel), while this peptide was very abundant in the His-tagged SLBP with the mutation of the cyclin binding site (Fig. 5A, right panel). Figure 5B shows the product ion spectrum acquired while this component was eluting. This MS/MS spectrum shows that the phosphoryl group is on S58 or T60, but not on T61, consistent with the phosphorylation of T61 being due to a cyclin/cdk.
FIG. 5.
LC-MS/MS analysis of His-tagged SLBP phosphorylation. (A) The purified His-tagged SLBP (Fig. 4C, lane 2) was digested with trypsin and subjected to mass spectrometric analysis. Wild-type SLBP (left) and the KRKL mutant SLBP (right) are shown. Extracted ion chromatograms of the product ion at m/z 527.6, which corresponds to the loss of a mass increment of 32.67 or H3PO4 from the triple-charged monophosphorylated peptide T13 of His-tagged SLBP at m/z 560.27. (B) LC-MS/MS spectrum of the tryptic peptide T13 from the His-tagged KRKL mutant SLBP, containing the SFTTP sequence, showing that the phosphoryl group is on either S58 or T60, but not T61. (C) Extracted ion chromatogram of the product ion at m/z 521.63 corresponding to the loss of 65.34 (two HPO3) from the triple-charged diphosphorylated SLBP peptide T13 (m/z 592.94) from the wild-type sample. (D) LC-MS/MS spectrum from the precursor ion at m/z 592.94 obtained at 70.65 min (panel C), confirming the presence of the diphosphorylation of the SLBP peptide T13 in the wild-type sample. The achieved fragment ions together with the MS/MS analysis from a synthetic SLBP T13 peptide phosphorylated at the equivalent position as T60 and T61 (data not shown) demonstrate that both threonine residues in the wild-type sample are phosphorylated. amu, atomic mass unit.
When we analyzed for the presence of the diphosphorylated peptide in the same samples, we detected the diphosphorylated peptide in the wild-type His-tagged protein (Fig. 5C) and did not detect any diphosphorylated peptide in the cyclin binding site mutant proteins (not shown). The sensitivity of detection for the diphosphorylated peptide was 10- to 20-fold lower than the monophosphorylated peptide using the synthetic peptides as standards, accounting for the relatively low signal to noise. Fragmentation analysis of the peptide was consistent with its assignment as the diphosphorylated peptide (Fig. 5D) and identical to that observed with the synthetic standard. Thus, in late S-phase cells treated with MG132, SLBP phosphorylated on both T60 and T61 accumulates, consistent with this being the signal for degradation. In the stable mutant SLBP, which has the cyclin binding site mutated, only T60 is phosphorylated, which is not sufficient to cause SLBP degradation.
Stabilization of SLBP does not affect histone mRNA degradation at the end of S phase.
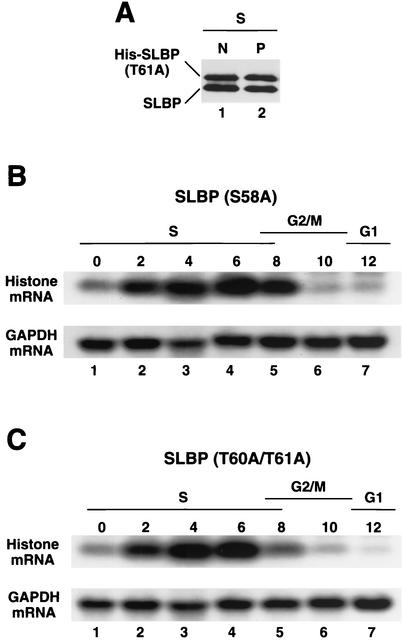
After histone pre-mRNA processing, SLBP accompanies the mRNA to the cytoplasm and is associated with the histone mRNA on polyribosomes (9, 16). Recombinant SLBPs with mutations or deletions in the amino terminus are fully active in processing in vitro (10), suggesting that they should substitute for the endogenous SLBP in histone mRNPs. To determine whether the T61A SLBP mutant that is stable at the end of S phase is associated with histone mRNAs in vivo, we fractionated S-phase cells into nuclear and polyribosomal fractions and analyzed each fraction for the two SLBPs. The ratio of the His-tagged SLBP to the endogenous SLBP was the same in each cell fraction (Fig. 6A), suggesting that it efficiently substitutes for the endogenous SLBP and was associated with histone mRNA on polyribosomes in S phase (Fig. 6A, lane 2). Identical results were obtained with the wild-type His-tagged SLBP (not shown) and the T60A/T61A SLBP.
FIG. 6.
Histone mRNA is degraded in the presence of a stable SLBP. (A) Cells transfected with the T61A SLBP were synchronized by a double-thymidine block, and the cells were harvested in S phase (4 h after release) and fractionated into nuclear (N) and polyribosomal (P) fractions. The amount of SLBP in each fraction was analyzed by Western blotting using the anti-SLBP antibody to detect both the His-tagged SLBP (His-SLBP) and endogenous SLBP. (B and C) HeLa cells transfected with the S58A mutant SLBP (B) and the T60A/T61A SLBP (C) were synchronized by a double-thymidine block. RNA was prepared from the blocked cells (lanes 1) and from cells at 2-h intervals after release from the block (lanes 2 to 7). Ten micrograms of total cell RNA was resolved by gel electrophoresis, transferred to a Nytran membrane, and probed with either the histone H3 gene (top panel) or the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA. The time after release and the cell cycle stage is indicated above each lane.
At the end of S phase, the histone mRNA is rapidly degraded at about the same time that SLBP is normally degraded (44), raising the possibility that the degradation of SLBP and histone mRNA are coupled. We determined whether histone mRNA metabolism was regulated normally in cells expressing the stable SLBP. Histone mRNA was degraded at the end of S phase in cells expressing both the S58A SLBP, which was degraded like the wild-type SLBP (Fig. 6B, lanes 4 to 7), and the T60A/T61A SLBP, which is stable (Fig. 6C, lanes 4 to 7). Thus, degradation of SLBP is not required for degradation of histone mRNA at the end of S phase. Similar results have been found for cells treated with inhibitors of DNA replication in S phase; histone mRNA is rapidly degraded while SLBP is stable (M. L. Whitfield and W. F. Marzluff, unpublished results).
SLBP is the only limiting factor for histone pre-mRNA processing in continuously cycling cells.
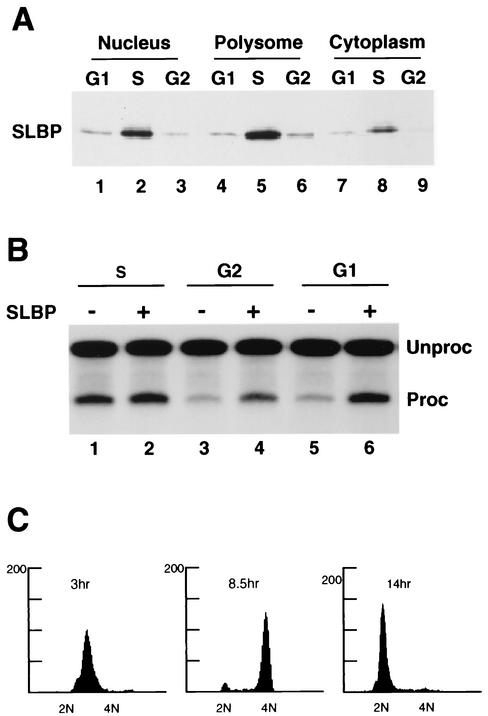
Histone pre-mRNA processing is activated as cells progress from G1 to S phase (17) and is likely also down-regulated at the end of S phase. The changes in SLBP levels that occur during the cell cycle might be solely responsible for this regulation. Since there are other factors required for histone pre-mRNA processing, it is also possible that additional factors are also regulated during the cell cycle. To determine whether SLBP is the major factor responsible for regulation of histone pre-mRNA processing, we prepared nuclear extracts from S-, G2-, and G1-phase cells after release of cells from the double-thymidine block. The degree of synchrony of these cultures was determined by fluorescence-activated cell sorter analysis (Fig. 7C). Western blot analysis demonstrated that there were significant amounts of SLBP present only in the S-phase cells and that as expected SLBP was located primarily in the nucleus and on the polyribosomes (Fig. 7A). Each nuclear extract was tested for the ability to process histone pre-mRNA and the ability of recombinant SLBP to stimulate histone pre-mRNA processing. Extracts prepared from S-phase cells (2 h after release from the double-thymidine block) processed histone H2a-614 pre-mRNA, and addition of recombinant SLBP increased the in vitro processing activity only about 50% (Fig. 7B, lanes 1 and 2). In contrast, extracts prepared from both G2- and G1-phase cells (8 and 14 h after release from the double-thymidine block, respectively) had very little processing activity (Fig. 7B, lanes 3 and 5). Addition of recombinant SLBP to either of these extracts resulted in a fivefold stimulation of the efficiency of processing, increasing the processing activity to close to the level of the S-phase extracts (Fig. 7B, lanes 4 and 6).
FIG. 7.
SLBP is the only essential histone pre-mRNA processing factor missing from G1 and G2 nuclear extracts. (A) HeLa cells were synchronized by a double-thymidine block, and equal cell equivalents of nuclear, polyribosomal, and cytosol fractions were prepared from S phase (3 h after release [lanes 2, 5, and 8]), G2 phase (8.5 h after release [lanes 3, 6, and 9]) and G1 phase (14 h after release [lanes 1, 4, and 7]) and analyzed for SLBP by Western blotting. (B) The nuclear extracts prepared from S-, G2-, and G1-phase cells were analyzed for processing activity using the H2a-614 pre-mRNA as a substrate. Recombinant SLBP was added to the extracts in lanes 2, 4, and 6 prior to addition of the pre-mRNA substrate. RNA was prepared from the reaction mixtures, and the pre-mRNA (Unproc) and processed mRNA (Proc) were resolved by gel electrophoresis. The results are representative of three independent preparations of synchronous cells. (C) Flow cytometric analysis of the cells used for preparation of the S-phase (3 h), G2-phase (8.5 h), and G1-phase (14 h) extracts, respectively. Chromosome content is shown on the x axis, and cell number is shown on the y axis.
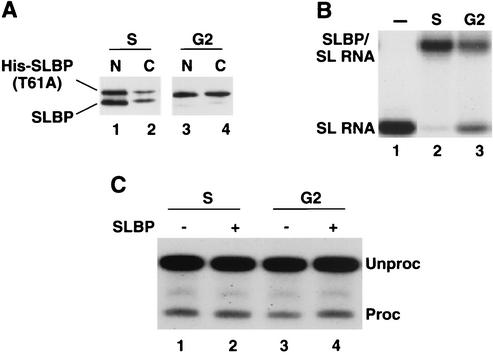
We also tested the processing activity in extracts from G2-phase cells expressing a stable SLBP. Cells expressing the T60A mutation were synchronized, and nuclear and cytoplasmic extracts were prepared from S-phase cells. Western blotting confirmed that the endogenous SLBP had been degraded in the G2-phase cells, which contained only the mutant SLBP (Fig. 8A). The mutant SLBP was present in both the nuclear and cytoplasmic fractions. In agreement with the results in Fig. 6A, the mutant and wild-type proteins were distributed similarly in the S-phase cells. In the G2-phase cells, there was a higher fraction of the protein in the cytoplasm than in the S-phase cells (Fig. 8A). The extracts from G2 cells were tested for SLBP binding activity using a mobility shift assay. There was reduced activity in the G2-phase extract compared with the S-phase extract (Fig. 8B) cells, in agreement with the lower amount of SLBP protein in the G2-phase nuclear extract (both because the wild-type protein has been degraded and because a larger fraction of the mutant protein is in the cytoplasm). We tested the S-phase and G2-phase extracts from cells expressing the T60A mutant for their ability to process histone pre-mRNA and for the ability of SLBP to stimulate processing. Both these extracts process histone pre-mRNA. The S-phase extract was slightly more efficient than the G2-phase extract, likely because of the reduced amount of SLBP in the G2-phase extract. However, the G2-phase extract was significantly more active than the G2-phase extract from wild-type cells (Fig. 7B, lane 3). Addition of recombinant SLBP had little effect on the activity of the S-phase extract and stimulated activity of the G2 extract about twofold, bringing it up to the level of the S-phase extract. Thus, the stable T60A SLBP in the G2-phase extract was functional in histone pre-mRNA processing. These results demonstrate that SLBP is the sole cell cycle-regulated factor required for histone pre-mRNA processing in vitro.
FIG. 8.
G2 extracts from cells expressing a stable SLBP are active in histone pre-mRNA processing. Cells stably expressing the T60A mutant SLBP were synchronized, and nuclear extracts and cytoplasm were prepared from S- and G2-phase cells. Equal amounts of protein were used in the assays for SLBP activity below. (A) Ten micrograms of nuclear (N) and cytoplasmic (C) proteins from S-phase (lanes 1 and 2) or G2-phase (lanes 3 and 4) cells were resolved by electrophoresis on an SDS-8% polyacrylamide gel, and SLBP was detected by Western blotting using antibody against the C-terminal peptide of SLBP. Note that the cytoplasm had 10 times more protein than the nuclear extract, so only 10% as many cell equivalents were analyzed in lanes 2 and 4. His-SLBP, His-tagged SLBP. (B) Equal amounts of nuclear extract from S-phase and G2-phase cells was incubated with a radiolabeled stem-loop (SL) (lane 1), and the complexes were resolved by native gel electrophoresis. (C) Equal amounts of S-phase (lanes 1 and 2) and G2-phase (lanes 3 and 4) cells were incubated with radiolabeled histone pre-mRNA, and the RNA products were analyzed by gel electrophoresis and detected by autoradiography. In lanes 2 and 4, recombinant SLBP was added to the extracts prior to addition of the substrate. The positions of pre-mRNA (Unproc) and processed mRNA (Proc) are shown to the right of the gel.
DISCUSSION
Phosphorylation of SLBP on T60 and T61 is required for SLBP degradation.
The levels of SLBP are regulated during the cell cycle. SLBP is up-regulated by a translational mechanism at the G1/S transition and is degraded at the end of S phase (17, 44). Treatment of cells in late S phase with the proteasome inhibitor MG132 prevents SLBP degradation at the end of S phase and results in the accumulation of a phosphorylated form of SLBP, suggesting that phosphorylation of SLBP may be the trigger for its degradation (44). Using a series of deletion mutants followed by site-directed mutation analysis, we found that mutation of either T60 or T61 to alanine stabilizes SLBP. T61 is immediately followed by a proline that is also essential for SLBP degradation, and hence SLBP is a possible substrate for a cyclin-dependent kinase. In support of this possibility, we found a potential cyclin binding site located 35 amino acids from the putative cyclin/cdk phosphorylation site. Mutation of this site to alanines also resulted in stabilization of SLBP at the end of S phase.
The degradation of several proteins is regulated during transitions from one phase of the cell cycle to the next. Two major complexes responsible for ubiquitination and subsequent degradation of proteins during the cell cycle have been identified: the anaphase-promoting complex (APC)/cyclosome and the SCF complex. The APC is activated in mitosis (30) and remains active through the next G1 phase (5). The APC recognizes a specific amino acid sequence, termed the destruction box, which is present in most proteins targeted for degradation by the APC. Since SLBP is degraded at the end of S phase or early G2 phase, it is unlikely that the APC is involved in the degradation of SLBP. The SCF complexes, a prototype of the Cullin/Roc1/2 complexes, are active throughout the cell cycle and are involved in degradation of specific proteins at the G1/S transition. They are responsible for the ubiquitination of S-phase inhibitors, Sic1 (12) and p27 (33), and the G1 cyclin, cyclin E (22, 29, 41). SCFs are also involved in protein degradation at the G2/M transition, for example for the degradation of the Wee1 kinase (28). The specificity of the SCF is provided by F-box proteins, which recognize the specific substrates. Since the F-box proteins recognize only phosphorylated substrates, synthesis of the F-box protein and/or phosphorylation of the substrate may be the regulatory step that triggers ubiquitination by SCF complexes.
The phosphorylation of both threonines 60 and 61 are required for SLBP degradation at the S/G2 border. We also identified a putative cyclin binding site, and mutation of the cyclin recognition motif, KRKL, stabilizes SLBP, providing additional evidence that phosphorylation of SLBP by a cyclin-cdk is required for SLBP degradation. SLBP containing this mutation is still phosphorylated at T60 (Fig. 5B), demonstrating that this phosphorylation at T60 either precedes the cyclin/cdk phosphorylation of T61 or can occur independently of this phosphorylation. We detected phosphorylation of both T60 and T61 in wild-type SLBP in late S-phase cells when the proteasome was inhibited (Fig. 5C). Since mutation of the SAVEE and SSSGSS sequences, which flank the cyclin binding site, does not affect SLBP degradation, it is unlikely that the mutation of KRKL affects SLBP degradation by disrupting its conformation. These results strongly argue that phosphorylation of threonines 60 and 61 is essential for SLBP degradation. Since the phosphorylation of SLBP is essential for its degradation, it is possible that SLBP is also a substrate of the SCF, with a specific F-box protein required for its degradation.
Since SLBP is degraded at the end of S phase or early G2 phase and cyclin B/cdc2 is activated at the beginning of mitosis, it is unlikely that phosphorylation of SLBP by cyclin B/cdc2 could play any role in SLBP degradation. A candidate cyclin- cdk that is active in late S phase is cyclin A/cdk2. The identity of the second kinase that phosphorylates T60 is not known. It is likely that these two threonines are phosphorylated independently (the phosphorylation of one site does not require previous phosphorylation of the other site), since the KRKL mutation blocks the phosphorylation of T61 but not T60. There are no precedents of phosphorylation of cdk sites requiring a neighboring phosphorylation. In preparations of recombinant SLBP expressed in baculovirus, some of the SLBP is phosphorylated on T61 and not T60 (our unpublished results with J. A. Erkmann), which is consistent with these phosphorylations being independent.
Preventing degradation of SLBP does not result in deregulation of histone mRNA.
All of the stably transfected SLBPs were expressed at similar levels to the endogenous SLBP, resulting in no more than two- to threefold-higher expression of total SLBP levels in the stably transfected cells in S phase. The His-tagged SLBPs function in histone pre-mRNA processing in vitro (10) and associate with the histone mRNP in vivo, as judged by association of the tagged SLBP with polyribosomes. The presence of stable SLBP at the end of S phase does not prevent the degradation of histone mRNA. This is consistent with our observation that SLBP is not degraded when cells are treated with inhibitors of DNA replication, although histone mRNA is rapidly degraded (M. L. Whitfield and W. F. Marzluff, unpublished results).
SLBP is the only regulated factor required for histone pre-mRNA processing.
Nuclear extracts prepared from exponentially growing cells are capable of processing synthetic histone pre-mRNAs in vitro. Processing requires that SLBP binds to the histone pre-mRNA. U7 snRNP then binds to the histone downstream element and its binding is stabilized by SLBP (8). There are likely other factors required for histone pre-mRNA processing that have not been well characterized (8, 10, 14). In vivo, histone pre-mRNA processing is regulated, both in cells arrested in G0/G1 (23, 40) and in continuously cycling cells (17). Since SLBP is required for efficient histone pre-mRNA processing, extracts prepared from G1- and G2-phase cells, which contain little SLBP, were essentially inactive in histone pre-mRNA processing. Addition of exogenous SLBP restored the processing activity of G1- and G2-phase nuclear extracts to levels similar to those in S-phase nuclear extracts, suggesting that SLBP is the only limiting factor for histone mRNA processing in both G2 and G1 phase. The SLBP that is stable remains active in histone pre-mRNA processing, since nuclear extracts prepared from cells in G2 phase expressing the T60/T61 stable mutant of SLBP are active in histone pre-mRNA processing (Fig. 8). Taken together, this evidence strongly suggests that SLBP is the sole essential cell cycle-regulated factor for histone pre-mRNA processing.
Two other possible mechanisms for the posttranscriptional regulation of histone pre-mRNA processing have been proposed: cell cycle regulation of a heat-labile factor (14, 23) or the cell cycle-regulated accessibility of the 5′ end of U7 snRNA (20). Our results demonstrate that the U7 snRNP present in the G1- or G2-phase extracts is active in processing. Indeed, there is a basal level of processing of the H2a-614 pre-mRNA in G2- and G1-phase nuclear extracts which is dependent on U7 snRNP and independent of SLBP (9, 10). If there is a heat-labile factor that is regulated in cells arrested in G0/G1, it is not regulated in continuously cycling cells. Many proteins necessary for cell cycle progression are down-regulated in cells arrested in G0/G1 but are not regulated in continuously cycling cells, since the proteins are relatively stable.
Additional events regulating histone mRNA accumulation.
Histone mRNAs are not present in G2 cells expressing a stable SLBP. Histone mRNAs are also not present in cells arrested at the G1/S border, even though these cells contain large amounts of SLBP, and extracts from these cells are active in histone pre-mRNA processing (unpublished results). One likely reason for the failure of histone mRNA to accumulate in these cells is that the mRNA is very unstable in the cytoplasm when there is not ongoing DNA replication. However, it is possible that there are also nuclear events that are regulated that result in histone mRNA being processed in the nucleus and degraded without being exported to the cytoplasm. If such a regulatory system exists and is cell cycle regulated, it could help ensure the rapid accumulation of histone mRNA as cells enter S phase and the cessation of production of cytoplasmic histone mRNA at the end of S phase.
Cell cycle signals regulate SLBP levels. Histone mRNAs and SLBP are both expressed at high levels only in S-phase cells. Since SLBP remains with the histone mRNA after histone pre-mRNA processing, SLBP is stoichiometrically required for accumulation of histone mRNA. However, histone mRNAs and SLBP are not regulated by the same signals and mechanisms. We previously observed that SLBP is present in cells blocked at the G1/S border, suggesting that SLBP starts to accumulate in late G1, prior to the accumulation of histone mRNA (44). Similarly, when S-phase cells are treated with inhibitors of DNA replication, histone mRNAs are degraded while SLBP is stable (unpublished results with M. L. Whitfield). Thus, all the evidence is consistent with histone mRNAs being regulated in response to changes in the rate of DNA synthesis, while SLBP is regulated by cell cycle signals. This suggests that the degradation of SLBP at the end of S phase inhibits histone pre-mRNA processing and functions to prevent further accumulation of histone mRNA prior to entry into the next cell cycle. Only when cells resynthesize SLBP as they approach S phase can they then accumulate histone mRNA.
There is no obvious phenotype in HeLa cells of persistent expression of SLBP through G2. Histone mRNA is degraded at the normal time with normal kinetics in these cells, so there will not be continued production of histone proteins during G2. Since the half-life of SLBP in S-phase cells is about 2 h and since the synthesis of SLBP is reduced at the end of S phase (L. Zheng, unpublished results) and during G1, due to regulation of translation of SLBP mRNA (44), there are not high levels of the stable SLBP throughout G1 in the cells expressing the stable SLBP. It is possible that the presence of the stable SLBP will result in precocious expression of some histone mRNA in G1 phase. However, in addition to the control of SLBP degradation, a higher percentage of the stable SLBP accumulates in the cytoplasm (Fig. 8B). This may be a secondary mechanism for stopping histone mRNA biosynthesis at the end of S phase.
The limitation of histone mRNA accumulation to S phase is likely to be important for proper chromatin function during development, although it may be less critical in a transformed cultured cell. It is becoming increasingly clear that there is a high level of specificity for assembly of variant core histones into nucleosomes, much of which may occur outside of S phase (for example, the H3cid protein, H3.3 protein, and H2a.X and H2a.Z proteins). These variants have specific functions (1, 2, 11, 19, 21, 34, 35). If large amounts of core histones are produced outside of S phase, then they could compete for specialized histone variants disrupting appropriate chromatin structure (26).
Exit from G1 and exit from mitosis both require degradation of specific critical proteins. Although SLBP degradation is not essential for the progression of the cell cycle, it is likely that there may be a group of proteins coordinately targeted for degradation as cells exit S phase and enter G2 phase and that some of these proteins are essential for cell cycle progression. Study of SLBP degradation will likely lead to further understanding of other proteins that are degraded at the end of S phase.
Acknowledgments
This work was supported in part by NIH grant GM29832 to W.F.M.
We thank Bob Duronio, Judy Erkmann, and Handan Kaygun for critical review of the manuscript.
REFERENCES
- 1.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Birchmeier, C., W. Folk, and M. L. Birnstiel. 1983. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3′ termini of sea urchin H2A mRNA. Cell 35:433-440. [DOI] [PubMed] [Google Scholar]
- 4.Bond, G. L., C. Prives, and J. L. Manley. 2000. Poly(A) polymerase phosphorylation is dependent on novel interactions with cyclins. Mol. Cell. Biol. 20:5310-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandeis, M., and T. Hunt. 1996. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 15:5280-5289. [PMC free article] [PubMed] [Google Scholar]
- 6.Cotten, M., O. Gick, A. Vasserot, G. Schaffner, and M. L. Birnstiel. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J. 7:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLisle, A. J., R. A. Graves, W. F. Marzluff, and L. F. Johnson. 1983. Regulation of histone mRNA production and stability in serum-stimulated mouse fibroblasts. Mol. Cell. Biol. 3:1920-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 9.Dominski, Z., J. Sumerel, R. J. Hanson, and W. F. Marzluff. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1:915-923. [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski, Z., L.-X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9:172-176. [DOI] [PubMed] [Google Scholar]
- 12.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 13.Gick, O., A. Krämer, W. Keller, and M. L. Birnstiel. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gick, O., A. Krämer, A. Vasserot, and M. L. Birnstiel. 1987. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. USA 84:8937-8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, C., D. M. Nelson, X. F. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson, R. J., J.-H. Sun, D. G. Willis, and W. F. Marzluff. 1996. Efficient extraction and partial purification of the polyribosomal-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35:2146-2156. [DOI] [PubMed] [Google Scholar]
- 17.Harris, M. E., R. Böhni, M. H. Schneiderman, L. Ramamurthy, D. Schümperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintz, N., H. L. Sive, and R. G. Roeder. 1983. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 3:539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff, S., K. Ahmad, J. S. Platero, and B. Van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann, I., and M. L. Birnstiel. 1990. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature 346:665-668. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, J. D., and M. A. Gorovsky. 2000. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28:3811-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koepp, D. M., L. K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J. W. Harper, and S. J. Elledge. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294:173-177. [DOI] [PubMed] [Google Scholar]
- 23.Lüscher, B., and D. Schümperli. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, F., A. Schaller, S. Eglite, D. Schümperli, and B. Müller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expr. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 26.Marzluff, W. F., and R. J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14:692-699. [DOI] [PubMed] [Google Scholar]
- 27.Marzluff, W. F., M. L. Whitfield, Z. Dominski, and Z.-F. Wang. 1997. Identification of the protein that interacts with the 3′ end of histone mRNA, p. 163-193. In J. D. Richter (ed.), mRNA formation and function. Academic Press, New York, N.Y.
- 28.Michael, W. M., and J. Newport. 1998. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science 282:1886-1889. [DOI] [PubMed] [Google Scholar]
- 29.Moberg, K. H., D. W. Bell, D. C. R. Wahrer, D. A. Haber, and I. K. Hariharan. 2001. Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413:311-316. [DOI] [PubMed] [Google Scholar]
- 30.Morgan, D. O. 1999. Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1:E47-E53. [DOI] [PubMed] [Google Scholar]
- 31.Mowry, K. L., and J. A. Steitz. 1987. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3′ end formation in vitro. Mol. Cell. Biol. 7:1663-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mowry, K. L., and J. A. Steitz. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238:1682-1687. [DOI] [PubMed] [Google Scholar]
- 33.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269:682-685. [DOI] [PubMed] [Google Scholar]
- 34.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 35.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 36.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massagué, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez, R., and W. F. Marzluff. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22:7093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulman, B. A., D. L. Lindstrom, and E. Harlow. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95:10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soldati, D., and D. Schümperli. 1988. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 8:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stauber, C., and D. Schümperli. 1988. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 16:9399-9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt, and S. I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413:316-322. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Z.-F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Z.-F., M. L. Whitfield, T. I. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 44.Whitfield, M. L., L.-X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]